Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Oligonol ameliorates liver function and brain function in the 5 × FAD mouse model: transcriptional and cellular analysis†
Danbi
Jo‡
ab,
Archana
Arjunan
a,
Seoyoon
Choi
ab,
Yoon Seok
Jung
a,
Jihyun
Park
cd,
Jihoon
Jo
e,
Oh Yoen
Kim
*cd and
Juhyun
Song
 *ab
*ab
aDepartment of Anatomy, Chonnam National University Medical School, Seoyangro 264, Hwasun 58128, Republic of Korea. E-mail: juhyunsong@chonnam.ac.kr; danbijo0818@gmail.com; sy20180433@gmail.com; yhemm@naver.com; archanaibms@gmail.com; Fax: +82-51-200-7353; Fax: +82-61-375-5834; Tel: +82-51-200-7326 Tel: +82-61-379-2706
bBiomedical Science Graduate Program (BMSGP), Chonnam National University, Seoyangro 264, Hwasun 58128, Republic of Korea
cDepartment of Food Science and Nutrition, Dong-A University, Nakdong-daero 550 beon-gil, Saha-gu, Busan, 49315, Republic of Korea. E-mail: jihyun6807@naver.com; oykim@dau.ac.kr
dDepartment of Health Sciences, Graduate School of Dong-A University, Nakdong-daero 550 beon-gil, Saha-gu, Busan, 49315, Republic of Korea
eDepartment of Biomedical Science, Chonnam National University Medical School, Seoyangro 264, Hwasun 58128, Republic of Korea. E-mail: Jo@jnu.ac.kr
First published on 16th October 2023
Abstract
Alzheimer's disease (AD) is a common neurodegenerative disease worldwide and is accompanied by memory deficits, personality changes, anxiety, depression, and social difficulties. For treatment of AD, many researchers have attempted to find medicinal resources with high effectiveness and without side effects. Oligonol is a low molecular weight polypeptide derived from lychee fruit extract. We investigated the effects of oligonol in 5 × FAD transgenic AD mice, which developed severe amyloid pathology, through behavioral tests (Barnes maze, marble burying, and nestle shredding) and molecular experiments. Oligonol treatment attenuated blood glucose levels and increased the antioxidant response in the livers of 5 × FAD mice. Moreover, the behavioral score data showed improvements in anxiety, depressive behavior, and cognitive impairment following a 2-month course of orally administered oligonol. Oligonol treatment not only altered the circulating levels of cytokines and adipokines in 5 × FAD mice, but also significantly enhanced the mRNA and protein levels of antioxidant enzymes and synaptic plasticity in the brain cortex and hippocampus. Therefore, we highlight the therapeutic potential of oligonol to attenuate neuropsychiatric problems and improve memory deficits in the early stage of AD.
1. Introduction
Alzheimer's disease (AD), a common neurodegenerative disorder, is characterized by chronic neuroinflammation, progressive accumulation of amyloid beta (Aβ) plaques, and neurofibrillary tangles.1 The pathogenesis of AD causes gradual cognitive decline and neuropsychiatric symptoms such as depressive mood and anxiety.2–5 Some studies have suggested that more than 50% of patients with AD have neuropsychiatric symptoms such as anxiety-like behaviors.6–9 Several studies have demonstrated that the AD brain has global damage in various regions, including the hippocampus, prefrontal cortex, limbic subcortical region, dentate gyrus, and para hippocampal region.10–12 The medial temporal lobe, including the cortex and hippocampus, is associated with memory function, and the amygdala is associated with anxiety, both of which exhibit slight atrophy and accumulation of Aβ plaque in the context of AD.13 Additionally, in AD, the brain exhibits severe oxidative stress, glial activation, neuronal cell death, and inflammation, leading to memory deficits.14,15Many attempts have been made to identify the potential of various natural plant products to ameliorate the pathogenesis of AD, with relatively low side effects. Polyphenols, a remarkable antioxidant that is abundant in various plant products (fruits and vegetables), play an important role in attenuating oxidative stress,16 improving network signaling between neurons and glia, and decreasing excessive accumulation of Aβ plaque and tau protein in the AD brain.17 Naringin from citrus fruits has been reported to have a neuroprotective effect in AD brain cerebellum by regulating tau phosphorylation and oxidative stress.18 Quercetin, a well-known flavonoid obtained from red berries, grapes, and onions, can increase mitochondrial biogenesis in neurons and protect neuronal cells against oxidative stress-induced free radicals.19 In addition, polyphenols obtained from conyza dioscoridis are known to attenuate AD pathologies,20 while others from resveratrol have been shown to enhance memory deficit in an AD mouse model.21
Recently, oligonol, a low-molecular-weight polyphenol derived from lychee fruit extract and containing catechin-type monomers and pro-anthocyanidin oligomers, has gained interest owing to its potential to attenuate cognition impairment in AD and aging models.22,23 Oliogonol has biologically beneficial effects, including anti-inflammation, anti-obesity, anti-cancer, and lipid regulation.24,25 Oral administration of oligonol at a high dose (100–200 mg per kg of body weight) in amyloid β (25–35)-induced AD model mice for 2 weeks has been found to improve memory deficit and cognition impairment.22 Moreover, in a senescence-accelerated prone mouse model (SAMP8), locomotive deficit was significantly improved and the severity of infection-induced inflammation was modulated following 36-week oligonol treatment (60 mg per kg of body weight), suggesting a benefit of oligonol in aging-associated diseases such as AD or Parkinson's disease.23
However, no previous study has identified the role of oligonol, a low-molecular-weight polyphenol, in metabolic conditions or cognitive function in an AD model with rapidly developing severe amyloid pathology. Therefore, in this study, we aimed to investigate whether oligonol influences liver function, cognitive decline, neuropsychiatric behaviors, and metabolic alterations in 5 × FAD mice, which rapidly develop severe amyloid pathology, a common mouse model of early stage AD.26 Our findings highlight the therapeutic potential of oligonol as a treatment for AD in terms of metabolic enhancement and cognitive improvement.
2. Materials and methods
2.1. Animal and experimental design
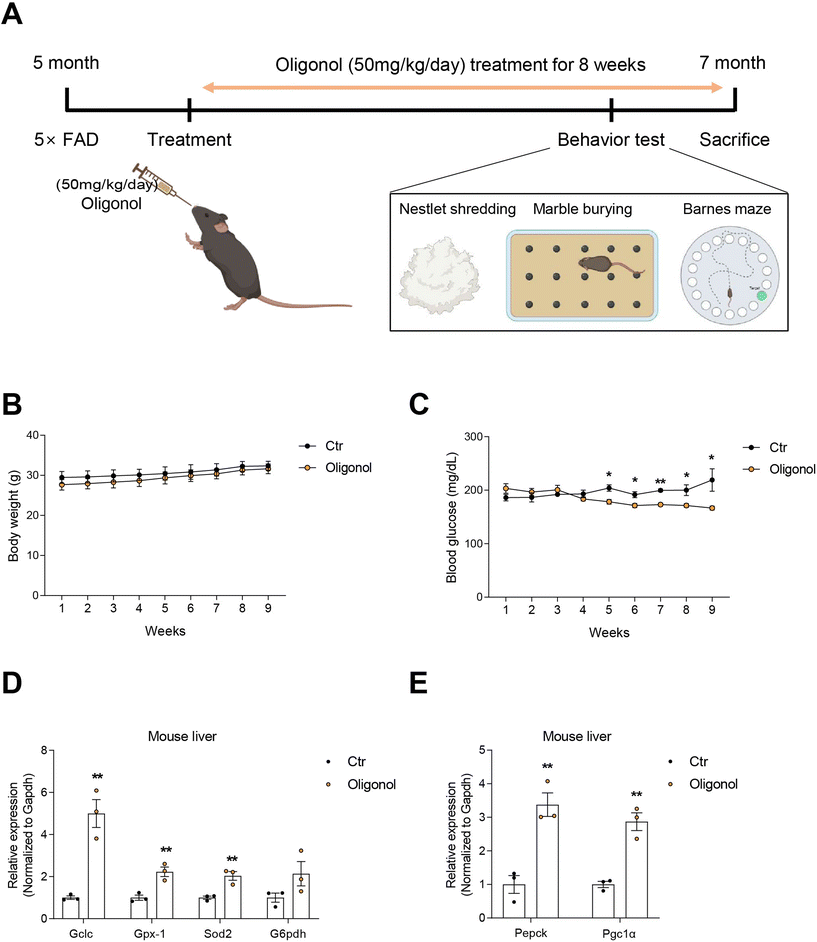
Five-month-old 5 × FAD male mice (n = 6) were housed in the Laboratory Animal Research Center, Chonnam National University (CNU), under a 16 h light/8 h dark cycle at 23 °C with 60% humidity and were given ad libitum access to food and water when the experimental procedures were conducted. Each animal was housed in a cage. The body weight and blood glucose levels of all mice were measured once a week. The 5 × FAD mice were subdivided into two groups: oligonol treatment (n = 3) and placebo (n = 3). The oligonol treatment groups were orally administered oligonol at a dose of 50 mg kg−1 day−1 as a suspension in drinking water daily for 8 weeks. The placebo group was given water in the same way as the oligonol treatment group (ESI Fig. 1† and Fig. 1A). The experiments were performed following the recommendations of the 96 guidelines for animal experiments established by the Animal Ethics Committee at CNU. The Animal Ethics Committee approved the protocol at CNU. At the 7th week of the experiments, animals were tested in the Barnes maze, marble burying test (MBT), and nestle shredding behavior test (NST), before being sacrificed at week 8.2.2. Barnes maze behavior test
The Barnes maze test was successfully performed on 5 × FAD mice to determine their spatial learning and memory. The Barnes maze (Jungdo Bio & Plant) is an opaque circular platform (diameter: 92 cm) with 20 equally spaced holes (diameter: 5 cm) located 2 cm from the edge. In a brightly lit environment, mice naturally seek the dark enclosed area provided by the black goal box (20 × 10 × 4 cm), which was located under the same escape hole throughout all trials. From the surface of the maze, the escape hole containing the goal box appears identical to the other 19 holes. The test was conducted in three phases: a habituation phase, lasting for 5 min (day 0); a training phase, lasting for 1–5 days; and a spatial reference memory phase, lasting for 7 days. In the training and spatial reference memory phases, the escape box was placed under one of the 20 holes. Initially, the mice were placed at the center of the maze, from which point, they explored the maze until they found the escape box. If the mice failed to enter the escape box within 240 s, the experimenter gently led them into the escape box. The mouse remained in the escape box for an additional 30 s before it was removed and returned to the home cage. To dissipate and disseminate odor cues for subsequent trials, the escape box, additional boxes, and maze surface were sprayed with 70% isopropyl alcohol and wiped non-systematically. The location of the escape box remained the same for each mouse during every trial of the training phase. The training was repeated twice a day, with a 30 min interval between each session. The performances of mice (latency: the time taken to escape box, and the number of incorrect entries) was recorded and averaged. The data were collected by a video camera located above the maze and were analyzed using ImageJ.272.3. Marble burying behavior test
The marble burying behavior test was used to assess the obsessive-compulsive behavior of 5 × FAD mice. Briefly, a polycarbonate cage (26 cm × 48 cm × 20 cm) was filled with bedding material to a depth of 5 cm and contained 20 standard glass marbles (15 mm diameter, 5.2 g in weight) in five rows of four marbles. The mice were allowed to explore the polycarbonate cage for 30 min. Water and food withheld during the experiment. Scoring was conducted when the rat buried the marble in two-thirds of its surface into the bedding material.282.4. Nestlet shredding behavior test
The polycarbonate cage (26 cm × 48 cm × 20 cm) was filled with bedding material, 1 g of nestlet (cotton bed) was kept, and the filter-top cover was placed on the cage. The mice were allowed to explore the cage for 30 min. Water and food were withheld during the experiment. Scoring was conducted by weighing the remaining unshredded nestle and dividing this weight by the starting weight to calculate the percentage of nestlets shredded.292.5. Quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Ambicon) according to the manufacturer's protocol. Total RNA from the liver, cortex, and hippocampus of 5 × FAD mice was converted to complementary DNA (cDNA) using random hexamers (Thermo Fisher Scientific) and RevertAid reverse transcriptases (Thermo Fisher Scientific). mRNA expression was measured by quantitative RT-PCR presented using the comparative CT method using the Power SYBR Green PCR master mix (Applied Biosystems) and the Step One Plus PCR system (Applied Biosystems). mRNA expression was normalized to the Gapdh primer set. The primer sequences of the mRNAs are listed in Table 1.| Mouse | ||
|---|---|---|
| Name | Forward sequence | Reverse sequence |
| Gclc | GTTATGGCTTTGAGTGCTGCAT | ATCACTCCCCAGCGACAATC |
| Gpx-1 | CCAGGAGAATGGCAAGAATGA | TCTCACCATTCACTTCGCACTT |
| Sod2 | TCTCACCATTCACTTCGCACTT | GGTGGCGTTGAGATTGTTCA |
| G6pdh | CTGGAACCGCATCATCGTGGAG | CCTGATGATCCCAAATTCATCAAAATAG |
| Pepck | AACTGTTGGCTGGCTCTC | GAACCTGGCGTTGAATGC |
| Pgc1α | TATGGAGTGACATAGAGTGTGCT | CCACTTCAATCCACCCAGAAAG |
| Fos | CTGTCCGTCTCTAGTGCCAAC | CCTCCTGACACGGTCTTCAC |
| Psd95 | GGTAACTCAGGTCTGGGCTTC | CACTGCAGCTGAATGGGTCA |
| Syp | ACATGCAAGGAACTGAGGGA | CCAGGTTCAGGAAGCCAAAC |
| Cyp2e1 | CTTTGCAGGAACAGAGACCA | ATGCACTACAGCGTCCATGA |
| Cyp4a10 | CAACTTGCCCATGATCACACA | CATCCTGCAGCTGATCCTTTC |
| Gapdh | AATGTGTCCGTCGTGGATCT | AGACAACCTGGTCCTCAGTG |
2.6. Western blotting
Proteins from the mice cortex and hippocampus were extracted in radioimmunoprecipitation assay buffer (RIPA, Translab) containing 1× phosphatase inhibitors and 1× protease inhibitors for 20 min on ice. Proteins were quantified using a bicinchoninic acid protein assay kit (BCA, Thermo Fisher Scientific) according to the manufacturer's protocol. Proteins (20 g) were loaded on a 10%–12% SDS-PAGE gel, and proteins were transferred onto a polyvinylidene fluoride membrane (PVDF, Millipore) activated by methanol. The membrane was blocked with 5% skim milk (BD Biosciences) or 5% bovine serum albumin (Thermo Fisher Scientific) for 1 h 30 min at room temperature. The membrane was incubated with primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) overnight at 4 °C. The membrane was incubated with horseradish peroxidase (HRP) secondary antibody (Santa Cruz, 1
1000 dilution) overnight at 4 °C. The membrane was incubated with horseradish peroxidase (HRP) secondary antibody (Santa Cruz, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution) for 2 h at room temperature. The membrane was detected using an enhanced chemiluminescence (ECL) solution (Thermo Fisher Scientific) and Fusion Solo software (Vilber). Protein expression was quantified using ImageJ software, and all protein expression levels were normalized to the GAPDH expression level. The following primary antibodies: brain-derived neurotrophic factor (BDNF) (Abcam, ab108319), Syp (Millipore, MAB368), c-Fos (Santa Cruz, sc-166940), glial fibrillary acidic protein (GFAP) (Santa Cruz, sc-33673), SR-2A (Santa Cruz, sc-166775), and GAPDH (Santa Cruz, sc-32233).
