Open Access Article
Open Access ArticleIn vivo administration of gut bacterial consortia replicates urolithin metabotypes A and B in a non-urolithin-producing rat model†
Carlos E.
Iglesias-Aguirre
 a,
Antonio
González-Sarrías
a,
Antonio
González-Sarrías
 a,
Adrián
Cortés-Martín
a,
Adrián
Cortés-Martín
 ab,
María
Romo-Vaquero
ab,
María
Romo-Vaquero
 a,
Leire
Osuna-Galisteo
a,
Leire
Osuna-Galisteo
 a,
José Joaquín
Cerón
c,
Juan Carlos
Espín
a,
José Joaquín
Cerón
c,
Juan Carlos
Espín
 a and
María Victoria
Selma
a and
María Victoria
Selma
 *a
*a
aLaboratory of Food & Health, Research Group on Quality, Safety and Bioactivity of Plant Foods, CEBAS-CSIC, Campus de Espinardo, Murcia 30100, Spain. E-mail: mvselma@cebas.csic.es
bAPC Microbiome Ireland & School of Microbiology, University College Cork, T12 YT20 Cork, Ireland
cInterdisciplinary Laboratory of Clinical Analysis, Interlab-UMU, Regional Campus of International Excellence Campus Mare Nostrum, University of Murcia, 30100 Espinardo, Murcia, Spain
First published on 14th February 2023
Abstract
Urolithin (Uro) production capacity and, consequently, at least partly, the health effects attributed to ellagitannin and ellagic acid consumption vary among individuals. The reason is that not all individuals have the gut bacterial ecology needed to produce the different Uro metabolites. Three human urolithin metabotypes (UM-A, UM-B, and UM-0) based on dissimilar Uro production profiles have been described in populations worldwide. Recently, the gut bacterial consortia involved in ellagic acid metabolism to yield the urolithin-producing metabotypes (UM-A and UM-B) in vitro have been identified. However, the ability of these bacterial consortia to customize urolithin production to mimic UM-A and UM-B in vivo is still unknown. In the present study, two bacterial consortia were assessed for their capacity to colonize the intestine of rats and convert UM-0 (Uro non-producers) animals into Uro-producers that mimic UM-A and UM-B, respectively. Two consortia of Uro-producing bacteria were orally administered to non-urolithin-producing Wistar rats for 4 weeks. Uro-producing bacterial strains effectively colonized the rats’ gut, and the ability to produce Uros was also effectively transferred. Bacterial strains were well tolerated. No changes in other gut bacteria, except Streptococcus reduction, or adverse effects on haematological and biochemical parameters were observed. Besides, two novel qPCR procedures were designed and successfully optimized to detect and quantify Ellagibacter and Enterocloster genera in faecal samples. These results suggest that the bacterial consortia are safe and could be potential probiotics for human trials, which is especially relevant for UM-0 individuals, who cannot produce bioactive Uros.
Introduction
For decades, the consumption of ellagitannins (ETs) and ellagic acid (EA) has been associated with numerous biological effects, including antioxidant, anticancer, anti-inflammatory, antibacterial, and anti-HIV replication activities.1 However, the absorption and bioavailability of ETs and EA are very poor. Previous human studies found that the urinary excretion of EA and an EA-O-glucuronide is less than 1% of the intake.2,3 Nevertheless, while ET and EA absorption is low, they are further metabolized by the colonic microbiota to urolithins (Uros), which are responsible, at least partially, for the beneficial effects of ET- and (or) EA-rich foods.1,4 First, EA is released through the hydrolysis of ET's ester bonds by the enzyme known as ellagitannase.5 EA undergoes further degradation, producing 6H-dibenzo[b,d]pyran-6-one derivatives called Uros. This cascade of reactions starts with a lactone ring cleavage by a lactonase enzyme, resulting in luteic acid, which is then decarboxylated to pentahydroxy-Uro (Uro-M5). Consecutive dehydroxylations convert it to tetrahydroxy-Uros (Uro-D, Uro-E, and Uro-M6) and trihydroxy-Uros (Uro-C, Uro-M7, and Uro-G), to finally yield dihydroxy-Uros (Uro-A and isoUro-A) and monohydroxy-Uro (Uro-B), the latter being generally detected when isoUro-A is also produced.1 The wide variation of Uro metabolic profiles detected in humans after consuming ET-rich foods indicates interindividual differences in the colonic microbiota responsible for ET degradation.6–8 The numerous possible interactions between the colonic bacteria and the hosting organism could explain the different metabolic fates of dietary (poly)phenols.6,9 This two-way interaction between gut microbiota and (poly)phenols has prompted the scientific community to cluster the population based on their metabolic phenotype (metabotype) to explain the differences in polyphenols’ effects.10,11 More specifically, according to the ET and EA metabolism, individuals can be stratified into Uro metabotypes (UMs) associated with the gut microbiome composition and functionality.7,8,12 Three human UMs (UM-A, UM-B, and UM-0) related to three different Uro production profiles have been described in Western and Eastern populations.13–15 In this regard, metabotype A (UM-A) individuals produce several intermediate Uros but only urolithin A (Uro-A), the main absorbed Uro in this UM, at the end of the microbial catabolic pathway. Metabotype B (UM-B) individuals produce some intermediate Uros and three final Uros, i.e., urolithin B (Uro-B), isourolithin A (IsoUro-A), and Uro-A, which are the main absorbed Uros in UM-B. Therefore, the intermediate Uros could act primarily in the intestine, while the final ones could have local and systemic effects.6 In contrast, individuals with metabotype 0 (UM-0) cannot produce these final Uros (only the precursor Urolithin-M5 has been detected so far, which is not absorbed in the gut). Differences in Uro profiles have been observed between UMs and along the large intestine, showing predominant Uro production in the distal colon region.16–19 Remarkably, the UM-0 percentage in Spanish and Chinese healthy volunteers is approximately 10% and could be even higher in the US population.13–15 The bacterial genera Gordonibacter and Ellagibacter have been identified as being capable of metabolizing EA into some Uros.20–23 However, it was found that some intermediate (for example, Uro-D, Uro-E, Uro-M7, and Uro-G) and final Uros (Uro-A and Uro-B) are not produced in pure cultures of these bacteria, which implies the need for other bacteria to complete the set of Uros that configure both UM-A and UM-B. Recently, the gut bacterial consortia involved in EA metabolism to yield the Uro-producing metabotypes (UM-A and UM-B) in vitro have been identified.24 Bacterial consortia containing Gordonibacter plus Enterocloster genera and Ellagibacter plus Enterocloster produced in vitro the urolithins associated with UM-A and UM-B, respectively.25 However, the ability of these bacterial consortia to replicate human Uro profiles associated with UM-A and UM-B in vivo is still unknown. Furthermore, the safety of consuming these Uro-producing bacteria as new probiotics is still uncertain, as it has not been previously tested in animals or humans. In the present study, the above bacterial consortia were assessed for their capacity to colonize the intestine of Uro-non-producing (UM-0-like) rats and convert them into Uro-producers mimicking UM-A and UM-B, respectively. The safety of these bacterial consortia was also evaluated. Besides, two novel real-time quantitative PCR (qPCR)-based procedures were developed to detect and quantify Ellagibacter and Enterocloster in faecal samples.Materials and methods
Chemical and reagents
Uros were chemically synthesized and purified by Villapharma Research S.L. (Parque Tecnológico de Fuente Álamo, Murcia, Spain). EA was purchased from Sigma-Aldrich (St Louis, MO, USA). Phosphate buffer saline (PBS) was obtained from Fisher Scientific (USA), while methanol, ethanol, and formic acid were from Panreac Química (Barcelona, Spain). Liquid chromatography-mass spectrometry (LC-MS) grade solvents were purchased from J.T. Baker (Deventer, The Netherlands). Ultrapure Millipore water (Bedford, MA, USA) was used throughout the study. All chemicals and reagents were of analytical grade.Preparation of bacterial consortia
