Open Access Article
Open Access ArticleDesigning an enzyme assembly line for green cascade processes using bio-orthogonal chemistry†
Li
Qiao‡
a,
Zhiyuan
Luo‡
ab,
Ru
Wang‡
c,
Xiaolin
Pei
 a,
Shujiao
Wu
c,
Haomin
Chen
a,
Tian
Xie
*bc,
Roger A.
Sheldon
a,
Shujiao
Wu
c,
Haomin
Chen
a,
Tian
Xie
*bc,
Roger A.
Sheldon
 *de and
Anming
Wang
*de and
Anming
Wang
 *a
*a
aKey Laboratory of Organosilicon Chemistry and Material Technology, Ministry of Education; College of Materials Chemistry and Chemical Engineering, Hangzhou Normal University, Hangzhou 311121, Zhejiang, China. E-mail: waming@hznu.edu.cn
bInnovation Research Institute of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China
cKey Laboratory of Elemene Class Anti-Cancer Medicines, Engineering Laboratory of Development of Chinese Medicines, and Collaborative Innovation Center of Chinese Medicines of Zhejiang Province, College of Pharmacy, Hangzhou Normal University, Hangzhou, Zhejiang 311121, China. E-mail: xbs@hznu.edu.cn
dMolecular Sciences Institute, School of Chemistry, University of the Witwatersrand, PO Wits. 2050, Johannesburg, South Africa. E-mail: roger.sheldon@wits.ac.za
eDepartment of Biotechnology, Section BOC, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands. E-mail: r.a.sheldon@tudelft.nl
First published on 16th August 2023
Abstract
Two non-canonical amino acids (ncAAs) with bio-orthogonal reactive groups, namely, p-azido-L-phenylalanine (p-AzF) and p-propargyloxy-L-phenylalanine (p-PaF), were genetically inserted into an aldo-keto reductase (AKR) and an alcohol dehydrogenase (ADH), respectively, at two preselected sites for each enzyme. The variants were expressed in the genome recoded bacterium Escherichia coli C321.ΔA. Supernatants of the individual cell lysates were subsequently mixed to produce orderly combi-crosslinked enzymes (O-CLEs) of AKR and ADH by co-polymerization of the two variants through their reactive bio-orthogonal groups. The site-specific cross-linked enzymes (S-CLEs) and cross-linked enzyme aggregates (CLEAs) were produced using dibenzocycloocta-4a,6a-diene-5,11-diyne (DBA) and glutaraldehyde as the crosslinking agent, respectively. The catalytic efficiencies of the O-CLEs, S-CLEs and combi-CLEAs were determined using the water soluble dihydro-4, 4-dimethyl-2, 3-furandione as a surrogate substrate in aqueous solution at 37 °C. The O-CLEs exhibited the highest catalytic efficiency (Kcat/KM = 11.36 S−1 mM−1) that was 4.24 and 22.27 times that of S-CLEs and combi-CLEAs, respectively. In the asymmetric cascade synthesis of (R)-1-(2-chlorophenyl) ethanol the product yield after 14 h using the O-CLEs, S-CLEs and the combi-CLEAs was 93%, 55% and 16%, respectively. Moreover, high activities and selectivity (ee > 99.99%) were maintained at high substrate concentrations in prolonged operation.
Introduction
The last two decades have witnessed continued growth of the use of biocatalysis for the green and sustainable manufacture of pharmaceuticals, fine chemicals and food.1–5 Enzymes are derived from readily available, inexpensive renewable resources and are biocompatible and biodegradable. Enzymatic reactions are performed at near ambient temperature and atmospheric pressure, in water as solvent. They avoid the need for functional-group activation, protection and de-protection steps, generate less waste and are, therefore, more cost-effective than conventional organic syntheses. Furthermore, they generally afford high chemo-, regio- and stereo-selectivities and the near-perfect enantioselectivities observed with highly engineered enzymes are unparalleled.In stark contrast with many chemo-catalytic reactions, enzymatic reactions involve roughly the same temperature and pressure. This facilitates the telescoping of multiple enzymatic steps into one-pot enzymatic cascades6 with additional economic and environmental benefits: fewer unit operations, less solvent and reactor volume and shorter cycle times that lead to higher volumetric and space time yields and less waste (lower E factors).7 Moreover, it circumvents the need for wasteful and costly separation and purification of intermediates and coupling of enzymatic steps drives equilibria towards product formation, thus enhancing overall yields.
In vivo a broad selection of complex molecules are synthesised, from simple precursors, in highly efficient metabolic pathways in microbial cell factories.8,9 This occurs in multi-enzyme cascade processes in which alignment of the individual enzymes in close proximity to each other enables highly efficient transfer of intermediates from the active site of one enzyme to that of the next enzyme, successfully competing with diffusion into the bulk solution. This process is referred to as substrate channeling.8,10 Much effort has been devoted in recent years, therefore, to the development of in vitro enzymatic cascade processes8,9,11–13 that emulate this magnificent orchestration of multiple enzymes in metabolic pathways, including the substrate channeling effect.8,10,14,15 Substrate channeling has been studied by biochemists for several decades16,17 but is now emerging as a novel approach to engineering highly efficient enzymatic cascade processes in vitro. Various strategies have been used to emulate substrate channeling: construction of fusion proteins,18,19 co-immobilisation20,21e.g. as combi-CLEAs,22 compartmentalisation,23,24 multidimensional scaffolding25 and the use of designer amphiphiles that accommodate both the enzyme and the substrate in close proximity in nanomicelles in water.26,27
However, spatial proximity is not the only feature that influences the overall reaction rates of multi-enzyme systems. The catalytic efficiency of metabolic pathways is highly dependent on precise control of the position and orientation of the individual enzymes in order to facilitate substrate transport between them.28,29 For example, substrate channeling in the bifunctional aldehyde-alcohol dehydrogenase enzyme, AdhE, the enzyme responsible for in vivo conversion of acetyl-CoA to ethanol via acetaldehyde using NADH as cofactor involves a helical arrangement of the two enzymes.30,31 Similarly, multienzyme assemblies for in vitro biocatalysis,32–34 involve orientation and alignment of reactants within confined spaces, usually with the active sites facing each other.