5000 dilution) for 2 h at room temperature. The membrane was detected using an enhanced chemiluminescence (ECL) solution (Thermo Fisher Scientific) and Fusion Solo software (Vilber). Protein expression was quantified using ImageJ software, and all protein expression levels were normalized to the GAPDH expression level. The following primary antibodies: brain-derived neurotrophic factor (BDNF) (Abcam, ab108319), Syp (Millipore, MAB368), c-Fos (Santa Cruz, sc-166940), glial fibrillary acidic protein (GFAP) (Santa Cruz, sc-33673), SR-2A (Santa Cruz, sc-166775), and GAPDH (Santa Cruz, sc-32233).
2.7. Detection of mouse cytokines and adipokines
A cytokine and adipokine array (R&D Systems, ARY006, and ARY013, USA) was performed according to the manufacturer's instructions using 5 × FAD mice. Blood samples of 5 × FAD mice were clotted at room temperature for 15 min and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to collect plasma. Subsequently, the plasma was diluted and incubated in a blocking solution containing the antibody detection cocktail for 1 h at room temperature, followed by incubating the membranes for cytokine and adipokine detection overnight at 4 °C. After incubating the membranes in streptavidin–horseradish peroxidase solution for 30 min at room temperature, the dot blots were visualized using an ECL solution and Fusion Solo software.
000 rpm for 10 min to collect plasma. Subsequently, the plasma was diluted and incubated in a blocking solution containing the antibody detection cocktail for 1 h at room temperature, followed by incubating the membranes for cytokine and adipokine detection overnight at 4 °C. After incubating the membranes in streptavidin–horseradish peroxidase solution for 30 min at room temperature, the dot blots were visualized using an ECL solution and Fusion Solo software.
2.8. Transcriptome analysis of RNA sequencing data
Total RNA from the cerebral hippocampus in three 5 × FAD mice models and three oligonol treated 5 × FAD mice models was extracted using TRIzol reagent (Thermo Fisher, MA, USA), and RNA integrity was verified using the Agilent 2100 BioAnalyzer (Agilent, CA, USA). The sequencing reads for each sample were mapped to the reference genome (Mus musculus GRCm39) by Kallisto (v0.46.1).30 The aligned results were applied to edgeR package31 to select differentially expressed genes.2.9. Functional analysis of significantly changed genes
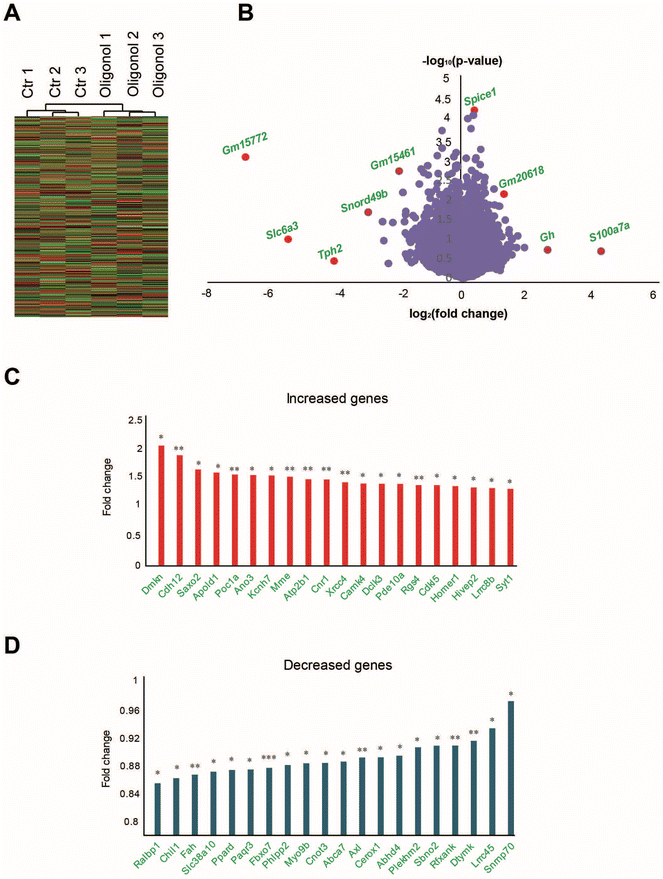
To select genes with significant changes in their expression in the oligonol-treated 5 × FAD group, we first selected transcripts with significantly different expression in the oligonol-treated 5 × FAD mouse data and the corresponding control group data. For this purpose, 577 genes with significant expression changes based on p-values 0.05 in the oligonol-treated 5 × FAD groups were selected. We distinguished genes in the volcano plots in Fig. 5A with genes based on p-values < 0.05. Statistically, among the genes with p-values < 0.05, the expression values of 328 genes were increased and those of 249 genes were decreased after oligonol treatment in 5 × FAD mice. Among the genes with a p-value < 0.05, we selected 20 increased and 20 decreased genes in the order of fold change. For Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using the Molecular Signatures Database,32 we selected 577 genes with p-values < 0.05 in the oligonol-treated 5 × FAD mouse model. The STRING (https://string-db.org) software program was used to screen interaction networks of the top 200 genes in order of decreasing p-values among genes with p-values <0.05. We selected only the networks with a minimum of two nodes.2.10. Statistical analysis
All data are presented as the group mean ± S.E.M. Statistical analysis was performed using an unpaired two-tailed t-test with Welch's correction in GraphPad Prism 8 (GraphPad Software Inc., USA). Data were considered significant at *p < 0.05, **p < 0.01, and ***p < 0.005 in the statistical analysis.3. Results
3.1. Oligonol treatment improved glucose metabolism and increased antioxidant gene expression in the livers of 5 × FAD mice
Oral administration of oligonol (50 mg kg−1 day−1) was administered to 5 × FAD mice at 5 months of age for 2 months (Fig. 1A) and the body weight and blood glucose level were checked (Fig. 1A). The body weights of both oligonol-treated and control 5 × FAD mice similarly increased over the 2-month period (Fig. 1B). The blood glucose levels in the oligonol-treated 5 × FAD mice gradually decreased from 4 weeks compared to those in the control 5 × FAD mice (Fig. 1C). The mRNA levels of antioxidant-related genes, such as glutamate-cysteine ligase catalytic (Gclc), glutathione peroxidase 1 (Gpx1), superoxide dismutase 2 (Sod2), and glucose 6-phosphate dehydrogenase (G6pdh), were significantly increased in the liver tissues of oligonol-treated 5 × FAD mice compared to the control 5 × FAD mice (Fig. 1D). Additionally, the mRNA levels of gluconeogenesis-related genes, such as phosphoenolpyruvate carboxykinase (Pepck) and peroxisome proliferator-activated receptor-c coactivator-1a (Pgc1α), were dramatically increased in the liver tissues of oligonol-treated 5 × FAD mice compared to control 5 × FAD mice (Fig. 1E).3.2. Oligonol treatment improved anxiety behavior and cognitive function in 5 × FAD mice
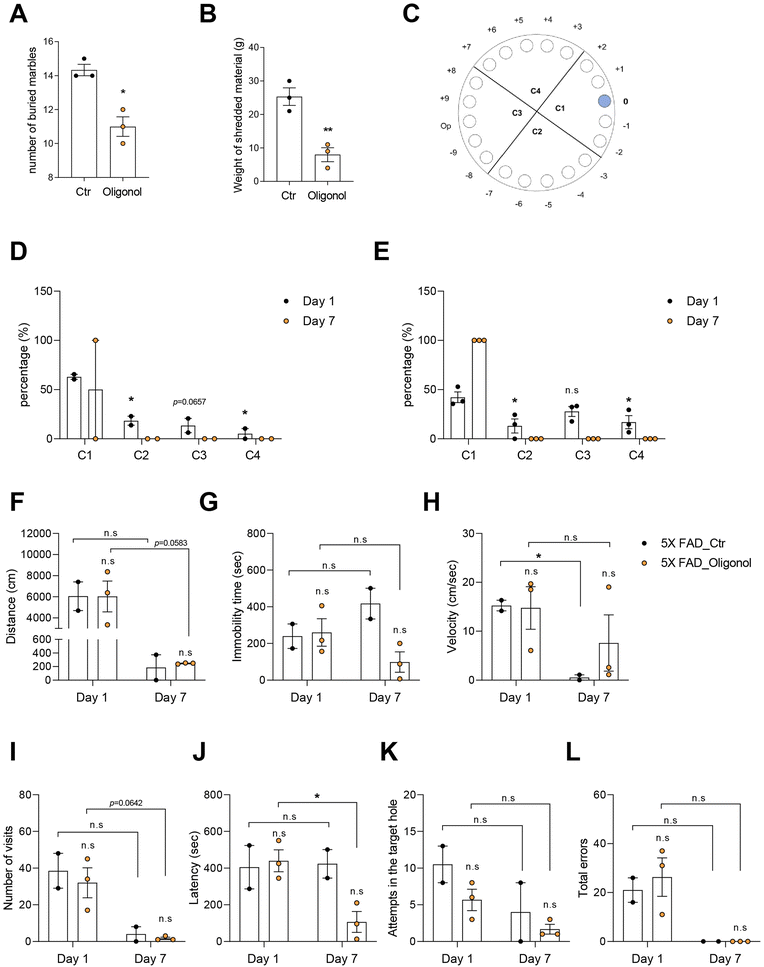
We conducted the MBT and NST to investigate the anxiety behavior, and the Barnes maze to examine the cognition in 5 × FAD mice after oral administration of oligonol (Fig. 2). The marble burying score, considered as the number of buried marbles, was significantly decreased in the oligonol-treated 5 × FAD mice (Fig. 2A). The nestle shredding score, considered as the weight of shredded material (g), was significantly reduced in oligonol-treated 5 × FAD mice (Fig. 2B). Fig. 2A and B show the suppressed anxiety in 5 × FAD mice as a result of oligonol treatment. We conducted the Barnes maze test to investigate the cognitive function in oligonol-treated 5 × FAD mice (Fig. 2D–L). Fig. 2C shows the quadrant region and target hole in the Barnes maze (Fig. 2C), while Fig. 2D and E show the percentage of each quadrant region visit on days 1 and 7 (Fig. 2D and E). Oligonol-treated 5 × FAD mice approached the C1 quadrant region containing the escape hole more frequently than the control 5 × FAD mice (Fig. 2D and E). Oligonol-treated 5 × FAD mice also showed a reduced total distance (Fig. 2F), immobility time (Fig. 2G), and velocity (Fig. 2H) to find escape holes on day 7. Additionally, oligonol-treated 5 × FAD mice demonstrated a reduced number of visits (Fig. 2I), decreased latency (Fig. 2J), reduced attempts in the target escape hole (Fig. 2K), and reduced total errors (Fig. 2L) in the fine target escape hole on day 7. These Barnes maze scores indicate that oligonol treatment improves cognitive function in 5 × FAD mice.3.3. Oligonol treatment alters the secretion of inflammatory cytokines and adipokines in the blood plasma of 5 × FAD mice
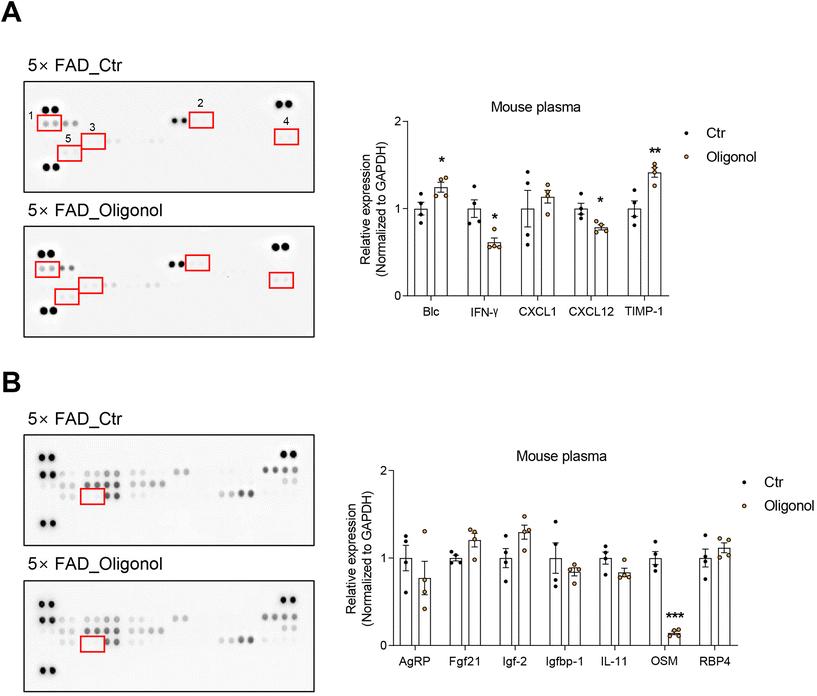
We detected various cytokines and adipokines in the blood plasma of 5 × FAD mice after oral administration of oligonol using cytokine and adipokine array kits (Fig. 3). The results revealed increased expression of cytokines, such as B lymphocyte chemoattractant (Blc) and tissue inhibitor of metalloproteinase-1 (TIMP-1), and reduced expression of interferon gamma (IFN-γ) and the chemokine ligand C–X–C motif chemokine ligand 12 (CXCL12) in the blood plasma of oligonol-treated 5 × FAD mice (Fig. 3A). We also found increased levels of adipokines, such as insulin-like growth factor 2 (IGF-2) and fibroblast growth factor 21 (FGF21), and dramatically reduced levels of oncostatin M (OSM) in the blood plasma of oligonol-treated 5 × FAD mice (Fig. 3B). These data indicate that oligonol influences the levels of cytokines and adipokines in the blood of 5 × FAD mice.3.4. Oligonol treatment reduces the expression of reactive oxygen species (ROS)-generating genes and increases the expression of neuronal function-related genes and serotonin receptors in the brains of 5 × FAD mice