Gordonibacter urolithinfaciens DSM 27213T, Ellagibacter isourolithinifaciens DSM 104140T, and Enterocloster bolteae CEBAS S4A9, all of them isolated and identified in our lab (CEBAS-CSIC, Spain), were anaerobically cultivated in 5 mL Wilkins-Chalgren anaerobe medium (WAM, Oxoid) tubes at 37 °C for 48 h in a Concept 400 anaerobic chamber (Baker Ruskin Technologies Ltd, Bridgend, South Wales, UK) and resuspended in PBS supplemented with 10% glycerol and 0.05% L-cysteine hydrochloride (PanReac Química, Barcelona, Spain). After anaerobic incubation, consortia were prepared.24,25 Briefly, “bacterial consortium A” contained G. urolithinfaciens and E. bolteae strains; “bacterial consortium B” contained E. isourolithinifaciens and E. bolteae cultures; and “control cocktail” contained sterile PBS with 10% glycerol and 0.05% L-cysteine hydrochloride. Before administration, all cocktails were distributed in 3.5 mL aliquots and kept at −80 °C.Animals, diets, and experimental design
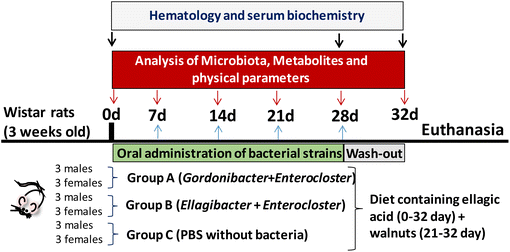
The experimental protocol (reference 624/2020) was approved by the local government, the Spanish National Research Council's Bioethics Committee (Madrid, Spain), and the Animal Experimentation Ethics Committee from the University of Murcia (Spain). The experiments followed the recommendations of the European Union regarding animal experimentation (Directive 2010/63/EU of the European Parliament and the Council on the protection of animals used for scientific purposes). Wistar rats (n = 18; 9 females and 9 males) weighing 240 ± 31 g were obtained from Envigo (Barcelona, Spain). This rat model was selected based on its inability to produce Uros observed in our preliminary studies in which faecal samples from different rat models were tested. Three groups (A, B, and C) were formed by randomly assigning 6 animals each (3 female and 3 male rats) per group (Fig. 1). Each rat group was housed in two different cages, divided by sex, in a room with a temperature-controlled environment (22 ± 2 °C) with 55 ± 10% relative humidity and a controlled light–dark cycle (12 h). The rats received a standard chow diet (Teklad, Barcelona, Spain) containing (%/100 g fresh weight) 14.3% proteins (corn gluten meal), 48% carbohydrates (wheat middlings, ground wheat, ground corn) and 4% fat (soybean oil), 2.9 kcal g−1 energy density, and 4.1% crude fibre, supplemented with EA powder ≥95% purity (3.6 mg per 100 g of chow). EA was mixed homogenously with ground standard feed, re-pelleted and lyophilized. The EA-enriched diet was stored away from moisture and light. The EA dose given to the rats in the diet was approximately 0.72 mg per rat per day (EA 1× diet), a human equivalent dose of 41 mg day−1.26 All groups were fed the EA 1× diet and tap water ad libitum throughout the experiment (5 weeks). Since the EA detected in faecal samples was low during the first week, some extra EA was added in the following days. At week 3, an extra dose of 1.5 mg EA per rat per day dissolved in water was orally administered by gavage to the animals (total = EA 3× diet), which was given only for 1 week. Finally, at week 4, ≈5 g walnuts per rat per day (another EA source) were added to the EA 1× diet and was maintained until the end of the study in order to evaluate the impact of the food matrix-containing EA on the bacteria's ability to produce Uros. Weight, food, and water intake were measured every day.The gut bacterial consortia were orally administered by gavage. Group A received 500 μL of bacterial cocktail A, Group B received 500 μL of bacterial cocktail B, and Group C (control) received 500 μL of PBS. Oral gavage was performed every 2 days during the first two weeks and every day during the following 2 weeks. In the last week (days 28 to 32), the animals continued with the same diet supplemented with EA and walnuts but without oral bacteria administration. At the end of the study, animals were sacrificed using a CO2 chamber.
Sampling procedures
Faecal samples were collected during the study for Uro analysis by UPLC-ESI-QTOF-MS/MS and gut microbiota by 16S rRNA gene sequencing and qPCR. Blood was extracted at different time points (baseline, day 28, and end of the study) for haematological and serum biochemical analyses. Blood samples were obtained from the tail vein (≈500 μL) and collected in heparin-containing tubes at the baseline and days 28 and 32 (Fig. 1). Plasma separation was performed immediately by centrifugation at 3000g for 10 min at 4 °C and frozen at −80 °C until further determination of serobiochemical variables. Liver, kidneys, and spleen were collected, weighed, and examined to detect any morphological differences at sacrifice.Design and optimization of the primers and probe for quantification of Ellagibacter and Enterocloster by qPCR
Sequences of the 16S rRNA genes of Ellagibacter isourolithinifaciens DSM 104140T and E. bolteae CEBAS S4A9 were obtained from the GenBank database (EMBL database). The sequences were aligned with the phylogenetically close bacterial genera using the Molecular Evolutionary Genetics Analysis software (MEGA 11) and inspected for regions of conserved and variable sequences to design the primers and probe to detect and quantify Ellagibacter and Enterocloster (Table 1). The primers and probe were designed and validated with the Primer Quest and Oligo Analyzer tools, respectively (Integrated DNA Technologies, Inc., Belgium) (https://eu.idtdna.com/pages). The TaqMan probe was labelled at the 5′ and 3′ ends with a 6-carboxy-fluorescein group (6FAM) and a blackberry quencher (BBQ), respectively. qPCR was performed using an ABI 7500 sequence detection system. The final concentrations of each primer and probe were 300 and 375 nM for Ellagibacter and 200 and 500 nM for Enterocloster. A conventional qPCR protocol was carried out for DNA amplification of Ellagibacter and Enterocloster, respectively. The ramping profile for Ellagibacter amplification was 1 cycle at 95 °C for 5 min, followed by 45 cycles of 95 °C for 15 s, 50 °C for 40 s, and 72 °C for 31 s. Finally, 1 cycle at 72 °C for 5 min was added. The ramping profile for Enterocloster amplification was 1 cycle at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 65 °C for 40 s, and 72 °C for 31 s. Finally, 1 cycle at 72 °C for 5 min was added.| Primer | Size | Oligonucleotide sequence 5′–3′ | T m (°C) | Location in the genea |
|---|---|---|---|---|
| a 16S ribosomal RNA gene. Tm, primer melting temperature. | ||||
| Ellagibacter Forward | 17 | GCTAGGTGTGGGGAAAC | 58.1 | 806–822 |
| Ellagibacter Reverse | 17 | CTCAAAGGAATTGACGG | 55.5 | 889–905 |
| Ellagibacter Probe | 14 | 6FAM-TACGGCGGCAACGC-BBQ | 61.5 | 870–883 |
| Enterocloster Forward | 20 | ACGTCCCAGTTCGGACTGTA | 65 | 1183–1202 |
| Enterocloster Reverse | 20 | GTTGCTGACTCCCATGGTGT | 64 | 1299–1318 |
| Enterocloster Probe | 24 | 6FAM-CAACCCGACTACACGAAGCTGGAA-BBQ | 68 | 1208–1231 |
Faecal DNA extraction and gut microbial analyses
DNA extraction from faecal samples was performed with the NucleoSpin® tissue DNA purification kit (Macherey-Nagel, Germany), according to the manufacturer's instructions with some modifications. DNA was quantified by fluorimetry (Qubit 3.0 – ThermoFisher Scientific™, UK), and the purity was measured from the absorbance ratio at a wavelength of 260/280 nm (A260/A280) (NanoDrop-ThermoFisher Scientific™, UK). Gut microbiota composition was determined by sequencing the V3 and V4 variable regions of the 16S rRNA gene following Illumina protocols (Illumina Inc., San Diego, CA, USA) with a read length of 2 × 300 bp paired-end run (MiSeq Reagent Kit v3, Illumina Inc.). Metagenomic sequencing was performed on a MiSeq-Illumina platform (FISABIO sequencing service, Spain). Data processing, chimeric sequence removal, sequence alignment, and 16S rRNA gene sequence clustering were performed as described elsewhere to obtain the taxonomic classification.8,27Gordonibacter DNA amplification was achieved in an ABI 7500 qPCR system as previously described.19,28 The Enterocloster and Ellagibacter-specific primers and probes designed in the present study were used following the qPCR protocol described above. Genomic DNA standard curves of Gordonibacter, Enterocloster, and Ellagibacter were used for their quantification. All the faecal samples were analysed in triplicate.Extraction, detection, and quantification of urolithins in faecal samples