The simplest examples of enzymatic cascades involve combinations of redox enzymes with co-factor recycling involving a second enzyme and a co-substrate, e.g. glucose dehydrogenase (GDH) with glucose35,36 or alcohol dehydrogenase (ADH) with isopropanol.36 Indeed, enzymatic reduction of prochiral ketones with highly engineered enzymes is the method of choice for the synthesis of enantiopure chiral alcohols as key building blocks for the industrial synthesis of a broad spectrum of pharmaceuticals.37–39
Cost-effective cofactor recycling is essential for commercial viability, especially for commodity chemicals40 and it can be achieved by carrier-free co-immobilisation of enzymes which can also increase the efficiency of cascade reactions through a crowding effect. Unfortunately, carrier-free immobilisation of enzymes can involve substantial activity losses owing to the random formation of covalent bonds between amine functionalities on the enzyme surface, or even in the active site, and the cross-linker.
A novel and effective method for avoiding this unwanted random formation of covalent bonds with reactive functional groups in the enzyme is through site-specific incorporation of suitable non-canonical amino acids, ncAAs, to enable controlled and precise crosslinking at specific, preselected sites. The first example of site-specific incorporation of ncAAs at pre-specified locations in proteins, thus enabling their subsequent covalent attachment to carriers through a single click reaction of the ncAA with surface functionalities on the carrier, was reported in 2013 by Bundy and co-workers.41 They noted that it constituted a minimally invasive method for covalent bioconjugation that enabled precise control of the immobilisation location. The methodology was demonstrated by attaching green fluorescent protein (GFP) to super-paramagnetic beads. The authors also noted that the technique could prove useful in biocatalysis applications.
The first demonstration of the methodology with an enzyme, was demonstrated in 2015 by Liu and Chen and co-workers42 who used genetic insertion of p-azidophenylalanine (p-AzF) into a laccase followed by immobilisation on a multi-walled carbon nanotube electrode. In 2018, Cha and Kwon43 described the immobilisation of enzymes directly from cell lysates by genetic insertion of a clickable ncAA, (p-AzF), using host cells with amber codon-free genomic DNAs, followed by immobilisation directly from cell supernatants onto alkyne-functionalised solid supports through a bioorthogonal azide–alkyne cycloaddition.
In 2020, we described the genetic insertion of five p-AzF units into an AKR.44 The 5-point variant was subsequently immobilised to form amorphous linear site-specific CLEs via a strain-promoted alkyne–azide cycloaddition (SPAAC) reaction with sym-dibenzo-1,5-cyclooctadiene-3,7-diyne (DBA) as a bifunctional cross-linker. However, the cofactor diffuses randomly between free or immobilised forms of the main and auxiliary enzymes. Therefore, the construction of ordered enzyme aggregates is necessary to explore how the spatially oriented arrangement of the active sites improve the efficiency of in vitro cascade reaction.
We subsequently extended this approach to the bio-orthogonal co-immobilisation of two-, three- and five-point p-AzF variants of an AKR with p-AzF variants of GDH or ADH.37 This was achieved by mixing the supernatant of the cell lysate of the AKR with that of the GDH or ADH followed by addition of the DBA cross-linker. The diyne cross-linker undergoes a copper-free click reaction with the azide groups on the two enzymes to afford amorphous AKR–GDH or AKR–ADH combi-CLEs. This can be compared with the glutaraldehyde cross-linker used in CLEA synthesis. The AKR-ADH combination catalysed the production of (S)-1-(2,6-dichloro-3-fluorophenyl) ethanol by asymmetric reduction of the corresponding prochiral ketone with an ee of >99.99%.
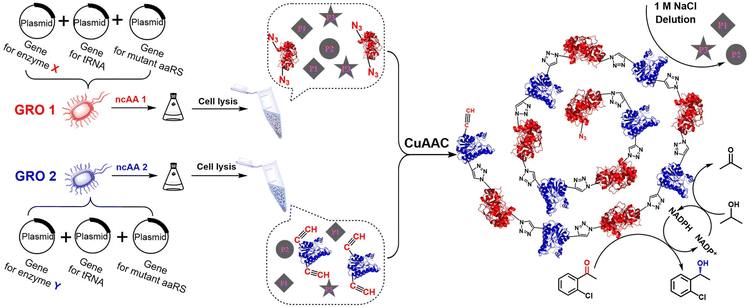
However, when both enzymes contain the same inserted ncAA, namely p-AzF, cross-linking with the DBA affords AKR–ADH combi-CLEs that are not exclusively –ADH–AKR–ADH–AKR– combinations but rather mixtures of possible combinations including, for example, AKR–AKR–AKR– or –ADH–ADH–ADH–. In order to generate linear orderly combi-crosslinked enzymes (O-CLEs) and avoid the separate aggregation of the ADH or AKR it is necessary to insert different ncAAs into the two enzymes so that they can only react with each other to afford only –AKR–ADH–AKR–ADH– combinations. We surmised that cross-linking of the two enzymes in an ordered –X–Y–X–Y– configuration would facilitate faster transfer of intermediates from one enzyme to the next enzyme in a cascade reaction, because of close spatial proximity, in emulation of the substrate channeling effect observed in metabolic pathways in vivo.
In the current article, we have followed this strategy by inserting p-AzF groups into the AKR and p-PaF into the ADH. Subsequent mixing of the two supernatants from the cell lysates results in the selective formation of orderly cross-linked enzymes (O-CLEs) in the –X–Y–X–Y– manner as precipitates by orthogonal ligation through Cu(I) catalysed click reaction of the azide and acetylene functions on the respective enzymes. Insertion of two types of ncAAs which subsequently undergo ligation via a click reaction also avoids the need for the very expensive DBA cross-linker. This one-step conversion of two enzymes, by mixing their cell lysates, into an ordered combi-CLE takes the methodology to a new level of sophistication.
