Open Access Article
Open Access ArticleHigh throughput laser ablation ICP-MS bioimaging of silver distribution in animal organisms and plant tissue after exposure to silver sulfide nanoparticles†
Gregor
Marolt
 a,
Sara
Novak
a,
Sara
Novak
 *b,
Anita Jemec
Kokalj
*b,
Anita Jemec
Kokalj
 b,
Iva
Talaber
b,
Veno
Kononenko
b,
Iva
Talaber
b,
Veno
Kononenko
 b,
Susana
Loureiro
b,
Susana
Loureiro
 c,
Zahra
Khodaparast
c,
Zahra
Khodaparast
 c,
Patrícia V.
Silva
c,
Patrícia V.
Silva
 c,
Martí Busquets
Fité
c,
Martí Busquets
Fité
 d,
Richard D.
Handy
d,
Richard D.
Handy
 e and
Damjana
Drobne
b
e and
Damjana
Drobne
b
aUniversity of Ljubljana, Faculty of Chemistry and Chemical Technology, Večna pot 113, Ljubljana, Slovenia
bUniversity of Ljubljana, Biotechnical Faculty, Department of Biology, Jamova 101, Ljubljana, Slovenia. E-mail: sara.novak@bf.uni-lj.si
cCESAM-Centre for Environmental and Marine Studies & Department of Biology, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal
dApplied Nanoparticles SL, C Alaba 88, 08018 Barcelona, Spain
eSchool of Biological and Marine Sciences, University of Plymouth, UK
First published on 10th October 2023
Abstract
Laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) is a method with high potential to visualize the distribution of elements in different samples, including a variety of organisms. This study aimed to demonstrate the broad application of LA-ICP-MS in nanomaterial-biota fate studies as a high throughput bioimaging tool and to broaden the choice of standard organism models in material-biota interaction research. In this study, operation parameters of LA-ICP-MS were optimised on one organism, woodlice Porcellio scaber. Subsequently, the scanning conditions were tested on a range of aquatic (Girardia tigrina, Lumbriculus variegatus, and Oncorhynchus mykiss), terrestrial organisms (Lumbricus rubellus, Porcellio scaber) and one plant (Triticum aestivum) upon exposure to Ag2S nanoparticles (NPs) and silver nitrate (AgNO3). Model organisms were exposed in aquatic or terrestrial mesocosm experiments where nominal concentrations of Ag were 10 μg Ag per L of water and 10 mg Ag per kg of soil, respectively. The results showed that both sample preparation and LA-ICP-MS imaging conditions, as optimized on the selected organism (65 μm laser diameter, scan rate 100 μm s−1, measuring duration 35 min), are applicable on different tissues. These LA-ICP-MS imaging conditions enable recognition of the main biological structures and biodistribution of elements of interest. By using fast-screening LA-ICP-MS, we confirmed the presence of Ag2S NPs on the body surface or in the gut lumen (adsorbed and retained), but not in other internal parts of organisms, which is consistent with our previous toxicokinetic studies. The presence of Ag was also confirmed in some parts of wheat roots. The advantage of this technique is the possibility of sequential use of fast- and slow-scanning steps to optimise the duration of analysis and data processing, whilst also improving cost-effectiveness without compromising the quality of results.
Introduction
The uptake, tissue distribution, and fate of internalized nanomaterials (NMs) result from complex interactions between the particles and organisms. Unless they dissolve, intact NMs are usually taken up across epithelia by endocytosis-related mechanisms, or in the case of pathology they presumably enter the organism at damaged surfaces.1 The concern is that the colloidal behaviour and reactivities of NMs are not the same as those of solutes, and consequently NMs may have different target organs and fates inside the organism compared to traditional chemicals. However, the target organs for NMs are poorly understood, especially across the different anatomies of biological taxa.The accumulation and fate of metals in organisms exposed to metal-based NMs have been the most studied. Traditional methods of metal analysis are used to analyse a whole tissue, an organ, or sometimes a whole organism to obtain the total metal concentration in the sample. This approach has been useful in developing toxicokinetic models of uptake and excretion of metals from NM exposures.2–4 However, the total metal concentration from a tissue digest does not identify if intact particles are present, and even the recent progress on single particle inductively coupled mass spectrometry (sp-ICP-MS) to identify particles in biological samples cannot identify the complex spatial pattern within an organ(s) in vivo. Different imaging techniques are available to complement these former approaches with spatial information on the location of the metal inside the organism.5,6 Among them, the laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) technique is becoming a primary analytical tool for imaging the distribution of metals as well as metal-based NPs in cell or tissue samples. Galazzi et al.7 and Doble et al.8 reviewed the contribution of ICP-MS-based platforms as tools for evaluating NP behavior in distinct biological samples.