Faecal samples (0.2–0.5 g) were extracted, in a proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, with a solution of MeOH/H2O (80/20), acidified with 0.1% HCl, and homogenised by vortexing for 2 min and shaking at 1500 rpm at room temperature for 10 min in a block heater. The suspension was centrifuged at 14
10, with a solution of MeOH/H2O (80/20), acidified with 0.1% HCl, and homogenised by vortexing for 2 min and shaking at 1500 rpm at room temperature for 10 min in a block heater. The suspension was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 10 min, and the supernatant was filtered through a 0.22 μm PVDF membrane filter (Millipore Corp., Bedford, MA) and diluted 3× with MeOH (0.1% formic acid) before injection in a UPLC-ESI-QTOF-MS system. A UPLC system (Agilent 1290 Infinity) coupled to a quadrupole time-of-flight (QTOF) LC/MS system (6550 Accurate-Mass) (Agilent Technologies, Waldbronn, Germany) was used to analyse the faecal metabolites using a gradient elution method as previously described.29,30 Briefly, separation was carried out in a Poroshell 120 EC-C18 reverse-phase column, using water and acetonitrile, both acidified with 0.1% formic acid, as mobile phases. The flow rate was 0.4 mL min−1, and the injection volume was 5 μL. The MassHunter Qualitative Analysis software (version B.10, Agilent Technologies, Waldbronn, Germany) was used for data processing. All metabolites were identified by direct comparison with standards and confirmed by their molecular mass. Calibration curves were obtained for EA and the different Uros with good linearity (R2 > 0.99).
000g at 4 °C for 10 min, and the supernatant was filtered through a 0.22 μm PVDF membrane filter (Millipore Corp., Bedford, MA) and diluted 3× with MeOH (0.1% formic acid) before injection in a UPLC-ESI-QTOF-MS system. A UPLC system (Agilent 1290 Infinity) coupled to a quadrupole time-of-flight (QTOF) LC/MS system (6550 Accurate-Mass) (Agilent Technologies, Waldbronn, Germany) was used to analyse the faecal metabolites using a gradient elution method as previously described.29,30 Briefly, separation was carried out in a Poroshell 120 EC-C18 reverse-phase column, using water and acetonitrile, both acidified with 0.1% formic acid, as mobile phases. The flow rate was 0.4 mL min−1, and the injection volume was 5 μL. The MassHunter Qualitative Analysis software (version B.10, Agilent Technologies, Waldbronn, Germany) was used for data processing. All metabolites were identified by direct comparison with standards and confirmed by their molecular mass. Calibration curves were obtained for EA and the different Uros with good linearity (R2 > 0.99).
Haematology and clinical chemistry
Haematological variables were determined in heparinized blood using an automated haematological analyser with specific software for rat blood samples (AVDIA 120, Siemens, Munich, Germany). The variables analysed were mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH), mean corpuscular haemoglobin concentration (MCHC), erythrocyte distribution (RDW), mean platelet volume (MPV), plateletcrit (PCT), platelet distribution width (PDW), mean platelet component (MPC), mean platelet mass (MPM), platelet count (PLT), reticulocyte haemoglobin content (CHr), and mean reticulocyte corpuscular volume (MCVr). Plasma biochemical variables, calcium (Ca), and phosphorus (P) were analysed using an Olympus AU400 chemistry and toxicology analyser (Beckman Coulter, California, USA). The biochemical variables were total proteins (PROT), albumin (ALBU), globulin (GLOB), creatinine (CREA), glucose (GLUC), cholesterol (CHOL), triglycerides (TRIGL), alkaline phosphatase (ALP), gamma glutamyl transpeptidase (GGT), aspartate aminotransferase (AST), and alanine aminotransferase (ALT). Finally, thyroxine (T4) levels were analysed using an IMMULITE® 1000 immunoassay system (Siemens).Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed with the SPSS software version 27.0 (SPSS Inc., Chicago, IL, USA), and differences with p < 0.05 were considered statistically significant. Data normality was evaluated with the Shapiro–Wilk test. Comparisons between different treatment groups were carried out using repeated measured analysis of variance (RM ANOVA) with Tukey's or Dunnett's T3 post hoc test, depending on whether the data followed a normal or non-normal distribution, respectively. Differences between males and females were explored using the paired Student's t-test, or Wilcoxon signed rank test when data were normally or non-normally distributed, respectively. Chi-square linear discriminant analysis effect size (LEfSe) was used to detect differential bacterial features between groups. Pearson correlation was used to analyse possible associations between variables in normal-distributed data, while Spearman correlation was applied in non-normal-distributed data. Data plots were performed using Sigma Plot 14.5 (Systat Software, San Jose, CA, USA).Results
Analysis of the specificity and sensitivity of the designed primers and probe for Ellagibacter and Enterocloster
The specificity of the Ellagibacter and Enterocloster primers and probe was confirmed by uploading their sequences on the sequence alignment program ProbeMatch.31 Results are shown in Tables 1S and 2S.† Subsequently, the specificity of the primers and probe was verified by qPCR using the optimised reaction conditions described in Materials and methods section. Representative bacterial species of the Eggerthellaceae and Lachnospiraceae families with the greatest similarity to Ellagibacter and Enterocloster in the 16S rRNA gene sequence were tested in addition to other closely related species frequently detected in the human intestine (Tables 1S and 2S†).
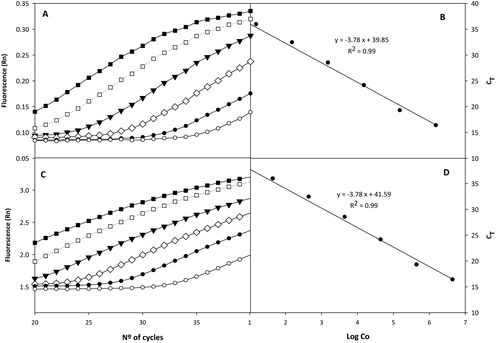
Fig. 2 shows the amplification plots corresponding to the standard quantification curves. CT values were plotted against bacterial concentration (Co) as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 genomic equivalents/reaction (6.25 × 10−6 ng, for haploid genome). The slope of the linear regression curve, calculated using genomic DNA, was −3.78, and the R2 value was greater than 0.99. Regarding the sensitivity of the qPCR system, 1.5 genome equivalents were needed as the minimum for a positive reaction using the TaqMan mode. The CT values obtained with the calibration curve of E. isourolithinifaciens and E. bolteae ranged from 15.5 ± 0.7 to 37.8 ± 0.7, while no DNA amplification was detected for the other strains tested. The quantification of Ellagibacter and Enterocloster in the rats’ faecal samples was performed with this method (qPCR), considering the mean of three replicates. The results were expressed as log
10 genomic equivalents/reaction (6.25 × 10−6 ng, for haploid genome). The slope of the linear regression curve, calculated using genomic DNA, was −3.78, and the R2 value was greater than 0.99. Regarding the sensitivity of the qPCR system, 1.5 genome equivalents were needed as the minimum for a positive reaction using the TaqMan mode. The CT values obtained with the calibration curve of E. isourolithinifaciens and E. bolteae ranged from 15.5 ± 0.7 to 37.8 ± 0.7, while no DNA amplification was detected for the other strains tested. The quantification of Ellagibacter and Enterocloster in the rats’ faecal samples was performed with this method (qPCR), considering the mean of three replicates. The results were expressed as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 bacteria per g of faeces and compared with the relative abundance values obtained by 16S rRNA sequencing. We observed a positive correlation between the results obtained by both methods in Ellagibacter (r = 0.660, p < 0.001) and Enterocloster (r = 0.459, p < 0.001) (data not shown).