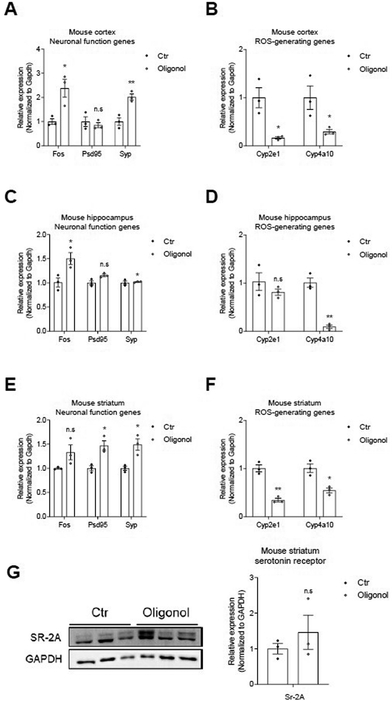
We next investigated the mRNA levels of neuronal function-related genes (c-FOS, PSD95, and SYP) and ROS generating genes (Cyp2e1 and Cyp4a10) in the cortex, hippocampus, and striatum regions of the brains of 5 × FAD mice oral administration (Fig. 4). We detected increased mRNA levels of PSD95 and SYP genes, as synaptic function-related markers, and c-Fos, as neuronal functional markers, in the mouse cortex (Fig. 4A), hippocampus (Fig. 4C), and striatum (Fig. 4E) of 5 × FAD mice after oligonol treatment (Fig. 4A, C, and E). We also observed reduced mRNA levels of Cyp2e1 and Cyp4a10 genes, as genes related to ROS generation, in the mouse cortex (Fig. 4B), hippocampus (Fig. 4D), and striatum (Fig. 4F) of 5 × FAD mice after oligonol treatment. Furthermore, we confirmed the protein levels of m-BDNF, Syp, c-Fos, and GFAP in the mouse cortex, hippocampus, and striatum of 5 × FAD mice following oligonol treatment (ESI Fig. 2†). We found an increased protein level pattern of GFAP, considered to indicate glial activation in the cortex (ESI Fig. 2A†), an increased protein level of c-Fos, considered to indicate neuronal activation in the hippocampus (ESI Fig. 2B†), and an increased protein level of c-Fos and SYP, considered to indicate neuronal activation and synaptic function in the striatum (ESI Fig. 2C†). Although these ESI Fig. 2† data are not significant, they reveal an increased pattern of proteins associated with neuronal function. Furthermore, we detected increased protein levels of serotonin receptors in the striatum region of 5 × FAD mice following oligonol treatment (Fig. 4G). Collectively, these findings show that oligonol treatment inhibits ROS generation and enhances synaptic plasticity, neuronal function, glial activation, and the expression of serotonin receptors in the brains of 5 × FAD mice.3.5. Transcriptome analysis of the hippocampus in oligonol-treated 5 × FAD mice
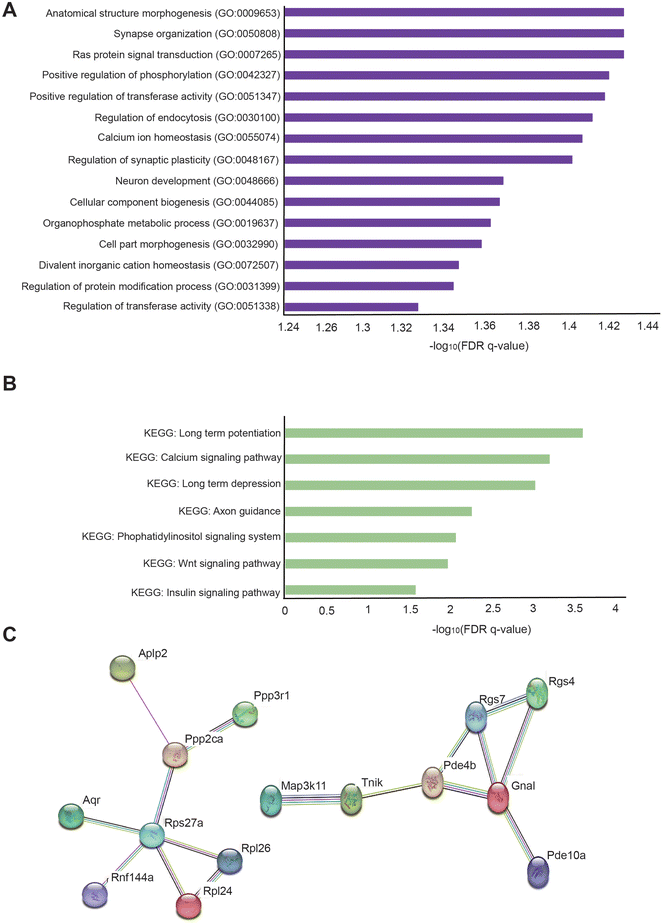
For transcriptome analysis of the hippocampus in oligonol-treated 5 × FAD mice, we conducted RNA sequencing of the total RNA in the hippocampus of three oligonol-treated 5 × FAD mice and three corresponding control 5 × FAD mice. First, we checked that the grouping between 5 × FAD group genes and oligonol treatment group genes was well performed (Fig. 5A). In the RNA sequencing data, the genes with exceptionally high expression levels and genes showing statistically significant changes in the oligonol-treated 5 × FAD group are shown in the volcano plot graph (Fig. 5B). Analysis of the hippocampus of an oligonol-treated 5 × FAD mouse revealed 328 genes with significantly increased expression and 249 genes with significantly decreased expression with a p-value of 0.05. As depicted in the volcano plot, the expression of Gm15772, Slc6a3, tryptophan hydroxylase 2 (Tph2), small nucleolar RNA, C/D Box 49B (Snord49b), Gm15461, spindle and centriole associated protein 1 (Spice1), Gm20618, growth hormone (Gh), and s100 calcium binding protein A7A (S100a7a) was significantly distinguished in the oligonol-treated 5 × FAD mouse hippocampus (Fig. 5B). Next, we identified fold changes in 20 genes with p-values < 0.05 in transcriptome data (Fig. 5C). Our results showed that fold-change-increased genes related to neuropathology were dermokine (Dmkn), cadmium-12 (Cdh12), stabilizer of axonemal microtubules 2 (Saxo2), apolipoprotein L domain-containing 1 (Apold1), POC1 centriolar protein A (Poc1a), anoctamin 3 (Ano3), potassium voltage-gated channel subfamily H member 7 (Kcnh7), membrane metallopentase (Mme), ATPase plasma membrane Ca2+ transporting 1 (Atp2b1), cannabinoid receptor 1 (Cnr1), X-ray repair cross complementing 4 (Xrcc4), calcium/calmodulin dependent protein kinase IV (Camk4), doublecortin-like kinase 3 (Dclk3), phosphodiesterase 10A (Pde10a), regulator of G protein signaling 4 (Rgs4), cyclin-dependent kinase-like 5 (Cdkl5), homer scaffold protein 1 (Homer1), HIVEP zinc finger 2 (Hivep2), leucine rich repeat-containing 8 VRAC subunit B (Lrrc8b), and synaptotagmin 1 (Syt1) genes in fold change order (Fig. 5C). We also checked the fold changes in 20 decreased genes with p-values < 0.05 in transcriptome data (Fig. 5D). Our results showed that the genes associated with decreased fold change in neuropathology were RalA binding protein 1 (Ralbp1), chitinase 3-like 1 (Chil1), fumarylacetoacetate hydrolase (Fah), solute carrier family 38 member 10 (Slc38a10), peroxisome proliferator activated receptor delta (Ppard), progestin and AdipoQ receptor family member 3 (Paqr3), F-Box protein 7 (Fbxo7), phospholipid phosphatase 2 (Phlpp2), myosin IXB (Myo9b), CCR4-NOT transcription complex subunit 3 (Cnot3), ATP-binding cassette A7 (Abca7), AXL receptor tyrosine kinase (Axl), cytoplasmic endogenous regulator of oxidative phosphorylation 1 (Cerox1), abhydrolase domain-containing 4, N-acyl phospholipase B (Abhd4), pleckstrin homology and RUN domain containing M2 (Plekhm2), strawberry notch homolog 2 (Sbno2), regulatory factor X associated ankyrin-containing protein (Rfxank), deoxythymidylate kinase (Dtymk), leucine rich repeat-containing 45 (Lrrc45), and small nuclear ribonucleoprotein U1 subunit 70 (Snrnp70) genes in fold change order (Fig. 5D). ESI Table 1† shows the 577 genes with p-value 0.05 or less genes (ESI Table 1†).Additionally, to identify the cellular pathways associated with genes significantly changed in the hippocampus of the oligonol-treated 5 × FAD mice, we performed GO analysis in The Molecular Signatures Database (MSigDB) with 577 genes with p-values < 0.05 (Fig. 6A). GO analysis showed that the most significantly enriched terms were anatomical structure morphogenesis, synapse organization, Ras protein signal transduction, positive regulation of phosphorylation, positive regulation of transferase activity, regulation of endocytosis, calcium ion homeostasis, regulation of synaptic plasticity, neuron development, cellular component biogenesis, organophosphate metabolic process, cell part morphogenesis, divalent inorganic cation homeostasis, regulation of protein modification process, and regulation of transferase activity (Fig. 6A). We also performed KEGG pathway analysis in MSigDB with 577 genes with p-values 0.05 (Fig. 6B). KEGG analysis showed that the most significant terms were long-term potentiation, calcium signaling pathway, long-term depression, axon guidance, phosphatidylinositol signaling system, Wnt signaling pathway, and insulin signaling pathway (Fig. 6B). Next, to analyze the protein networks affected in the oligonol-treated 5 × FAD mouse hippocampus, we selected 200 genes in order of low p-values among genes with p-values < 0.05. We then analyzed the STRING network analysis database using selected genes33 (Fig. 6C). The protein interaction network obtained from the STRING database for the genes that were changed in the oligonol-treated 5 × FAD group is shown in Fig. 6C. Interestingly, the network containing the APLP2 protein, related to the amyloid precursor protein, the RGS7 protein, related to the GTPase activating protein in the brain, the PDE4B and RGS4 proteins, related to schizophrenia, and the TNIK protein, related to postsynaptic density of glutamatergic synapse and neuropsychiatric disorders were selected after STRING analysis (Fig. 6C).