The basic principle of LA-ICP-MS is that the upper layer of the dry sample (whole organism or lyophilized slices of tissue sections) is ablated (i.e., vapourised or sublimed) by energy from a laser and the ions/atoms given off are then detected by inductively coupled plasma mass spectrometry.9,10 For a graphical representation of the selected elemental distributions in the sample, the data from LA-ICP-MS measurements are processed and converted into maps. The quality of the maps depends mostly on the laser diameter (LD), scanning speed (SS), and dwell time (DT). In addition, LA-ICP-MS allows scanning either large tissue sections or small spot sizes for a detailed analysis.11 LA-ICP-MS is a very sensitive method with detection limits of imaging in the μg g−1 to sub μg g−1 range and with a lateral resolution between 5 μm and 200 μm.12 In addition to providing elemental tissue distribution, LA-ICP-MS can be combined with a single particle (SP) counting technique and can provide indirect visualization of the number and size of NPs at a tissue level together with discrimination between ions and particles.13 Namely, in LA-SP-ICP-MS metals in ionic forms behave differently upon ablation and can therefore be separated in post-processing.14 An important advantage of using LA-ICP-MS is a relatively simple sample preparation. The gold standard in tissue preparation for LA-ICP-MS is a routine histological method of chemical fixation and paraffin embedding (FFPE).15
LA-ICP-MS has already been applied to different biological samples to study the presence of NPs in cells or their distribution in tissues. For example, Au NPs have been investigated in a mouse leukemic monocyte macrophage cell line, and Ag NPs in alveolar macrophages. SeCd/ZnS quantum dot or Au and Ag NP distribution was studied in fibroblasts.6,16,17 At the organ level, Au NPs were inspected in the mouse liver.18 Doped Fe-oxide particles have been studied in zebrafish larvae and in mouse models showing atherosclerotic plaques.19–21 There are also some reports of elemental imaging in plants exposed to NPs.8,22 Yamashita et al. showed the size and position of Ag and Au nanoparticles in onion cells, and Metarapi et al. examined the NP distribution in the roots of Ag+ exposed plants.14,23 However, much less data exists for invertebrates such as Annelida, Mollusca, and Arthropoda or complex vertebrate samples when NMs are studied. The exceptions are two studies by Böhme et al.20,24 who provided evidence on the applicability of LA-ICP-MS to study the crustacean Daphnia magna and zebrafish Danio rerio larvae exposed to engineered nanoparticles. In the study from 2015, they found Al2O3, Ag, and Au NMs attached to D. rerio larvae chorion and in the gut of D. magna.20 In the follow-up study they showed differences in the distribution of Ag, Au, CuO, and ZnO NPs.24
The technique has also been used for studying the elemental distribution profile of organisms25 or imaging different trace elements and isotopes in different biological samples.26,27 However, to fully explore the potential of LA-ICP-MS as a reliable tool for biological samples, the selection of optimal conditions for (multi)elemental fast or slow mode mapping is needed.28
In the present study, the application of LA-ICP-MS was tested for a fast screening of the presence and distribution of selected elements, in our case primarily Ag and also some other elements commonly present in biological samples, in different animal and plant tissue samples after exposure to silver sulfide NPs (Ag2S NPs) in terrestrial or aquatic mesocosm experiments. Ag NPs are among the most commonly used NPs in commercial products, and their production is still on the upswing. Therefore, their deposition in the environment is inevitable, and they are expected to enter the soil as one of the main repositories.29 When Ag NPs enter the soil, the majority are transformed into poorly soluble Ag sulfide NPs (Ag2S NPs); however they are also available as Ag NPs as well as Ag ions.4,30 The distribution of Ag NPs in biota as well as their biokinetics have been studied extensively.4,31,32
We tested the hypothesis that the Ag distribution pattern in Ag2S NP and AgNO3 exposed organisms is different. The work here was organized into three steps: (a) method optimization in a single-species experiment with the terrestrial isopod Porcellio scaber, then (b) testing the parameters in an environmentally realistic set-up on the same organism, and finally (c) testing selected scanning parameters and sample preparation on a wide range of biological samples [terrestrial species: earthworms (Lumbricus rubellus), crustaceans (Porcellio scaber), and plants (Triticum aestivum L.) and aquatic species: planarians (Girardia tigrina), blackworms (Lumbriculus variegatus), and the rainbow trout intestine (Oncorhynchus mykiss)] to find the best compromise between the scanning speed and image resolution. The data were also used to show the utility of LA-ICP-MS for the fast screening of the metal/NM content in organisms with different anatomy and biological matrices either as a basis for other more advanced techniques or to confirm the outcomes of other approaches (i.e., toxicokinetic studies).
