Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Solvent-free and ball mill-free catalytic C–H methylation†
Matic
Hribersek
a,
Carolina
Méndez-Gálvez
a,
Martin
Huber
a,
Paul J.
Gates
 b,
Patrick
Shakari
c,
Ayan
Samanta
b,
Patrick
Shakari
c,
Ayan
Samanta
 c and
Lukasz T.
Pilarski
c and
Lukasz T.
Pilarski
 *a
*a
aDepartment of Chemistry – BMC, Uppsala University, Box 576, 75123, Uppsala, Sweden. E-mail: lukasz.pilarski@kemi.uu.se; Web: https://pilarski.group
bSchool of Chemistry, University of Bristol, Cantock's Close, Clifton, Bristol, BS8 1TS, UK
cDepartment of Chemistry – Ångström Laboratories, Box 538, 75121, Uppsala, Sweden
First published on 12th September 2023
Abstract
An expedient, mechanochemical, operationally simple protocol is reported for the Rh-catalysed C–H methylation of (hetero)arenes under solvent-free conditions without the use of a ball mill. Reagent mixing and activation are delivered using simple pestle-and-mortar grinding and subsequent heating, providing access to the same sustainability benefits as ball milling without the need for specialised equipment. Calculated E-factors are identical to those of ball milling and 5–25 times lower than for solution based conditions. The C–H methylation displays complete regioselectivity and good functional group tolerance. Reaction mixture analyses using scanning electron microscopy and differential scanning calorimetry are described.
Introduction
Growing concern with the environmental impact of chemical synthesis has revitalised interest in mechanochemistry – the promotion of reactivity through mechanical action, such as milling or grinding.1 Mechanochemical synthesis can enhance sustainability in various ways,1a including – most prominently – decreasing reliance on hazardous, toxic or environmentally harmful solvents. In pharmaceutical production alone, bulk solvent constitutes an estimated eighty-five percent of generated waste by mass.2 Other advantages of mechanochemistry include reduced reaction times and temperatures, the prospects of conducting ordinarily air-sensitive transformations under aerobic conditions,3 and even routes to new products via otherwise inaccessible mechanistic pathways.4 Significant advances have been made recently in the mechanistic understanding of mechanochemical reactions5 and IUPAC recently recognized mechanochemistry as one of the top ten “chemical innovations that will change our world”.6Meanwhile, developments in C–H functionalisation methodology continue apace, providing previously impossible transformations and streamlining access to valuable molecules through the direct substitution of classically ‘inert’ C–H bonds.7
New approaches to catalytic C–H methylation rank amongst the most sought-after8 because the introduction of a methyl group to bioactive molecules can improve their activity, lipophilicity, metabolic stability, pharmacokinetics and pharmacodynamics – benefits collectively termed the ‘magic methyl effect’.9
Combining the advantages of mechanochemistry and C–H functionalisation has garnered increasing interest.10 As part of our ongoing work in these areas,8h,11 we developed a mechanochemical Rh-catalysed C–H methylation carried out in an automated ball mill.8h Our approach avoided the toxic solvents (e.g. carcinogenic 1,2-DCE) previous C–H methylations were reported to require, and some of which have been effectively regulated into obsolescence.12 Other advantages of our mechanochemical protocol included dramatically reduced reaction times (from a typical 18 h to 0.5–2 h), and the possibility of late-stage C–H methylation of biologically active molecules.13
Importantly, however, automated ball milling can present up-front costs and is not yet a mainstream practice in reaction development for most laboratories. Notably, reports describing the advantages of ball milling in organic methodology very rarely consider pestle-and-mortar grinding as an accessible, low-cost alternative to deliver solvent-free reaction mixing and activation.
Some solvent-free C–H functionalisations that do not use ball milling have been reported,14 but many involve heating neat mixtures that include one or more liquid reagents used in large excess, some of which presumably act as de facto solvents.
Here, we describe an approach to catalytic C–H methylation where reagent mixing and activation energy are delivered without the use of a ball mill, but instead by very simple and brief pestle-and mortar grinding15 and subsequent heating of neat mixtures under air. This approach offers several benefits: (1) it affords the green credentials of a solvent-free, mechanochemical approach (i.e. exclusion of toxic solvents, reduction in waste, low temperature and short reaction times) in an accessible, low-cost way; (2) it allows for a comparison between automated ball milling and the cheaper alternative, which is largely absent from the literature.
We describe the applicability of the ‘grind-and-heat’ approach to diversely substituted substrates and provide some initial insights into the role played by the grinding and heating stages of our procedure obtained from Scanning Electron Microscopy (SEM) and Differential Scanning Calorimetry (DSC).