4. Discussion
Here, we investigated the role of oligonol in metabolic condition and cognitive function in 5 × FAD mice, which together with 3TG mice, is a widely used AD mouse model.34,35 In particular, the 5 × FAD mouse is an AD model that rapidly develops severe amyloid pathologies within 2–4 months after birth.35 During the oral administration of oligonol to 5 × FAD mice, we checked the body weight and blood glucose levels every week until the day of sacrifice. We observed a reduction in blood glucose levels in the oligonol-treated 5 × FAD mice compared to control mice. A previous study mentioned that oligonol improved insulin sensitivity and glucose uptake.36 We previously confirmed the positive regulatory role of oligonol in glucose metabolism in AD mice. We have also detected increased mRNA levels of antioxidant-related genes, such as Gclc,37Gpx1,38Sod2,39 and G6pdh,40 and nuclear factor erythroid-2-related factor 2 (Nrf2)41 in the livers of 5 × FAD mice (Fig. 1). Increased expression of these genes indicates that oligonol treatment causes an increased antioxidant response in the liver and reduced liver damage in AD mice.42 In addition, the increased levels of Pepck and Pgc1α genes observed in our study are associated with gluconeogenesis through the activation of PI3K/AKT signaling.43,44PGC-1α regulates the mitochondrial catabolic capacity of the cell and boosts the expression of antioxidant enzymes against damage conditions.45–47 These findings indicate that oligonol promotes the antioxidant response and plays a cellular protective role in the livers of AD mice.In this study, we performed three behavior tests (Barnes maze test, BMT, and NST). The Barnes maze test, as one of the major behavior tests, is used to assess spatial memory and learning function more accurately than previous tests such as the Morris water maze.48–52 Several studies have used the Barnes maze to investigate learning and memory deficits in AD model mice.53,54 The results of the Barnes maze test suggest that oligonol enhances spatial memory in the AD brain. MBT is used to investigate anxiolytic behavior55 and obsessive–compulsive behavior.56 Wild-type mice usually dig in the ground to search for food, bury harmful things, and create safe nurseries for their babies.28 Some studies have attempted to investigate the behavioral and psychological symptoms of dementia, such as anxiety, in AD mouse models using the MBT.34,57–59Considering the MBT scores observed in our study, oligonol may alleviate anxiety-like psychiatric problems in the AD brain. Additionally, the NST is a behavior test to assess psychiatric diseases such as anxiety, compulsive-like behaviors, distress, and pain.60–62 Some studies have identified brain functional deficits and brain damage using the NST.63,64 Our results suggest that oligonol improves anxiety, compulsive-like behavior, and distress in the AD brain. The data shown in Fig. 4G confirm the increased expression of the serotonin receptor SR-2A in the striatum. The serotonin receptor 5-HT 2a is commonly expressed on neurons and glia in the cortex, nucleus accumbens, claustrum, olfactory bulb, limbic system, striatum including the amygdala, and basal ganglia.65,66 Several studies have mentioned that reduced serotonin 5-HT 2a receptors are associated with object memory loss, spatial memory loss,67,68 and reduced long term memory69 in AD. Other studies have reported that reductions in serotonin 5-HT 2a receptors lead to impaired fear memory, mental disorders, depression and anxiety.70–73 Serotonin 5-HT 2a receptors colocalize with PSD-95 protein74 and influence dendritic spine maturation and synapse stabilization.75 Serotonin 5-HT 2a receptor signaling also induces intercellular signaling pathway activation, including ERK and tyrosine kinase signaling in neurons.76 Accordingly, our results indicate that oligonol enhances the serotonergic system in the brains of 5 × FAD mice and improves anxiety, spatial memory, and depression in the AD brain.
Subsequently, we detected the cytokine and adipokine levels in the blood plasma of 5 × FAD mice. Our data revealed that oligonol treatment increased the levels of Blc (CXCL13), related to T cell activity and THE immune response,77 and I-TAC, a chemokine that regulates the activity of T cells,78 in the blood plasma of 5 × FAD mice. Oligonol treatment also reduced the levels of INF-γ and IL-27, which serve to heighten the immune and inflammatory responses by T cell polarization, T-cell proliferation, and T cell activation79,80 in the plasma of 5 × FAD mice. Additionally, the level of oncostatin M, an IL-6 family member that accelerates severe inflammation,81 was dramatically reduced in the blood plasma of 5 × FAD mice following oral administration of oligonol. In contrast, the levels of IGF-2, an anti-inflammatory regulator and enhancer of neuronal density,82 and FGF21, an anti-inflammatory regulator,83 were considerably increased in the plasma of 5 × FAD mice following oral administration of oligonol. Based on the data obtained for cytokines and myokines, we speculate that oligonol treatment suppresses the inflammatory response and contributes to the immune system in AD mice.
Furthermore, we confirmed increased mRNA levels and protein level patterns of synapse-related genes, including c-Fos, PSD95, and SYP, in the cortex, hippocampus, and striatum of 5 × FAD mice as a result of oligonol treatment. Overall, we hypothesize that oligonol treatment contributes to the expression of neuronal activation and synaptic plasticity in the brains of 5 × FAD mice. c-Fos, as an index of neuronal activity, has been reported to be associated with several neurological issues, including anxiety, depression, fear memory, and spatial memory.84–87 PSD95, as a postsynaptic density protein, regulates the localization of various neurotransmitter receptors,88,89 clustering of ion channels,90 and synaptic plasticity.91 SYP, as a presynaptic density protein, regulates neuronal networks and affects cognitive function,92,93 and depressive like behavior and anxiety.94 Considering this evidence, we consider that oligonol serves to modulate synapse plasticity and neuronal activity, which is involved in neuropathological issues.
We also observed that the mRNA levels of Cyp2e1 and Cyp4a10 genes related to ROS generation in mitochondria95,96 were decreased in the brain cortex, hippocampus, and striatum of 5 × FAD mice treated with oligonol. This may indicate that oligonol suppresses ROS generation in the cortex, hippocampus, and striatum of the AD brain. Decreased ROS generation in the cortex and hippocampus is linked to enhanced long-term potentiation,97 improved learning and memory function,98 reduced neuronal apoptosis,99 the inhibition of blood–brain barrier (BBB) breakdown,100 and reduced pain.101 Increased ROS production in the striatum induces apoptosis,102 hyperglycemia,103 Impaired dopamine release and synaptic plasticity.104 Our data suggest that oligonol can restore dysfunctions caused by ROS production, suggesting that the expression of genes related to ROS generation was reduced by oligonol treatment in the striatum. Our western blot data showed an increased protein level of GFAP in the cortex of 5 × FAD mice after oligonol treatment. Increased GFAP levels are related to astrocyte reactivity,105 neurogenesis,106 attenuation of depressive like behavior,107 and neuronal cell survival108 through autophagy in the brain.
The volcano graph demonstrates the distinguishing genes of the dopamine transporter Slc6a3 gene, related to neuropsychiatric diseases,109,110Tph2, related to serotonergic neuronal cell regulation,111,112 and Gh, related to inflammation.113 In addition, our transcriptome data showed 20 increased genes, including Cdh12, related to the upregulation of neurite growth;114Apold1, associated with angiogenesis and blood brain permeability;115,116Poc1a, related to the regulation of neuronal spindle function;117Kcnh7, associated with the downregulation of schizophrenia;118Atp2b, related to the protection of γ-aminobutyric acid (GABA)ergic neurons;119Cnr1, related to mood disorder and dopaminergic neurotransmission;120,121Camp4, related to the upregulation of memory formation improvement;122Dclk3, associated with neuroprotective function in the dentate gyrus in hippocampal formation and Huntington's disease;123Pde10a, related to regulation of the dopamine system in neurons;124Rgs4, associated with neurotransmitter transmission, psychiatric disorders such as depression and autism, and cognition;125–127Cdkl5, related to the downregulation of seizure and motor dysfunction;128Homer1, related to the downregulation of epilepsy, autism, addiction, schizophrenia, and depression;128Hivep2, related to the regulation of dopaminergic neurons;129 and Syt1, related to the regulation of neurotransmitter release130 in the 5 × FAD mouse hippocampus after oligonol treatment.
We also found 20 decreased genes, including Ralbp1, related to mitochondrial function and decreased neuroinflammation;131,132Slc38a10, related to the upregulation of cell survival and the homeostasis of neurotransmitters such as GABA, dopamine, serotonin, and glutamate;133Ppard, associated with the protection of axonal injury and cell growth and the regulation of immune response;134Paqr3, related to the regulation of energy metabolism, and Golgi apparatus;135Fbxo7, related to the upregulation of pyramidal neuronal function with Pink1;136,137Myo9b, related to the maintenance of the dendrite morphology of neurons;138Abca7, associated with the downregulation of the phagocytosis process in glia, and the process of Alzheimer's disease;139,140Sbno2, related to the downregulation of interleukin-6 secretion, and the suppression of inflammation;141,142 and Snrnp70, related to the inhibition of amyotrophic lateral sclerosis and motoneuronal degeneration143,144 in the 5 × FAD mouse hippocampus after oligonol treatment.
Considering our transcriptome analysis data, we found enriched GO terms, including calcium signaling, which is promoted by synaptic activity;145 the regulation of endocytosis, which modulates the hippocampal synapse strength146 and neuronal survival;147 and RAS protein signaling for the regulation of hippocampal neurogenesis and cognition148 in the 5 × FAD mouse hippocampus after oligonol treatment. We also observed significant KEGG terms such as long-term potentiation, long-term depression which are linked to neurotransmitter secretion, synaptic plasticity, neuronal circuit, learning and memory function;149–151 axon guidance, which is related to improved hippocampal circuit and synaptogenic activity;152 Wnt signaling, which is related to hippocampal neurogenesis;153 and insulin signaling, which can regulate energy homeostasis, cognition, and mood154,155 in the 5 × FAD mouse hippocampus after oligonol treatment.
Our STRING protein network software data showed the interaction between Ppp2ca, related to neuropsychiatric diseases;156 Aplp2, associated with amyloid beta peptide processing;157 and Rps27a, affecting microglia activation.158 Additionally, Tnik, related to cognition and postsynaptic formation;159 Rgs7, associated with synaptic plasticity and memory formation;160 Pde4b, related to the inhibition of memory acquisition;161 and Pde10a, related to the secretion of dopamine by neurons.124
In summary, oral oligonol administration may contribute to the activation of the antioxidant response pathway, reduce ROS generation, modulate glucose metabolism and cytokine and adipokine production, increase glial reactivity, activate synaptic plasticity and neuronal function, and increase serotonin receptor expression in AD. These consequences may lead to stable mood conditions and improved cognitive function in the AD brain. Taken together, our results suggest that oligonol has therapeutic potential to ameliorate liver function and metabolic function and attenuate neuropathological and neuropsychiatric issues such as anxiety and cognitive decline in AD.
Consent for publication
All authors have reviewed the manuscript and provided consent for publication.Ethics approval and consent to participate
All animal procedures were performed in accordance with protocols approved by the recommendations of the 96 guidance for animal experiments established by the Animal Ethics Committee at Chonnam National University (CNU). The Animal Ethics Committee approved the protocol at the CNU.Abbreviations
| AD | Alzheimer's disease |
| Aβ | Amyloid beta |
| SAMP8 | Senescence-accelerated prone mice |
| MBT | Marble burying test |
| NST | Nestlet shredding behavior test |
| cDNA | Complementary DNA |
| Gclc | Glutamate-cysteine ligase catalytic |
| Gpx1 | Glutathione peroxidase 1 |
| Sod2 | Superoxide dismutase 2 |
| G6pdh | Glucose 6-phosphate dehydrogenase |
| Pepck | Phosphoenolpyruvate carboxykinase |
| Pgc1α | Peroxisome proliferator-activated receptor-c coactivator-1a |
| Blc | B lymphocyte chemoattractant |
| TIMP-1 | Metalloproteinase-1 |
| IFN-γ | Interferon gamma |
| CXCL12 | Chemokine ligand C–X–C motif chemokine ligand 12 |
| IGF-2 | Insulin-like growth factor 2 |
| FGF21 | Fibroblast growth factor 21 |
| OSM | Oncostatin M |
| ROS | Reactive oxygen species |
| Tph2 | Tryptophan hydroxylase 2 |
| Snord49b | Small Nucleolar RNA: C/D Box 49B |
| Spice1 | Spindle and centriole associated protein 1 |
| Gh | Growth hormone |
| S100a7a | s100 calcium binding protein A7A |
| Dmkn | Dermokine |
| Cdh12 | Cadherin-12 |
| Saxo2 | Stabilizer of axonemal microtubules 2 |
| Apold1 | Apolipoprotein L domain-containing 1 |
| Poc1a | POC1 centriolar protein A |
| Ano3 | Anoctamin 3 |
| Kcnh7 | Potassium voltage-gated channel subfamily H member 7 |
| Mme | Membrane metalloendopeptidase |
| Atp2b1 | ATPase plasma membrane Ca2+ Transporting 1 |
| Cnr1 | Cannabinoid receptor 1 |
| Xrcc4 | X-ray repair cross complementing 4 |
| Camk4 | Calcium/calmodulin dependent protein kinase IV |
| Dclk3 | Doublecortin-like kinase 3 |
| Pde10a | Phosphodiesterase 10A |
| Rgs4 | Regulator of G protein signaling 4 |
| Cdkl5 | Cyclin-dependent kinase-like 5 |
| Homer1 | Homer scaffold protein 1 |
| Hivep2 | HIVEP zinc finger 2 |
| Lrrc8b | Leucine rich repeat-containing 8 VRAC subunit B |
| Syt1 | Synaptotagmin 1 |
| Ralbp1 | RalA binding protein 1 |
| Chil1 | Chitinase 3-like 1 |
| Fah | Fumarylacetoacetate hydrolase |
| Slc38a10 | Solute carrier family 38 member 10 |
| Ppard | Peroxisome proliferator activated receptor delta |
| Paqr3 | Progestin and adipoQ receptor family member 3 |
| Fbxo7 | F-box protein 7 |
| Phlpp2 | Phospholipid phosphatase 2 |
| Myo9b | Myosin IXB |
| Cnot3 | CCR4-NOT transcription complex subunit 3 |
| Abca7 | ATP-binding cassette A7 |
| Axl | AXL Receptor tyrosine kinase |
| Cerox1 | Cytoplasmic endogenous regulator of oxidative phosphorylation 1 |
| Abhd4 | Abhydrolase domain-containing 4: N-acyl phospholipase B |
| Plekhm2 | Pleckstrin homology and RUN domain containing M2 |
| Sbno2 | Strawberry notch homolog 2 |
| Rfxank | Regulatory factor X associated ankyrin containing protein |
| Dtymk | Deoxythymidylate kinase |
| Lrrc45 | Leucine rich repeat containing 45 |
| Snrnp70 | Small nuclear ribonucleoprotein U1 subunit 70 |
| Nrf2 | Nuclear factor erythroid-2-related factor 2 |
| GABA | γ-Aminobutyric acid |
| BBB | Blood brain barrier |
| ALS | Amyotrophic lateral sclerosis |
Author contributions
Conceptualization and data curation: Oh Yoen Kim and Juhyun Song; formal analysis, investigation, and methodology: Danbi Jo, Archana Arjunan, Seoyoon Choi, Yoon Seok Jung, Jihyun Park, Oh Yoen Kim, and Juhyun Song; validation and visualization: Danbi Jo, Oh Yoen Kim, and Juhyun Song; writing – original draft: Danbi Jo, Oh Yoen Kim and Juhyun Song; writing – review and editing: Juhyun Song; funding acquisition and supervision: Oh Yoen Kim and Juhyun Song. All authors have read and approved the final manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF grant 2022R1A2C1006125 to Juhyun Song and 2022R1A2C1010398 to Oh Yoen Kim).References
- D. J. Selkoe, Alzheimer's disease: genes, proteins, and therapy, Physiol. Rev., 2001, 81, 741–766 CrossRef CAS PubMed.