Methods
Nanoparticle characterization
For the single-species tests with woodlice, polyvinylpyrrolidone (PVP) coated sulfidised silver nanoparticle (Ag2S NP) colloids with a mean size 20 nm were used in the study. Particles were provided by Applied Nanoparticles (Barcelona, Spain). Detailed particle characteristics are provided in the ESI† (Fig. S1). A colloidal suspension of Ag2S NPs (also provided by Applied Nanoparticles) was used for the terrestrial and aquatic mesocosm experiments. The Ag2S NPs were also PVP-coated with a reported size by the manufacturer of 20.4 ± 11.9 nm (mean ± S.D., n = 613). Detailed information on the characteristics of these Ag2S NP colloids is described by Peixoto et al.30 Silver nitrate (AgNO3) was used as a metal salt control (Sigma-Aldrich, 99% purity, CAS 7761–88–8, Germany).Single-species test and optimization of LA-ICP-MS
The first step of the LA-ICP-MS imaging optimization process was performed on paraffin cross sections of woodlice exposed to AgNO3, at 500 mg Ag per kg of soil as high concentrations of silver were expected to be found in the digestive tissue. The selected operating parameters were further tested on slices from animals exposed to Ag2S NPs with a concentration of 10 mg of Ag per kg of soil or an unexposed control soil. The variables tested in the LA-ICP-MS optimization phase are described in Table 1. Four unstained slices of the same animal from each exposure group (control, AgNO3, and Ag2S NPs) were used for selecting optimal measuring parameters and to assess the within-animal variation (precision) of the technique.
| Abbreviation | Laser ablation | |
|---|---|---|
| Wavelength | 193 nm | |
| Laser ablation chamber | Fast washout 2-volume HelEx II cell | |
| Ablation mode | Raster (bidirectional line scanning) | |
| BD (=BS) | Beam diameter (=beam size); laser beam spot diameter | 65 μm or 35 μm circle |
| Line length | 2.0–4.0 mm | |
| Sample dimensions (=sample size) | approx. 2 × 4 mm2 or 3.0 × 4.6 mm2 | |
| No. of line scans | 30–100 | |
| Fluence | 1.0 J cm−2 | |
| RR | Repetition rate | 10 Hz |
| SS | Scanning speed (=scan rate) | 10, 100, 200, and 500 μm s−1 |
| He carrier gas flow rate | 0.5 L min−1 (cell); 0.3 L min−1 (cup) | |
| Ar make-up flow rate | 0.8 L min−1 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| ICP-MS | ||
| RF power (W) | 1500 W | |
| Isotopes measured | 34S, 35Cl, 44Ca, 47Ti, 63Cu, 66Zn, and 107Ag | |
| AT | Acquisition time/mass | 0.05–0.10 s |
| DT | Sample period/point (dwell time) | 0.5 s |
| Point/mass | 1 | |
| Measurement mode | Time-resolved TRA | |
| Ar plasma gas flow rate (L min−1) | 15 L min−1 | |
| Ar auxillary gas flow rate (L min−1) | 0.8 L min−1 | |
The instrument for LA-ICP-MS has a laser ablation system (193 nm ArF* excimer; Analyte G2, Teledyne Photon Machines Inc., Bozeman, MT), with a 2-volume ablation cell (HelEx II; He carrier gas flow rate, cell = 0.5 L min−1 and cup = 0.3 L min−1). The LA unit was interfaced with a quadrupole ICP-MS (Agilent 7900, Agilent Technologies, Palo Alto-CA, USA); Ar make-up gas was added before the ICP torch (0.8 L min−1) and MS operation was in time-resolved mode, measuring one point per mass, acquiring the following isotope masses for all analysed samples: 34S, 35Cl, 44Ca, 63Cu, 66Zn, and 107Ag. In addition to Ag, these elements were chosen to be measured as they outline the anatomy or are present at specific locations (digestive glands) and thus serve as reference points in the elemental maps. The scan direction in all experiments was horizontal and bidirectional, i.e. exchanging the directions from left to right and right to left, alternately. The lines of odd numbers were recorded in the direction left-to-right, while the lines of even numbers were recorded in the opposite direction, right-to-left, in order to reduce the total analysis time.
LA-ICP-MS analyses of terrestrial and aquatic organisms exposed to Ag2S NPs and AgNO3 in a mesocosm set-up
For the aquatic organisms, an indoor modular mesocosm system was used to simulate a stream environment. For a detailed description of the experimental design, see Clark et al.34 Briefly, the system was composed of 36 artificial rivers made of glass arranged in triplicate, and four sets of triplicate were assigned to controls, Ag2S NPs, and AgNO3. The bottom of each stream was covered by a layer of sediment mixed with ground alder leaves, and streams were filled with an enriched artificial pond water (APW) medium. The following species were introduced in each artificial stream: G. tigrina (planarian), Physa acuta (snail), Chironomus riparius (midge larvae), L. variegatus (blackworm), D. magna (water flea), and the rainbow trout O. mykiss (daphnids and fish were kept separately in submerged plastic chambers). The water was spiked daily to reach a nominal concentration of 10 μg Ag per L of water, and the system operated in recirculation mode. The experiment lasted 14 days, and destructive sampling was performed for all organisms on days 2, 7, and 14 (organisms were also sampled at time 0). Fish were dissected into the intestine, liver, gill, muscle, and remaining carcass.