Results & discussion
Optimisation
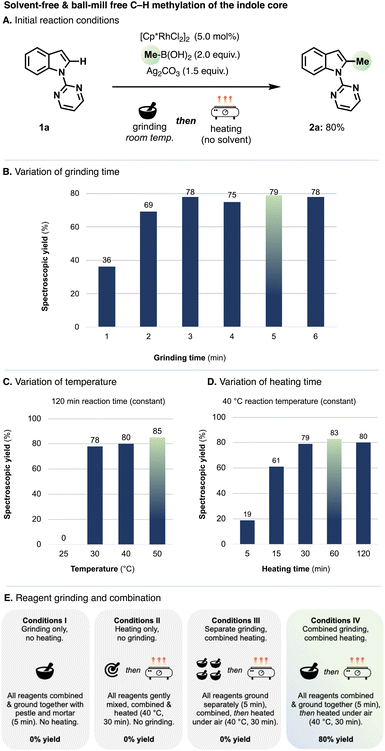
Initially, we tested the prospect of replacing a ball mill's reagent mixing and energy delivery using an agate pestle and mortar in which all reaction components were combined and ground manually for 1–5 min, and from which they were then transferred into a glass vial and heated conventionally under air, without solvent, liquid additives, stirring or exclusion of air (‘grind-and-heat’ conditions). We selected Rh-catalysed oxidative C–H methylation of 1-pyrimidylindole (1a) as a ‘workhorse’ reaction to test our approach.16 In an initial reaction using 1a, [Cp*RhCl2]2 as the catalyst precursor, MeB(OH)2 as the methyl source and Ag2CO3 as oxidant, all reaction components were ground manually for 5 min, then heated at 40 °C for 30 min, giving the corresponding C2-methylated compound 2a in 80% yield (Fig. 1A). In a separate run of reactions (Fig. 1B), we compared the effect of the grinding time (1–5 min) on the yield. Longer grinding times correlated with greater yields (after heating); grinding for more than 3 min returned 2a in yields >70%, with 5 min giving 79% and thus confirming the reproducibility of the reaction in Fig. 1A. Grinding times >5 min gave no further advantage.Fig. 1C shows the effect of temperature variations for the heating portion of our protocol, for which all reagents were ground together (for 5 min) and heated for 120 min. No conversion of 1a to 2a took place at or below 25 °C. In the reaction run at 30 °C, 2a formed in 78% yield, in 80% yield for the reaction performed at 40 °C and 85% yield for the reaction heated to 50 °C. Fig. 1D delineates the effect of heating duration. Extending the heating period to longer than 30 min gave only a small increase in yield (79% after 30 min, vs. 83% after 60 min). Magnetic stirring of the reaction mixture during the heating step had no effect on yields.
As under our ball milling conditions,8h amongst a variety of oxidants tested, Ag2CO3 gave the highest yields and only AgI salts (Ag2CO3, Ag2O, AgF and AgOAc) resulted in any conversion to methylated products; other common oxidants, including Cu(OAc)2, Cu(TFA)2, MnO2 and Fe2O3 proved ineffective (see SI for more details). We obtained 2a in 81% spectroscopic yield upon lowering the Ag2CO3 loading to 1.2 equiv. Other Rh salts, including RhCl3·H2O, [RhCl(cod)]2 and [RhCl(C2H4)2]2 gave no conversion to 2a when used instead of [Cp*RhCl2]2 (see Table S4† for more details), although cyclometalated complexes were catalytically active (see below).
Conditions I–IV (Fig. 1E) summarise variations in how the reagents were ground and heated. Grinding the starting materials, catalyst and oxidant only (i.e., excluding the heating step) gave no conversion of 1a to 2a (Conditions I). Replacing the grinding step with gentle mixing of the starting materials, catalyst and oxidant (without grinding), and then heating the resulting mixture at 40 °C for 30 min resulted in no formation of 2a (Conditions II). Grinding the reagents, catalyst and oxidant separately for 5 min, then combining them and heating at 40 °C for 30 min also resulted in no conversion (Conditions III). Only combined grinding of the reagents, catalyst and oxidant for 5 min, and subsequent heating, gave 2a in good yield (Conditions IV, 80%).
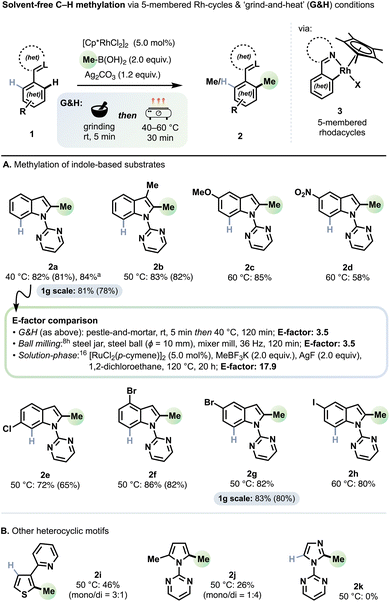
Scope of C–H methylation
Under the optimised conditions described above, a variety of decorated 1-pyrimidyl indoles (1a–h) underwent efficient C2−H methylation to give derivatives 2a–h with total reaction times of 35 min and optimal reaction temperatures in the 40–60 °C range (Scheme 1).Generally, for these conditions, we observed the evolution of ethane gas arising from competitive oxidative homocoupling of MeB(OH)2 (see ESI† for more information). Increasing reaction temperatures above 70 °C gave lower yields due to an acceleration of the homocoupling. A methyl group at the indole C3 position did not provide enough steric hindrance to inhibit the methylation (product 2b). Good tolerance towards electron-donating groups (e.g., –OMe, product 2c) and electron-withdrawing groups (e.g., –NO2 product 2d) on the indole's benzenoid ring was observed. The superior performance of electron-rich substrates is consistent with the outcome of reactions performed under ball milling conditions.8h Substrates with chloro- (product 2e), bromo- (2f–2g) and iodo- (2h) groups, each of which can serve as useful synthetic handles for subsequent derivatisation, gave generally very good yields.
C2–H Methylation of thiophene (2i) and pyrrole (2j) rings also proved viable, whereas the imidazole (2k) core did not participate in the reaction. Substrates with oxygen-based directing groups (1-acetylindole and N-methyl-acetanilide) gave no conversion.