Results and discussion
Preparation and characterisation of O-CLEs of AKR and ADH enzymes
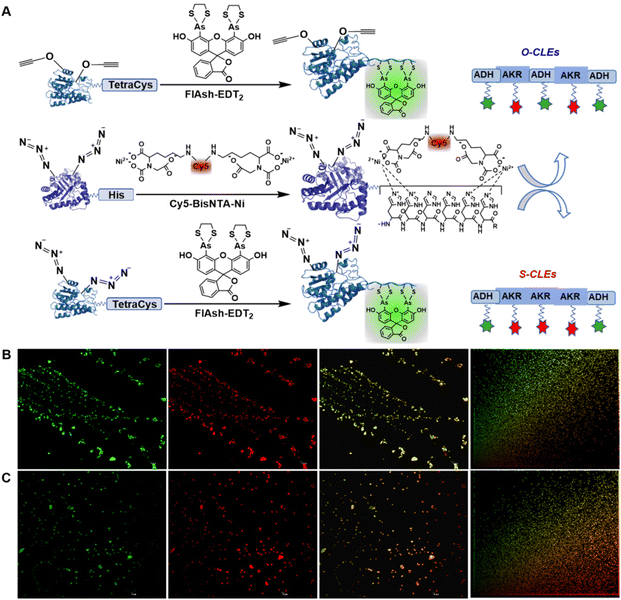
In the present work, we report the incorporation of ncAAs containing azide and alkyne groups into an aldo-keto reductase (AKR) and an alcohol dehydrogenase (ADH), respectively, at preselected sites. Each mutation site is kept away from the catalytic triad but close to the catalytic pocket in order to achieve vectorial biocatalysis,45,46 Positioning of the ncAAs on the protein surface facilitates subsequent cross-linking and the overall enhanced efficiency is an important stimulus for operation in continuous flow.47The engineered bacterium used for the expression of the enzyme proteins containing ncAAs was E. coli C321.ΔA. All known UAG stop codons in E. coli C321.ΔA had been replaced by synonymous UAA codons, which allowed the deletion of release factor 1 and redistribution of UAG translation function.48 UAG was no longer a stop codon, but a blank codon that allowed to reintroduce UAG codons and orthogonal translation machinery to permit efficient and site-specific incorporation of ncAAs into proteins.48 After insertion of the ncAAs, the two enzymes with two functional groups were conjugated in the mixed cell lysates, in a controllable –X–Y–X–Y–X– manner as in a string of rosary beads, to afford ordered cross-linked enzymes (O-CLEs) as shown in Fig. 1. The O-CLEs were characterized using Confocal Laser Scanning Microscopy (CLSM) with two kinds of fluorescent probes (Fig. 2). The morphology of the O-CLEs was characterized using SEM and cross-linkages were verified using FT-IR and MS.
 | ||
| Fig. 1 Schematic illustration of cross-linked enzymes of AKR (X) and ADH (Y) using cell lysates and bio-orthogonal click chemistry under consective microwave irradiation. | ||
In order to promote substrate channeling the two enzymes were cross-linked in an orderly fashion by mixing the supernatants of the respective cell lysates, using bio-orthogonal click chemistry between the two p-azido-L-phenylalanine (p-AzF) ncAAs on the AKR and the two p-propargyloxy-L-phenylalanines (p-PaFs) on the ADH. Based on structure analysis using Swiss-PdbViewe software, we selected two sets of mutation site pairs including 49(Y)–266(E), and 138(Q)–215(E) of the AKR to produce AKR49266 and AKR138215, and 155(Y)–189(Y) and 3(R)–251(Q) of ADH to produce ADH155189 and ADH3251, respectively. The two mutation sites in each set are in an opposite position but all of them are exposed on the surface of the protein.
The E. coli C321.ΔA eliminates the tag-stopping function and successfully inserts ncAAs at preselected sites.48 The expressed target enzymes were analysed by SDS-PAGE (Fig. S5†). The apparent molecular weights of the AKR and ADH variants were approximately 32.0 kDa and 27.5 kDa, respectively. MALDI-TOF-MS was used to determine the molecular weight of the purified enzyme, and the result was consistent with the theoretical molecular weight of the multipoint variants inserted with p-AzF and p-PaF (Fig. S6†).
When the two supernatants of the cell lysates, containing the corresponding target enzymes, were mixed a precipitate was formed and was separated by centrifugation and washed with 1.0 M NaCl solution. Similarly, site-specific cross-linked enzymes (S-CLEs) acting as a control group were obtained by inserting only p-AzF into all the preselected sites in both enzymes and linking them with dibenzocycloocta-4a,6a-diene-5,11-diyne (DBA) as the cross-linker (Fig. S2B†).
The covalent linkage in (O/S)-CLEs was confirmed using FT-IR and MS. Bio-orthogonal click chemistry afforded N-containing heterocycles, which were characterised by an unsaturated carbon–carbon bond tensile vibration in the 2900 cm−1 region and a C–H tensile vibration in the 2800 cm−1 region.49 No similar bands were observed prior to cross-linking, which effectively verified the covalent linking of the enzyme variants in the (O/S)-CLEs (Fig. S10†). To further demonstrate covalent linking in O-CLEs, a mass spectrometric investigation of the acid hydrolysate was carried out. As shown in Fig. S11a,† the ion peak of 425 in the mass spectrogram is consistent with the cross-linked structure [M + H+] formed in O-CLEs. The ion peak of 612 in Fig. S11b† corresponds to the cross-linked structure in S-CLEs, thus verifying the ordered and disordered cross-linking modes of O-CLEs and S-CLEs, respectively.
In order to study their morphology, the obtained (O/S)-CLEs were characterised using SEM. As Fig. S7† shows, orderly CLEs present a banded structure. However, AKR and ADH variants, and the mode of cross-linking of the two enzymes could not be distinguished by scanning electron microscopy. However, this could be addressed by statistical colocalization analysis50–53 of the data obtained in the experiments with Image Pro Plus software where the cross-linked enzymes were characterized by CLSM using red and green dyes.