The selected organisms or tissues from both mesocosm studies were immediately preserved in Carnoy B fixative and histological sections were prepared as described by Lešer et al. Sections (8 μm) were stained with eosin and haematoxylin in order to define the organism anatomy/morphology and some were left unstained for the LA-ICP-MS measurements, as in the initial isopod study.33 For each exposure group (control, NPs, or ionic control) one or two slices per animal were measured.
Results and discussion
In the work presented here, the LA-ICP-MS scanning parameters were optimized on laboratory animals exposed to insoluble Ag2S NPs, or AgNO3, and then tested in realistic aquatic and terrestrial mesocosm exposures on a range of biota with different anatomies and tissue matrices.Optimization of scanning conditions on AgNO3 exposed P. scaber in a single-species set-up
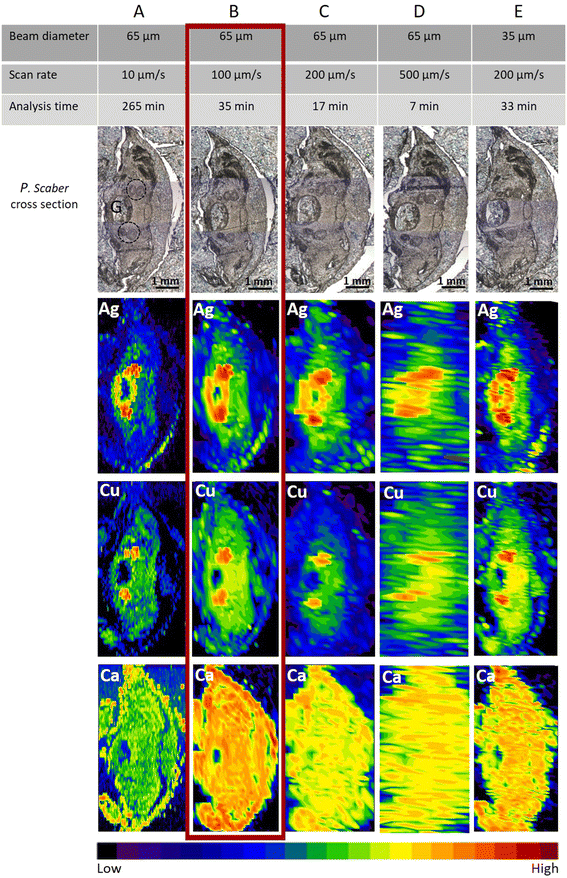
No mortality was observed in the control or any of the exposed groups in the single species tests with woodlice exposed to AgNO3. The optimal fast screening LA-ICP-MS conditions were a compromise between image quality, laser diameter, and scan speed. The “image quality” means that one can identify the main anatomical structures. The main anatomical structure of the woodlice where ingested and accumulated material is expected, is the digestive system (Fig. S2†). By knowing this, we could optimise the measurement parameters on animals exposed to the high concentration of AgNO3 (500 mg of Ag per kg of soil). In our study, apart from the element of interest, Ag, elements such as Cu and Ca were also measured to identify animal's internal morphology. The distribution of Ca outlines the animal’s body and appendages. The distribution of Cu serves to identify digestive glands with metal-storing cells that accumulate different metals when they are present in bioavailable form in crustaceans' food.29,35As expected, with a constant laser beam diameter (65 μm circle), the best lateral resolution was achieved using the slowest scan rate (10 μm s−1) (Fig. 1A). However, the time of analysis for a typical sample size (approx 10 mm2) took more than 4.4 h using 10 μm s−1 scan speed and 65 μm beam diameter (∼25 min mm−2). This is not feasible for studies with large numbers of samples where the presence/absence of an element of interest in organs is the first piece of information of interest. Therefore slow LA-ICP-MS scanning should be applied only in cases where more detailed information is needed, as slow LA-ICP-MS scanning remarkably increases the resolution.
When the scan rate was increased by 10 times (i.e., 100 μm s−1), the time for measuring one sample was reduced significantly to approx. 35 minutes (∼2.5 min mm−2). As seen in Fig. 1B, the resolution is still sufficient, and distinct biological structures of interest could be clearly seen as well as metal-storing cells with accumulated Ag. In Fig. 1C and D, due to a further increase in the scan rate to 200 μm s−1 and 500 μm s−1, respectively, the images were not of sufficient quality to define the Ag and Cu locations. However, the presence of Ag in the sample could still be confirmed despite the scan time being reasonably short, i.e., 17 min and 7 min, respectively. These latter conditions were therefore deemed satisfactory for getting information on the presence or absence of elements, but not for identifying the exact tissue location.