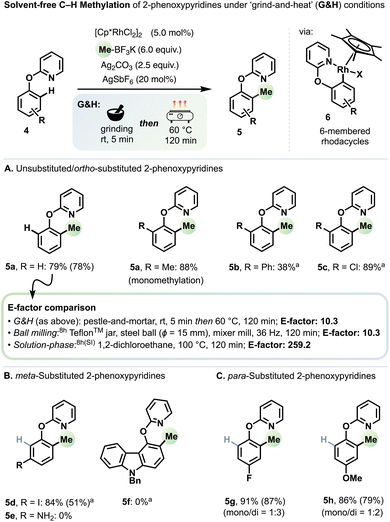
2-Phenoxypyridines (4, Scheme 2) occur in various biologically active molecules as well as luminescent materials,17 and can also serve as masked phenols18 or as pseudohalides in transition metal catalysis.19 In Rh-catalysed ortho-directed C–H methylation (to form products 5), 2-phenoxypyridines react via six-membered rhodacyclic intermediates (6).20 As under ball milling conditions,8h optimised yields for 5 relied on a switch of methyl source from MeB(OH)2 to MeBF3K,21 an increase of its loading to 3–6 equivalents and the inclusion of sub-stoichiometric AgSbF6. We attribute the need for these changes to the greater difficulty of transmetalation22 or reductive elimination from six-membered rhodacyclic intermediates.8h In all cases, increasing the reaction temperature to 60 °C and the duration to 120 min proved beneficial. Generally, methylated products were obtained in very good or excellent yields from unsubstituted (5a) as well ortho- (5a–c), meta- (5d), and para-substituted (5g,h) substrates (Scheme 2). Exceptions included the primary amine 5e and carbazole derivative 5f.
E-Factors and sustainability
A foremost sustainability credential of mechanochemistry is its avoidance of bulk solvents as reaction media, which greatly reduces waste and helps side-step key environmental and human health hazards.1a Practically all advances in mechanochemical C–H functionalisation methodology rely on automated ball milling.10bSchemes 1 and 2 include a comparison of E-factors (the mass ratio of waste/product)23 for the syntheses of representative products 2a and 5a, respectively, each under three sets of conditions: ‘grind-and-heat’ (G&H), automated ball milling, and in solution.
For both 2a and 5a, our ‘grind-and-heat’ protocol and automated ball milling gave identical E-factors: 3.5 for 2a and 10.3 for 5a. We calculated the highest yielding solution-based system reported for generating 2avia C–H methylation to have an E-factor of 17.9.16 Thus, both solvent-free approaches reduce the E-factor more than fivefold for 2a. For 5a, the highest yielding solution-based system has an E-factor of 259.2.8h Therefore, in this case, both solvent-free approaches offer a twenty-five-fold improvement. It is additionally noteworthy that for both 2a and 5a (and their analogues) the previous highest yielding reported solution-based systems used 1,2-DCE as solvent, a potent carcinogen that has been banned for all practical purposes in some jurisdictions.12a
By these metrics, both ‘grind-and-heat’ and automated ball milling significantly outperform their solution-based counterparts in sustainability terms. However, we stress that, in contrast to automated ball milling, ‘grind-and-heat’ conditions require no specialised equipment. Thus, the sustainability benefits of solvent-free, mechanochemical catalytic C–H functionalisation may be much more widely and cheaply accessible than might be commonly assumed. This is an important and overlooked aspect of solvent-free C–H functionalisation methodology and we anticipate its extension to many other reaction types.
Late-stage functionalization (LSF)
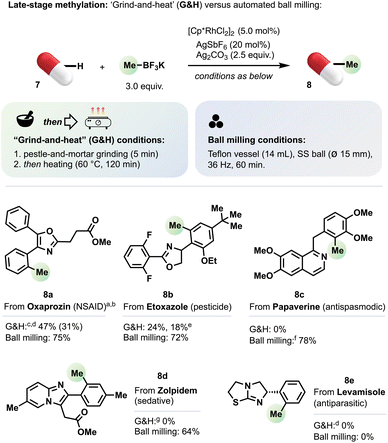
Research into strategies for late-stage functionalization (LSF) of bioactive compounds, natural products and advanced intermediates is presently a ‘hot topic’.10c,13b–d,24 LSF can streamline access to derivatives of valuable molecules by reducing significantly the need for their de novo synthesis. Thus, LSF can accelerate the exploration of chemical space and, for example, expedite drug development.13d,24,25 We reported previously that several biologically active molecules (or their close relatives) underwent efficient late stage C–H methylation under conditions based on automated ball milling.8h We sought to compare this with the ‘grind-and-heat’ approach.Under ‘grind-and-heat’ conditions, methylated Oxaprozin (8a) and Etoxazole (8b) derivatives formed in 47% and 24% yield, respectively (Scheme 3), somewhat lower than for automated ball milling.8h Although the ‘grind-and-heat’ regime was not effective for Papaverine (8c), Zolpidem derivative 8d or Levamisole (8e), the amounts of 8a and 8b we obtained would be generally considered useful for physiochemical and biological testing.