10 bacteria per g of faeces and compared with the relative abundance values obtained by 16S rRNA sequencing. We observed a positive correlation between the results obtained by both methods in Ellagibacter (r = 0.660, p < 0.001) and Enterocloster (r = 0.459, p < 0.001) (data not shown).
Effect of orally administered bacterial consortia on the in vivo production of urolithins
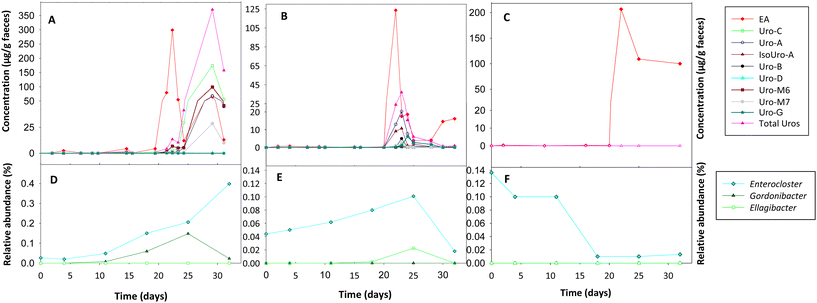
The Uro analysis in faeces showed that initially (T0), the rats did not produce Uros after ingesting EA powder (Fig. 3A–C). However, after administering the bacterial consortia A (Fig. 3A) and B (Fig. 3B), rats became capable of producing Uros. During the first three weeks, when the rats from groups A and B only ingested EA powder, the Uro concentration (range: 0–1.43 μg g−1) and that of their EA precursor (range: 0.07–4.54 μg g−1) in faeces was low. However, from day 21, when the consumption of walnuts started as a second EA source, there was a very significant increase in faecal EA (Fig. 3A–C) and Uro production in groups A (Fig. 3A) and B (Fig. 3B). Moreover, differences in the Uro profile were observed between groups A and B. In group A, fecal Uro-C, Uro-M6, Uro-M7, and the final metabolite Uro-A were predominant. In contrast, in group B, the three final Uros (Uro-A, Uro-B, and IsoUro-A) were present, whereas the intermediates were scarce. However, a higher faecal concentration of the recently described Uro-G was detected in group B than in group A. In addition, the Uro-B concentration was lower than that of Uro-A and IsoUro-A in group B. Furthermore, Uro-A and IsoUro-A, but not Uro-B, were produced in all rats of group B (Fig. 3B). When walnuts were added to the diet of control rats (group C) on day 21, a higher faecal concentration of EA was also observed than before walnut consumption, where the faecal EA concentration was very low (Fig. 3C). However, neither EA powder intake nor walnut consumption promoted the production of Uros in the gut of control rats (i.e., with no bacterial consortia).Intestinal colonization of orally administered bacteria
Initially, before rats of groups A and B started the consumption of the bacterial consortia, Gordonibacter or Ellagibacter were under the limit of detection in the faecal samples when metagenomic sequencing analysis was performed using the MiSeq-Illumina platform (Fig. 3D–F). When qPCR was used, Gordonibacter levels in rats were low (<5.8 log bacteria per g faeces), and Ellagibacter levels were under the limit of detection (data not shown). In contrast, Enterocloster was detected using the MiSeq-Illumina and qPCR in the faeces of rats from all groups (A, B, and C) at the baseline (Fig. 3D–F). The relative abundance (mean ± SD) of Enterocloster in groups A, B, and C was 0.03 ± 0.02, 0.04 ± 0.06, and 0.13 ± 0.17%, respectively, but without significant differences (p = 0.570). The administration of the bacterial consortia A and B resulted in the appearance and gradual increase of Gordonibacter and Ellagibacter in the corresponding group A and B faeces, respectively (Fig. 3D and E). Furthermore, faecal Enterocloster levels increased throughout the study in groups A and B (Fig. 3D and E). However, LEfSe analysis showed scarce differences in other gut bacterial groups when metagenomic sequencing data of groups A and B were compared to those of the control group (Fig. 4A and B). Interestingly, a reduction of the Streptococcus genus and Streptococcaceae family was observed in groups A and B compared to the control group (Fig. 4A and B). Five days after stopping the oral administration of the bacterial consortia (end date of the study), a reduction in the abundance of Ellagibacter and Enterocloster was detected in group B, which also resulted in a significant decrease in Uro production (Fig. 3B and E). In group A, a reduction in the abundance of Gordonibacter but not Enterocloster was also observed (Fig. 3A), which resulted in a decrease in Uro concentration but was less pronounced than in group B (Fig. 3D and E). In the control group, Ellagibacter was under the limit of detection throughout the study, and Enterocloster and Gordonibacter remained at very low levels (Fig. 3F).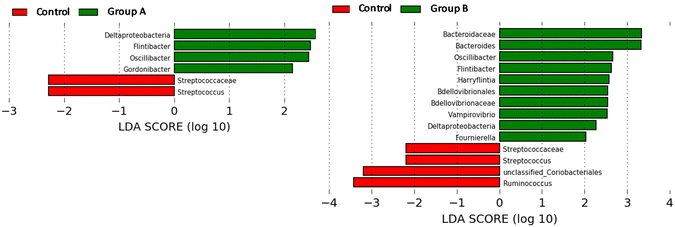 | ||
| Fig. 4 Linear discriminant analysis (LDA) effect size (LEfSe) plot of taxonomic biomarkers identified in the gut microbiome of rats after administering the gut bacterial consortia for 4 weeks. | ||
Study of the toxicity of the orally administered gut bacteria
Potential side effects from ingestion of the bacterial consortia were investigated. No adverse effects on growth, weight gain, food intake, and vital organs were observed after 4 weeks of oral administration by gavage of bacterial consortia in rats from groups A and B compared to the control group. However, male rats gained 107.4 ± 2.4 g, while female rats gained less weight (25.7 ± 5 g) at the end of the study without differences compared to the control group. Food consumption by cage was 63.1 ± 1.4 g for male rats and 33.8 ± 1.8 g for female rats. However, water intake was 136.1 ± 37.2 g and 76.0 ± 12.6 g, by male and female rats by cage respectively. Haematological differences were observed between males and females in some variables, such as erythrocyte distribution (RDW, p = 0.005), leucocytes (p = 0.005), platelet distribution width (PDW, p = 0.005), and reticulocyte haemoglobin content (CHr, p = 0.013) (Table 2). Still, no differences were observed for any variables between groups that consumed the bacterial consortia (groups A and B) and the control group (group C). The biochemical values were also analysed, and differences were detected only between males and females in some variables such as the creatinine index (p < 0.001), phosphorus (p = 0.007), alkaline phosphatase (ALP) (p = 0.003), and thyroxine (T4) (p = 0.002) (Table 3). However, no differences were observed for any of the variables between the groups that consumed the bacterial consortia (groups A and B) and the control group (group C).