To further investigate the order of –AKR–ADH–AKR–ADH– (–X–Y–X–Y– manner) in the O-CLEs, a labelling strategy using a short peptide tag and a complementary recognition pair of small molecular probes was applied.54 With AKR-49–266 and ADH-155–189, for example, when the (O/S)-CLEs were mixed together with two fluorescent probes (-Cy5-Bisnta-Ni and Flash-EDT2-), the His-Tag in the AKR variant was specifically bound to the Cy5-Bisnta-Ni55 resulting in red fluorescence in the wavelength range of 770–622 nm. Similarly, the Cys-Cys-Pro-Gly-Cys-Cys peptide segment56 in ADH bound to Flash-EDT2 resulting in green fluorescence in the wavelength range of 577–492 nm (Fig. 2A). Co-localization fluorescence analysis afforded scatter plots of fluorescence intensity of red and green channels. The overall linear diagonal distribution in the O-CLEs diagram, indicates a strong co-localization relationship between the two enzyme molecules, and the fluorescence intensity of the two enzymes was almost the same. However, in the S-CLEs group diagram, the basic linear distribution was close to the red channel, indicating that there was a certain co-localization relationship between the two enzymes, but the correlation was not strong.57
In addition, Pearson's correlation coefficient (PCC) and Manders’ overlap coefficient (MOC)53 are 0.990747 and 0.993085, respectively, for the O-CLEs (Fig. 2B) whereas they are 0.765394 and 0.698419, respectively, for the S-CLEs (Fig. 2C). This consist with O-CLEs exhibiting uniform red and green fluorescence patterns, and the combined image in the full wavelength exhibits a uniform yellow colour image.50 These fluorescence and copositioning relation results are consistent with the AKR and ADH in the O-CLEs, in contrast with those in the S-CLEs, being cross-linked in sequence with just one ADH between two AKR enzyme molecules. Similar CLSM image was also detected in the O- and S-CLEs formed by AKR138215 and ADH3251 (Fig. S8 and 9†).
To verify the order of AKR and ADH on O-CLEs, a SpyTag/SpyCatcher system was employed to increase the identification of CLEs under transmission electron microscopy (TEM) by fusing proteins at one of its protein components. First, a SpyTag (SpT) was fused at N-terminal of AKR49266 of O-CLEs, and a SpyCatcher (SpC) was fused at N-terminal of wild AKR. They were subsequently mixed to form the new CLEs via the specific coupling of SpyCatcher and SpyTag.58 As the characterization in Fig. S13,† the bulgy protein in the new CLEs verified the success bio-orthogonal connection of SpT-AKR49266 with SpC-AKR. This demonstrates the formation of O-CLEs in –X(X)–Y–X(X)–Y– manner, and further indicates that the order of AKR49266 and ADH155189 in the dual enzyme O-CLEs as –X–Y–X–Y– manner.
Enzymatic enantioselective synthesis of the chiral alcohol using cross-linked enzyme preparations
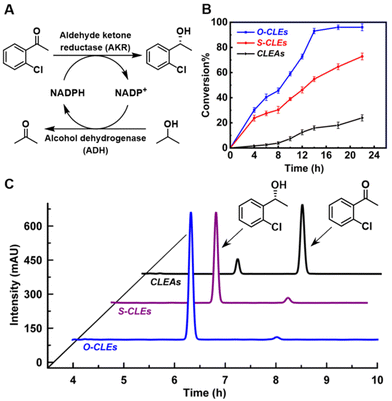
Prior to evaluating the enantioselective synthesis of the chiral alcohol, (R)-1-(2-chlorophenyl) ethanol, a comparative analysis of cofactor (NADPH) regeneration efficiency and reducing activity was carried out (Fig. S14A and B†). The results showed that the cross-linked enzymes prepared by ordered cross-linking (O-CLEs) had better reducing activity and a higher NADPH production rate than those prepared by fixed point crosslinking (S-CLEs) and random crosslinking (CLEAs), obtained by glutaraldehyde mediated cross-linking of pure AKR and ADH proteins (Fig. S2A†). In general, in the co-immobilisation of multiple enzymes, cofactor regeneration is faster than that in the freely diffusing system, and the cofactor regeneration efficiency increases with decreasing distance between the two enzymes.59 The initial production rate of NADPH using NADP+ as substrate and ADH in the cross-linked enzymes as catalyst provides a measure of the efficiency of the cascade reaction.60,61In the apparent kinetic analysis of O-CLEs, S-CLEs and CLEAs (Table 1) based on the determination of enzyme activities (Fig. S4†), ordered cross-linking (O-CLEs) showed lower KM values and better catalytic efficiency than solely site-specific disordered cross-linking (S-CLEs) and random cross-linking (CLEAs), consistent with the enzymes in O-CLEs having a stronger affinity for the substrate.62 Moreover, in the cross-linking of AKR49266–ADH155189, the Kcat/KM value of O-CLEs was 4.24 times that of S-CLEs and 22.27 times that of CLEAs (see Table 1). The enhanced rate observed with the O-CLEs can be rationalised as being a consequence of the ordered arrangement facilitating the movement of the substrate and intermediates between the two enzymes.63,64 Thus, in the aligned assembly of AKR and ADH in the –X–Y–X–Y– manner alignment impedes diffusion of the intermediates into the bulk solution65,66 and reduces the transient time,10 resulting in a higher reaction rate through substrate chanelling.46,67 This is analogous to that observed in enzymatic cascades in vivo, such as the natural polyketide synthase module,68,69 In addition, intensified artificial vectorial catalysis in the close alignment of AKR and ADH can also cause macromolecular crowding70,71 and induce conformational changes that directly affect the structure of active sites in favour of increasing affinity for substrates.72 In these crowded environments the conformational flexibility of enzymes and protein dynamics critical to enzyme catalysis73 can be altered.