Interest in the mechanochemical synthesis and reactivity of coordination and organometallic complexes has also experienced recent growth.3a–c,10d,j,26 We previously reported that rhodacyclic complexes of the types 3 or 6 can be generated efficiently under ball milling conditions and that they are competent precatalysts for oxidative C–H methylation.8h Under ‘grind-and-heat’ conditions, rhodacycle 3a (X = Cl) formed in 28% yield from 1a and [Cp*RhCl2]2 (see Table S1†). Complex 3a catalysed the C–H methylation of indole derivative 1a to give 2a in 84% yield (Scheme 1) and the C–H methylation of 7b to give 8b in 18% yield (Scheme 3). Hernandez and co-workers recently described the formation and role of rhodacycles in mechanochemical C–H functionalisation.26b,c
Scanning electron microscopy (SEM)
Scanning Electron Microscopy (SEM) allowed a comparison of morphological changes in the reaction mixtures corresponding to different steps of the ‘grind-and-heat’ C–H methylation of 1a (Fig. 2).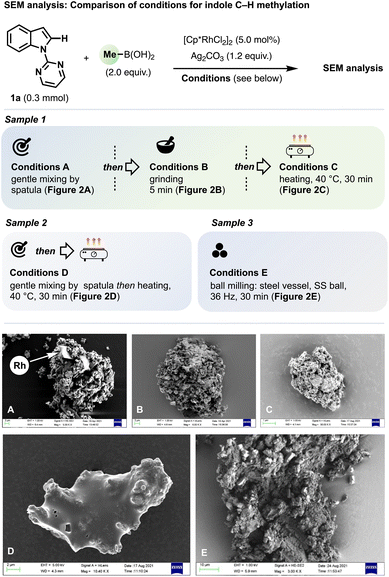 | ||
| Fig. 2 Comparison of reaction mixtures for the Rh-catalyzed C–H methylation of 1a using different conditions using scanning electron microscopy (SEM). | ||
For Sample 1, all the reagents were first mixed gently (Conditions A), then subjected to pestle-and-mortar grinding (Conditions B) and, finally, heated (Conditions C). An SEM image was recorded after each step (Fig. 2A–C). After gentle mixing only (Conditions A), SEM imaging revealed fine particles aggregated around larger crystalline fragments approximately 10 μm in diameter (Fig. 2A). Elemental analysis using Energy Dispersive X-ray Spectroscopy (EDS) confirmed only the larger fragments contained Rh, and these were there therefore assigned as crystals of [Cp*RhCl2]2. After 5 min of manual pestle-and-mortar grinding (Conditions B), SEM revealed the sample to be significantly more homogenised and compacted (Fig. 2B) and X-ray elemental analysis revealed uniform distribution of Rh-, Cl- and Ag-containing particles throughout the sample. After heating (Conditions C), Sample 1 retained its homogeneity but acquired a porous appearance (Fig. 2C), which we ascribe to the background oxidative ethane formation discussed above.
For Sample 2, all reagents were gently mixed by spatula only and heated directly (Conditions D), by-passing the pestle-and-mortar grinding stage (this corresponds to Conditions II in Fig. 1E). In this case, the SEM image (Fig. 2D) showed smooth, coated particles for which X-Ray elemental analysis indicated very even nitrogen distribution across the surface. We assign this to a coating of 1a, which we measured to have a melting temperature range starting at 35 °C, as determined by Differential Scanning Calorimetry (DSC). The smooth appearance is evidence that the background oxidative ethane formation did not occur. As the pestle-and-mortar grinding step proved necessary for successful C–H methylation without a ball mill (Fig. 1E), the present result indicates substrate melting alone does not ensure successful C–H methylation under ‘grind-and-heat’ conditions.
For Sample 3, all reagents were loaded into a 14 mL stainless steel milling vessel with a ball bearing (ϕ = 15 mm) and subjected to automated ball milling at 36 Hz for 30 min (Conditions E). This produced the most homogenized and compressed sample of the set (Fig. 2E). The absence of the porosity seen in Sample 3 is ascribed to the continuous mechanical action of the mill even after complete consumption of MeB(OH)2.
To give 2a in 82% yield (Scheme 1) using a 5.0 mol% loading of [Cp*RhCl2]2, each Rh centre must, on average, interact with 8.2 equivalents of substrate. The results above suggest that sufficient sample homogenisation by pestle-and-mortar grinding (Fig. 2B) combined with substrate mobility (e.g., from melting, Fig. 2D) can enable this for some, even all-solid solvent-free reaction mixtures. For such reactions, therefore, the more thorough mixing offered by ball milling is not needed.
By contrast, ‘grind-and-heat’ conditions did not provide 8d from Zolpidem analogue 7d. For the corresponding Sample 4 (Fig. 3F), SEM analysis revealed homogenised but sharp-edged, brittle-looking particles lacking the evidence of ethane evolution or substrate melting observed for 1a (cf.Fig. 2C and D). However, 8d formed in 64% yield using ball milling (Scheme 3). The corresponding SEM image (Sample 5, Fig. 3G) and elemental X-Ray analysis showed a very homogeneous distribution of all elements in a mixture coated onto flakes of Teflon originating from the milling vessel. The melting point of 7d (120 °C) is significantly above the reaction temperature of both the ‘grind-and-heat’ conditions we used (40 °C) and the temperatures the Teflon vessels (which rose to 50–90 °C during milling). These results suggest continuous reagent mixing is achieved through the mechanical action of ball bearings