- Y. E. Geda, L. S. Schneider, L. N. Gitlin, D. S. Miller, G. S. Smith, J. Bell, J. Evans, M. Lee, A. Porsteinsson, K. L. Lanctot, P. B. Rosenberg, D. L. Sultzer, P. T. Francis, H. Brodaty, P. P. Padala, C. U. Onyike, L. A. Ortiz, S. Ancoli-Israel, D. L. Bliwise, J. L. Martin, M. V. Vitiello, K. Yaffe, P. C. Zee, N. Herrmann, R. A. Sweet, C. Ballard, N. A. Khin, C. Alfaro, P. S. Murray, S. Schultz, C. G. Lyketsos and I. Neuropsychiatric, Syndromes Professional Interest Area of, Neuropsychiatric symptoms in Alzheimer's disease: past progress and anticipation of the future, Alzheimers Dement., 2013, 9, 602–608 CrossRef PubMed.
- L. Ferretti, S. M. McCurry, R. Logsdon, L. Gibbons and L. Teri, Anxiety and Alzheimer's disease, J. Geriatr. Psychiatry Neurol., 2001, 14, 52–58 CrossRef CAS PubMed.
- C. G. Lyketsos, O. Lopez, B. Jones, A. L. Fitzpatrick, J. Breitner and S. DeKosky, Prevalence of neuropsychiatric symptoms in dementia and mild cognitive impairment: results from the cardiovascular health study, J. Am. Med. Assoc., 2002, 288, 1475–1483 CrossRef PubMed.
- S. Babri, G. Mohaddes, I. Feizi, A. Mohammadnia, A. Niapour, A. Alihemmati and M. Amani, Effect of troxerutin on synaptic plasticity of hippocampal dentate gyrus neurons in a beta-amyloid model of Alzheimer's disease: an electrophysiological study, Eur. J. Pharmacol., 2014, 732, 19–25 CrossRef CAS PubMed.
- C. Ballard and A. Corbett, Management of neuropsychiatric symptoms in people with dementia, CNS Drugs, 2010, 24, 729–739 Search PubMed.
- A. J. Petkus, C. A. Reynolds, J. L. Wetherell, W. S. Kremen, N. L. Pedersen and M. Gatz, Anxiety is associated with increased risk of dementia in older Swedish twins, Alzheimers Dement., 2016, 12, 399–406 CrossRef PubMed.
- D. Gallagher, R. Coen, D. Kilroy, K. Belinski, I. Bruce, D. Coakley, B. Walsh, C. Cunningham and B. A. Lawlor, Anxiety and behavioural disturbance as markers of prodromal Alzheimer's disease in patients with mild cognitive impairment, Int. J. Geriatr. Psychiatry, 2011, 26, 166–172 CrossRef PubMed.
- J. N. Perusini and M. S. Fanselow, Neurobehavioral perspectives on the distinction between fear and anxiety, Learn. Mem., 2015, 22, 417–425 CrossRef CAS PubMed.
- J. P. Aggleton, A. Pralus, A. J. Nelson and M. Hornberger, Thalamic pathology and memory loss in early Alzheimer's disease: moving the focus from the medial temporal lobe to Papez circuit, Brain, 2016, 139, 1877–1890 CrossRef PubMed.
- D. R. Thal, U. Rub, M. Orantes and H. Braak, Phases of A beta-deposition in the human brain and its relevance for the development of AD, Neurology, 2002, 58, 1791–1800 CrossRef PubMed.
- G. W. Van Hoesen, J. C. Augustinack, J. Dierking, S. J. Redman and R. Thangavel, The parahippocampal gyrus in Alzheimer's disease. Clinical and preclinical neuroanatomical correlates, Ann. N. Y. Acad. Sci., 2000, 911, 254–274 CrossRef CAS PubMed.
- G. A. Carlesimo, F. Piras, M. D. Orfei, M. Iorio, C. Caltagirone and G. Spalletta, Atrophy of presubiculum and subiculum is the earliest hippocampal anatomical marker of Alzheimer's disease, Alzheimers Dement., 2015, 1, 24–32 Search PubMed.
- J. Zhang, Y. F. Zhen, P. B. C. Ren, L. G. Song, W. N. Kong, T. M. Shao, X. Li and X. Q. Chai, Salidroside attenuates beta amyloid-induced cognitive deficits via modulating oxidative stress and inflammatory mediators in rat hippocampus, Behav. Brain Res., 2013, 244, 70–81 CrossRef CAS PubMed.
- A. Gella and N. Durany, Oxidative stress in Alzheimer disease, Cell Adhes. Migr., 2009, 3, 88–93 CrossRef PubMed.
- A. Basli, S. Soulet, N. Chaher, J. M. Merillon, M. Chibane, J. P. Monti and T. Richard, Wine polyphenols: potential agents in neuroprotection, Oxid. Med. Cell. Longevity, 2012, 2012, 805762 Search PubMed.
- A. Dal-Pan, S. Dudonne, P. Bourassa, M. Bourdoulous, C. Tremblay, Y. Desjardins, F. Calon and C. Neurophenols, Cognitive-Enhancing Effects of a Polyphenols-Rich Extract from Fruits without Changes in Neuropathology in an Animal Model of Alzheimer's Disease, J. Alzheimer's Dis., 2017, 55, 115–135 CAS.
- H. M. Hassan, M. R. Elnagar, E. Abdelrazik, M. R. Mahdi, E. Hamza, E. M. Elattar, E. M. ElNashar, M. A. Alghamdi, Z. Al-Qahtani, K. M. Al-Khater, R. A. Aldahhan and E. L. M, Neuroprotective effect of naringin against cerebellar changes in Alzheimer's disease through modulation of autophagy, oxidative stress and tau expression: An experimental study, Front. Neuroanat., 2022, 16, 1012422 CrossRef CAS PubMed.
- C. L. Ho, N. J. Kao, C. I. Lin, T. L. Cross and S. H. Lin, Quercetin Increases Mitochondrial Biogenesis and Reduces Free Radicals in Neuronal SH-SY5Y Cells, Nutrients, 2022, 14, 3310 CrossRef CAS PubMed.
- A. A. Gomaa, H. S. M. Farghaly, R. M. Makboul, A. M. Hussien and M. A. Nicola, Polyphenols from Conyza dioscoridis (L.) ameliorate Alzheimer's disease- like alterations through multi-targeting activities in two animal models, BMC Complementary Med. Ther., 2022, 22, 288 CrossRef CAS PubMed.
- Y. D. Wei, X. X. Chen, L. J. Yang, X. R. Gao, Q. R. Xia, C. C. Qi and J. F. Ge, Resveratrol ameliorates learning and memory impairments induced by bilateral hippocampal injection of streptozotocin in mice, Neurochem. Int., 2022, 159, 105385 CrossRef CAS PubMed.
- Y. Y. Choi, T. Maeda, H. Fujii, T. Yokozawa, H. Y. Kim, E. J. Cho and T. Shibamoto, Oligonol improves memory and cognition under an amyloid beta(25-35)-induced Alzheimer's mouse model, Nutr. Res., 2014, 34, 595–603 CrossRef CAS PubMed.
- T. Sakurai, K. Kitadate, H. Nishioka, H. Fujii, J. Ogasawara, T. Kizaki, S. Sato, T. Fujiwara, K. Akagawa, T. Izawa and H. Ohno, Oligomerised lychee fruit-derived polyphenol attenuates cognitive impairment in senescence-accelerated mice and endoplasmic reticulum stress in neuronal cells, Br. J. Nutr., 2013, 110, 1549–1558 CrossRef CAS PubMed.
- J. S. Noh, C. H. Park and T. Yokozawa, Treatment with oligonol, a low-molecular polyphenol derived from lychee fruit, attenuates diabetes-induced hepatic damage through regulation of oxidative stress and lipid metabolism, Br. J. Nutr., 2011, 106, 1013–1022 CrossRef CAS PubMed.
- R. Yamanishi, E. Yoshigai, T. Okuyama, M. Mori, H. Murase, T. Machida, T. Okumura and M. Nishizawa, The anti-inflammatory effects of flavanol-rich lychee fruit extract in rat hepatocytes, PLoS One, 2014, 9, e93818 CrossRef PubMed.
- M. Zhang, L. Zhong, X. Han, G. Xiong, D. Xu, S. Zhang, H. Cheng, K. Chiu and Y. Xu, Brain and Retinal Abnormalities in the 5xFAD Mouse Model of Alzheimer's Disease at Early Stages, Front. Neurosci., 2021, 15, 681831 CrossRef PubMed.
- K. E. Bernstein, Y. Koronyo, B. C. Salumbides, J. Sheyn, L. Pelissier, D. H. Lopes, K. H. Shah, E. A. Bernstein, D. T. Fuchs, J. J. Yu, M. Pham, K. L. Black, X. Z. Shen, S. Fuchs and M. Koronyo-Hamaoui, Angiotensin-converting enzyme overexpression in myelomonocytes prevents Alzheimer's-like cognitive decline, J. Clin. Invest., 2014, 124, 1000–1012 CrossRef CAS PubMed.
- R. M. Deacon, Digging and marble burying in mice: simple methods for in vivo identification of biological impacts, Nat. Protoc., 2006, 1, 122–124 CrossRef CAS PubMed.
- S. L. Thompson, A. C. Welch, E. V. Ho, J. M. Bessa, C. Portugal-Nunes, M. Morais, J. W. Young, J. A. Knowles and S. C. Dulawa, Btbd3 expression regulates compulsive-like and exploratory behaviors in mice, Transl. Psychiatry, 2019, 9, 222 CrossRef PubMed.
- N. L. Bray, H. Pimentel, P. Melsted and L. Pachter, Near-optimal probabilistic RNA-seq quantification, Nat. Biotechnol., 2016, 34, 525–527 CrossRef CAS PubMed.
- M. D. Robinson, D. J. McCarthy and G. K. Smyth, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 2010, 26, 139–140 CrossRef CAS PubMed.
- A. Liberzon, A. Subramanian, R. Pinchback, H. Thorvaldsdottir, P. Tamayo and J. P. Mesirov, Molecular signatures database (MSigDB) 3.0, Bioinformatics, 2011, 27, 1739–1740 CrossRef CAS PubMed.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen and C. V. Mering, STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- S. Oddo, A. Caccamo, J. D. Shepherd, M. P. Murphy, T. E. Golde, R. Kayed, R. Metherate, M. P. Mattson, Y. Akbari and F. M. LaFerla, Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction, Neuron, 2003, 39, 409–421 CrossRef CAS PubMed.
- H. Oakley, S. L. Cole, S. Logan, E. Maus, P. Shao, J. Craft, A. Guillozet-Bongaarts, M. Ohno, J. Disterhoft, L. Van Eldik, R. Berry and R. Vassar, Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation, J. Neurosci., 2006, 26, 10129–10140 CrossRef CAS PubMed.
- H. K. Bhakta, P. Paudel, H. Fujii, A. Sato, C. H. Park, T. Yokozawa, H. A. Jung and J. S. Choi, Oligonol promotes glucose uptake by modulating the insulin signaling pathway in insulin-resistant HepG2 cells via inhibiting protein tyrosine phosphatase 1B, Arch. Pharmacal Res., 2017, 40, 1314–1327 CrossRef CAS PubMed.
- M. Mani, S. Khaghani, T. Gol Mohammadi, Z. Zamani, K. Azadmanesh, R. Meshkani, P. Pasalar and E. Mostafavi, Activation of Nrf2-Antioxidant Response Element Mediated Glutamate Cysteine Ligase Expression in Hepatoma Cell line by Homocysteine, Hepatitis Mon., 2013, 13, e8394 Search PubMed.
- E. N. Terry, P. H. Gann, R. Molokie, M. Deininger and A. M. Diamond, Changes in the activity of the GPx-1 anti-oxidant selenoenzyme in mononuclear cells following imatinib treatment, Leuk. Res., 2011, 35, 831–833 CrossRef CAS PubMed.
- V. Srivastava, B. Buzas, R. Momenan, G. Oroszi, A. J. Pulay, M. A. Enoch, D. W. Hommer and D. Goldman, Association of SOD2, a mitochondrial antioxidant enzyme, with gray matter volume shrinkage in alcoholics, Neuropsychopharmacology, 2010, 35, 1120–1128 CrossRef CAS PubMed.
- Y. Zhang, Y. Xu, W. Lu, J. Li, S. Yu, E. J. Brown, B. Z. Stanger, J. D. Rabinowitz and X. Yang, G6PD-mediated increase in de novo NADP(+) biosynthesis promotes antioxidant defense and tumor metastasis, Sci. Adv., 2022, 8, eabo0404 CrossRef CAS PubMed.
- Z. Zhang, J. Qu, C. Zheng, P. Zhang, W. Zhou, W. Cui, X. Mo, L. Li, L. Xu and J. Gao, Nrf2 antioxidant pathway suppresses Numb-mediated epithelial-mesenchymal transition during pulmonary fibrosis, Cell Death Dis., 2018, 9, 83 CrossRef PubMed.
- J. A. Godoy-Lugo, M. A. Thorwald, D. A. Mendez, R. Rodriguez, D. Nakano, A. Nishiyama and R. M. Ortiz, Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade, Int. J. Mol. Sci., 2022, 23, 10897 CrossRef CAS PubMed.