| Enzyme mutations used for cross-linking | Cross-linking manner | K M (mM) | K cat (S−1) | K cat/KM |
|---|---|---|---|---|
| a The concentration of the dihydro-4, 4-dimethyl-2, 3-furandione surrogate substrate was in the range 0.065 mM to 15.6 mM. | ||||
| AKR49266 and ADH155189 | CLEAs | 1.34 ± 0.13 | 0.68 ± 0.01 | 0.51 |
| S-CLEs | 0.44 ± 0.05 | 1.18 ± 0.03 | 2.68 | |
| O-CLEs | 0.11 ± 0.01 | 1.25 ± 0.01 | 11.36 | |
In the enantioselective synthesis of (R)-1-(2-chlorophenyl) ethanol from the corresponding ketone (Fig. 3A) the product was obtained in 99.99% ee. As shown in Fig. 3B, the yield of (R)-1-(2-chlorophenyl) ethanol obtained with the O-CLEs was 93% after 14 h, compared with 55% with the S-CLEs and 16% with the CLEAs. Previous work showed that CLEAs produced using glutaraldehyde lost some of their original stereoselectivity with a reduced ee value of the product (72%).74 This decrease in activity and enantioselectivity may result from the random and unstable nature of glutaraldehyde cross-linking21 which results in disordered polymers75 and unfavourable structural changes in the enzyme.60
The simple one-step preparation and purification of O/S-CLEs using cell lysate supernatants preserves the enzyme activity observed with whole cells and avoids tedious purification.76 The O-CLEs with just one ADH between two AKR enzymes have the additional advantage of localizing a succession of sites inside a channel where it is easier to exchange intermediates between active sites and hence reduce the loss of unstable intermediates or cofactors by diffusion.77 Furthermore, the shorter distance between the enzymes leads to an increase in catalytic activity, presumably through substrate channeling.59 The turnover frequency (TOF) in the catalytic cascade showed a decline over 30 h (Fig. S15†) from 2.02 to 1.75 for O-CLEs, 1.96 to 1.5 for S-CLEs and 0.98 to 0.57 for CLEAs, respectively, indicating higher stability of O-CLEs. This may be due to the more stable enzyme structure in O-CLEs, resulting from the bio-orthogonal chemical linkages.
Docking analysis for the interaction of AKR and ADH in O-CLEs
To further explore the interaction and possible spatial orientation of AKR and ADH in orderly cross-linked enzymes, docking prediction of protein–protein complexes was carried out. After the mutants AKR-49–266 and ADH-155–189 are sequentially linked by ncAAs, each AKR protein is tightly linked to two ADHs. Therefore, when the substrate NADP+ leaves the active site of AKR, it can easily enter the active site of the ADH, thus minimising unnecessary diffusion of intermediates into the bulk solution. This reduces the transient time and effectively compartmentalises reaction intermediates10,78 and increases the cascade reaction efficiency. In addition, the close proximity of the active sites of the AKR and ADH facilitates shuttling of NADPH and NADP+ between them via substrate channeling79,80 (Fig. 4).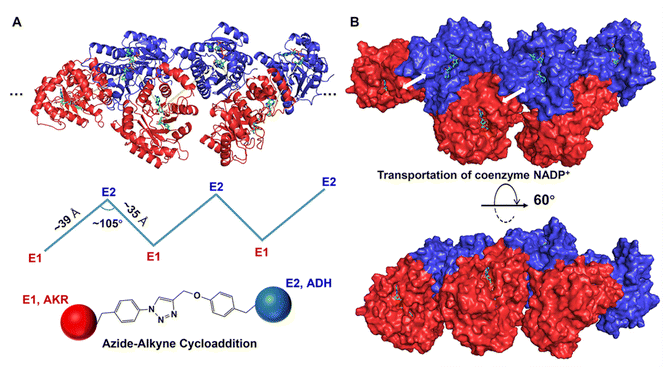 | ||
| Fig. 4 Docking simulation diagram of double enzyme crosslinking (A, docking site AKR-266–ADH-155; B, docking site AKR-49–ADH-189). | ||
In order to further examine the cross-linked protein–protein complex, the AKR and ADH were docked in an orderly manner using the protein–protein algorithm ZDOCK. The rigid body protein–protein docking program ZDOCK uses a fast Fourier transform algorithm to achieve an efficient global docking search on a three-dimensional grid and scores using a combination of shape complementarity, electrostatics and statistical potential terms.81 In addition, the docking results show that the distance between the active sites of the AKR and ADH in O-CLEs was shortened to an amazing 3.4 nm (34 Å) with an angle of 105° and the formed NADP+/NADPH substrate channel is favourable for their transport between enzymes.59 Indeed, inter-enzyme distance and rotational orientation were the primary factors affecting the efficiency of the model enzyme system on a scaffold surface.
In short, maximum enhancement in the transfer of the substrate is achieved when the enzymes are assembled with minimum inter-enzyme separation and suitable cross-linking sites oriented towards each other.82 Our results using a docking model of the varying dual enzyme orientation suggest that the geometric details from the three dimensional structures of the enzymes used are important in designing an enzyme assembly line.83,84 Orderly cross-linking and tight linkage of the two enzymes with a short inter-enzyme distance in O-CLEs facilitates the transfer of intermediates, thus increasing the throughput and product yield in the cascade.85
Conclusions
Here, CLEAs constitute a different concept. They involve the precipitation of the enzyme as physical aggregates followed by crosslinking of the enzymes within these aggregates using a cross-linker such as glutaraldehyde. The aggregation process in this way is disordered and uncontrolled. In contrast, CLEs involve the direct formation of cross-linked enzymes as precipitates through a (co-)polymerisation process in which the active functional groups on the individual enzymes (monomers), such as azide and acetylene functional groups, react with each other. An insoluble copolymer is formed. This cross-linked polymerisation of enzymes is characterised by controlled orientation or binding sites. In the text, we use the word ‘CLEs’ to mean cross-linked enzymes in general. It refers to both O-CLEs and S-CLEs.In conclusion, we have prepared novel ordered cross-linked enzymes (O-CLEs), with retention of activity and near-perfect enantioselectivity, through the use of bio-orthogonal click chemistry to enable precision cross-linking of two p-PaF and two p-AzF residues, genetically introduced at preselected sites in an ADH and AKR, respectively. This highly selective co-polymerisation process involved mixing supernatants from individual cell lysates derived from expression of the respective ADH and AKR variants and subsequent fermentation. The enzymes are assembled in a controllable –X–Y–X–Y–X– manner with regard to both spatial position and orientation.