When using the fast-screening, ∼4 × 103 data points are produced to analyse a typical sample size (approx. 3 × 5 mm2). This is 5–6 orders of magnitude lower than that in the studies by Yamashita et al. and Materapi et al., which used the single particle (LA-SP-ICP-MS) technique for elemental analyses.14,23 These authors reported an accumulation of ∼2 × 107 data points for the analysis of a 0.5 × 0.5 mm2 sample area, corresponding to ∼1.2 × 109 data points if the same sample size as in our study was analysed.14,23 In our work, we have collected substantially fewer experimental data points, which is mainly due to the significantly longer dwell time of the fast-screening method (500 ms), but the resolution quality is still at a sufficient level to study the presence of elements of interest (Ca, Cu for outlining the major biological structures and Ag as an element of interest) in different organisms.
When the laser diameter was smaller (35 μm) and a medium scan rate (200 μm s−1) was applied (Fig. 1E), the lateral resolution of images on Ag distribution in the sample (size 3.0 × 4.6 mm2) was comparable with the resolution seen in Fig. 1B and also the time of the analysis (33 min), corresponding to ∼2.4 min mm−2, was comparable. However, the obtained resolution for Cu and Ca was slightly lower in the case of Fig. 1E. Therefore, the settings applied as seen in Fig. 1B (65 μm laser beam diameter and 100 μm s−1 scan rate) were selected as optimal scanning parameters for a fast screening (35 minutes) and used for analyses of the Ag presence in all other selected organisms exposed in the two mesocosm experiments.
Testing the optimized scanning conditions in single-species tests with P.scaber exposed to an environmentally relevant concentration of Ag2S NPs
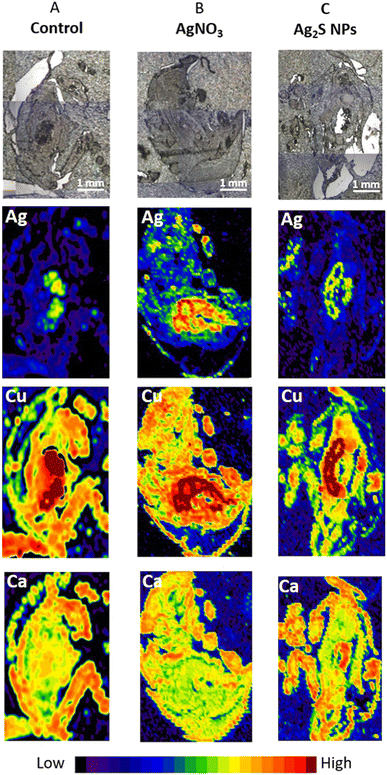
After selecting the optimal LA-ICP-MS imaging parameters on P. scaber exposed to high concentrations of AgNO3 (500 mg Ag per kg of soil) these parameters were used to test the sensitivity of the method on P. scaber exposed to environmentally relevant concentrations of Ag2S NPs or AgNO3 with a final concentration of 10 mg Ag per kg of soil (Fig. 2). From the maps presented in Fig. 2A, it is evident that some Ag was also present in the digestive glands of control animals, but not in the area of the gut. Our previous work has shown that control isopods have some background of Ag.4 In the case of exposure to AgNO3 a strong Ag signal in the area of digestive glands indicates that a much higher amount of silver was present in the digestive system than in the gut indicating accumulation of Ag in digestive glands (Fig. 2B). As expected, Cu is present in the digestive glands (dark red colour). In the case of Ag2S NP exposure, Ag was also detected in digestive glands and also in the gut confirming the consumption of Ag2S NPs (Fig. 2C).Application of the optimized LC-ICP-MS scanning conditions to organisms exposed to Ag2S NPs and AgNO3 in aquatic and terrestrial mesocosm experiments
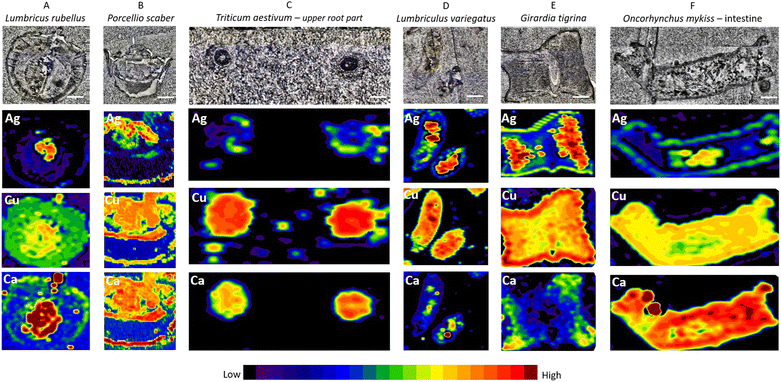
The cross sections of whole earthworms (Fig. 3A), whole isopods (Fig. 3B), and upper parts of wheat roots (Fig. 3C) as well as aquatic organisms – longitudinal sections of blackworm (Fig. 3D), planarian cross section (Fig. 3E), and longitudinal sections of the fish intestine (Fig. 3F) exposed to Ag2S NPs and AgNO3 (exposure concentration 10 mg Ag per kg of soil and 10 μg Ag per L of water) were inspected by LA-ICP-MS using parameters defined in a previous step and described in detail above (Fig. 3E). In Fig. 3, the measurements on the seventh day of exposure for all organisms are presented.We provide evidence that anatomically very different biological samples could be prepared for LA-ICP-MS analyses following the protocol for conventional histological sample preparation (i.e., chemical fixation and paraffin embedding). It turned out that by using Carnoy B fixative, major/important biological structures in all samples were sufficiently preserved. A unified sample preparation approach is therefore very welcome when anatomically different organisms are needed to be processed in a relatively short time, such as specimen collections from complex large-scale experiments such as mesocosms or field sampling. In addition, conventional histological sample preparation allows differences in the localization of silver to be identified after exposure to Ag2S NPs and AgNO3 or in non-exposed organisms.