- Y. C. Kuo, I. Y. Chen, S. C. Chang, S. C. Wu, T. M. Hung, P. H. Lee, K. Shimotohno and M. F. Chang, Hepatitis C virus NS5A protein enhances gluconeogenesis through upregulation of Akt-/JNK-PEPCK signalling pathways, Liver Int., 2014, 34, 1358–1368 CrossRef CAS PubMed.
- B. Onken, N. Kalinava and M. Driscoll, Gluconeogenesis and PEPCK are critical components of healthy aging and dietary restriction life extension, PLoS Genet., 2020, 16, e1008982 CrossRef CAS PubMed.
- I. Valle, A. Alvarez-Barrientos, E. Arza, S. Lamas and M. Monsalve, PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells, Cardiovasc. Res., 2005, 66, 562–573 CrossRef CAS PubMed.
- A. M. Tormos, S. Perez-Garrido, R. Talens-Visconti, A. R. Nebreda and J. Sastre, Long term p38-a deficiency up-regulates antioxidant enzymes through compensatory NF-?B activation, Free Radicals Biol. Med., 2014, 75(Suppl 1), S52 CrossRef PubMed.
- J. Iacovelli, G. C. Rowe, A. Khadka, D. Diaz-Aguilar, C. Spencer, Z. Arany and M. Saint-Geniez, PGC-1alpha Induces Human RPE Oxidative Metabolism and Antioxidant Capacity, Invest. Ophthalmol. Visual Sci., 2016, 57, 1038–1051 CrossRef CAS PubMed.
- Y. Suzuki and I. Imayoshi, Network analysis of exploratory behaviors of mice in a spatial learning and memory task, PLoS One, 2017, 12, e0180789 CrossRef PubMed.
- T. Illouz, R. Madar and E. Okun, A modified Barnes maze for an accurate assessment of spatial learning in mice, J. Neurosci. Methods, 2020, 334, 108579 CrossRef PubMed.
- C. A. Barnes, Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat, J. Comp. Physiol. Psychol., 1979, 93, 74–104 CAS.
- F. P. Varodayan, H. Sidhu, M. Kreifeldt, M. Roberto and C. Contet, Morphological and functional evidence of increased excitatory signaling in the prelimbic cortex during ethanol withdrawal, Neuropharmacology, 2018, 133, 470–480 CrossRef CAS PubMed.
- K. Gawel, E. Gibula, M. Marszalek-Grabska, J. Filarowska and J. H. Kotlinska, Assessment of spatial learning and memory in the Barnes maze task in rodents-methodological consideration, Naunyn Schmiedebergs Arch. Pharmacol., 2019, 392, 1–18 CrossRef CAS PubMed.
- A. Attar, T. Liu, W. T. Chan, J. Hayes, M. Nejad, K. Lei and G. Bitan, A shortened Barnes maze protocol reveals memory deficits at 4-months of age in the triple-transgenic mouse model of Alzheimer's disease, PLoS One, 2013, 8, e80355 CrossRef PubMed.
- T. P. O'Leary and R. E. Brown, Visuo-spatial learning and memory deficits on the Barnes maze in the 16-month-old APPswe/PS1dE9 mouse model of Alzheimer's disease, Behav. Brain Res., 2009, 201, 120–127 CrossRef PubMed.
- K. Njung'e and S. L. Handley, Evaluation of marble-burying behavior as a model of anxiety, Pharmacol., Biochem. Behav., 1991, 38, 63–67 CrossRef PubMed.
- G. T. Taylor, S. Lerch and S. Chourbaji, Marble burying as compulsive behaviors in male and female mice, Acta Neurobiol. Exp., 2017, 77, 254–260 CrossRef.
- C. L. Broekkamp, H. W. Rijk, D. Joly-Gelouin and K. L. Lloyd, Major tranquillizers can be distinguished from minor tranquillizers on the basis of effects on marble burying and swim-induced grooming in mice, Eur. J. Pharmacol., 1986, 126, 223–229 CrossRef CAS PubMed.
- V. Torres-Lista, S. Lopez-Pousa and L. Gimenez-Llort, Marble-burying is enhanced in 3xTg-AD mice, can be reversed by risperidone and it is modulable by handling, Behav. Processes, 2015, 116, 69–74 CrossRef PubMed.
- V. Torres-Lista and L. Gimenez-Llort, Impairment of nesting behaviour in 3xTg-AD mice, Behav. Brain Res., 2013, 247, 153–157 CrossRef PubMed.
- F. Dorninger, G. Zeitler and J. Berger, Nestlet Shredding and Nest Building Tests to Assess Features of Psychiatric Disorders in Mice, Bio-Protoc., 2020, 10, e3863 CAS.
- P. Jirkof, Burrowing and nest building behavior as indicators of well-being in mice, J. Neurosci. Methods, 2014, 234, 139–146 CrossRef PubMed.
- M. Angoa-Perez, M. J. Kane, D. I. Briggs, D. M. Francescutti and D. M. Kuhn, Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice, J. Visualized Exp., 2013, 50978, DOI:10.3791/50978.
- T. N. Sager, J. Kirchhoff, A. Mork, J. Van Beek, K. Thirstrup, M. Didriksen and J. B. Lauridsen, Nest building performance following MPTP toxicity in mice, Behav. Brain Res., 2010, 208, 444–449 CrossRef CAS PubMed.
- R. M. Deacon, A. Croucher and J. N. Rawlins, Hippocampal cytotoxic lesion effects on species-typical behaviours in mice, Behav. Brain Res., 2002, 132, 203–213 CrossRef PubMed.
- J. Hannon and D. Hoyer, Molecular biology of 5-HT receptors, Behav. Brain Res., 2008, 195, 198–213 CrossRef CAS PubMed.
- C. Bombardi, Neuronal localization of the 5-HT2 receptor family in the amygdaloid complex, Front. Pharmacol., 2014, 5, 68 Search PubMed.
- J. Versijpt, K. J. Van Laere, F. Dumont, D. Decoo, M. Vandecapelle, P. Santens, I. Goethals, K. Audenaert, G. Slegers, R. A. Dierckx and J. Korf, Imaging of the 5-HT2A system: age-, gender-, and Alzheimer's disease-related findings, Neurobiol. Aging, 2003, 24, 553–561 CrossRef CAS PubMed.
- S. Khaliq, S. Haider, S. P. Ahmed, T. Perveen and D. J. Haleem, Relationship of brain tryptophan and serotonin in improving cognitive performance in rats, Pak. J. Pharm. Sci., 2006, 19, 11–15 CAS.
- L. Marner, V. G. Frokjaer, J. Kalbitzer, S. Lehel, K. Madsen, W. F. Baare, G. M. Knudsen and S. G. Hasselbalch, Loss of serotonin 2A receptors exceeds loss of serotonergic projections in early Alzheimer's disease: a combined [11C]DASB and [18F]altanserin-PET study, Neurobiol. Aging, 2012, 33, 479–487 CrossRef CAS PubMed.
- D. Sierra-Mercado, N. Padilla-Coreano and G. J. Quirk, Dissociable roles of prelimbic and infralimbic cortices, ventral hippocampus, and basolateral amygdala in the expression and extinction of conditioned fear, Neuropsychopharmacology, 2011, 36, 529–538 CrossRef PubMed.
- G. Zhang, H. N. Asgeirsdottir, S. J. Cohen, A. H. Munchow, M. P. Barrera and R. W. Stackman Jr., Stimulation of serotonin 2A receptors facilitates consolidation and extinction of fear memory in C57BL/6J mice, Neuropharmacology, 2013, 64, 403–413 CrossRef CAS PubMed.
- G. J. Quirk, D. Pare, R. Richardson, C. Herry, M. H. Monfils, D. Schiller and A. Vicentic, Erasing fear memories with extinction training, J. Neurosci., 2010, 30, 14993–14997 CrossRef CAS PubMed.
- C. S. Grob, A. L. Danforth, G. S. Chopra, M. Hagerty, C. R. McKay, A. L. Halberstadt and G. R. Greer, Pilot study of psilocybin treatment for anxiety in patients with advanced-stage cancer, Arch. Gen. Psychiatry, 2011, 68, 71–78 CrossRef CAS PubMed.
- K. A. Jones, D. P. Srivastava, J. A. Allen, R. T. Strachan, B. L. Roth and P. Penzes, Rapid modulation of spine morphology by the 5-HT2A serotonin receptor through kalirin-7 signaling, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 19575–19580 CrossRef CAS PubMed.
- Z. Xia, J. A. Gray, B. A. Compton-Toth and B. L. Roth, A direct interaction of PSD-95 with 5-HT2A serotonin receptors regulates receptor trafficking and signal transduction, J. Biol. Chem., 2003, 278, 21901–21908 CrossRef CAS PubMed.
- M. J. Millan, P. Marin, J. Bockaert and C. Mannoury la Cour, Signaling at G-protein-coupled serotonin receptors: recent advances and future research directions, Trends Pharmacol. Sci., 2008, 29, 454–464 CrossRef CAS PubMed.
- Y. Yan, J. Wang, B. Qu, Y. Zhang, Y. Wei, H. Liu and C. Wu, CXCL13 and TH1/Th2 cytokines in the serum and cerebrospinal fluid of neurosyphilis patients, Medicine, 2017, 96, e8850 CrossRef CAS PubMed.
- K. E. Cole, C. A. Strick, T. J. Paradis, K. T. Ogborne, M. Loetscher, R. P. Gladue, W. Lin, J. G. Boyd, B. Moser, D. E. Wood, B. G. Sahagan and K. Neote, Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3, J. Exp. Med., 1998, 187, 2009–2021 CrossRef CAS PubMed.
- Z. Beizavi, M. Zohouri, M. Asadipour and A. Ghaderi, IL-27, a pleiotropic cytokine for fine-tuning the immune response in cancer, Int. Rev. Immunol., 2021, 40, 319–329 CrossRef CAS PubMed.
- L. B. Ivashkiv, IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy, Nat. Rev. Immunol., 2018, 18, 545–558 CrossRef CAS PubMed.
- N. R. West, A. N. Hegazy, B. M. J. Owens, S. J. Bullers, B. Linggi, S. Buonocore, M. Coccia, D. Gortz, S. This, K. Stockenhuber, J. Pott, M. Friedrich, G. Ryzhakov, F. Baribaud, C. Brodmerkel, C. Cieluch, N. Rahman, G. Muller-Newen, R. J. Owens, A. A. Kuhl, K. J. Maloy, S. E. Plevy, Oxford IBD Cohort Investigators, S. Keshav, S. P. L. Travis and F. Powrie, Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease, Nat. Med., 2017, 23, 579–589 CrossRef CAS PubMed.
- F. Vafaee, A. Zarifkar, M. Emamghoreishi, M. R. Namavar, S. Shirzad, H. Ghazavi and V. Mahdavizadeh, Insulin-Like Growth Factor 2 (IGF-2) Regulates Neuronal Density and IGF-2 Distribution Following Hippocampal Intracerebral Hemorrhage, J. Stroke Cerebrovasc. Dis., 2020, 29, 105128 CrossRef PubMed.
- H. Jia, J. Cheng, Q. Zhou, J. Peng, Y. Pan and H. Han, Fibroblast growth factor 21 attenuates inflammation and oxidative stress in atherosclerotic rat via enhancing the Nrf1-ARE signaling pathway, Int. J. Clin. Exp. Pathol., 2018, 11, 1308–1317 Search PubMed.
- M. H. Qiu, M. C. Chen, Z. L. Huang and J. Lu, Neuronal activity (c-Fos) delineating interactions of the cerebral cortex and basal ganglia, Front. Neuroanat., 2014, 8, 13 Search PubMed.
- E. Knapska and S. Maren, Reciprocal patterns of c-Fos expression in the medial prefrontal cortex and amygdala after extinction and renewal of conditioned fear, Learn. Mem., 2009, 16, 486–493 CrossRef PubMed.
- Y. Sun, H. Zhu, R. Cheng, Z. Tang and M. Zhang, Outer membrane protein Amuc_1100 of Akkermansia muciniphila alleviates antibiotic-induced anxiety and depression-like behavior in mice, Physiol. Behav., 2023, 258, 114023 CrossRef CAS PubMed.
- C. L. Ge, W. Chen, L. N. Zhang, Y. H. Ai, Y. Zou and Q. Y. Peng, Hippocampus-prefrontal cortex inputs modulate spatial learning and memory in a mouse model of sepsis induced by cecal ligation puncture, CNS Neurosci. Ther., 2023, 29, 390–401 CrossRef CAS PubMed.
- K. S. Yoo, K. Lee, J. Y. Oh, H. Lee, H. Park, Y. S. Park and H. K. Kim, Postsynaptic density protein 95 (PSD-95) is transported by KIF5 to dendritic regions, Mol. Brain, 2019, 12, 97 CrossRef CAS PubMed.
- M. J. Broadhead, M. H. Horrocks, F. Zhu, L. Muresan, R. Benavides-Piccione, J. DeFelipe, D. Fricker, M. V. Kopanitsa, R. R. Duncan, D. Klenerman, N. H. Komiyama, S. F. Lee and S. G. Grant, PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits, Sci. Rep., 2016, 6, 24626 CrossRef CAS PubMed.
- K. S. Christopherson, N. T. Sweeney, S. E. Craven, R. Kang, D. El-Husseini Ael and D. S. Bredt, Lipid- and protein-mediated multimerization of PSD-95: implications for receptor clustering and assembly of synaptic protein networks, J. Cell Sci., 2003, 116, 3213–3219 CrossRef CAS PubMed.
- A. Yoshii and M. Constantine-Paton, Postsynaptic localization of PSD-95 is regulated by all three pathways downstream of TrkB signaling, Front. Synaptic Neurosci., 2014, 6, 6 CAS.