| Haematocrit (%) | Erythrocytes (×106 cells per μL) | Haemoglobin (g dL−1) | MCV (fL) | MCH (pg) | MCHC (g dL−1) | RDW (%) | Leucocytes (×103 cells per μL) | Neutrophils (%) | Lymphocytes (%) | Monocytes (%) | Eosinophils (%) | Basophils (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Females | 38.0 ± 4.6 | 7.1 ± 0.7 | 11.3 ± 1.9 | 54.0 ± 1.9 | 15.5 ± 1.4 | 33.3 ± 0.4 | 10.6 ± 0.6 a | 4.0 ± 1.4 a | 45.3 ± 8.6 | 45.9 ± 10.3 | 0.3 ± 0.2 | 8.5 ± 2.0 | 0.0 ± 0.1 |
| Control | 35.3 ± 4.5 | 6.8 ± 0.8 | 10.3 ± 2.1 | 52.5 ± 1.6 | 14.5 ± 1.6 | 33.4 ± 0.4 | 10.6 ± 0.3 | 3.0 ± 0.6 | 47.4 ± 12.0 | 43.4 ± 13.1 | 0.2 ± 0.2 | 8.9 ± 2.2 | 0.0 ± 0.0 |
| Group A | 39.7 ± 6.5 | 7.2 ± 1.2 | 11.7 ± 2.5 | 54.8 ± 2.1 | 15.6 ± 1.2 | 33.5 ± 0.5 | 11.1 ± 0.7 | 4.0 ± 0.9 | 46.9 ± 4.0 | 44.3 ± 6.3 | 0.3 ± 0.2 | 8.4 ± 2.1 | 0.1 ± 0.1 |
| Group B | 39.0 ± 2.6 | 7.2 ± 0.4 | 12.0 ± 1.0 | 54.8 ± 1.5 | 16.5 ± 0.9 | 33.2 ± 0.4 | 10.3 ± 0.5 | 4.9 ± 2.0 | 41.5 ± 10.4 | 50.0 ± 13.2 | 0.3 ± 0.4 | 8.1 ± 2.4 | 0.1 ± 0.1 |
| Males | 38.1 ± 5.3 | 7.2 ± 0.9 | 10.6 ± 1.9 | 53.0 ± 1.2 | 14.6 ± 1.3 | 33.4 ± 0.2 | 11.3 ± 0.3 b | 5.9 ± 1.2 b | 36.3 ± 9.9 | 56.5 ± 11.7 | 0.6 ± 0.6 | 6.5 ± 2.0 | 0.1 ± 0.1 |
| Control | 33.0 ± 3.6 | 6.3 ± 0.5 | 9.0 ± 2.0 | 52.1 ± 1.8 | 14.2 ± 1.9 | 33.5 ± 0.2 | 11.2 ± 0.2 | 5.7 ± 0.8 | 34.2 ± 9.0 | 56.8 ± 11.9 | 1.3 ± 0.7 | 7.6 ± 2.6 | 0.0 ± 0.0 |
| Group A | 41.0 ± 6.2 | 7.6 ± 1.3 | 11.3 ± 2.1 | 53.7 ± 0.7 | 14.3 ± 0.6 | 33.3 ± 0.3 | 11.5 ± 0.2 | 7.3 ± 0.9 | 34.2 ± 16.2 | 60.2 ± 18.5 | 0.3 ± 0.2 | 5.1 ± 2.2 | 0.1 ± 0.2 |
| Group B | 40.3 ± 1.5 | 7.7 ± 0.2 | 11.7 ± 0.6 | 53.2 ± 0.9 | 15.3 ± 1.2 | 33.3 ± 0.2 | 11.4 ± 0.5 | 4.9 ± 0.4 | 40.5 ± 3.9 | 52.5 ± 4.8 | 0.3 ± 0.2 | 6.7 ± 0.8 | 0.1 ± 0.1 |
| Platelets (×103 cells per μL) | MPV (fL) | PCT (%) | PDW (%) | MPC (g dL−1) | MPM (pg) | Large PLT (×103 cells per μL) | Reticulocytes (%) | CHr (pg) | MCVr (fL) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Values are presented as the mean ± SD of 3 replicates. Values with different letters within each column are significantly different (p < 0.05). Dunnett's T3 post hoc test was applied in monocytes and basophils because data did not follow normal distribution. Tukey's post hoc test was applied for the rest of variables because data followed normal distribution. MCV: mean corpuscular volume; MCH: mean corpuscular haemoglobin; MCHC: mean corpuscular haemoglobin concentration; RDW: erythrocyte distribution; MPV: mean platelet volume; PCT: plateletcrit; PDW: platelet distribution width; MPC: mean platelet component; MPM: mean platelet mass; PLT: platelet count; CHr: reticulocyte haemoglobin content; MCVr: mean reticulocyte corpuscular volume. | ||||||||||
| Females | 880.0 ± 90.7 | 7.5 ± 0.4 | 0.6 ± 0.1 | 56.4 ± 6.5 a | 20.1 ± 1.1 | 1.3 ± 0.5 | 8.9 ± 7.8 | 1.4 ± 0.4 | 17.2 ± 0.5 a | 56.9 ± 1.7 |
| Control | 956.7 ± 75.7 | 7.1 ± 0.2 | 0.7 ± 0.1 | 51.9 ± 2.1 | 20.5 ± 1.1 | 1.3 ± 0.0 | 3.3 ± 5.8 | 1.1 ± 0.3 | 16.8 ± 0.6 | 55.2 ± 1.7 |
| Group A | 853.3 ± 196.6 | 7.5 ± 0.4 | 0.6 ± 0.2 | 56.9 ± 7.5 | 20.2 ± 1.5 | 1.3 ± 0.1 | 6.7 ± 5.8 | 1.6 ± 0.5 | 17.4 ± 0.1 | 58.0 ± 1.5 |
| Group B | 830.0 ± 17.3 | 7.9 ± 0.2 | 0.7 ± 0.1 | 60.4 ± 7.4 | 19.5 ± 0.4 | 1.3 ± 0.0 | 16.7 ± 5.8 | 1.4 ± 0.4 | 17.±0.2 | 57.6 ± 0.3 |
| Males | 864.4 ± 123.4 | 7.6 ± 0.4 | 0.6 ± 0.1 | 65.7 ± 5.5 b | 20.0 ± 0.6 | 1.3 ± 0.4 | 12.2 ± 4.4 | 1.7 ± 0.6 | 16.6 ± 0.5 b | 57.1 ± 1.7 |
| Control | 760.0 ± 55.7 | 7.8 ± 0.4 | 0.6 ± 0.0 | 66.8 ± 9.5 | 19.8 ± 0.3 | 1.3 ± 0.1 | 13.3 ± 5.8 | 1.5 ± 0.8 | 16.2 ± 0.4 | 56.5 ± 1.0 |
| Group A | 903.3 ± 153.1 | 7.5 ± 0.4 | 0.7 ± 0.1 | 63.3 ± 3.7 | 20.3 ± 0.8 | 1.3 ± 0.0 | 10.0 ± 0.0 | 1.6 ± 0.6 | 16.8 ± 0.2 | 58.3 ± 2.5 |
| Group B | 930.0 ± 96.4 | 7.6 ± 0.6 | 0.7 ± 0.1 | 67.1 ± 1.7 | 19.9 ± 0.6 | 1.3 ± 0.0 | 13.3 ± 5.8 | 2.1 ± 0.3 | 16.8 ± 0.6 | 56.6 ± 1.4 |
| PROT (g dL−1) | ALBU (g dL−1) | GLOB (g dL−1) | CREA (mg L−1) | UREA (mg dL−1) | GLUC (mg dL−1) | CHOL (mg dL−1) | TRIGL (mg dL−1) | Ca (mg dL−1) | P (mg dL−1) | ALP (IU L−1) | GGT (IU L−1) | AST (IU L−1) | ALT (IU L−1) | T4 (μg dL−1) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Values are presented as the mean ± SD of 3 replicates. Values with different letters within each column are significantly different (p < 0.05). Dunnett's T3 post hoc test was applied in albumin, urea and alanine aminotransferase because data did not follow normal distribution. Tukey's post hoc test was applied for the rest of variables because data followed normal distribution. PROT, total proteins; ALBU, albumin; GLOB, globulin; CREA, creatinine; GLUC, glucose; CHOL, cholesterol; TRIGL, triglycerides; Ca, calcium; P, phosphorus; ALP, alkaline phosphatase; GGT, gamma glutamyl transpeptidase; AST, aspartate aminotransferase; ALT, alanine aminotransferase; T4, thyroxine; IU L−1, international units per litre. | |||||||||||||||