All inspected tissues from different organisms were prepared following the same protocol as described in the methods. On the stained sections, the major biological structures could be defined (Fig. S3†), demonstrating that the tissues were sufficiently preserved. In the case of small invertebrates, the whole-body cross section could be analysed, while for larger organisms, distinct organs or plant parts with the expected NP presence were isolated, fixed, and then inspected by LA-ICP-MS (i.e., plant roots and fish intestines). The scanned size of the samples was approx. 2 × 4 mm2.
The aim of measuring other elements also in parallel to the element of interest with LA-ICP-MS was to contribute to the identification of major anatomical features in model tissues. The advantage of the method is that the time of measurement is not prolonged when multiple elements present in the sample are analysed (i.e. simultaneous elemental analysis). To present the results we have chosen two elements commonly present in all biological tissues, Ca and Cu.
The results show that fast-screening LA-ICP-MS settings (65 μm laser diameter and scan rate 100 μm s−1), which enable a short measuring time (35 min), are sufficient to obtain enough lateral resolution to locate the main biological structures and biodistribution of elements of interest on the elemental maps.
Elevated concentrations of Ag indicated by a strong signal (red color on images, see Fig. 3, second row) were present only in the specific areas recognized as the gut (confirmed on the histological images – Fig. S3†) of all invertebrate organisms (Fig. 3A, B, D and E), wheat root parts (Fig. 3C) and the lumen of the fish intestine (Fig. 3F). This is likely explained by the adsorption of Ag after Ag2S NP exposure on the body surface, or as a result of ingested or precipitated materials present in the gut lumen. P. scaber did not accumulate Ag and G. tigrina showed little Ag accumulation upon Ag2S NP exposures in the mesocosm tests, which can be the result of eventual excretion of Ag present in the gut lumen, with no Ag internalisation. Adsorption of NMs on the body surface has previously been documented, for example in daphnids.30,36 These results are in agreement with the literature reporting that passive translocation of NMs through invertebrate integuments is very unlikely if the barrier is intact.2,3,31,37 In the case of plants (T. aestivum L.), we confirmed the NP translocation from soil to roots and potentially to other body parts as reported by other authors.32,38 In the case of O. mykiss, the resolution assured by the fast screening could not be distinguished if Ag is present in the gut lumen or gut epithelium; here also slow LA-ICP-MS scanning of the same region should be provided to answer this question.
It is crucial to emphasize that Ag was measured also in all controls; therefore a background signal was present in all samples (Fig. S4†). When the model organisms were exposed to AgNO3, elevated concentrations of Ag could also be seen in other body parts, not only the digestive system as in the case of exposure to NPs, as illustrated in Fig. 2 in the case of isopods and Fig. S5† in earthworms (represented as an example).
These findings were also confirmed in the study by Clark et al.34 where total metal concentrations in the organs of the fish reflected the lower bioavailability of Ag2S compared to AgNO3. Clear differences in Ag biokinetics were also shown in the study by Talaber et al.4 where Ag bioaccumulation after exposure of two soil invertebrate species to pristine Ag NPs, ionic Ag and Ag2S NPs (same as those used in our study) depended on the Ag form, soil type and test organism.
In conclusion, the present study confirmed that optimized LA-ICP-MS analyses provide Ag elemental distribution in invertebrate, vertebrate and plant samples exposed to AgNO3 or Ag2S NPs. By complementing this information with knowledge on the anatomy and physiology of organisms, the fate of materials/ions in the body or organ can be revealed. This confirms that LA-ICP-MS analyses can provide spatial information about the locations of material assimilation inside the body or on its surface. To our knowledge, this is the first time that the LA-ICP-MS method has been applied to an environmentally relevant mesocosm study and there are no field studies reported for NMs using the LA-ICP-MS method for detection in organisms. However, the present study paves the way for ecological fieldwork, first by demonstrating the method on an organism (isopod) under controlled laboratory conditions and then on a range of organisms in the mesocosm study. Crucially, despite complex environmental matrices such as soil, or sandy sediment in the aquatic mesocosm, these did not interfere with the measurements. Arguably, if organisms are carefully rinsed after collection to remove excess media and quickly preserved in fixative, then this method will have application on field-collected organisms as well. Clearly, fixed samples from mesocosms, or perhaps fieldwork, could be used for LA-ICP-MS to study the fate of metal-based (nano)materials in biota or samples could be stored for an indefinite time for the purpose of future investigations.