even for all-solid reaction mixtures under milling conditions, whereas ‘grind-and-heat’ C–H methylation requires sufficient sample homogenisation first but also depends on additional reagent mobility, e.g., afforded through a substrate's melting window starting below the reaction temperature. We confirmed using DSC that successful C–H methylation under ‘grind-and-heat’ conditions requires either the substrate or reaction mixtures (after grinding) to have a melting temperature range beginning below the reaction temperature or, otherwise, a glass transition temperature, Tg, lower than the reaction temperature.27 We note that these properties and associated reaction outcomes may also depend on co-crystal formation during the grinding step; Hernández and co-workers recently described the formation of [Cp*RhCl2]2/substrate co-crystals prior to C–H rhodation under ball milling conditions.26b,c
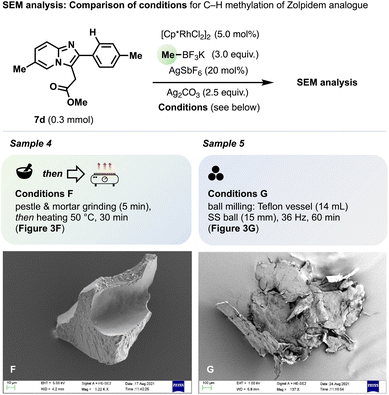 | ||
| Fig. 3 Comparison by scanning electron microscopy (SEM) of ‘grind-and-heat’ versus ball milling conditions for the attempted C–H methylation of 7d. | ||
Conclusions
We have demonstrated that selective and efficient Rh-catalysed C–H methylation is viable under solvent-free and ball mill-free conditions – even for some all-solid reaction mixtures – using a simple ‘grind-and-heat’ protocol. This approach boasts broad functional group tolerance and can be applied to some more complex, biologically active molecules. Whilst it is not a full replacement for automated ball milling, we suspect a ‘grind-and-heat’ approach is likely to work for a large range of mechanochemical reactions for which only ball milling conditions have been reported previously. The ‘grind-and-heat’ approach offers essentially identical sustainability benefits to automated ball milling, but it is operationally simpler and cheaper, as it requires no specialised equipment. Thus, it may become a route through which greener, solvent-free reactions become more often considered as increasing emphasis is placed on sustainable synthetic method development.28Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Swedish Research Council (Vetenskapsrådet, dnr 2019-05424), Carl Tryggers Stiftelse (CTS 21:1495) and the Erasmus Programme for funding. We are grateful to Dr Jannica Heinrichs for help with SEM, as well as Dr Christine Dyrager and Dr Johanna Larsson for manuscript proof-reading. This study made use of the NMR Uppsala infrastructure, which is funded by the Department of Chemistry – BMC and the Disciplinary Domain of Medicine and Pharmacy.References
- (a) K. J. Ardila-Fierro and J. G. Hernández, ChemSusChem, 2021, 14, 2145–2162 CrossRef CAS PubMed; (b) T. Friščić, C. Mottillo and H. M. Titi, Angew. Chem., Int. Ed., 2020, 59, 1018–1029 CrossRef PubMed; (c) J.-L. Do and T. Friščić, ACS Cent. Sci., 2017, 3, 13–19 CrossRef CAS PubMed; (d) J. L. Howard, Q. Cao and D. L. Browne, Chem. Sci., 2018, 9, 3080–3094 RSC; (e) J. Andersen and J. Mack, Green Chem., 2018, 20, 1435–1443 RSC; (f) C. Xu, S. De, A. M. Balu, M. Ojeda and R. Luque, Chem. Commun., 2015, 51, 6698–6713 RSC; (g) S. Hwang, S. Grätz and L. Borchardt, Chem. Commun., 2022, 581661–1671 Search PubMed; (h) M. Leonardi, M. Villacampa and J. C. Menéndez, Chem. Sci., 2018, 9, 2042–2064 RSC; (i) J. A. Leitch and D. L. Browne, Chem. – Eur. J., 2021, 27, 9721–9726 CrossRef CAS PubMed; (j) K. Kubota and H. Ito, Trends Chem., 2020, 2, 1066–1081 CrossRef CAS; (k) R. R. A. Bolt, J. A. Leitch, A. C. Jones, W. I. Nicholson and D. L. Browne, Chem. Soc. Rev., 2022, 514243–4260 Search PubMed; (l) L. Gonnet, C. B. Lennox, J.-L. Do, I. Malvestiti, S. G. Koenig, K. Nagapudi and T. Friščić, Angew. Chem., Int. Ed., 2022, e202115030 CAS; (m) R. Laskar, T. Pal, T. Bhattacharya, S. Maiti, M. Akita and D. Maiti, Green Chem., 2022, 24, 2296–2320 RSC.
- (a) C. J. Clarke, W.-C. Tu, O. Levers, A. Bröhl and J. P. Hallett, Chem. Rev., 2018, 118, 747–800 CrossRef CAS PubMed; (b) C. Jimenez-Gonzalez, C. S. Ponder, Q. B. Broxterman and J. B. Manley, Org. Process Res. Dev., 2011, 15, 912–917 CrossRef CAS.
- (a) V. S. Pfennig, R. C. Villella, J. Nikodemus and C. Bolm, Angew. Chem., Int. Ed., 2022, 61, e202116514 CrossRef CAS PubMed; (b) R. Takahashi, A. Hu, P. Gao, Y. Gao, Y. Pang, T. Seo, J. Jiang, S. Maeda, H. Takaya, K. Kubota and H. Ito, Nat. Commun., 2021, 12, 6691 CrossRef CAS PubMed; (c) K. Kubota, R. Takahashi and H. Ito, Chem. Sci., 2019, 10, 5837–5842 RSC; (d) Y. Pang, T. Ishiyama, K. Kubota and H. Ito, Chem. – Eur. J., 2019, 25, 4654–4659 CrossRef CAS PubMed.
- (a) J. G. Hernández and C. Bolm, J. Org. Chem., 2017, 82, 4007–4019 CrossRef PubMed; (b) M. E. McFadden and M. J. Robb, J. Am. Chem. Soc., 2021, 143, 7925–7929 CrossRef CAS PubMed; (c) E. Boldyreva, Chem. Soc. Rev., 2013, 42, 7719–7738 RSC; (d) S. Lukin, L. S. Germann, T. Friščić and I. Halasz, Acc. Chem. Res., 2022, 55, 1262–1277 CrossRef CAS PubMed.