- Y. Li, H. Wang, Y. Gao, R. Zhang, Q. Liu, W. Xie, Z. Liu, D. Geng and L. Wang, Circ-Vps41 positively modulates Syp and its overexpression improves memory ability in aging mice, Front. Mol. Neurosci., 2022, 15, 1037912 CrossRef CAS PubMed.
- A. K. Jyothi, B. Thotakura, S. C. Priyadarshini, S. Patil, M. S. Poojari and M. Subramanian, Paternal stress alters synaptic density and expression of GAP-43, GRIN1, M1 and SYP genes in the hippocampus and cortex of offspring of stress-induced male rats, Morphologie, 2022, 107, 67–79 CrossRef PubMed.
- A. Karimani, N. Ramezani, A. Afkhami Goli, M. H. Nazem Shirazi, H. Nourani and A. M. Jafari, Subchronic neurotoxicity of diazinon in albino mice: Impact of oxidative stress, AChE activity, and gene expression disturbances in the cerebral cortex and hippocampus on mood, spatial learning, and memory function, Toxicol. Rep., 2021, 8, 1280–1288 CrossRef CAS PubMed.
- X. M. Qi, L. L. Miao, Y. Cai, L. K. Gong and J. Ren, ROS generated by CYP450, especially CYP2E1, mediate mitochondrial dysfunction induced by tetrandrine in rat hepatocytes, Acta Pharmacol. Sin., 2013, 34, 1229–1236 CrossRef CAS PubMed.
- A. Veith and B. Moorthy, Role of Cytochrome P450s in the Generation and Metabolism of Reactive Oxygen Species, Curr. Opin. Toxicol., 2018, 7, 44–51 CrossRef PubMed.
- L. T. Knapp and E. Klann, Role of reactive oxygen species in hippocampal long-term potentiation: contributory or inhibitory?, J. Neurosci. Res., 2002, 70, 1–7 CrossRef CAS PubMed.
- P. Liu, J. Wang, S. Peng, D. Zhang, L. Zhuang, C. Liu, Y. Zhang and X. Shi, Suppression of phosphodiesterase IV enzyme by roflumilast ameliorates cognitive dysfunction in aged rats after sevoflurane anaesthesia via PKA-CREB and MEK/ERK pathways, Eur. J. Neurosci., 2022, 56, 4317–4332 CrossRef CAS PubMed.
- X. X. Xu, R. X. Shi, Y. Fu, J. L. Wang, X. Tong, S. Q. Zhang, N. Wang, M. X. Li, Y. Tong, W. Wang, M. He, B. Y. Liu, G. L. Chen and F. Guo, Neuronal nitric oxide synthase/reactive oxygen species pathway is involved in apoptosis and pyroptosis in epilepsy, Neural Regener. Res., 2023, 18, 1277–1285 CrossRef PubMed.
- L. F. Liu, Y. Hu, Y. N. Liu, D. W. Shi, C. Liu, X. Da, S. H. Zhu, Q. Y. Zhu, J. Q. Zhang and G. H. Xu, Reactive oxygen species contribute to delirium-like behavior by activating CypA/MMP9 signaling and inducing blood-brain barrier impairment in aged mice following anesthesia and surgery, Front. Aging Neurosci., 2022, 14, 1021129 CrossRef CAS PubMed.
- S. S. Schattauer, A. Bedini, F. Summers, A. Reilly-Treat, M. M. Andrews, B. B. Land and C. Chavkin, Reactive oxygen species (ROS) generation is stimulated by kappa opioid receptor activation through phosphorylated c-Jun N-terminal kinase and inhibited by p38 mitogen-activated protein kinase (MAPK) activation, J. Biol. Chem., 2019, 294, 16884–16896 CrossRef CAS PubMed.
- K. J. Cho, H. W. Kim, S. Y. Cheon, J. E. Lee and G. W. Kim, Apoptosis signal-regulating kinase-1 aggravates ROS-mediated striatal degeneration in 3-nitropropionic acid-infused mice, Biochem. Biophys. Res. Commun., 2013, 441, 280–285 CrossRef CAS PubMed.
- L. Amador-Alvarado, T. Montiel and L. Massieu, Differential production of reactive oxygen species in distinct brain regions of hypoglycemic mice, Metab. Brain Dis., 2014, 29, 711–719 CrossRef CAS PubMed.
- T. Kitada, A. Pisani, D. R. Porter, H. Yamaguchi, A. Tscherter, G. Martella, P. Bonsi, C. Zhang, E. N. Pothos and J. Shen, Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11441–11446 CrossRef CAS PubMed.
- A. S. Gravesteijn, H. Beckerman, E. A. Willemse, H. E. Hulst, B. A. de Jong, C. E. Teunissen and V. de Groot, Brain-derived neurotrophic factor, neurofilament light and glial fibrillary acidic protein do not change in response to aerobic training in people with MS-related fatigue - a secondary analysis of a randomized controlled trial, Mult. Scler. Relat. Disord., 2022, 70, 104489 CrossRef PubMed.
- A. D. Garcia, N. B. Doan, T. Imura, T. G. Bush and M. V. Sofroniew, GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain, Nat. Neurosci., 2004, 7, 1233–1241 CrossRef CAS PubMed.
- C. Li, Y. Zhu, Y. Wu, M. Fu, Y. Wu, Y. Wu, Y. Qiu, H. Zhang and M. Ding, Oridonin Alleviates LPS-Induced Depression by Inhibiting NLRP3 Inflammasome via Activation of Autophagy, Front. Med., 2021, 8, 813047 CrossRef PubMed.
- Y. Ying, G. Xiang, M. Chen, J. Ye, Q. Wu, H. Dou, S. Sheng and S. Zhu, Gelatine nanostructured lipid carrier encapsulated FGF15 inhibits autophagy and improves recovery in spinal cord injury, Cell Death Discovery, 2020, 6, 137 CrossRef CAS PubMed.
- C. Habak, A. Noreau, A. Nagano-Saito, B. Mejia-Constain, C. Degroot, A. P. Strafella, S. Chouinard, A. L. Lafontaine, G. A. Rouleau and O. Monchi, Dopamine transporter SLC6A3 genotype affects cortico-striatal activity of set-shifts in Parkinson's disease, Brain, 2014, 137, 3025–3035 CrossRef PubMed.
- M. E. A. Reith, S. Kortagere, C. E. Wiers, H. Sun, M. A. Kurian, A. Galli, N. D. Volkow and Z. Lin, The dopamine transporter gene SLC6A3: multidisease risks, Mol. Psychiatry, 2022, 27, 1031–1046 CrossRef CAS PubMed.
- D. J. Walther, J. U. Peter, S. Bashammakh, H. Hortnagl, M. Voits, H. Fink and M. Bader, Synthesis of serotonin by a second tryptophan hydroxylase isoform, Science, 2003, 299, 76 CrossRef CAS PubMed.
- L. Gutknecht, N. Araragi, S. Merker, J. Waider, F. M. Sommerlandt, B. Mlinar, G. Baccini, U. Mayer, F. Proft, M. Hamon, A. G. Schmitt, R. Corradetti, L. Lanfumey and K. P. Lesch, Impacts of brain serotonin deficiency following Tph2 inactivation on development and raphe neuron serotonergic specification, PLoS One, 2012, 7, e43157 CrossRef CAS PubMed.
- F. Wasinski, M. R. Tavares, D. O. Gusmao, E. O. List, J. J. Kopchick, G. A. Alves, R. Frazao and J. Donato Jr., Central growth hormone action regulates neuroglial and proinflammatory markers in the hypothalamus of male mice, Neurosci. Lett., 2023, 806, 137236 CrossRef CAS PubMed.
- B. Guo, M. Qi, S. Huang, R. Zhuo, W. Zhang, Y. Zhang, M. Xu, M. Liu, T. Guan and Y. Liu, Cadherin-12 Regulates Neurite Outgrowth Through the PKA/Rac1/Cdc42 Pathway in Cortical Neurons, Front. Cell Dev. Biol., 2021, 9, 768970 CrossRef PubMed.
- J. B. Regard, S. Scheek, T. Borbiev, A. A. Lanahan, A. Schneider, A. M. Demetriades, H. Hiemisch, C. A. Barnes, A. D. Verin and P. F. Worley, Verge: a novel vascular early response gene, J. Neurosci., 2004, 24, 4092–4103 CrossRef CAS PubMed.
- Z. Fan, R. Ardicoglu, A. A. Batavia, R. Rust, L. von Ziegler, R. Waag, J. Zhang, T. Desgeorges, O. Sturman, H. Dang, R. Weber, M. Roszkowski, A. E. Moor, M. E. Schwab, P. L. Germain, J. Bohacek and K. De Bock, The vascular gene Apold1 is dispensable for normal development but controls angiogenesis under pathological conditions, Angiogenesis, 2023, 26, 385–407 CrossRef CAS PubMed.
- J. H. Chen, M. Segni, F. Payne, I. Huang-Doran, A. Sleigh, C. Adams, U. K. Consortium, D. B. Savage, S. O'Rahilly, R. K. Semple and I. Barroso, Truncation of POC1A associated with short stature and extreme insulin resistance, J. Mol. Endocrinol., 2015, 55, 147–158 CAS.
- X. Wang, Y. Su, H. Yan, Z. Huang, Y. Huang and W. Yue, Association Study of KCNH7 Polymorphisms and Individual Responses to Risperidone Treatment in Schizophrenia, Front. Psychiatry, 2019, 10, 633 CrossRef PubMed.
- J. Gao, Z. Qin, X. Qu, S. Wu, X. Xie, C. Liang and J. Liu, Endogenous neuroprotective mechanism of ATP2B1 in transcriptional regulation of ischemic preconditioning, Am. J. Transl. Res., 2021, 13, 1170–1183 CAS.
- A. A. Sultan, M. K. Dimick, C. C. Zai, J. L. Kennedy, B. J. MacIntosh and B. I. Goldstein, The association of CNR1 genetic variants with resting-state functional connectivity in youth bipolar disorder, Eur. Neuropsychopharmacol., 2023, 71, 41–54 CrossRef CAS PubMed.
- C. Leukel, D. Schumann, R. Kalisch, T. Sommer and N. Bunzeck, Dopamine Related Genes Differentially Affect Declarative Long-Term Memory in Healthy Humans, Front. Behav. Neurosci., 2020, 14, 539725 CrossRef CAS PubMed.
- H. Fukushima, R. Maeda, R. Suzuki, A. Suzuki, M. Nomoto, H. Toyoda, L. J. Wu, H. Xu, M. G. Zhao, K. Ueda, A. Kitamoto, N. Mamiya, T. Yoshida, S. Homma, S. Masushige, M. Zhuo and S. Kida, Upregulation of calcium/calmodulin-dependent protein kinase IV improves memory formation and rescues memory loss with aging, J. Neurosci., 2008, 28, 9910–9919 CrossRef CAS PubMed.
- L. Galvan, L. Francelle, M. C. Gaillard, L. de Longprez, M. A. Carrillo-de Sauvage, G. Liot, K. Cambon, L. Stimmer, S. Luccantoni, J. Flament, J. Valette, M. de Chaldee, G. Auregan, M. Guillermier, C. Josephine, F. Petit, C. Jan, M. Jarrige, N. Dufour, G. Bonvento, S. Humbert, F. Saudou, P. Hantraye, K. Merienne, A. P. Bemelmans, A. L. Perrier, N. Deglon and E. Brouillet, The striatal kinase DCLK3 produces neuroprotection against mutant huntingtin, Brain, 2018, 141, 1434–1454 CrossRef PubMed.
- R. Bonate, G. Kurek, M. Hrabak, S. Patterson, F. Padovan-Neto, A. R. West and H. Steiner, Phosphodiesterase 10A (PDE10A): Regulator of Dopamine Agonist-Induced Gene Expression in the Striatum, Cells, 2022, 11, 2214 CrossRef CAS PubMed.
- E. Schwarz, A gene-based review of RGS4 as a putative risk gene for psychiatric illness, Am. J. Med. Genet., Part B, 2018, 177, 267–273 CrossRef PubMed.
- M. Stratinaki, A. Varidaki, V. Mitsi, S. Ghose, J. Magida, C. Dias, S. J. Russo, V. Vialou, B. J. Caldarone, C. A. Tamminga, E. J. Nestler and V. Zachariou, Regulator of G protein signaling 4 [corrected] is a crucial modulator of antidepressant drug action in depression and neuropathic pain models, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 8254–8259 CrossRef CAS PubMed.
- C. D. Paspalas, L. D. Selemon and A. F. Arnsten, Mapping the regulator of G protein signaling 4 (RGS4): presynaptic and postsynaptic substrates for neuroregulation in prefrontal cortex, Cereb. Cortex, 2009, 19, 2145–2155 CrossRef PubMed.
- P. D. Negraes, C. A. Trujillo, N. K. Yu, W. Wu, H. Yao, N. Liang, J. D. Lautz, E. Kwok, D. McClatchy, J. Diedrich, S. M. de Bartolome, J. Truong, R. Szeto, T. Tran, R. H. Herai, S. E. P. Smith, G. G. Haddad, J. R. Yates 3rd and A. R. Muotri, Altered network and rescue of human neurons derived from individuals with early-onset genetic epilepsy, Mol. Psychiatry, 2021, 26, 7047–7068 CrossRef PubMed.
- J. Zhao, C. Chen, R. L. Bell, H. Qing and Z. Lin, Identification of HIVEP2 as a dopaminergic transcription factor related to substance use disorders in rats and humans, Transl. Psychiatry, 2019, 9, 247 CrossRef PubMed.