| Females | 6.2 ± 0.4 | 3.0 ± 0.3 | 3.7 ± 0.7 | 6.4 ± 0.4 a | 34.4 ± 6.7 | 151.45 ± 72.7 | 74.35 ± 7.5 | 62.6 ± 31.2 | 9.8 ± 0.8 | 8.7 ± 1.9 a | 254.9 ± 95.8 a | 0.1 ± 0.1 | 239.3 ± 184.9 | 53.4 ± 26.2 | 3.7 ± 0.7 a |
| Control | 6.2 ± 1.7 | 2.9 ± 0.4 | 3.3 ± 1.3 | 6.3 ± 0.3 | 31.6 ± 11.3 | 115.5 ± 9.4 | 73.9 ± 9.9 | 55.3 ± 34.3 | 9.4 ± 0.7 | 7.5 ± 1.6 | 207.2 ± 131.5 | 0.1 ± 0.0 | 273.9 ± 0165.7 | 40.1 ± 20.7 | 3.6 ± 1.1 |
| Group A | 7.2 ± 0.5 | 3.2 ± 0.3 | 4.0 ± 0.3 | 6.8 ± 0.2 | 35.0 ± 5.4 | 206.8 ± 116.1 | 75.6 ± 7.1 | 59.9 ± 30.5 | 10.4 ± 0.5 | 10.4 ± 0.9 | 236.2 ± 60.6 | 0.3 ± 0.3 | 303.5 ± 287.6 | 55.4 ± 32.9 | 3.6 ± 0.2 |
| Group B | 6.8 ± 0.4 | 2.9 ± 0.4 | 3.9 ± 0.1 | 6.2 ± 0.4 | 36.5 ± 2.3 | 132.1 ± 23.1 | 73.6 ± 8.7 | 72.7 ± 39.4 | 9.5 ± 0.8 | 8.1 ± 2.1 | 321.5 ± 71.8 | 0.1 ± 0.0 | 140.5 ± 63.4 | 64.9 ± 27.5 | 4.1 ± 0.9 |
| Males | 6.3 ± 0.1 | 2.2 ± 0.2 | 4.1 ± 0.2 | 5.7 ± 0.2 b | 35.9 ± 3.9 | 161.5 ± 159.1 | 93.6 ± 13.1 | 98.9 ± 45.5 | 9.3 ± 0.6 | 10.6 ± 2.2 b | 437.3 ± 118. b | 0.1 ± 0.1 | 424.4 ± 274.4 | 55.3 ± 22.2 | 4.1 ± 0.9 b |
| Control | 6.2 ± 0.3 | 2.1 ± 0.3 | 4.1 ± 0.2 | 5.6 ± 0.2 | 34.9 ± 3.5 | 261.7 ± 278.5 | 98.1 ± 18.4 | 114.8 ± 0.8 | 9.4 ± 1.1 | 10.0 ± 1.9 | 453.9 ± 158.3 | 0.2 ± 0.2 | 407.7 ± 341.6 | 62.4 ± 14.9 | 3.7 ± 1.6 |
| Group A | 6.4 ± 0.1 | 2.3 ± 0.2 | 4.2 ± 0.2 | 5.8 ± 0.3 | 34.2 ± 3.6 | 124.4 ± 19.6 | 96.1 ± 11.8 | 84.4 ± 50.6 | 9.2 ± 0.3 | 10.3 ± 2.1 | 360.7 ± 52.5 | 0.1 ± 0.0 | 387.8 ± 276.9 | 56.2 ± 37.1 | 4.0 ± 0.3 |
| Group B | 6.3 ± 0.1 | 2.1 ± 0.1 | 4.1 ± 0.1 | 5.7 ± 0.0 | 38.8 ± 4.5 | 98.5 ± 13.6 | 86.5 ± 9.8 | 97.5 ± 3.4 | 9.4 ± 0.2 | 11.5 ± 3.0 | 497.3 ± 116.0 | 0.1 ± 0.0 | 477.6 ± 317.9 | 47.4 ± 14.0 | 4.4 ± 0.7 |
Discussion
In the last decade, there has been growing interest in ET and EA-rich plant foods, including pomegranate and walnuts, for their potential protective effects against chronic degenerative diseases such as cancer, cardiovascular and neurodegenerative diseases (Alzheimer's and Parkinson's diseases).32,33 However, ET and EA bioavailability and absorption rate are very low, and they are further metabolised by the gut microbiota to produce Uros. Thus, in the last few decades, many preclinical studies have suggested that Uros might be the main contributors to the health benefits of consuming ET-rich sources.1 In parallel, several studies have been performed to identify the bacterial groups that are able to metabolise EA into different types of Uros. Recently, the gut bacterial consortia involved in EA metabolism to yield the Uro-producing metabotypes (UM-A and UM-B) in vitro have been identified.25 In the present study, we develop two qPCR-based methods to detect two bacterial genera of these consortia (Ellagibacter and Enterocloster) in a specific, sensitive and cost-efficient manner. Developing a qPCR-based method to detect Gordonibacter was unnecessary because it was previously developed and tested in human faecal samples.19,28 As depicted in the ESI Tables 1S and 2S,† the system was accurate and suitable for detecting the Ellagibacter and Enterocloster genera specifically, as it excluded neighbour species and other bacteria commonly detected in the intestine. In faecal samples, Ellagibacter and Enterocloster quantification using this qPCR method correlated well with the results obtained by 16S rRNA sequencing. Hence, this qPCR method for Ellagibacter and Enterocloster quantification can be considered a specific, efficient, and feasible methodology.The present study investigated the viability of Uro-producing bacteria isolated from the human gut in a non-Uro-producing rat model for the first time. We confirmed here and in other parallel studies that Wistar rats are a suitable model of non-Uro-producing mammals (UM-0-like animal model). However, this should be verified by further studies since the rodents’ gut microbiota of a strain can also differ depending on the animal supplier. The present study confirms the essential role that Gordonibacter and (or) Ellagibacter play in the initial steps of the Uro production pathway. Even though the Enterocloster genus was detected in the control rats’ faeces throughout the study, it was insufficient to lead the animals to produce Uros. This result is in line with previous studies which correlated the presence of Gordonibacter and (or) Ellagibacter in the human gut with Uro production.8,19 Overall, the fact that Gordonibacter, Ellagibacter, and Enterocloster increased their abundance in faeces and successfully reproduced the production of Uros in vivo proves that all or some of the ingested bacteria reached the intestine and remained alive. We observed a higher amount of Uro-A than IsoUro-A and Uro-B in group B. Consequently, in this group, the production of Uro-A was more favoured over IsoUro-A and Uro-B production, replicating the profile of many individuals from UM-B. This Uro production profile has also been described recently in a different approach with a pseudo-germ-free mouse model after transplanting daily the faecal microbiota of different UM donors for 4 weeks, which could be a valuable model to explore the health effects of EA and ETs.34 However, our study has shown that consuming the bacterial consortia A (Gordonibacter + Enterocloster) and B (Ellagibacter + Enterocloster) is enough to produce the intestinal colonization of Uro-producing bacteria in a non-Uro-producing rat model, where the continued consumption of ETs was not sufficient to modify the UM-0 towards a Uro-producing metabotype (UM-A or UM-B).