Author contributions
Conceptualization: G. M, S. N., D. D., and S. L.; methodology: G. M, S. N., S. L., D. D., M.-B.-F., and R. D. H.; investigation: G. M, S. N., V. K., I. T., S. L., Z. K., P. V.-S., M. B.-F., and R. D. H.; data curation: G. M., S. N., V. K., I. T., S. L., Z. K., P. V-S, M. B.-F., R. D. H., and D. D.; writing—original draft preparation: G. M., S. N., D. D., and A. J.-K.; writing—review and editing: G. M., A. J.-K., S. N., S. L., Z. K., P. V.-S., R. D. H., and D. D.; funding acquisition: D. D.; EU H2020 project NanoFASE. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded by the EU H2020 project NanoFASE (Nanomaterial Fate and Speciation in the Environment; grant no. 646002) and supported by the Slovenian Research Agency (ARRS) through research programmes P1-0153 and P1-0184. Thanks are due to Fundação para a Ciência e a Tecnologia, Ministério da Ciência, Tecnologia e Ensino Superior (FCT/MCTES) for the financial support to CESAM (UIDP/50017/2020 + UIDB/50017/2020 + LA/P/0094/2020) through national funds. PVS was supported by a doctoral grant (SFRH/BD/52571/2014) from FCT. ZK was supported by a doctoral grant (BD/UI88/7260/2015). Thanks are also due to NanoFASE partners which provided the model organisms for mesocosm experiments.References
- R. D. Handy and G. Al-Bairuty, Ecotoxicology of Nanoparticles in Aquatic Systems, 2019, pp. 156–168 Search PubMed.
- E. J Petersen, M. Mortimer, R. M. Burgess, R. Handy, S. Hanna, K. T. Ho and P. Holden, Environ. Sci.: Nano, 2019, 6(6), 1619–1656 RSC.
- N. W Van Den Brink, A. J. Kokalj, P. V. Silva, E. Lahive, K. Norrfors, M. Baccaro and C. A. Van Gestel, Environ. Sci.: Nano, 2019, 6(7), 1985–2001 RSC.
- I. Talaber, C. A Van Gestel, A. J. Kokalj, G. Marolt, S. Novak, P. Zidar and D. Drobne, Environ. Sci.: Nano, 2020, 7(9), 2735–2746 RSC.
- Z. Qin, J. A. Caruso, B. Lai, A. Matusch and J. S. Becker, Metallomics, 2011, 3(1), 28–37 CrossRef CAS PubMed.
- D. Drescher, C. Giesen, H. Traub, U. Panne, J. Kneipp and N. Jakubowski, Anal. Chem., 2012, 84(22), 9684–9688 CrossRef CAS PubMed.
- R. M. Galazzi, K. Chacón-Madrid, D. C. Freitas, L. F. da Costa and M. A. Arruda, Rapid Commun. Mass Spectrom., 2020, 34, e8726 CrossRef CAS.
- P. A. Doble, R. G. de Vega, D. J. Bishop, D. J. Hare and D. Clases, Chem. Rev., 2021, 121(19), 11769–11822 CrossRef CAS.
- D. Günther and B. Hattendorf, Trends Anal. Chem., 2005, 24(3), 255–265 CrossRef.
- D. Clases and R. Gonzalez de Vega, Anal. Bioanal. Chem., 2022, 414, 7363–7386 CrossRef CAS PubMed.
- C. M. Ackerman, P. K. Weber, T. Xiao, B. Thai, T. J. Kuo, E. Zhang and C. J. Chang, Metallomics, 2018, 10(3), 474–485 CrossRef CAS PubMed.
- J. S. Becker, M. Zoriy, J. S. Becker, J. Dobrowolska and A. Matusch, J. Anal. At. Spectrom., 2007, 22(7), 736–744 RSC.
- D. Metarapi and J. T. van Elteren, J. Anal. At. Spectrom., 2020, 35(4), 784–793 RSC.
- D. Metarapi, J. T. van Elteren, M. Šala, K. Vogel-Mikuš, I. Arčon, V. S. Šelih and S. B. Hočevar, Environ. Sci.: Nano, 2021, 8(3), 647–656 RSC.
- M. Bonta, S. Török, B. Hegedus, B. Döme and A. Limbeck, Anal. Bioanal. Chem., 2017, 409, 1805–1814 CrossRef CAS PubMed.
- M. Wang, L. N. Zheng, B. Wang, H. Q. Chen, Y. L. Zhao, Z. F. Chai, H. J. Reid, B. L. Sharp and W. Y. Feng, Anal. Chem., 2014, 86, 10252–10256 CrossRef CAS PubMed.