- (a) E. Boldyreva, Nat. Chem., 2022, 14, 10–12 CrossRef CAS PubMed; (b) S. Lukin, T. Stolar, M. Tireli, M. V. Blanco, D. Babic, T. Friscic, K. Uzarevic and I. Halasz, Chem. – Eur. J., 2017, 23, 13941–13949 CrossRef CAS PubMed; (c) J. M. Andersen and J. Mack, Chem. Sci., 2017, 8, 5447–5453 RSC; (d) X. Ma, W. Yuan, S. E. J. Bell and S. L. James, Chem. Commun., 2014, 50, 1585–1587 RSC; (e) G. I. Lampronti, A. A. L. Michalchuk, P. P. Mazzeo, A. M. Belenguer, J. K. M. Sanders, A. Bacchi and F. Emmerling, Nat. Commun., 2021, 12, 6134 CrossRef CAS PubMed; (f) A. A. L. Michalchuk and F. Emmerling, Angew. Chem., Int. Ed., 2022, 61, e202117270 CrossRef CAS PubMed; (g) S. Lukin, L. S. Germann, T. Friščić and I. Halasz, Acc. Chem. Res., 2022, 55, 1262–1277 CrossRef CAS PubMed; (h) C. Leroy, S. Mittelette, G. Félix, N. Fabregue, J. Špačková, P. Gaveau, T.-X. Métro and D. Laurencin, Chem. Sci., 2022 10.1039/D2SC01496C.
- F. Gomollón-Bel, Chem. Int., 2019, 41, 12–17 Search PubMed.
- (a) T. Dalton, T. Faber and F. Glorius, ACS Cent. Sci., 2021, 7, 245–261 CrossRef CAS PubMed; (b) T. Rogge, N. Kaplaneris, N. Chatani, J. Kim, S. Chang, B. Punji, L. L. Schafer, D. G. Musaev, J. Wencel-Delord, C. A. Roberts, R. Sarpong, Z. E. Wilson, M. A. Brimble, M. J. Johansson and L. Ackermann, Nat. Rev. Methods Primers, 2021, 1, 43 CrossRef CAS; (c) B. Liu, A. M. Romine, C. Z. Rubel, K. M. Engle and B.-F. Shi, Chem. Rev., 2021, 121, 14957–15074 CrossRef CAS PubMed; (d) S. Vásquez-Céspedes, X. Wang and F. Glorius, ACS Catal., 2018, 8, 242–257 CrossRef.
- For recent reviews see: (a) D. Aynetdinova, M. C. Callens, H. B. Hicks, C. Y. X. Poh, B. D. A. Shennan, A. M. Boyd, Z. H. Lim, J. A. Leitch and D. J. Dixon, Chem. Soc. Rev., 2021, 50, 5517–5563 RSC; (b) N.-D. Mao, Y. Ye, X.-T. Zhuo, X.-Y. Ye and T. Xie, Tetrahedron, 2021, 96, 132402 CrossRef CAS. For a selection of recent studies see: (c) E. Weis, M. A. Hayes, M. J. Johansson and B. Martín-Matute, iScience, 2021, 24, 102467 CrossRef CAS PubMed; (d) L. F. T. Novaes, J. S. K. Ho, K. Mao, K. Liu, M. Tanwar, M. Neurock, E. Villemure, J. A. Terrett and S. Lin, J. Am. Chem. Soc., 2022, 144(3), 1187–1197 CrossRef CAS PubMed; (e) A. Vasilopoulos, S. W. Krska and S. S. Stahl, Science, 2021, 372, 398–403 CrossRef CAS PubMed; (f) K. Feng, R. E. Quevedo, J. T. Kohrt, M. S. Oderinde, U. Reilly and M. C. White, Nature, 2020, 580, 621–627 CrossRef CAS PubMed; (g) S. D. Friis, M. J. Johansson and L. Ackermann, Nat. Chem., 2020, 12, 511–519 CrossRef CAS PubMed; (h) S. Ni, M. Hribersek, S. K. Baddigam, F. J. L. Ingner, A. Orthaber, P. J. Gates and L. T. Pilarski, Angew. Chem., Int. Ed., 2021, 60, 6660–6666 CrossRef CAS PubMed; (i) Q. Gao, Y. Shang, F. Song, J. Ye, Z.-S. Liu, L. Li, H.-G. Cheng and Q. Zhou, J. Am. Chem. Soc., 2019, 141, 15986–15993 CrossRef CAS PubMed; (j) R. Kumar, R. Sharma, R. Kumar and U. Sharma, Org. Lett., 2020, 22, 305–309 CrossRef CAS PubMed; (k) F. Serpier, F. Pan, W. S. Ham, J. Jacq, C. Genicot and T. Ritter, Angew. Chem., Int. Ed., 2018, 57, 10697–10701 CrossRef CAS PubMed; (l) J. Jin and D. W. C. MacMillan, Nature, 2015, 525, 87–90 CrossRef CAS PubMed; (m) A. Hogg, M. Wheatley, P. Domingo-Legarda, A. Carral-Menoyo, N. Cottam and I. Larrosa, JACS Au, 2022, 2, 2529–2538 CrossRef CAS PubMed.
- (a) H. Schönherr and T. Cernak, Angew. Chem., Int. Ed., 2013, 52, 12256–12267 CrossRef PubMed; (b) S. Sun and J. Fu, Bioorg. Med. Chem. Lett., 2018, 28, 3283–3289 CrossRef CAS PubMed; (c) E. J. Barreiro, A. E. Kümmerle and C. A. M. Fraga, Chem. Rev., 2011, 111, 5215–5246 CrossRef CAS PubMed.