On the other hand, data showed that when an EA powder-rich diet was supplemented with walnuts, faecal EA concentration and the production of Uros significantly increased in both groups A (Fig. 3A) and B (Fig. 3B). This indicates that the matrix of the food containing EA impacts its bioavailability in the intestine and, therefore, the bacteria's ability to produce Uros. Thus, not all dietary sources of EA are equally appropriate to produce Uros at the intestinal level. Furthermore, walnuts were highly suitable for the production of Uros at the intestinal level in humans compared to the intake of pomegranate juice and EA-rich extracts.19,35 Nevertheless, in the control group, even though the faecal concentration of EA increased after adding walnuts to the diet, it was not enough to modulate the rats’ intestinal microbiota to produce Uros (Fig. 4A). These results agree with previous studies on humans, in which consuming foods rich in EA did not turn UM-0 individuals into Uro-producers, probably due to the absence or minimal levels of Uro-producing bacteria in their gut microbiota.11,36 Overall, our results suggest that the modulation of the relative abundance of bacteria such as Gordonibacter, Ellagibacter, and Enterocloster could modify the UMs of individuals. Indeed, continuous consumption of bacterial consortia containing Gordonibacter plus Enterocloster (in group A rats) or Ellagibacter plus Enterocloster (in group B rats) resulted in intestinal colonization of Uro-producing bacteria in a non-Uro-producing rat model, allowing the metabotype of rats to change from UM-0 to UM-A or UM-B. Additionally, the safety of these orally administered gut bacterial consortia in rats for 1 month was investigated with no adverse effects or modulation of other bacterial groups compared to control rats (Tables 2 and 3). However, more research is needed to confirm the impact of Uro-producing consortia consumption on the host's health and safety before being considered as possible next-generation probiotics to customise Uro production in humans.
Conclusions
In addition to other dietary polyphenol-transforming bacteria, the Uro-producing gut bacterial consortia described in this study could have potential as novel probiotics, especially relevant in UM-0 individuals, who cannot produce bioactive Uros. Moreover, compared to chemical synthesis, metabolite production by microorganisms is often more convenient from a health and environmental point of view. Diseases previously treated only by synthetic medicines are progressively relying more and more on bacterial secondary metabolites, such as antibiotics and antitumoral, cholesterol-lowering, and immunomodulation agents. However, further research should be carried out to demonstrate these emerging notions.Author contributions
Conceptualization and design: M.V.S.; experiment execution: C.E.I.-A.; A.C.M.; A.G.S.; methodology and software: C.E.I.-A.; M.R.S.; L.O.; J.J.C. writing: M.V.S.; critical review of the manuscript for important intellectual content: J.C.E., A.G.S., A.C.M. and M.V.S. All authors have read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been funded by the project PID2019-103914RB-I00 from the Ministry of Science and Innovation (MICIN, AEI/10.13039/501100011033, Spain) and Projects 21647/PDC/21 and 20880/PI/18 (Fundación Séneca de la Región de Murcia, Spain). C. E. I.-A. is the holder of a predoctoral grant from MICIN (grant number FPU18/03961).References
- R. García-Villalba, J. A. Giménez-Bastida, A. Cortés-Martín, M. Á. Ávila-Gálvez, F. A. Tomás-Barberán, M. V. Selma, J. C. Espín and A. González-Sarrías, Urolithins: a comprehensive update on their metabolism, bioactivity, and associated gut microbiota, Mol. Nutr. Food Res., 2022, 66, 1–22 CrossRef PubMed.
- R. González-Barrio, G. Borges, W. Mullen and A. Crozier, Bioavailability of anthocyanins and ellagitannins following consumption of raspberries by healthy humans and subjects with an ileostomy, J. Agric. Food Chem., 2010, 58, 3933–3939 CrossRef PubMed.
- A. González-Sarrías, R. García-Villalba, M. Á. Núñez-Sánchez, J. Tomé-Carneiro, P. Zafrilla, J. Mulero, F. A. Tomás-Barberán and J. C. Espín, Identifying the limits for ellagic acid bioavailability: A crossover pharmacokinetic study in healthy volunteers after consumption of pomegranate extracts, J. Funct. Foods, 2015, 19, 225–235 CrossRef.
- B. Cerdá, F. A. Tomás-Barberán and J. C. Espín, Metabolism of Antioxidant and Chemopreventive Ellagitannins from Strawberries, Raspberries, Walnuts, and Oak-Aged Wine in Humans: Identification of Biomarkers and Individual Variability, J. Agric. Food Chem., 2005, 53, 227–235 CrossRef PubMed.
- J. A. Ascacio-Valdés, A. F. Aguilera-Carbó, J. J. Buenrostro, A. Prado-Barragán, R. Rodríguez-Herrera and C. N. Aguilar, The complete biodegradation pathway of ellagitannins by Aspergillus niger in solid-state fermentation, J. Basic Microbiol., 2016, 56, 329–336 CrossRef PubMed.
- C. E. Iglesias-Aguirre, A. Cortés-Martín, M. Á. Ávila-Gálvez, J. A. Gimenez-Bastida, M. V. Selma, A. Gonzalez-Sarrias and J. C. Espin, Main drivers of (poly)phenols effects on human health: metabolites production and (or) gut microbiota-associated metabotypes?, Food Funct., 2021, 12, 10324–10355 RSC.
- F. A. Tomás-Barberán, R. García-Villalba, A. González-Sarrías, M. V. Selma and J. C. Espín, Ellagic acid metabolism by human gut microbiota: consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status, J. Agric. Food Chem., 2014, 62, 6535–6538 CrossRef PubMed.
- M. Romo-Vaquero, A. Cortés-Martín, V. Loria-Kohen, A. Ramírez-de-Molina, I. García-Mantrana, M. C. Collado, J. C. Espín and M. V. Selma, Deciphering the Human Gut Microbiome of Urolithin Metabotypes: Association with Enterotypes and Potential Cardiometabolic Health Implications, Mol. Nutr. Food Res., 2019, 63, 1800958 CrossRef PubMed.
- F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, Interactions of gut microbiota with dietary polyphenols and consequences to human health, Curr. Opin. Clin. Nutr. Metab. Care, 2016, 19, 471–476 CrossRef.
- T. Meroño, G. Peron, G. Gargari, R. González-Domínguez, A. Miñarro, E. Vegas-Lozano, N. Hidalgo-Liberona, C. Del Bo’, S. Bernardi, P. A. Kroon, B. Carrieri, A. Cherubini, P. Riso, S. Guglielmetti and C. Andrés-Lacueva, The relevance of urolithins-based metabotyping for assessing the effects of a polyphenol-rich dietary intervention on intestinal permeability: A post-hoc analysis of the MaPLE trial, Food Res. Int., 2022, 159, 111632 CrossRef PubMed.
- A. González-Sarrías, R. García-Villalba, M. Romo-Vaquero, C. Alasalvar, A. Örem, P. Zafrilla, F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, Clustering according to urolithin metabotype explains the interindividual variability in the improvement of cardiovascular risk biomarkers in overweight-obese individuals consuming pomegranate: A randomized clinical trial, Mol. Nutr. Food Res., 2017, 61, 1600830 CrossRef PubMed.
- A. Cortés-Martín, M. V. Selma, F. A. Tomás-Barberán, A. González-Sarrías and J. C. Espín, Where to Look into the Puzzle of Polyphenols and Health? The Postbiotics and Gut Microbiota Associated with Human Metabotypes, Mol. Nutr. Food Res., 2020, 64, 1900952 CrossRef PubMed.