- O. Reifschneider, A. Vennemann, G. Buzanich, M. Radtke, U. Reinholz, H. Riesemeier, J. Hogeback, C. Koppen, M. Grossgarten, M. Sperling, M. Wiemann and U. Karst, Chem. Res. Toxicol., 2020, 33, 1250–1255 Search PubMed.
- Q. Li, Z. Wang, J. Mo, G. Zhang, Y. Chen and C. Huang, Sci. Rep., 2017, 7(1), 2965 Search PubMed.
- S. Zarco-Fernandez, A. M. Coto-Garcia, R. Munoz-Olivas, J. Sanz-Landaluze, S. Rainieri and C. Camara, Chemosphere, 2016, 148, 328–335 CrossRef CAS PubMed.
- S. Böhme, H. J. Stärk, D. Kühnel and T. Reemtsma, Anal. Bioanal. Chem., 2015, 407, 5477–5485 CrossRef PubMed.
- C. Scharlach, L. Müller, S. Wagner, Y. Kobayashi, H. Kratz, M. Ebert and E. Schellenberger, J. Biomed. Nanotechnol., 2016, 12(5), 1001–1010 CrossRef CAS PubMed.
- V. M. Neves, G. M. Heidrich, E. S. Rodrigues, M. S. P. Enders, E. I. Muller, F. T. Nicoloso, H. W. Pereira de Carvalho and V. L. Dressler, Environ. Sci. Technol., 2019, 53, 10827–10834 CrossRef CAS PubMed.
- S. Yamashita, Y. Yoshikuni, H. Obayashi, T. Suzuki, T. Green and T. Hirata, Anal. Chem., 2019, 91(7), 4544–4551 CrossRef CAS PubMed.
- S. Böhme, M. Baccaro, M. Schmidt, A. Potthoff, H. J. Stärk, T. Reemtsmae and D. Kühnel, Environ. Sci.: Nano, 2017, 4, 1005 RSC.
- S. J. M. Van Malderen, B. Laforce, T. Van Acker, C. Nys, M. De Rijcke, R. de Rycke, M. De Bruyne, M. N. Boone, K. De Schamphelaere, O. Borovinskaya, B. De Samber, L. Vincze and F. Vanhaecke, Anal. Chem., 2017, 89(7), 4161–4168 CrossRef CAS PubMed.
- J. S. Becker, J. Mass Spectrom., 2013, 48(2), 255–268 CrossRef PubMed.
- W. X. Wang, Crit. Rev. Environ. Sci. Technol., 2022, 52(19), 3384–3414 CrossRef CAS.
- J. T. Van Elteren, V. S. Šelih and M. Šala, J. Anal. At. Spectrom., 2019, 34(9), 1919–1931 RSC.
- J. He, D. Wang and D. Zhou, Sci. Total Environ., 2019, 648, 102–108 CrossRef CAS PubMed.
- S. Peixoto, Z. Khodaparast, G. Cornelis, E. Lahive, A. G. Etxabe, M. Baccaro and V. Puntes, Ecotoxicol. Environ. Saf., 2020, 206, 111405 CrossRef CAS PubMed.
- Z. Khodaparast, C. A. van Gestel, A. G. Papadiamantis, S. F. Gonçalves, I. Lynch and S. Loureiro, Sci. Total Environ., 2021, 777, 146071 CrossRef CAS PubMed.
- P. S. Tourinho, C. A. van Gestel, A. J. Morgan, P. Kille, C. Svendsen, K. Jurkschat and S. Loureiro, Ecotoxicology, 2016, 25, 267–278 CrossRef CAS PubMed.
- V. Lešer, D. Drobne, B. Vilhar, A. Kladnik, N. Žnidaršič and J. Štrus, Zoology, 2008, 111(6), 419–432 CrossRef PubMed.
- N. Clark, J. Vassallo, P. V. Silva, A. R. R. Silva, M. Baccaro, N. Medvešček and R. D. Handy, Sci. Total Environ., 2022, 850, 157912 CrossRef CAS PubMed.
- A. Bibic, D. Drobne and J. Strus, Bull. Environ. Contam. Toxicol., 1997, 5(58), 814–821 Search PubMed.
- S. Novak, A. J Kokalj, M. Hočevar, M. Godec and D. Drobne, Ecotoxicol. Environ. Saf., 2018, 152, 61–66 CrossRef CAS PubMed.
- M. Van der Zande, A. J. Kokalj, D. Spurgeon, S. Loureiro, P. V. Silva, Z. Khodaparast, D. Drobne, N. J. Clark, N. W. Van den Brink, M. Baccaro, C. A. Van Gestel, H. Bouwmeester and R. D. Handy, Environ. Sci.: Nano, 2020, 7(7), 1874–1898 RSC.
- E. Kranjc, D. Mazej, M. Regvar, D. Drobne and M. Remškar, Environ. Sci.: Nano, 2018, 5(2), 520–532 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ja00223c |
| This journal is © The Royal Society of Chemistry 2023 |