Their proximity and alignment facilitate transfer of reactive intermediates between the two active sites, thereby hindering competing diffusion into the bulk solution observed with mixtures of the corresponding free or immobilised enzymes. This so-called substrate channeling results in an increase in the overall catalytic efficiency of enzymes in cascade processes in emulation of metabolic pathways in vivo.
Moreover, formation of the O-CLEs by simply mixing the two supernatants to precipitate the assembled ADH-AKR rosary in a seamless combination of enzyme purification and immobilisation into one unit operation provides an assembly line for green biocatalytic cascade processes. We expect, therefore, that the methodology will find applications in a variety of multi-enzyme and chemo-enzymatic cascade processes, in particular for use in continuous operation (biocatalysis in flow).47,86–88
Author contributions
Li Qiao: investigation, data curation, visualization, and writing – review & editing. Zhiyuan Luo: investigation, formal analysis, and writing – original draft. Ru Wang: investigation, methodology, data curation – original draft. The above authors contributed equally to this work. Xiaolin Pei: theoretical calculation and writing – original draft. Shujiao Wu and Haomin Chen: validation – original draft. Tian Xie: supervision, project administration. Anming Wang: conceptualization and supervision, visualization, funding acquisition, project administration, and writing – review & editing. Roger A. Sheldon: supervision and coordination–review, writing & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Natural Science Foundation of China (22078079), the Natural Science Foundation of Zhejiang Province (LY18B060009, LY22B060003), and the Program for Postgraduates in Innovation Practice and Service for Locality” in HZNU (2022).References
- A. R. Alcántara, P. Domínguez de María, J. A. Littlechild, M. Schürmann, R. A. Sheldon and R. Wohlgemuth, ChemSusChem, 2022, 15, e202102709 Search PubMed.
- A. Alcántara, Catalysts, 2019, 9, 792 CrossRef.
- J. P. Adams, M. J. B. Brown, A. Diaz-Rodriguez, R. C. Lloyd and G.-D. Roiban, Adv. Synth. Catal., 2019, 361, 2421–2432 CAS.
- M. D. Truppo, ACS Med. Chem. Lett., 2017, 8, 476–480 CrossRef CAS PubMed.
- R. A. Sheldon and D. Brady, ACS Sustainable Chem. Eng., 2021, 9, 8032–8052 CrossRef CAS.
- R. A. Sheldon, Enzyme-Catalyzed Cascade Reactions, Wiley-VCH, Weinheim, 2008 Search PubMed.
- A. Bruggink, R. Schoevaart and T. Kieboom, Org. Process Res. Dev., 2003, 7, 622–640 CrossRef CAS.
- A. I. Benitez-Mateos, D. R. Padrosa and F. Paradisi, Nat. Chem., 2022, 14, 489–499 CrossRef CAS PubMed.
- J. H. Schrittwieser, S. Velikogne, M. Hall and W. Kroutil, Chem. Rev., 2018, 118, 270–348 CrossRef CAS PubMed.
- G. R. Welch and F. H. Gaertner, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 4218–4222 CrossRef CAS PubMed.
- C. An and K. M. Maloney, Curr. Opin. Green Sustainable Chem., 2022, 34, 100591 CrossRef CAS.
- Y. Zhou, S. Wu and U. T. Bornscheuer, ChemComm, 2021, 57, 10661–10674 RSC.
- K. Rosenthal, U. T. Bornscheuer and S. Lütz, Angew. Chem., Int. Ed., 2022, 61, e202208358 CrossRef CAS PubMed.
- I. Wheeldon, S. D. Minteer, S. Banta, S. C. Barton, P. Atanassov and M. Sigman, Nat. Chem., 2016, 8, 299–309 CrossRef CAS PubMed.
- F. Manea, V. G. Garda, B. Rad and C. M. Ajo-Franklin, Biotechnol. Bioeng., 2020, 117, 912–923 CrossRef CAS PubMed.
- I. Wheeldon, S. D. Minteer, S. Banta, S. C. Barton, P. Atanassov and M. Sigman, Nat. Chem., 2016, 8, 299–309 CrossRef CAS PubMed.
- G. R. Welch and F. H. Gaertner, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 4218–4222 CrossRef CAS PubMed.
- K. Yang, F. Li, Y. Qiao, Q. Zhou, Z. Hu, Y. He, Y. Yan, L. Xu, C. Madzak and J. Yan, ACS Sustainable Chem. Eng., 2018, 6, 17035–17043 CrossRef CAS.
- N. C. Dubey and B. P. Tripathi, ACS Appl. Bio Mater., 2021, 4, 1077–1114 CrossRef CAS PubMed.
- E. T. Hwang and S. Lee, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS.
- D. M. Liu, J. Chen and Y. P. Shi, TrAC, Trends Anal. Chem., 2018, 102, 332–342 CrossRef CAS.
- R. A. Sheldon, Catalysts, 2019, 9, 261 CrossRef CAS.
- T. Thomik, I. Wittig, J. Y. Choe, E. Boles and M. Oreb, Nat. Chem. Biol., 2017, 13, 1158–1163 CrossRef CAS PubMed.
- L. Zhang, J. Shi, Z. Jiang, Y. Jiang, S. Qiao, J. Li, R. Wang, R. Meng, Y. Zhu and Y. Zheng, Green Chem., 2011, 13, 300–306 RSC.
- Z. Liu, S. Cao, M. Liu, W. Kang and J. Xia, ACS Nano, 2019, 13, 11343–11352 CrossRef CAS PubMed.
- J. Dussart-Gautheret, J. Yu, K. Ganesh, G. Rajendra, F. Gallou and B. H. Lipshutz, Green Chem., 2022, 24, 6172–6178 RSC.