- A. Cortés-Martín, R. García-Villalba, A. González-Sarrías, M. Romo-Vaquero, V. Loria-Kohen, A. Ramírez-De-Molina, F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, The gut microbiota urolithin metabotypes revisited: the human metabolism of ellagic acid is mainly determined by aging, Food Funct., 2018, 9, 4100–4106 RSC.
- W. Xian, S. Yang, Y. Deng, Y. Yang, C. Chen, W. Li and R. Yang, Distribution of Urolithins Metabotypes in Healthy Chinese Youth: Difference in Gut Microbiota and Predicted Metabolic Pathways, J. Agric. Food Chem., 2021, 69, 13055–13065 CrossRef CAS PubMed.
- A. Singh, D. D'Amico, P. A. Andreux, G. Dunngalvin, T. Kern, W. Blanco-Bose, J. Auwerx, P. Aebischer and C. Rinsch, Direct supplementation with Urolithin A overcomes limitations of dietary exposure and gut microbiome variability in healthy adults to achieve consistent levels across the population, Eur. J. Clin. Nutr., 2022, 76, 297–308 CrossRef CAS.
- R. García-Villalba, H. Vissenaekens, J. Pitart, M. Romo-Vaquero, J. C. Espín, C. Grootaert, M. V. Selma, K. Raes, G. Smagghe, S. Possemiers, J. Van Camp and F. A. Tomas-Barberan, Gastrointestinal simulation model TWIN-SHIME shows differences between human urolithin-metabotypes in gut microbiota composition, pomegranate polyphenol metabolism, and transport along the intestinal tract, J. Agric. Food Chem., 2017, 65, 5480–5493 CrossRef PubMed.
- R. García-Villalba, D. Beltrán, J. C. Espín, M. V. Selma and F. A. Tomás-Barberán, Time course production of urolithins from ellagic acid by human gut microbiota, J. Agric. Food Chem., 2013, 61, 8797–8806 CrossRef PubMed.
- R. García-Villalba, M. V. Selma, J. C. Espín and F. A. Tomás-Barberán, Identification of Novel Urolithin Metabolites in Human Feces and Urine after the Intake of a Pomegranate Extract, J. Agric. Food Chem., 2019, 67, 11099–11107 CrossRef PubMed.
- M. Romo-Vaquero, R. García-Villalba, A. González-Sarrías, D. Beltrán, F. A. Tomás-Barberán, J. C. Espín and M. V. Selma, Interindividual variability in the human metabolism of ellagic acid: Contribution of Gordonibacter to urolithin production, J. Funct. Foods, 2015, 17, 785–791 CrossRef CAS.
- D. Beltrán, M. Romo-Vaquero, J. C. Espín, F. A. Tomás-Barberán and M. V. Selma, Ellagibacter isourolithinifaciens gen. Nov., sp. Nov., a new member of the family Eggerthellaceae, isolated from human gut, Int. J. Syst. Evol. Microbiol., 2018, 68, 1707–1712 CrossRef.
- M. V. Selma, D. Beltrán, M. C. Luna, M. Romo-Vaquero, R. García-Villalba, A. Mira, J. C. Espín and F. A. Tomás-Barberán, Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid, Front. Microbiol., 2017, 8, 1521 CrossRef PubMed.
- M. V. Selma, F. A. Tomas-Barberan, D. Beltran, R. Garcia-Villalba and J. C. Espin, Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut, Int. J. Syst. Evol. Microbiol., 2014, 64, 2346–2352 CrossRef CAS PubMed.
- M. V. Selma, D. Beltrán, R. García-Villalba, J. C. Espín and F. A. Tomás-Barberán, Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter, species, Food Funct., 2014, 5, 1779–1784 RSC.
- M. V. Selma, F. A. Tomás-Barberán, D. Beltran, R. García-Villalba and J. C. Espín, Eur. Pat, EP22382999, 2022 Search PubMed.
- C. E. Iglesias-Aguirre, R. García-Villalba, D. Beltrán, M. D. Frutos, J. C. Espín, F. A. Tomás-Barberán and M. V. Selma, The Gut Bacterial Consortia Involved in Ellagic Acid Metabolism to Yield Human Urolithin Metabotypes Revealed, J. Agric. Food Chem., 2023, 71 DOI:10.1021/acs.jafc.2c08889.
- S. Reagan-Shaw, M. Nihal and N. Ahmad, Dose translation from animal to human studies revisited, FASEB J., 2008, 22, 659–661 CrossRef CAS.
- M. Romo-Vaquero, E. Fernández-Villalba, A. L. Gil-Martinez, L. Cuenca-Bermejo, J. C. Espín, M. T. Herrero and M. V. Selma, Urolithins: potential biomarkers of gut dysbiosis and disease stage in Parkinson's patients, Food Funct., 2022, 13, 6306–6316 RSC.
- A. Cortés-Martín, R. García-Villalba, I. García-Mantrana, A. Rodríguez-Varela, M. Romo-Vaquero, M. C. Collado, F. A. Tomás-Barberán, J. C. Espín and M. V. Selma, Urolithins in Human Breast Milk after Walnut Intake and Kinetics of Gordonibacter, Colonization in Newly Born: The Role of Mothers’ Urolithin Metabotypes, J. Agric. Food Chem., 2020, 68, 12606–12616 CrossRef PubMed.
- R. García-Villalba, J. C. Espín and F. A. Tomás-Barberán, Chromatographic and spectroscopic characterization of urolithins for their determination in biological samples after the intake of foods containing ellagitannins and ellagic acid., J. Chromatogr. A, 2016, 1428, 162–175 CrossRef PubMed.
- A. Cortés-Martín, M. Romo-Vaquero, I. García-Mantrana, A. Rodríguez-Varela, M. C. Collado, J. C. Espín and M. V. Selma, Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum, Nutrients, 2019, 11, 2079 CrossRef PubMed.
- B. L. Maidak, J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt and J. M. Tiedje, The RDP-II (Ribosomal Database Project), Nucleic Acids Res., 2001, 29, 173–174 CrossRef CAS PubMed.
- C. Sánchez-González, C. J. Ciudad, V. Noé and M. Izquierdo-Pulido, Health benefits of walnut polyphenols: An exploration beyond their lipid profile, Crit. Rev. Food Sci. Nutr., 2017, 57, 3373–3383 CrossRef PubMed.
- J. A. Giménez-Bastida, M. Á. Ávila-Gálvez, J. C. Espín and A. González-Sarrías, Evidence for health properties of pomegranate juices and extracts beyond nutrition: A critical systematic review of human studies, Trends Food Sci. Technol., 2021, 114, 410–423 CrossRef.
- W. Xian, S. Yang, Y. Deng, Y. Yang, Z. Tan, W. Li and R. Yang, Potential of Establishing the Corresponding Human Microbial Community in Pseudo Germ-Free Mice through Fecal Microbe Transfer from Three Urolithin Metabotypes, J. Agric. Food Chem., 2022, 70, 9388–9398 CrossRef CAS PubMed.
- J. Long, Y. Guo, J. Yang, S. M. Henning, R. P. Lee, A. Rasmussen, L. Zhang, Q. Y. Lu, D. Heber and Z. Li, Bioavailability and bioactivity of free ellagic acid compared to pomegranate juice, Food Funct., 2019, 10, 6582–6588 RSC.
- A. Cortés-Martín, C. E. Iglesias-Aguirre, A. Meoro, M. V. Selma and J. C. Espín, Pharmacological Therapy Determines the Gut Microbiota Modulation by a Pomegranate Extract Nutraceutical in Metabolic Syndrome: A Randomized Clinical Trial, Mol. Nutr. Food Res., 2021, 65, 2001048 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo03957e |
| This journal is © The Royal Society of Chemistry 2023 |