- (a) J. G. Hernández, Chem. – Eur. J., 2017, 23, 17157–17165 CrossRef PubMed; (b) S. Zhao, Y. Li, C. Liu and Y. Zhao, Tetrahedron Lett., 2018, 59, 317–324 CrossRef CAS; (c) X. J. Yang, C. Y. Wu, W. K. Su and J. B. Yu, Eur. J. Org. Chem., 2022, e202101440 CrossRef CAS; (d) H. Cheng, J. G. Hernández and C. Bolm, Adv. Synth. Catal., 2018, 360, 1800–1804 CrossRef CAS; (e) S. R. Yetra, Z. Shen, H. Wang and L. Ackermann, Beilstein J. Org. Chem., 2018, 14, 1546–1553 CrossRef CAS PubMed; (f) G. N. Hermann and C. Bolm, ACS Catal., 2017, 7, 4592–4596 CrossRef CAS; (g) G. N. Hermann, P. Becker and C. Bolm, Angew. Chem., Int. Ed., 2015, 54, 7414–7417 CrossRef CAS PubMed; (h) G. N. Hermann, M. T. Unruh, S.-H. Jung, M. Krings and C. Bolm, Angew. Chem., Int. Ed., 2018, 57, 10723–10727 CrossRef CAS PubMed; (i) G. N. Hermann, C. L. Jung and C. Bolm, Green Chem., 2017, 19, 2520–2523 RSC; (j) J. G. Hernández and C. Bolm, Chem. Commun., 2015, 51, 12582–12584 RSC; (k) G. N. Hermann, P. Becker and C. Bolm, Angew. Chem., Int. Ed., 2016, 55, 3781–3784 CrossRef CAS PubMed; (l) H. Cheng, J. G. Hernández and C. Bolm, Org. Lett., 2017, 19, 6284–6287 CrossRef CAS PubMed; (m) M. Juribašić, K. Užarević, D. Gracin and M. Ćurić, Chem. Commun., 2014, 50, 10287–10290 RSC; (n) S.-J. Lou, Y.-J. Mao, D.-Q. Xu, J.-Q. He, Q. Chen and Z.-Y. Xu, ACS Catal., 2016, 6, 3890–3894 CrossRef CAS; (o) K.-Y. Jia, J.-B. Yu, Z.-J. Jiang and W.-K. Su, J. Org. Chem., 2016, 81, 6049–6055 CrossRef CAS PubMed; (p) T. Gensch, M. N. Hopkinson, F. Glorius and J. Wencel-Delord, Chem. Soc. Rev., 2016, 45, 2900–2936 RSC; (q) X. Lu, Y. Bai, J. Qin, N. Wang, Y. Wu and F. Zhong, ACS Sustainable Chem. Eng., 2021, 9, 1684–1691 CrossRef CAS; (r) P. Shi, Y. Tu, D. Kong, P. Wu, D. Ma and C. Bolm, Angew. Chem., Int. Ed., 2022, 61, e202204874 CrossRef CAS PubMed; (s) S. Min, B. Park, J. Nedsaengtip and S. Hyeok Hong, Adv. Synth. Catal., 2022, 364, 1975–1981 CrossRef CAS.
- F. J. L. Ingner, Z. X. Giustra, S. Novosedlik, A. Orthaber, P. J. Gates, C. Dyrager and L. T. Pilarski, Green Chem., 2020, 22, 5648–5655 RSC.
- (a) J. Sherwood, Angew. Chem., Int. Ed., 2018, 57, 14286–14290 CrossRef CAS PubMed; (b) A. Jordan, C. G. J. Hall, L. R. Thorp and H. F. Sneddon, Chem. Rev., 2022, 122, 6749–6794 CrossRef CAS PubMed.
- (a) L. Zhang and T. Ritter, J. Am. Chem. Soc., 2022, 144, 2399–2414 CrossRef CAS PubMed; (b) B. Hong, T. Luo and X. Lei, ACS Cent. Sci., 2020, 6, 622–635 CrossRef CAS PubMed; (c) J. Börgel and T. Ritter, Chem, 2020, 6, 1877–1887 CrossRef; (d) M. Moir, J. J. Danon, T. A. Reekie and M. Kassiou, Expert Opin. Drug Discovery, 2019, 14, 1137–1149 CrossRef CAS PubMed; (e) R. Jana, H. M. Begam and E. Dinda, Chem. Commun., 2021, 57, 10842–10866 RSC.
- For selected examples see: (a) N. Zappimbulso, M. A. M. Capozzi, A. Porcheddu, G. M. Farinola and A. Punzi, ChemSusChem, 2021, 14, 1363–1369 CrossRef CAS PubMed; (b) T. W. von Zuben, G. Cariello Silva and A. G. Salles, Green Chem., 2021, 23, 6361–6365 RSC; (c) B. S. Gore, J.-H. Lin and J.-J. Wang, Green Chem., 2021, 23, 4144–4149 RSC; (d) F. Zhao, Q. Tan, D. Wang and G.-J. Deng, Green Chem., 2020, 22, 427–432 RSC; (e) C. Teja and F. R. Nawaz Khan, Org. Lett., 2020, 22, 1726–1730 CrossRef CAS PubMed; (f) E. de Oliveira Lima Filho and I. Malvestiti, ACS Omega, 2020, 5, 33329–33339 CrossRef CAS PubMed; (g) A. Punzi, M. A. M. Capozzi, S. Di Noja, R. Ragni, N. Zappimbulso and G. M. Farinola, J. Org. Chem., 2018, 83, 9312–9321 CrossRef CAS PubMed; (h) S. Firoozi, M. Hosseini-Sarvari and M. Koohgard, Green Chem., 2018, 20, 5540–5549 RSC; (i) F. G. Cirujano, M. Stalpaert and D. E. D. Vos, Green Chem., 2018, 20, 2481–2485 RSC; (j) R. A. Jagtap, V. Soni and B. Punji, ChemSusChem, 2017, 10, 2242–2248 CrossRef CAS PubMed; (k) S. Yaragorla, G. Singh and R. Dada, Tetrahedron Lett., 2015, 56, 5924–5929 CrossRef CAS; (l) S. Shome and S. P. Singh, Eur. J. Org. Chem., 2015, 6025–6032 CrossRef CAS; (m) S. Bensaid and H. Doucet, ChemSusChem, 2012, 5, 1559–1567 CrossRef CAS PubMed; (n) T. Wang, R.-d. Ma, L. Liu and Z.-p. Zhan, Green Chem., 2010, 12, 1576–1579 RSC; (o) M. Ghobrial, K. Harhammer, M. D. Mihovilovic and M. Schnürch, Chem. Commun., 2010, 46, 8836–8838 RSC; (p) R. B. Bedford, C. J. Mitchell and R. L. Webster, Chem. Commun., 2010, 46, 3095–3097 RSC; (q) R. B. Bedford, J. U. Engelhart, M. F. Haddow, C. J. Mitchell and R. L. Webster, Dalton Trans., 2010, 39, 10464–10472 RSC.