- M. Cortes-Clerget, N. Akporji, J. Zhou, F. Gao, P. Guo, M. Parmentier, F. Gallou, J.-Y. Berthon and B. H. Lipshutz, Nat. Commun., 2019, 10, 2169 CrossRef PubMed.
- C. M. Agapakis, P. M. Boyle and P. A. Silver, Nat. Chem. Biol., 2012, 8, 527–535 CrossRef CAS PubMed.
- D. F. Savage, B. Afonso, A. H. Chen and P. A. Silver, Science, 2010, 327, 1258–1261 CrossRef CAS PubMed.
- G. Kim, J. Yang, J. Jang, J.-S. Choi, A. J. Roe, O. Byron, C. Seok and J.-J. Song, Commun. Biol., 2020, 3, 298 CrossRef CAS PubMed.
- P. Pony, C. Rapisarda, L. Terradot, E. Marza and R. Fronzes, Nat. Commun., 2020, 11, 1426 CrossRef CAS PubMed.
- J. Fu, Y. R. Yang, S. Dhakal, Z. Zhao, M. Liu, T. Zhang, N. G. Walter and H. Yan, Nat. Protoc., 2016, 11, 2243–2273 CrossRef CAS PubMed.
- J. Fu, Y. R. Yang, A. Johnson-Buck, M. Liu, Y. Liu, N. G. Walter, N. W. Woodbury and H. Yan, Nat. Nanotechnol., 2014, 9, 531–536 CrossRef CAS PubMed.
- D. L. Bates, M. J. Danson, G. Hale, E. A. Hooper and R. N. Perham, Nature, 1977, 268, 313–316 CrossRef CAS PubMed.
- X. Zhong, Y. Ma, X. Zhang, J. Zhang, B. Wu, F. Hollmann and Y. Wang, Mol. Catal., 2022, 521, 112188 CrossRef CAS.
- E. Ricca, B. Brucher and J. H. Schrittwieser, Adv. Synth. Catal., 2011, 353, 2239–2262 CrossRef CAS.
- R. Wang, J. Zhang, Z. Luo, T. Xie, Q. Xiao, X. Pei and A. Wang, Int. J. Biol. Macromol., 2022, 205, 682–691 CrossRef CAS PubMed.
- J. Nazor, J. Liu and G. Huisman, Curr. Opin. Biotechnol., 2021, 69, 182–190 CrossRef CAS PubMed.
- F. Hollmann, I. Arends and D. Holtmann, Green Chem., 2011, 13, 2285–2314 RSC.
- J. U. Bowie, S. Sherkhanov, T. P. Korman, M. A. Valliere, P. H. Opgenorth and H. Liu, Trends Biotechnol., 2020, 38, 766–778 CrossRef CAS PubMed.
- M. T. Smith, J. C. Wu, C. T. Varner and B. C. Bundy, Biotechnol. Prog., 2013, 29, 247–254 CrossRef CAS PubMed.
- D. L. Guan, Y. Kurra, W. S. Liu and Z. L. Chen, Chem. Commun., 2015, 51, 2522–2525 RSC.
- J. Cha and I. Kwon, Biotechnol. J., 2018, 13, 1700739 CrossRef PubMed.
- H. Li, R. Wang, A. Wang, J. Zhang, Y. Yin, X. Pei and P. Zhang, ACS Sustainable Chem. Eng., 2020, 8, 6466–6478 CrossRef CAS.
- P. D. Mitchell, Biosci. Rep., 2004, 24, 386–435 CrossRef PubMed.
- A. L. Semrau, P. M. Stanley, D. Huber, M. Schuster, B. Albada, H. Zuilhof, M. Cokoja and R. A. Fischer, Angew. Chem., Int. Ed., 2022, 61, e202115100 CAS.
- A. P. Mattey, G. J. Ford, J. Citoler, C. Baldwin, J. R. Marshall, R. B. Palmer, M. Thompson, N. J. Turner, S. C. Cosgrove and S. L. Flitsch, Angew. Chem., Int. Ed., 2021, 60, 18660–18665 CrossRef CAS PubMed.
- M. J. Lajoie, A. J. Rovner, D. B. Goodman, H. R. Aerni, A. D. Haimovich, G. Kuznetsov, J. A. Mercer, H. H. Wang, P. A. Carr, J. A. Mosberg, N. Rohland, P. G. Schultz, J. M. Jacobson, J. Rinehart, G. M. Church and F. J. Isaacs, Science, 2013, 342, 357–360 CrossRef CAS PubMed.
- H. Jornvall, Cell. Mol. Life Sci., 2008, 65, 3875–3878 CrossRef PubMed.
- K. W. Dunn, M. M. Kamocka and J. H. McDonald, Am. J. Physiol.: Cell Physiol., 2011, 300, C723–C742 CrossRef CAS PubMed.
- J. Jonkman, C. M. Brown, G. D. Wright, K. I. Anderson and A. J. North, Nat. Protoc., 2020, 15, 1585–1611 CrossRef CAS PubMed.
- K. Chung, J. Wallace, S.-Y. Kim, S. Kalyanasundaram, A. S. Andalman, T. J. Davidson, J. J. Mirzabekov, K. A. Zalocusky, J. Mattis, A. K. Denisin, S. Pak, H. Bernstein, C. Ramakrishnan, L. Grosenick, V. Gradinaru and K. Deisseroth, Nature, 2013, 497, 332–337 CrossRef CAS PubMed.
- E. M. M. Manders, F. J. Verbeek and J. A. Aten, J. Microsc., 1993, 169, 375–382 CrossRef CAS PubMed.
- Y. Takaoka, A. Ojida and I. Hamachi, Angew. Chem., Int. Ed., 2013, 52, 4088–4106 CrossRef CAS PubMed.
- V. Roullier, S. Clarke, C. You, F. Pinaud, G. Gouzer, D. Schaible, V. Marchi-Artzner, J. Piehler and M. Dahan, Nano Lett., 2009, 9, 1228–1234 CrossRef CAS PubMed.