- H. Melland, F. Bumbak, A. Kolesnik-Taylor, E. Ng-Cordell, A. John, P. Constantinou, S. Joss, M. Larsen, C. Fagerberg, L. W. Laulund, J. Thies, F. Emslie, M. Willemsen, T. Kleefstra, R. Pfundt, R. Barrick, R. Chang, L. Loong, M. Alfadhel, J. van der Smagt, M. Nizon, M. A. Kurian, D. J. Scott, J. J. Ziarek, S. L. Gordon and K. Baker, Expanding the genotype and phenotype spectrum of SYT1-associated neurodevelopmental disorder, Genet. Med., 2022, 24, 880–893 CrossRef CAS PubMed.
- S. Awasthi, A. Hindle, N. A. Sawant, M. George, M. Vijayan, S. Kshirsagar, H. Morton, L. E. Bunquin, P. T. Palade, J. J. Lawrence, H. Khan, C. Bose, P. H. Reddy and S. P. Singh, RALBP1 in Oxidative Stress and Mitochondrial Dysfunction in Alzheimer’s Disease, Cells, 2021, 10, 3113 CrossRef CAS PubMed.
- B. Bennani-Baiti, S. Toegel, H. Viernstein, E. Urban, C. R. Noe and I. M. Bennani-Baiti, Inflammation Modulates RLIP76/RALBP1 Electrophile-Glutathione Conjugate Transporter and Housekeeping Genes in Human Blood-Brain Barrier Endothelial Cells, PLoS One, 2015, 10, e0139101 CrossRef PubMed.
- R. Tripathi, T. Aggarwal and R. Fredriksson, SLC38A10 Transporter Plays a Role in Cell Survival Under Oxidative Stress and Glutamate Toxicity, Front. Mol. Biosci., 2021, 8, 671865 CrossRef CAS PubMed.
- E. R. Doroshenko, P. C. Drohomyrecky, A. Gower, H. Whetstone, L. S. Cahill, M. Ganguly, S. Spring, T. J. Yi, J. G. Sled and S. E. Dunn, Peroxisome Proliferator-Activated Receptor-delta Deficiency in Microglia Results in Exacerbated Axonal Injury and Tissue Loss in Experimental Autoimmune Encephalomyelitis, Front. Immunol., 2021, 12, 570425 CrossRef CAS PubMed.
- L. Wang, X. Wang, Z. Li, T. Xia, L. Zhu, B. Liu, Y. Zhang, F. Xiao, Y. Pan, Y. Liu, F. Guo and Y. Chen, PAQR3 has modulatory roles in obesity, energy metabolism, and leptin signaling, Endocrinology, 2013, 154, 4525–4535 CrossRef CAS PubMed.
- T. Huang, L. Fang, R. He, H. Weng, X. Chen, Q. Ye and D. Qu, Fbxo7 and Pink1 play a reciprocal role in regulating their protein levels, Aging, 2020, 13, 77–88 CrossRef PubMed.
- M. Correa-Vela, V. Lupo, M. Montpeyo, P. Sancho, A. Marce-Grau, J. Hernandez-Vara, A. Darling, A. Jenkins, S. Fernandez-Rodriguez, C. Tello, L. Ramirez-Jimenez, B. Perez, A. Sanchez-Montanez, A. Macaya, M. J. Sobrido, M. Martinez-Vicente, B. Perez-Duenas and C. Espinos, Impaired proteasome activity and neurodegeneration with brain iron accumulation in FBXO7 defect, Ann. Clin. Transl. Neurol., 2020, 7, 1436–1442 CrossRef CAS PubMed.
- H. Long, X. Zhu, P. Yang, Q. Gao, Y. Chen and L. Ma, Myo9b and RICS modulate dendritic morphology of cortical neurons, Cereb. Cortex, 2013, 23, 71–79 CrossRef PubMed.
- P. Hollingworth, D. Harold, R. Sims, A. Gerrish, J. C. Lambert, M. M. Carrasquillo, R. Abraham, M. L. Hamshere, J. S. Pahwa, V. Moskvina, K. Dowzell, N. Jones, A. Stretton, C. Thomas, A. Richards, D. Ivanov, C. Widdowson, J. Chapman, S. Lovestone, J. Powell, P. Proitsi, M. K. Lupton, C. Brayne, D. C. Rubinsztein, M. Gill, B. Lawlor, A. Lynch, K. S. Brown, P. A. Passmore, D. Craig, B. McGuinness, S. Todd, C. Holmes, D. Mann, A. D. Smith, H. Beaumont, D. Warden, G. Wilcock, S. Love, P. G. Kehoe, N. M. Hooper, E. R. Vardy, J. Hardy, S. Mead, N. C. Fox, M. Rossor, J. Collinge, W. Maier, F. Jessen, E. Ruther, B. Schurmann, R. Heun, H. Kolsch, H. van den Bussche, I. Heuser, J. Kornhuber, J. Wiltfang, M. Dichgans, L. Frolich, H. Hampel, J. Gallacher, M. Hull, D. Rujescu, I. Giegling, A. M. Goate, J. S. Kauwe, C. Cruchaga, P. Nowotny, J. C. Morris, K. Mayo, K. Sleegers, K. Bettens, S. Engelborghs, P. P. De Deyn, C. Van Broeckhoven, G. Livingston, N. J. Bass, H. Gurling, A. McQuillin, R. Gwilliam, P. Deloukas, A. Al-Chalabi, C. E. Shaw, M. Tsolaki, A. B. Singleton, R. Guerreiro, T. W. Muhleisen, M. M. Nothen, S. Moebus, K. H. Jockel, N. Klopp, H. E. Wichmann, V. S. Pankratz, S. B. Sando, J. O. Aasly, M. Barcikowska, Z. K. Wszolek, D. W. Dickson, N. R. Graff-Radford, R. C. Petersen, Alzheimer's Disease Neuroimaging Initiative, C. M. van Duijn, M. M. Breteler, M. A. Ikram, A. L. DeStefano, A. L. Fitzpatrick, O. Lopez, L. J. Launer, S. Seshadri, C. Consortium, C. Berr, D. Campion, J. Epelbaum, J. F. Dartigues, C. Tzourio, A. Alperovitch, M. Lathrop, E. Consortium, T. M. Feulner, P. Friedrich, C. Riehle, M. Krawczak, S. Schreiber, M. Mayhaus, S. Nicolhaus, S. Wagenpfeil, S. Steinberg, H. Stefansson, K. Stefansson, J. Snaedal, S. Bjornsson, P. V. Jonsson, V. Chouraki, B. Genier-Boley, M. Hiltunen, H. Soininen, O. Combarros, D. Zelenika, M. Delepine, M. J. Bullido, F. Pasquier, I. Mateo, A. Frank-Garcia, E. Porcellini, O. Hanon, E. Coto, V. Alvarez, P. Bosco, G. Siciliano, M. Mancuso, F. Panza, V. Solfrizzi, B. Nacmias, S. Sorbi, P. Bossu, P. Piccardi, B. Arosio, G. Annoni, D. Seripa, A. Pilotto, E. Scarpini, D. Galimberti, A. Brice, D. Hannequin, F. Licastro, L. Jones, P. A. Holmans, T. Jonsson, M. Riemenschneider, K. Morgan, S. G. Younkin, M. J. Owen, M. O'Donovan, P. Amouyel and J. Williams, Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease, Nat. Genet., 2011, 43, 429–435 CrossRef CAS PubMed.
- S. Dib, J. Pahnke and F. Gosselet, Role of ABCA7 in Human Health and in Alzheimer’s Disease, Int. J. Mol. Sci., 2021, 22, 4603 CrossRef CAS PubMed.
- T. E. Syme, M. Grill, E. Hayashida, B. Viengkhou, I. L. Campbell and M. J. Hofer, Strawberry notch homolog 2 regulates the response to interleukin-6 in the central nervous system, J. Neuroinflammation, 2022, 19, 126 CrossRef CAS PubMed.
- M. Grill, T. E. Syme, A. L. Nocon, A. Z. Lu, D. Hancock, S. Rose-John and I. L. Campbell, Strawberry notch homolog 2 is a novel inflammatory response factor predominantly but not exclusively expressed by astrocytes in the central nervous system, Glia, 2015, 63, 1738–1752 CrossRef PubMed.
- T. Nakaya, A specific gene-splicing alteration in the SNRNP70 gene as a hallmark of an ALS subtype, Gene, 2022, 818, 146203 CrossRef CAS PubMed.
- T. Nakaya, Dissection of FUS domains involved in regulation of SnRNP70 gene expression, FEBS Lett., 2020, 594, 3518–3529 CrossRef CAS PubMed.
- H. Bading, Nuclear calcium signalling in the regulation of brain function, Nat. Rev. Neurosci., 2013, 14, 593–608 CrossRef CAS PubMed.
- B. Granseth and L. Lagnado, The role of endocytosis in regulating the strength of hippocampal synapses, J. Physiol., 2008, 586, 5969–5982 CrossRef CAS PubMed.
- M. Overhoff, E. De Bruyckere and N. L. Kononenko, Mechanisms of neuronal survival safeguarded by endocytosis and autophagy, J. Neurochem., 2021, 157, 263–296 CrossRef CAS PubMed.
- M. Manns, O. Leske, S. Gottfried, Z. Bichler, P. Lafenetre, P. Wahle and R. Heumann, Role of neuronal ras activity in adult hippocampal neurogenesis and cognition, Front. Neurosci., 2011, 5, 18 Search PubMed.
- J. R. Whitlock, A. J. Heynen, M. G. Shuler and M. F. Bear, Learning induces long-term potentiation in the hippocampus, Science, 2006, 313, 1093–1097 CrossRef CAS PubMed.
- Y. Ge, Z. Dong, R. C. Bagot, J. G. Howland, A. G. Phillips, T. P. Wong and Y. T. Wang, Hippocampal long-term depression is required for the consolidation of spatial memory, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16697–16702 CrossRef CAS PubMed.
- T. Sumi and K. Harada, Mechanism underlying hippocampal long-term potentiation and depression based on competition between endocytosis and exocytosis of AMPA receptors, Sci. Rep., 2020, 10, 14711 CrossRef CAS PubMed.
- H. Blockus, S. V. Rolotti, M. Szoboszlay, E. Peze-Heidsieck, T. Ming, A. Schroeder, N. Apostolo, K. M. Vennekens, P. S. Katsamba, F. Bahna, S. Mannepalli, G. Ahlsen, B. Honig, L. Shapiro, J. de Wit, A. Losonczy and F. Polleux, Synaptogenic activity of the axon guidance molecule Robo2 underlies hippocampal circuit function, Cell Rep., 2021, 37, 109828 CrossRef CAS PubMed.
- S. B. Arredondo, D. Valenzuela-Bezanilla, M. D. Mardones and L. Varela-Nallar, Role of Wnt Signaling in Adult Hippocampal Neurogenesis in Health and Disease, Front. Cell Dev. Biol., 2020, 8, 860 CrossRef PubMed.
- M. Soto, W. Cai, M. Konishi and C. R. Kahn, Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 6379–6384 CrossRef CAS PubMed.
- A. Kleinridders, H. A. Ferris, W. Cai and C. R. Kahn, Insulin action in brain regulates systemic metabolism and brain function, Diabetes, 2014, 63, 2232–2243 CrossRef PubMed.
- S. Reynhout, S. Jansen, D. Haesen, S. van Belle, S. A. de Munnik, E. Bongers, J. H. Schieving, C. Marcelis, J. Amiel, M. Rio, H. McLaughlin, R. Ladda, S. Sell, M. Kriek, C. Peeters-Scholte, P. A. Terhal, K. L. van Gassen, N. Verbeek, S. Henry, J. Scott Schwoerer, S. Malik, N. Revencu, C. R. Ferreira, E. Macnamara, H. M. H. Braakman, E. Brimble, M. R. Z. Ruzhnikov, M. Wagner, P. Harrer, D. Wieczorek, A. Kuechler, B. Tziperman, O. Barel, B. B. A. de Vries, C. T. Gordon, V. Janssens and L. Vissers, De Novo Mutations Affecting the Catalytic Calpha Subunit of PP2A, PPP2CA, Cause Syndromic Intellectual Disability Resembling Other PP2A-Related Neurodevelopmental Disorders, Am. J. Hum. Genet., 2019, 104, 139–156 CrossRef CAS PubMed.
- T. Fanutza, D. Del Prete, M. J. Ford, P. E. Castillo and L. D'Adamio, APP and APLP2 interact with the synaptic release machinery and facilitate transmitter release at hippocampal synapses, eLife, 2015, 4, e09743 CrossRef PubMed.
- N. Khayer, M. Mirzaie, S. A. Marashi and M. Jalessi, Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases, PLoS One, 2020, 15, e0239219 CrossRef CAS PubMed.
- M. P. Coba, N. H. Komiyama, J. Nithianantharajah, M. V. Kopanitsa, T. Indersmitten, N. G. Skene, E. J. Tuck, D. G. Fricker, K. A. Elsegood, L. E. Stanford, N. O. Afinowi, L. M. Saksida, T. J. Bussey, T. J. O'Dell and S. G. Grant, TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function, J. Neurosci., 2012, 32, 13987–13999 CrossRef CAS PubMed.
- O. Ostrovskaya, K. Xie, I. Masuho, A. Fajardo-Serrano, R. Lujan, K. Wickman and K. A. Martemyanov, RGS7/Gbeta5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling, eLife, 2014, 3, e02053 CrossRef PubMed.
- A. McGirr, T. V. Lipina, H. S. Mun, J. Georgiou, A. H. Al-Amri, E. Ng, D. Zhai, C. Elliott, R. T. Cameron, J. G. Mullins, F. Liu, G. S. Baillie, S. J. Clapcote and J. C. Roder, Specific Inhibition of Phosphodiesterase-4B Results in Anxiolysis and Facilitates Memory Acquisition, Neuropsychopharmacology, 2017, 42, 1178 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo03451h |
| ‡ These authors contributed equally to this study as corresponding authors. |
| This journal is © The Royal Society of Chemistry 2023 |