- (a) L. Liu, J. Lin, M. Pang, H. Jin, X. Yu and S. Wang, Org. Lett., 2022, 24, 1146–1151 CrossRef CAS PubMed; (b) G. Pisanò and C. S. J. Cazin, ACS Sustainable Chem. Eng., 2021, 9, 9625–9631 CrossRef.
- For a related solution-based C2–H methylation of indoles see: M. D. L. Tonin, D. Zell, V. Müller and L. Ackermann, Synthesis, 2017, 49, 127–134 CAS.
- (a) W. Zhang, J. Ma, G.-J. Liu, X.-Y. Liu, J. Fan and L.-S. Liao, J. Mater. Chem. C, 2017, 5, 9496–9503 RSC; (b) G. E. Norby, C.-D. Park, B. O'Brien, G. Li, L. Huang and J. Li, Org. Electron., 2016, 37, 163–168 CrossRef CAS; (c) E. Turner, N. Bakken and J. Li, Inorg. Chem., 2013, 52, 7344–7351 CrossRef CAS PubMed.
- (a) Q. Gou, X. Tan, M. Zhang, M. Ran, T. Yuan, S. He, L. Zhou, T. Cao and F. Luo, Org. Lett., 2020, 22, 1966–1971 CrossRef CAS PubMed; (b) L. Zhang, Y. Wang, Y. Shi, Y. Wu, J. Lan, W. Ma and J. You, ACS Catal., 2019, 9, 5358–5364 CrossRef CAS; (c) C. Chen, Y. Pan, H. Zhao, X. Xu, Z. Luo, L. Cao, S. Xi, H. Li and L. Xu, Org. Lett., 2018, 20, 6799–6803 CrossRef CAS PubMed; (d) D.-W. Yin and G. Liu, J. Org. Chem., 2018, 83, 3987–4001 CrossRef CAS PubMed; (e) Y. Wu, W. Li, L. Jiang, L. Zhang, J. Lan and J. You, Chem. Sci., 2018, 9, 6878–6882 RSC; (f) S. Yang, B. Feng and Y. Yang, J. Org. Chem., 2017, 82, 12430–12438 CrossRef CAS PubMed; (g) G. Tan, Q. You and J. You, ACS Catal., 2018, 8, 8709–8714 CrossRef CAS; (h) D. C. McAteer, E. Javed, L. Huo and S. Huo, Org. Lett., 2017, 19, 1606–1609 CrossRef CAS PubMed.
- (a) X. Zeng, Y. Zhang, Z. Liu, S. Geng, Y. He and Z. Feng, Org. Lett., 2020, 22, 2950–2955 CrossRef CAS PubMed; (b) Y.-Y. Kong and Z.-X. Wang, Adv. Synth. Catal., 2019, 361, 5440–5448 CrossRef CAS; (c) J. Li and Z.-X. Wang, Chem. Commun., 2018, 54, 2138–2141 RSC.
- For a recent discussion of metalacyclic complexes in C–H functionalisation see: J. M. Zakis, T. Smejkal and J. Wencel-Delord, Chem. Commun., 2022, 58, 483–490 RSC.
- Y. Chen, Chem. – Eur. J., 2019, 25, 3405–3439 CrossRef CAS PubMed.
- K. Osakada and Y. Nishihara, Dalton Trans., 2022, 51, 777–796 RSC.
- (a) R. A. Sheldon, ACS Sustainable Chem. Eng., 2018, 6, 32–48 CrossRef CAS; (b) R. A. Sheldon, Green Chem., 2023, 25, 1704–1728 RSC.
- (a) J. Wencel-Delord and F. Glorius, Nat. Chem., 2013, 5, 369–375 CrossRef CAS PubMed; (b) X. J. Yang, C. Y. Wu, W. K. Su and J. B. Yu, Eur. J. Org. Chem., 2022, e202101440 CrossRef CAS.
- T. Cernak, K. D. Dykstra, S. Tyagarajan, P. Vachal and S. W. Krska, Chem. Soc. Rev., 2016, 45, 546–576 RSC.
- (a) Y. Liu, F. Z. Liu and K. Yan, Angew. Chem., Int. Ed., 2022, 61, e202116980 CrossRef CAS PubMed; (b) K. J. Ardila-Fierro, M. Rubcic and J. G. Hernandez, Chem. – Eur. J., 2022, 28, e202200737 CrossRef CAS PubMed; (c) J. G. Hernández, K. J. Ardila-Fierro, S. Gómez, T. Stolar, M. Rubčić, E. Topić, C. Z. Hadad and A. Restrepo, Chem. – Eur. J., 2023, e202301290 Search PubMed.
- The latter was observed only with Etoxazole (7b).
- (a) J. B. Zimmerman, P. T. Anastas, H. C. Erythropel and W. Leitner, Science, 2020, 367, 397–400 CrossRef CAS PubMed; (b) J. L. Tyler, F. Katzenburg and F. Glorius, Chem. Sci., 2023, 14, 7408–7410 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3gc02411c |
| This journal is © The Royal Society of Chemistry 2023 |