- S. R. Adams, R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis and R. Y. Tsien, J. Am. Chem. Soc., 2002, 124, 6063–6076 CrossRef CAS PubMed.
- P. G. Penarrubia, X. F. Ruiz and J. Galvez, IEEE Trans. Image Process., 2005, 14, 1151–1158 Search PubMed.
- B. Zakeri, J. O. Fierer, E. Celik, E. C. Chittock, U. Schwarz-Linek, V. T. Moy and M. Howarth, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E690–E697 CrossRef CAS PubMed.
- T. A. Ngo, E. Nakata, M. Saimura and T. Morii, J. Am. Chem. Soc., 2016, 138, 3012–3021 CrossRef CAS PubMed.
- A. M. Wang, F. K. Zhang, F. F. Chen, M. Z. Wang, H. F. Li, Z. W. Zeng, T. A. Xie and Z. M. Chen, Korean J. Chem. Eng., 2011, 28, 1090–1095 CrossRef CAS.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305 CrossRef CAS PubMed.
- J. U. Bowie, S. Sherkhanov, T. P. Korman, M. A. Valliere, P. H. Opgenorth and H. J. Liu, Trends Biotechnol., 2020, 38, 766–778 CrossRef CAS PubMed.
- D. L. Zhu, S. S. Ao, H. H. Deng, M. Wang, C. Q. Qin, J. Zhang, Y. R. Jia, P. Ye and H. G. Ni, ACS Appl. Mater. Interfaces, 2019, 11, 33581–33588 CrossRef CAS PubMed.
- A. Badoei-dalfard, Z. Karami and S. Malekabadi, Bioresour. Technol., 2019, 278, 473–476 CrossRef CAS PubMed.
- W. J. Keenleyside and C. Whitfield, J. Biol. Chem., 1996, 271, 28581–28592 CrossRef CAS PubMed.
- P. K. Ajikumar, W.-H. Xiao, K. E. J. Tyo, Y. Wang, F. Simeon, E. Leonard, O. Mucha, T. H. Phon, B. Pfeifer and G. Stephanopoulos, Science, 2010, 330, 70–74 CrossRef CAS PubMed.
- X. Lian, Y. P. Chen, T. F. Liu and H. C. Zhou, Chem. Sci., 2016, 7, 6969–6973 RSC.
- F. Cao, L.-F. Ma, L.-S. Hu, C.-X. Xu, X. Chen, Z.-J. Zhan, Q.-W. Zhao and X.-M. Mao, Angew. Chem., Int. Ed., 2023, 62, e202214814 CAS.
- L. Chen, X. Wei and Y. Matsuda, J. Am. Chem. Soc., 2022, 144, 19225–19230 CrossRef CAS PubMed.
- D. Balchin, M. Hayer-Hartl and F. U. Hartl, Science, 2016, 353, aac4354 CrossRef PubMed.
- S. L. Speer, C. J. Stewart, L. Sapir, D. Harries and G. J. Pielak, Annu. Rev. Biophys., 2022, 51, 267–300 CrossRef PubMed.
- C. Echeverria and R. Kapral, Phys. Chem. Chem. Phys., 2015, 17, 29243–29250 RSC.
- M. Jiang and Z. H. Guo, J. Am. Chem. Soc., 2007, 129, 730–731 CrossRef CAS PubMed.
- R. Wang, J. Zhang, Z. Luo, T. Xie, Q. Xiao, X. Pei and A. Wang, Int. J. Biol. Macromol., 2022, 205, 682–691 CrossRef CAS PubMed.
- N. R. Mohamad, N. H. C. Marzuki, N. A. Buang, F. Huyop and R. A. Wahab, Biotechnol. Biotechnol. Equip., 2015, 29, 205–220 CrossRef CAS PubMed.
- S. Naseer, O. Y. Jie, X. Chen, S. J. Pu, Y. T. Guo, X. Zhang, D. L. Li and C. L. Yang, Int. J. Biol. Macromol., 2020, 154, 1490–1495 CrossRef CAS PubMed.
- E. Ricca, B. Brucher and J. H. Schrittwieser, Adv. Synth. Catal., 2011, 353, 2239–2262 CrossRef CAS.
- K. S. Rabe, J. Mueller, M. Skoupi and C. M. Niemeyer, Angew. Chem., Int. Ed., 2017, 56, 13574–13589 CrossRef CAS PubMed.
- E. W. Miles, S. Rhee and D. R. Davies, J. Biol. Chem., 1999, 274, 12193–12196 CrossRef CAS PubMed.
- P. Bauler, G. Huber, T. Leyh and J. A. McCammon, J. Phys. Chem. Lett., 2010, 1, 1332–1335 CrossRef CAS PubMed.
- B. G. Pierce, K. Wiehe, H. Hwang, B. H. Kim, T. Vreven and Z. P. Weng, Bioinformatics, 2014, 30, 1771–1773 CrossRef CAS PubMed.
- C. C. Roberts and C.-E. A. Chang, J. Chem. Theory Comput., 2015, 11, 286–292 CrossRef CAS PubMed.
- J. L. Smith and D. H. Sherman, Science, 2008, 321, 1304–1305 CrossRef CAS PubMed.
- P. F. Leadlay, Nature, 2014, 510, 482–483 CrossRef CAS PubMed.
- P. Bauler, G. Huber, T. Leyh and J. A. McCammon, J. Phys. Chem. Lett., 2010, 1, 1332–1335 CrossRef CAS PubMed.
- M. P. Thompson, I. Penafiel, S. C. Cosgrove and N. J. Turner, Org. Process Res. Dev., 2019, 23, 9–18 CrossRef CAS.
- P. Fernandes and C. C. C. R. de Carvalho, Processes, 2021, 9, 225 CrossRef CAS.
- L. L. Martin, T. Peschke, F. Venturoni and S. Mostarda, Curr. Opin. Green Sustainable Chem., 2020, 25, 100350 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3gc01898a |
| ‡ They contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |