Open Access Article
Open Access ArticleRutin attenuates D-galactose-induced oxidative stress in rats’ brain and liver: molecular docking and experimental approaches†
Shaimaa M.
Saafan
a,
Shymaa A.
Mohamed
b,
Ahmed E.
Noreldin
c,
Fayza A.
El Tedawy
a,
Yaser H. A.
Elewa
de,
Reda S.
Fadly
a,
Soad Khalil
Al Jaouni
 f,
Ali H.
El-Far
f,
Ali H.
El-Far
 *g and
Abdelwahab A.
Alsenosy
*g and
Abdelwahab A.
Alsenosy
 g
g
aDepartment of Biochemistry, Animal Health Research Institute (AHRI), Damanhour Branch, Agriculture Research Centre (ARC), Damanhour 22511, Egypt. E-mail: saafanshaimaa@gmail.com; Fayzaeltedawy@gmail.com; dr_redafadly@yahoo.com
bMolecular Biology, Molecular Biology Unit, Medical Technology Centre, Medical Research Institute, Alexandria University, Egypt. E-mail: shymaa.abdullah@alexu.edu.eg
cDepartment of Histology and Cytology, Faculty of Veterinary Medicine, Damanhour University, Damanhour 22511, Egypt. E-mail: ahmed.elsayed@damanhour.edu.eg
dDepartment of Histology, Faculty of Veterinary Medicine, Zagazig University, Zagazig 44519, Egypt
eLaboratory of Anatomy, Faculty of Veterinary Medicine, Basic Veterinary Sciences, Hokkaido University, Sapporo 060-0818, Japan. E-mail: y-elewa@vetmed.hokudai.ac.jp
fDepartment of Hematology/Pediatric Oncology, Yousef Abdulatif Jameel Scientific Chair of Prophetic Medicine Application, Faculty of Medicine, King Abdulaziz University, Jeddah 21589, Saudi Arabia. E-mail: saljaouni@kau.edu.sa
gDepartment of Biochemistry, Faculty of Veterinary Medicine, Damanhour University, Damanhour 22511, Egypt. E-mail: ali.elfar@damanhour.edu.eg; dr_alsenosy_2010@yahoo.com
First published on 31st March 2023
Abstract
Oxidative stress results from the imbalance between reactive oxygen species (ROS) production and antioxidant defence and is primarily involved in aging. The current study investigated the antioxidant activity of rutin in aging in rats induced by D-galactose (D-gal) for 42 days. Rutin was orally used at doses of 50 and 100 mg kg−1 daily. Results showed that D-gal induced oxidative alterations in the brain and liver recognized via upregulation of aging and oxidative markers. In contrast, rutin ameliorated the oxidative stress induced by D-gal by enhancing antioxidant markers such as superoxide dismutase-1, glutathione peroxidase-1, and glutathione S-transferase-α. Also, rutin significantly decreased the accumulation of β-galactosidase and reduced the expression of p53, p21, Bcl-2-associated X protein (Bax), caspase-3 (CASP3), and mammalian target of rapamycin (mTOR) in brain and hepatic tissues. Rutin potentially attenuated these aging-related oxidative alterations in a dose-dependent manner. Moreover, rutin markedly reduced the increased immunohistochemical expression of β-galactosidase, 8-hydroxy-2′-deoxyguanosine, calcium-binding adapter molecule 1, glial fibrillary acidic protein, Bax, and interleukin-6 and significantly increased Bcl2, synaptophysin, and Ki67. Furthermore, a molecular docking study revealed that rutin exhibited high affinity to rat and human caspases, PI3K/AKT/mTOR, and the IL-6 receptor. Finally, we can conclude that rutin supplementation can be a promising natural protective compound that could delay aging and maintain health.
1. Introduction
Aging is associated with numerous morphological and functional changes in the body. Oxidative stress and inflammation are the main mechanisms involved in the aging process that may cause severe cellular injuries.1 Reactive oxygen species (ROS) are produced by living organisms due to normal cellular metabolism and environmental factors. ROS are highly reactive molecules and can damage cell structures such as carbohydrates, nucleic acids, lipids, and proteins and alter their functions.2 The shift in the balance between oxidants and antioxidants in favor of oxidants is termed oxidative stress.3The liver and the brain are the two most crucial organs, and ROS assault throughout aging causes both of these organs to steadily deteriorate.4 There is a relationship between the functional status of the liver and brain; hepatic encephalopathy results from the insufficiency of the liver to remove toxins from the blood, leading to brain dysfunction.5 Furthermore, exposure to ROS increases the incidence of several liver diseases throughout the life-span, including aging.6
Chronic experimental D-galactose (D-gal) exposure induces premature aging similar to natural aging in rodents.7–10D-gal is largely metabolised in the liver, which also plays a crucial detoxifying role. Additionally, this organ is regularly exposed to ROS due to its rapid respiratory rate. Second, despite the fact that the brain has the most complex tissue of any organ in the body and needs a lot of oxygen and energy to operate normally, it only has a small concentration of antioxidant enzymes.11
In nature, D-gal is a reducing sugar that can be metabolized at normal concentrations. However, at high levels, D-gal can be converted into aldose and hydroperoxide under the catalysis of galactose oxidase, resulting in the generation of a superoxide anion and oxygen-derived free radicals.12D-gal also reacts with the free amines of amino acids in proteins and peptides both in vivo and in vitro to form advanced glycation end products (AGEs), which cause ROS accumulation.13,14 Oxidative stress-mediated DNA damage caused alterations in the synaptic structure, and memory maintenance and learning abilities were also demonstrated in mouse models.15,16
Since ancient times, medicinal plants have been an integral part of traditional medicine for the discovery of new drug leads.17–26 Rutin (3,3′,4′,5,7-pentahydroxyflavone-3-rhamnoglucoside) is a flavonol abundant in plants such as passionflower, buckwheat, tea, and apple. The name ‘rutin’ comes from the plant Ruta graveolens, which contains rutin. Chemically, it is a glycoside comprising the flavonol aglycone quercetin and the disaccharide rutinose. The 4-oxo group and the 2,3 double bond in the C ring in rutin may be related to its neuroprotective action.27 Polyhydroxylated substitutions on rings A and B, a 2,3-double bond, a free 3-hydroxyl substitution, and a 4-keto moiety confer antiperoxidative properties to rutin.28 It has demonstrated many pharmacological activities, including antioxidant, neuroprotective, cytoprotective, vasoprotective, anticarcinogenic, and cardioprotective effects.29–31 The present experiment aims to assess the anti-aging protective effect of rutin in experimentally induced aging by D-gal in rats' brain and liver.
2. Materials and methods
2.1. Ethics statement
All methods are reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org) and approved (DMU/VetMed-2023/012) by the Faculty of Veterinary Medicine Ethics Committee, Damanhour University, Egypt.2.2. Experimental design
Fifty healthy male Wistar rats weighing (90–110 g B.W.), were purchased from the Medical Research Institute of Alexandria University, Egypt. The animals were kept in metal cages under controlled environmental conditions with optimum temperature (23 ± 2), humidity (55 ± 5), and dark/light cycle (12 hours) with free access to basal diet (ESI Table 1†) and drinking water. All animals were housed for one week before the experiment to acclimate and ensure average growth and behavior.Rats were randomly assigned into five groups containing 10 rats each. In the control group, rats were subcutaneously injected with physiological saline solution (0.9%) daily. In the vehicle group, rats were injected subcutaneously with physiological saline solution (0.9%) and orally supplemented with corn oil daily. In the D-gal (Sigma-Aldrich Co., Missouri, USA) group, rats were injected subcutaneously with 200 mg of D-gal per kg body weight (B.W.) dissolved in saline solution daily and supplemented orally with corn oil.32 However, in the D-gal + RU50 group, rats were injected subcutaneously with 200 mg of D-gal per kg B.W. dissolved in saline solution daily and supplemented orally with 50 mg of rutin (Sigma-Aldrich Co.) per kg B.W.33 In the D-gal + RU100 group, rats were injected subcutaneously with 200 mg of D-gal per kg B.W. dissolved in saline solution daily and supplemented orally with 100 mg of rutin per kg B.W.34 dissolved in corn oil. The experimental period lasted for 42 days.
2.3. Sampling
On day 42, the animals were anesthetized with isoflurane inhalation and euthanized by cervical dislocation. The brain (cerebrum, hippocampus, and cerebellum) and liver (right lobe) were immediately dissected, rinsed with chilled normal saline 0.9%, and divided into three parts; the first was used for histopathological and immunohistochemical examination (fixed in neutral buffered formalin, 10%) and the other two parts were stored at −80 °C until further analysis. The first part was used for studying mRNA expression of p53, p21, Bcl-2-associated X protein (Bax), B-cell lymphoma-2 (Bcl2), caspase-3 (CASP3), mammalian target of rapamycin (mTOR), and β-actin by RT-PCR. The second part was used for the determination of superoxidase dismutase 1 (SOD1), glutathione peroxidase-1 (GPx-1), and glutathione S-transferase-α (GST-α) by ELISA.2.4. Histopathological assessment
The conventional paraffin embedding technique was performed to process the fixed specimens according to Bancroft and Layton.35 Four μm thick sections from brain and liver samples were stained using hematoxylin and eosin (H and E) as described by Bancroft and Layton35 and periodic acid Schiff (PAS) in liver sections according to Layton and Bancroft.36 Semiquantitative scoring of brain and liver lesions was calculated according to Gibson-Corley et al.37 Briefly, lesions in 10 fields randomly chosen from each rat's slide were obtained and averaged. The lesions were scored in a blinded way [score scale: 0 = normal; 1 ≤ 25%; 2 = 26–50%; 3 = 51–75%; 4 = 76–100%].2.5. Immunohistochemical assessment
Antibodies, sources, working dilutions, and methods for antigen retrieval are listed in the ESI Table 2.† The immunohistochemical technique was investigated according to the method described by Noreldin et al.38 Micrographs of the sections were taken with a digital camera (Leica EC3, Leica, Germany) connected to a microscope (Leica DM500). The ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to quantify immunostaining intensities. Vis et al. (2000) reported that the inverse means density was determined in 10 randomly chosen fields from various sections of five rats in each group.2.6. Antioxidant status assessment
Oxidative stress and antioxidant biomarkers were analyzed in the brain and liver homogenates (20% (w/v)) using cooled 0.1 M phosphate buffered saline. SOD1 (cat. no. ER0332), GPx-1 (cat. no. ER0274), and GST-α (cat. no. ER1022) protein levels were determined using ELISA kits (FineTest, Wuhan, Hubei, China). The Bradford method was followed to determine protein levels in all samples.392.7. Gene expression assessment by real-time polymerase chain reaction (RT-PCR)
According to the manufacturer's kit, total RNA was extracted from the tissue samples using the easy-spin kit for total RNA extraction (INTRON Biotechnology, Korea). The purities and concentration of RNA were measured by a nanodrop spectrophotometer (Genway Nanodrop, Germany). 1 μg of RNA (260/280 ratio = 1.8–2.0) was used for the transcription of cDNA using the RT-Premix kit (INTRON Biotechnology). 2 μl of RT product was mixed with 10 μl of SYBR-Green master mix (INTRON, Biotechnology, Korea) and 0.5 mM of each forward and reverse primer (ESI Table 3†) and nuclease-free water to make a final volume of 20 μl. All reactions were performed on a 7500 Applied Biosystems, USA, under the following conditions: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 58 °C for 15 °C, and 72 °C for 30 s. Relative expression of mRNA was normalized to β-actin as a housekeeper gene. The fold changes of mRNA expression were calculated by the 2−ΔΔCt method described by Livak and Schmittgen.402.8. Molecular docking
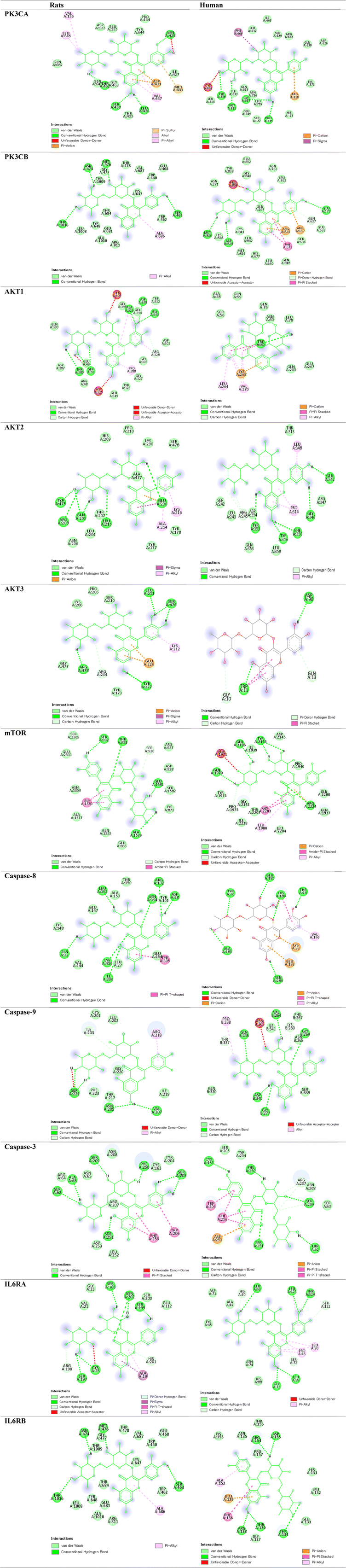
Molecular docking assessment was performed to determine the binding affinity (pKi) of rutin toward phosphatidylinositol-4,5-bisphosphate 3-kinase-catalytic subunit alpha (PK3CA), phosphatidylinositol-4,5-bisphosphate 3-kinase-catalytic subunit beta (PK3CB), protein kinase B1 (AKT1), AKT2, AKT3, mammalian target of rapamycin (mTOR), caspase-8, caspase-9, caspase-3, interleukin 6 receptor-alpha (IL6RA), and interleukin 6 receptor-beta (IL6RB) proteins in rats and humans.Rat PK3CA (AF-A0A0G2K344), PK3CB (AF-Q9Z1L0), AKT1 (AF-P47196), AKT2 (AF-P47197), AKT3 (AF-Q63484), mTOR (AF-P42346), caspase-8 (AF-Q9JHX4), caspase-9 (AF-A0A0G2K3V0), caspase-3 (AF-P55213), IL6RA (AF-G3V8T6), IL6RB (AF-P40190), and human PK3CB (AF-P42338) were retrieved from the AlphaFold protein structure database (https://alphafold.ebi.ac.uk/). Human PK3CA (3HIZ), AKT1 (6S9X), AKT2 (1GZO), AKT3 (2X18), mTOR (4JT6), caspase-8 (1I4E), caspase-9 (1NW9), caspase-3 (1NMQ), IL6RA (1N26), and IL6RB (1P9M) were obtained from the RCSB protein data bank (https://www.rcsb.org/).
The binding affinity (pKi) and the ligand efficiency of rutin and target proteins were determined using the InstaDock software.41 In addition, visualization of ligands–proteins interactions was visualized using the Discovery Studio Visualizer software (https://discover.3ds.com/discovery-studio-visualizer-download).
2.9. Statistical analysis
A one-way ANOVA with Tukey's post hoc multiple range tests was used for data analysis using GraphPad Prism v.5 (https://www.graphpad.com/) (GraphPad, San Diego, CA, USA). All declarations of significance were based on P < 0.05.3. Results
3.1. Histopathology
The histopathological investigation of the cerebrum of rats in the control and vehicle groups showed normal histoarchitecture of the neurons and neuropil (ESI Fig. 1A and B†). On the other hand, the cerebrum of D-gal-treated rats revealed necrotic neurons and congested blood vessels (ESI Fig. 1C†). However, the cerebrum of the D-gal + RU50-treated rats showed improved cerebral neurons and neuropil with minimal necrotic neurons (ESI Fig. 1D†). Moreover, the D-gal + RU100-treated rats exhibited a quite normal cerebral architecture (ESI Fig. 1E†). ESI Fig. 1F† represents a significant decrease in the cerebral necrotic score in the D-gal + RU50 and D-gal + RU100 groups compared with that of the D-gal group. Also, the D-gal + RU100 rats exhibited a significant reduction in necrotic score compared with those in the D-gal + RU50 group.The histopathologic examination of the hippocampus of rats in the control and vehicle groups showed normal hippocampal structures of the granule cell layer and molecular layer of the dentate gyrus (ESI Fig. 2A and B†). However, the D-gal-treated rats showed necrotic neurons of the dentate gyrus with a small number and layers of neurons with a disordered arrangement (ESI Fig. 2C†). Rats treated with D-gal + RU50 and D-gal + RU100 revealed an improvement in the dentate gyrus morphology, with a few degenerated neurons (ESI Fig. 2D and E†). Hippocampal necrotic scores were significantly decreased in the D-gal + RU50 and D-gal + RU100 compared with those of the D-gal. Also, the D-gal + RU100 group exhibited a significant reduction in the necrotic score compared with the D-gal + RU50 group (ESI Fig. 2F†).
Upon examining the cerebellum of rats in the control and vehicle groups, we detected the normal cerebellar architecture of the molecular cell, Purkinje cell, and granule cell layers (ESI Fig. 3A and B†). D-gal-treated rats showed a complete loss or necrotic nuclei of the Purkinje cell layer with a focal loss of neurons of the granule cell layer (ESI Fig. 3C†). However, in the D-gal + RU50 group, rats revealed a better histologic cerebellar structure with a few pyknotic Purkinje cells (ESI Fig. 3D†). Furthermore, D-gal + RU100-treated rats have a nearly normal histologic cerebellar architecture with minimal degenerated Purkinje cells (ESI Fig. 3E†). Cerebellar necrotic scores were significantly decreased in the D-gal + RU50 and D-gal + RU100 groups compared with those of the D-gal group. Also, the D-gal + RU100 rats exhibited a significant reduction in the necrotic score compared with the D-gal + RU50 rats (ESI Fig. 3F†).
No histopathologic hepatic lesions were detected in the control and vehicle groups (ESI Fig. 4A and B†). However, hepatic samples isolated from the D-gal group showed dilated and congested central vein, hydropic degeneration, and accumulated inflammatory cells (ESI Fig. 4C†). Rats treated with D-gal + RU50 revealed an improved hepatic architecture with a few pyknotic nuclei (ESI Fig. 4D†). Furthermore, D-gal + RU100-treated rats exhibited a relatively normal hepatic arch similar to the control group (ESI Fig. 4E†). Semi-quantitative statistical analysis of hepatic lesion scores revealed that the D-gal group had a markedly increased level of hepatic vacuolation score compared to the rats in the control group. However, the D-gal + RU50 and D-gal + RU100 groups showed a significant reduction in the hepatic lesion score (ESI Fig. 4F†).
The negative control and vehicle groups had normal content and distribution of glycogen inside hepatocytes (ESI Fig. 5A and B†). On the other hand, hepatic sections of the D-gal group revealed a low amount and distribution of glycogen (ESI Fig. 5C†). The glycogen amount and distribution in the D-gal + RU50 and D-gal + RU100-treated rats had increased in a dose-dependent manner (ESI Fig. 5D and E†). The D-gal group revealed a marked reduction in glycogen distribution compared with the control and negative vehicle groups. Moreover, rats protected against D-gal using RU50 and RU100 exhibited an even glycogen distribution in all hepatic lobules (ESI Fig. 5F†).
3.2. Immunohistochemistry
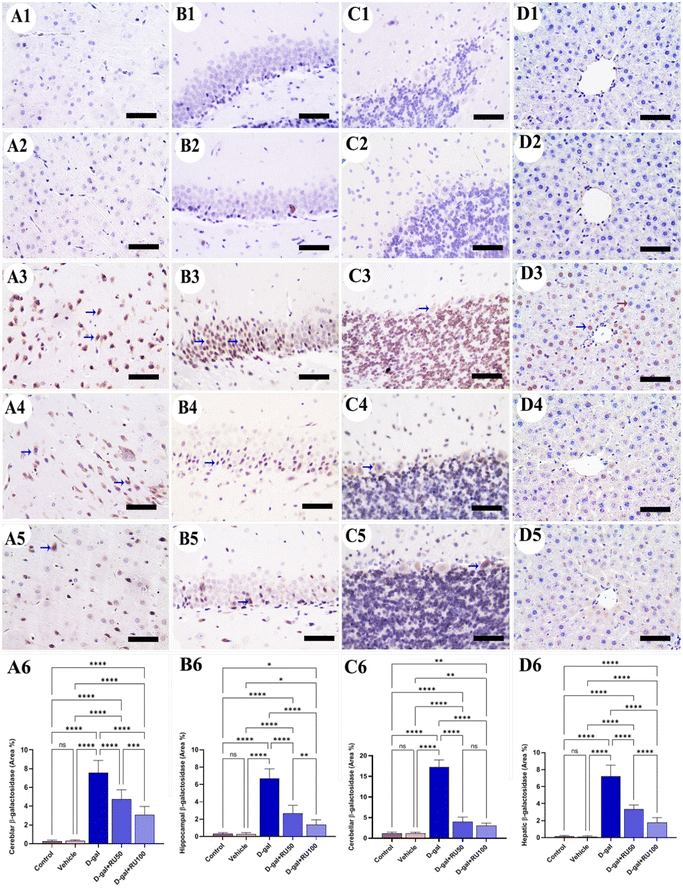
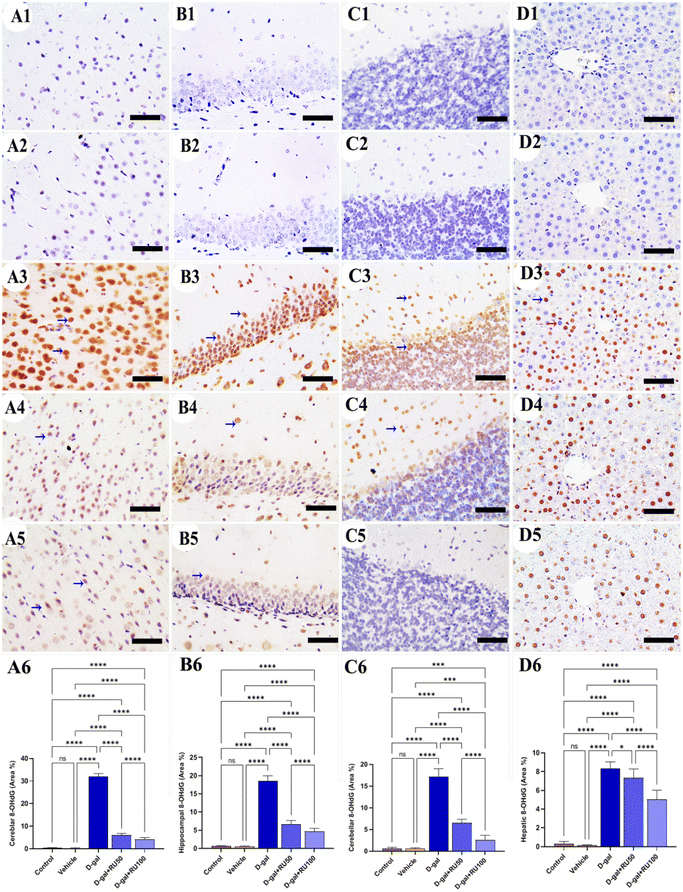
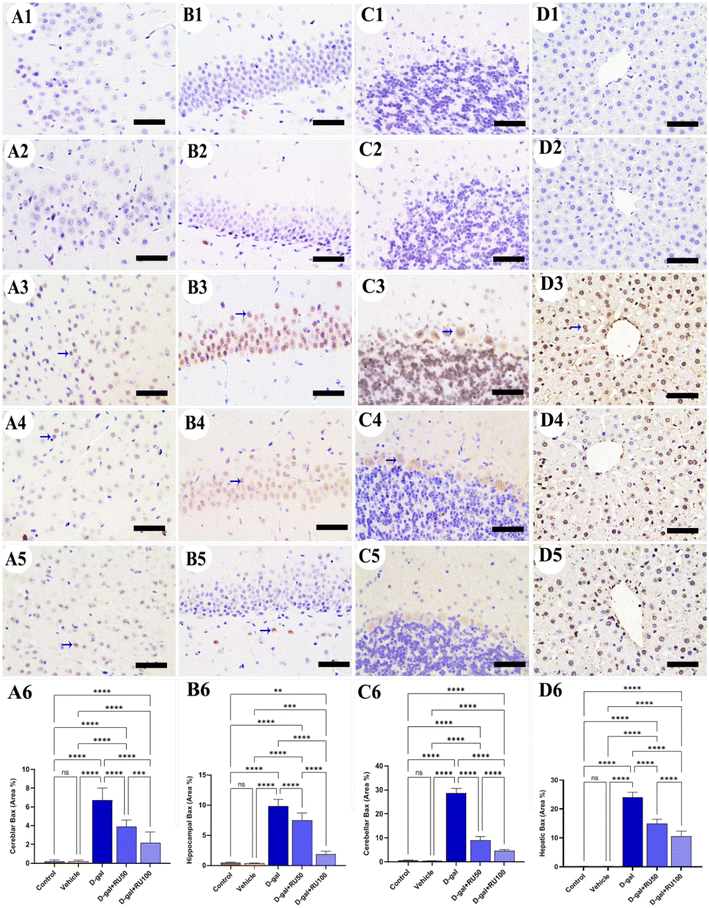
The investigation of β-galactosidase immunohistochemical expression in the control and vehicle groups showed a negative immune reaction in the cerebrum (Fig. 1A1 and A2), hippocampus (Fig. 1B1 and B2), cerebellum (Fig. 1C1 and C2), and liver (Fig. 1D1 and D2) respectively. However, the D-gal-group showed many β-galactosidase immune reactive nuclei in all brain regions and the liver (Fig. 1A3, B3, C3, and D3). On the other hand, the group treated with D-gal + RU50 showed a lower distribution of nuclei showing the β-galactosidase reaction than the D-gal alone group (Fig. 1A4, B4, C4, and D4). Moreover, the D-gal + RU100 group revealed the lowest distribution of nuclei showing the β-galactosidase reaction (Fig. 1A5, B5, C5, and D5). The nonparametric quantitative analysis for the area percentage of nuclei showing the β-galactosidase immunohistochemical reaction revealed a markedly high expression in the D-gal group compared with the control rats. This expression was significantly reduced in the D-gal + RU50 and D-gal + RU100 groups (Fig. 1A6, B6, C6, and D6).No immunohistochemical reaction of 8-hydroxy-2′-deoxyguanosine (8-OHdG) could be detected in the cerebrum (Fig. 2A1 and A2), hippocampus (Fig. 2B1 and B2), cerebellum (Fig. 2C1 and C2), and liver (Fig. 2D1 and D2) of control and vehicle groups. The D-gal-treated rats revealed the highest distribution of nuclei showing 8-OHdG immunoreaction (Fig. 2A3, B3, C3, and D3). However, the D-gal + RU50-supplemented rats exhibited a smaller distribution of nuclei showing the 8-OHdG immunoreaction than the D-gal alone group (Fig. 2A4, B4, C4, and D4). In addition, the D-gal + RU100 group exhibited the smallest expression of nuclei showing the 8-OHdG immunoreaction in all brain compartments and liver (Fig. 2A5, B5, C5, and D5). The nonparametric quantitative analysis for the area percentage of nuclei showing 8-OHdG immunohistochemical reaction reveals a significantly increased expression in D-gal-treated rats in comparison with control rats. This expression was markedly lowered in the D-gal + RU50 and D-gal + RU100 groups (Fig. 2A6, B6, C6, and D6).
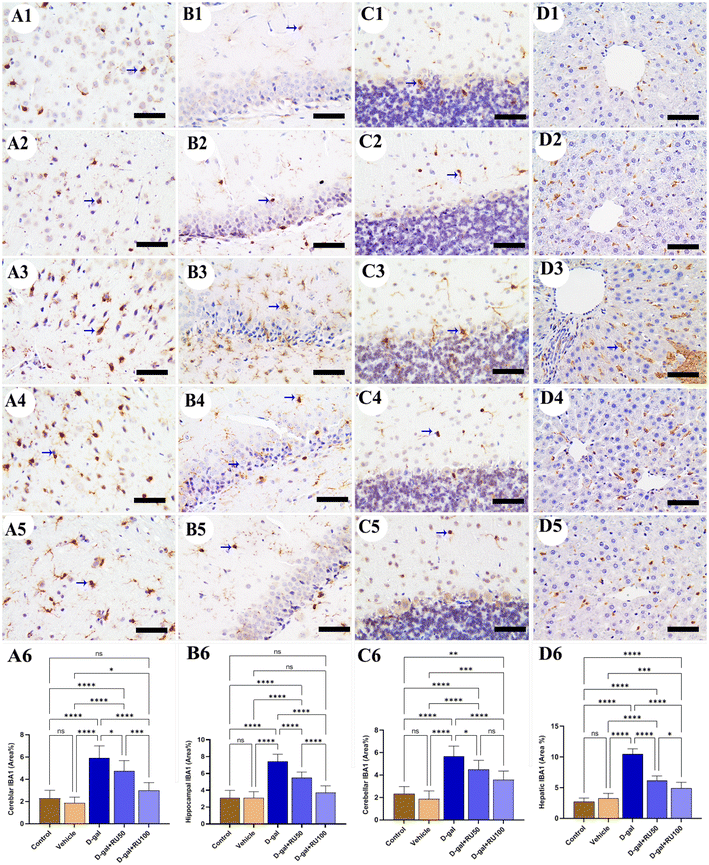
The detection of cells showing calcium-binding adapter molecule 1 (IBA1) immunoreaction in the control and vehicle groups revealed the lowest number in the cerebrum (Fig. 3A1 and A2), hippocampus (Fig. 3B1 and B2), cerebellum (Fig. 3C1 and C2), and liver (Fig. 3D1 and D2). However, the rats in the D-gal group exhibited the highest microglia distribution in all brain compartments and Kupffer cells in the liver (Fig. 3A3, B3, C3, and D3). On the other hand, rats in the D-gal + RU50 group showed lower microglia and Kupffer cell distribution than those in the D-gal group (Fig. 3A4, B4, C4, and D4). Furthermore, the D-gal + RU100 group revealed the lowest microglia and Kupffer cell number among all treated groups (Fig. 3A5, B5, C5, and D5). The nonparametric quantitative analysis for the area percentage of cells showing IBA1 immunohistochemical reaction showed a markedly high immunohistochemical reaction in the D-gal-treated rats compared with the control rats. This expression was significantly decreased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 3A6, B6, C6, and D6).
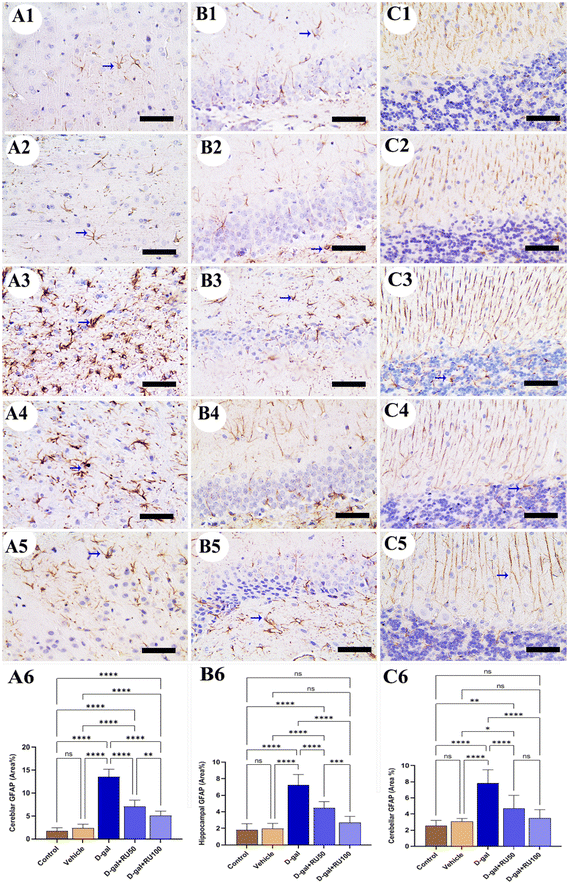
Regarding the immunohistochemical expression of the glial fibrillary acidic protein (GFAP) in rat brain, a low distribution of astrocytes was detected in the cerebrum (Fig. 4A1 and A2), hippocampus (Fig. 4B1 and B2), and cerebellum (Fig. 4C1 and C2), respectively. In contrast, in the D-gal group, rats revealed the highest astrocyte number in all brain regions (Fig. 4A3, B3, and C3). The D-gal + RU50 group showed lower astrocyte distribution than the D-gal group (Fig. 4A4, B4, and C4). Moreover, rats treated with D-gal + RU100 revealed the lowest astrocyte distribution than all groups (Fig. 4A5, B5, and C5). The nonparametric quantitative analysis for the area percentage of GFAP expression in astrocytes showed a markedly high immunohistochemical GFAP reaction in the D-gal group compared with the control group. This reaction was markedly decreased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 4A6, B6, and C6).
The examination of Bax immunohistochemical expression in the control and vehicle groups revealed negative to mild reactions in the cerebrum (Fig. 5A1 and A2), hippocampus (Fig. 5B1 and B2), cerebellum (Fig. 5C1 and C2), and liver (Fig. 5D1 and D2). On the other hand, brain and liver specimens obtained from rats in the D-gal group showed massive Bax expression in the nuclei of neurons and hepatocytes (Fig. 5A3, B3, C3, and D3). At the same time, the D-gal + RU50 rats exhibited decreased Bax expression than the D-gal group (Fig. 5A4, B4, C4, and D4). Furthermore, the D-gal + RU100 rats showed the lowest Bax expression among all groups (Fig. 5A5, B5, C5, and D5). The nonparametric quantitative analysis for the area percentage of Bax expression showed a markedly high Bax expression in the D-gal group compared with the control group. This reaction was markedly reduced in the D-gal + RU50 and D-gal + RU100 groups (Fig. 5A6, B6, C6, and D6).
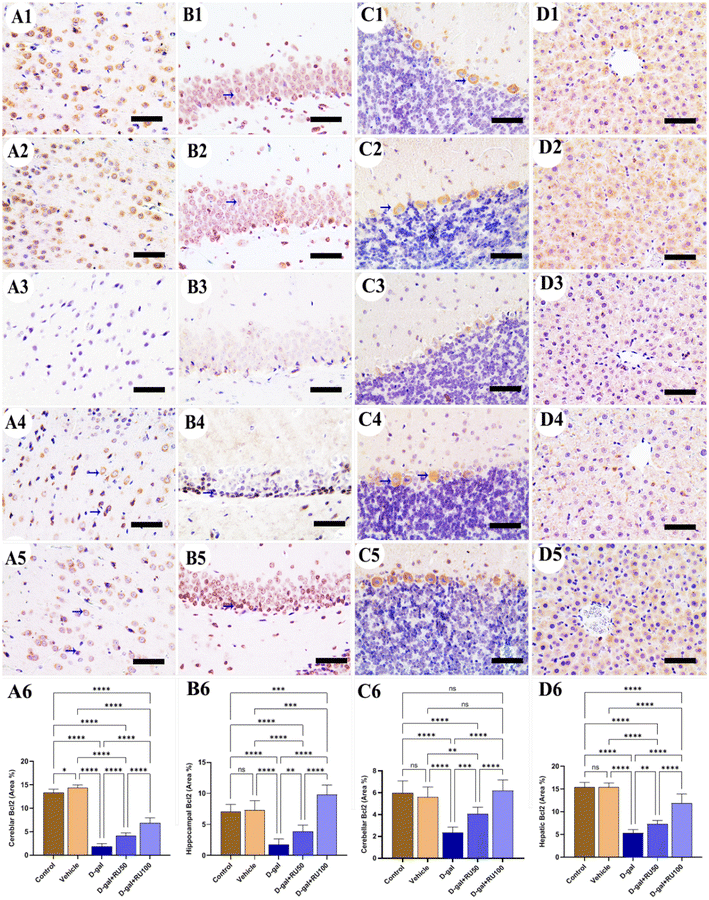
Concerning the studying of Bcl2 expression in the control and vehicle groups, high expression was seen in the cerebrum (Fig. 6A1 and A2), hippocampus (Fig. 6B1 and B2), cerebellum (Fig. 6C1 and C2), and liver (Fig. 6D1 and D2). However, treating rats with D-gal lowered the distribution of Bcl2 in all brain compartments and the liver (Fig. 6A3, B3, C3, and D3). On the other hand, the expression of Bcl2 increased in the D-gal + RU50 group more than in rats treated with D-gal alone (Fig. 6A4, B4, C4, and D4). Moreover, Bcl2 expression was the highest in the D-gal + RU100 group among all treated groups (Fig. 6A5, B5, C5, and D5). The nonparametric quantitative analysis for the area percentage of Bcl2 expression revealed a markedly decreased Bcl2 expression in the D-gal group compared with control groups. This reaction was markedly increased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 6A6, B6, C6, and D6).
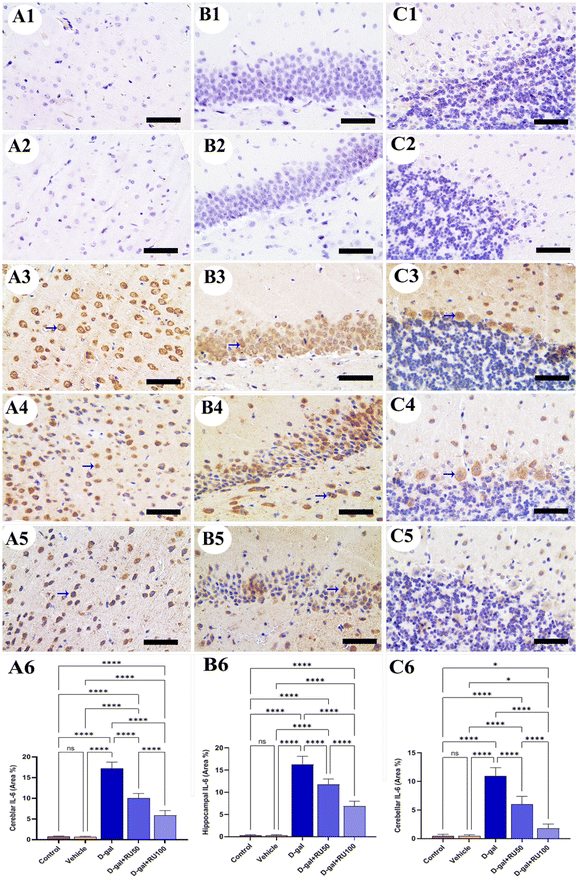
In the control and vehicle groups, no expression for interleukin-6 (IL-6) could be detected in the cerebrum (Fig. 7A1 and A2), hippocampus (Fig. 7B1 and B2), and cerebellum (Fig. 7C1 and C2). Otherwise, the rats exposed to D-gal showed extensive IL-6 expression in all brain locations (Fig. 7A3, B3, and C3), which was reduced in the samples isolated from rats treated with D-gal + RU50 (Fig. 7A4, B4, and C4). IL-6 expression was the lowest in the D-gal + RU100 group among all treated groups (Fig. 7A5, B5, and C5). The nonparametric quantitative analysis for the area percentage of IL-6 expression showed a markedly high IL-6 expression in the D-gal group compared with the control groups. This reaction was markedly decreased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 7A6, B6, and C6).
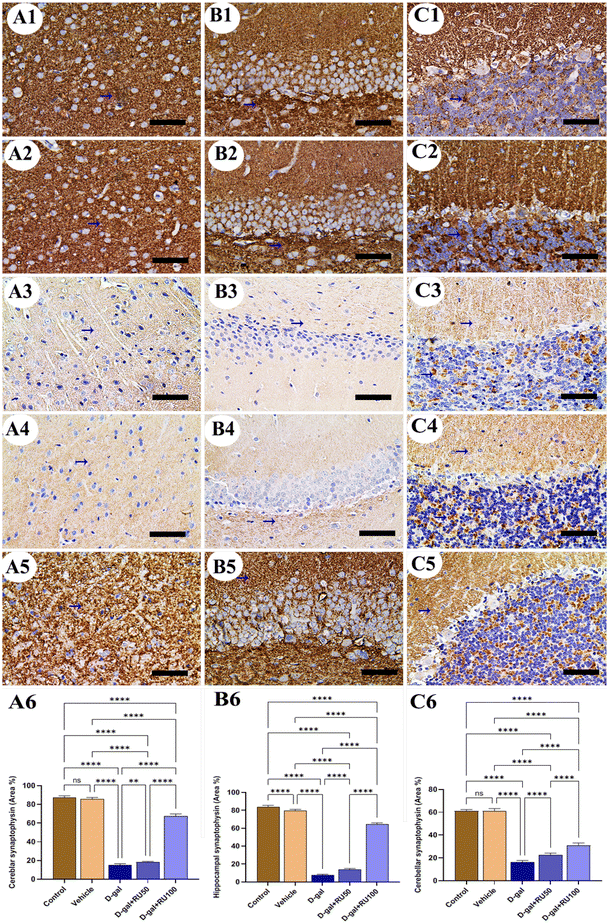
Synaptophysin expression in rats of the control and vehicle groups was intense and normal in the cerebrum (Fig. 8A1 and A2), hippocampus (Fig. 8B1 and B2), and cerebellum (Fig. 8C1 and C2). However, synaptophysin expression was the lowest in the brain of rats in the D-gal group (Fig. 8A3, B3, and C3). In contrast, synaptophysin expression improved in the D-gal + RU50 group more than in the D-gal group (Fig. 8A4, B4, and C4). Interestingly, the best improvement in synaptophysin expression of all groups was detected in the D-gal + RU100 group (Fig. 8A5, B5, and C5). The nonparametric quantitative analysis for the area percentage of the synaptophysin expression revealed a marked reduced synaptophysin expression in the D-gal group compared with the control group. This reaction was markedly increased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 8A6, B6, and C6).
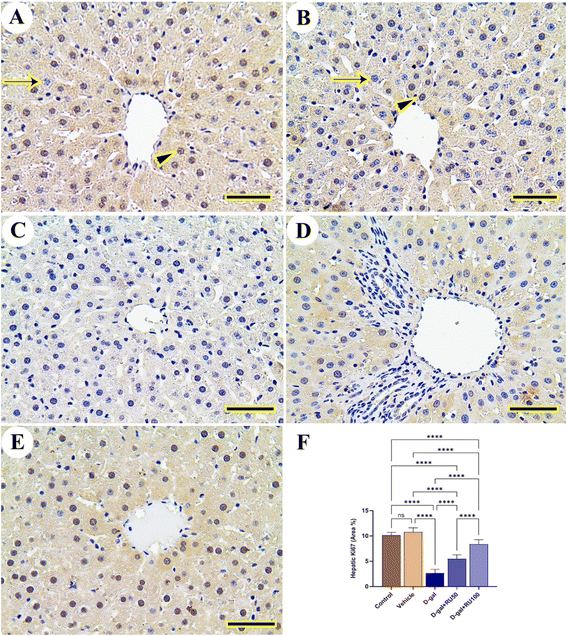
The exploration of hepatic tissue for Ki67 expression in the control and vehicle groups displayed extensive expression in nuclei of hepatocytes with mild reaction in the cytoplasm (Fig. 9A and B). However, this extensive reaction was decreased in the D-gal group (Fig. 9C). The D-gal + RU50 group showed mild restoration for Ki67 expression in hepatocytes nuclei (Fig. 9D), which increased extensively in the D-gal + RU100 group (Fig. 9E). The nonparametric quantitative analysis for the area percentage of Ki67 expression revealed a marked decreased Ki67 expression in the D-gal group compared to control groups. This reaction was markedly increased in the D-gal + RU50 and D-gal + RU100 groups (Fig. 9F).
3.3. Antioxidant status
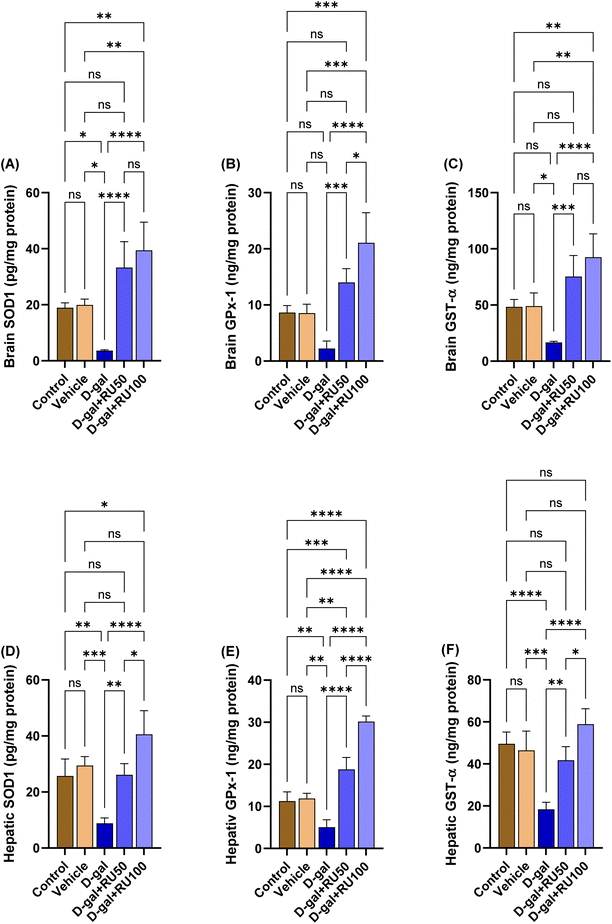
The data in Fig. 10A show a significant (P < 0.05) decrease in brain SOD1 concentration in the D-gal group compared with the control and the vehicle groups, whereas in the D-gal + RU50 and D-gal + RU100 groups, the brain SOD1 concentration was significantly (P < 0.0001) increased compared with the D-gal group. Its concentration was significantly increased in the D-gal + RU100 (P < 0.01) group compared with the control group; in addition, its concentration was significantly (P < 0.01) increased in the D-gal + RU100 group compared with the vehicle group.The data in Fig. 10B and C show a significant (P < 0.05) decrease in brain GPx-1 and GST-α concentrations, respectively in the D-gal group compared with the control and the vehicle groups, while in the D-gal + RU50 (P < 0.001) and D-gal + RU100 (P < 0.0001) groups, the brain GPx-1 concentration was significantly increased compared with the D-gal group. In addition, GPx-1 concentration was significantly increased in the D-gal + RU100 group compared with the control and vehicle groups (P < 0.001) and D-gal + RU50 group (P < 0.05). Furthermore, GST-α concentration was significantly (P < 0.01) increased in the D-gal + RU100 group compared with the control and vehicle groups.
The levels of hepatic SOD1, GPx-1, and GST-α are shown in Fig. 10D, E, and F, respectively. Their levels were significantly (P < 0.01, P < 0.01, and P < 0.001, respectively) decreased in the D-gal group. The D-gal group treated with rutin showed significant increases in SOD1, GPx-1, and GST-α in the D-gal + RU50 and D-gal + RU100 groups in a dose-dependent manner.
3.4. mRNA expression
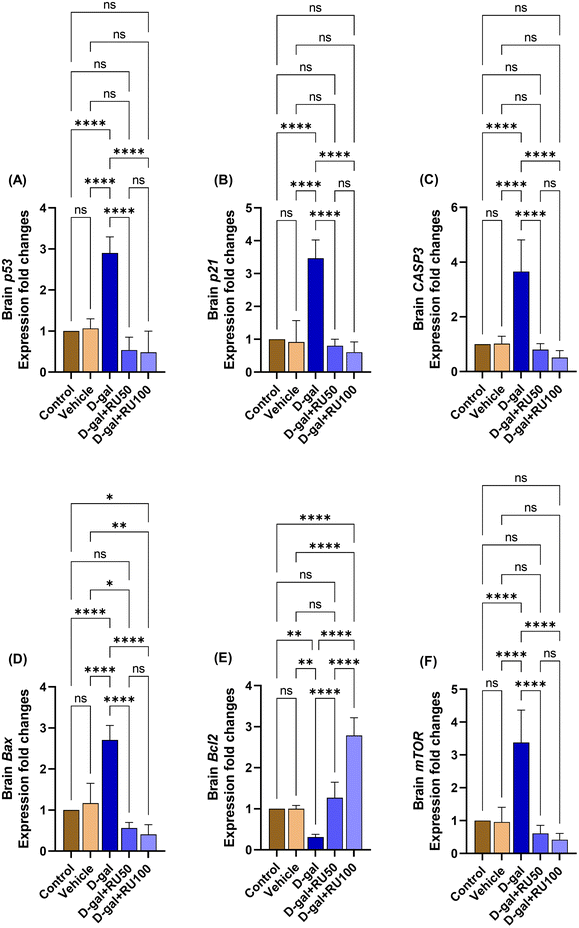
The brain p53 and p21 mRNA expression (Fig. 11A and B, respectively) in the D-gal group was significantly increased (P < 0.0001) compared with the control and vehicle groups. In the D-gal + RU50 and D-gal + RU100 groups, p53 mRNA expression was significantly decreased (P < 0.0001) compared with the D-gal group. In addition, p53 expression levels in the D-gal + RU100 were non significantly reduced compared with the vehicle group. Also, the brain p21 mRNA expression was significantly decreased (P < 0.0001) in the D-gal + RU50 and D-gal + RU100 groups compared with the D-gal group.The brain CASP3 mRNA expression in the D-gal group was significantly increased (P < 0.0001) compared with the control and vehicle groups. In contrast, in the D-gal + RU50 and D-gal + RU100 groups, their levels showed a significant decrease (P < 0.0001) compared with the D-gal group (Fig. 11C).
The brain Bax mRNA expression in the D-gal group was significantly increased (P < 0.0001) compared with the control and vehicle groups. On the other hand, the brain Bax mRNA expression was significantly decreased (P < 0.0001) in the D-gal + RU50 and D-gal + RU100 groups compared with the D-gal group. In comparison with the vehicle group, the brain Bax mRNA expression was significantly decreased in the D-gal + RU50 (P < 0.05) and D-gal + RU100 (P < 0.01) groups. At the same time, its expression was significantly decreased in the D-gal + RU100 (P < 0.05) compared with the control group (Fig. 11D).
The brain Bcl2 mRNA expression in the D-gal group was significantly decreased (P < 0.01) compared with the control and the vehicle groups. On the other hand, the brain Bcl2 mRNA expression was significantly increased (P < 0.0001) in the D-gal + RU50 and the D-gal + RU100 groups compared with the D-gal group. In addition, its expression was significantly (P < 0.0001) increased in the D-gal + RU100 group compared with the control group, the vehicle group, and the D-gal + RU50 group (Fig. 11E).
The brain mTOR mRNA expression in the D-gal group was significantly increased (P < 0.0001) compared with the control and the vehicle groups. On the other hand, its expression was significantly decreased (P < 0.0001) in the D-gal + RU50 and D-gal + RU100 groups compared with the D-gal group (Fig. 11F).
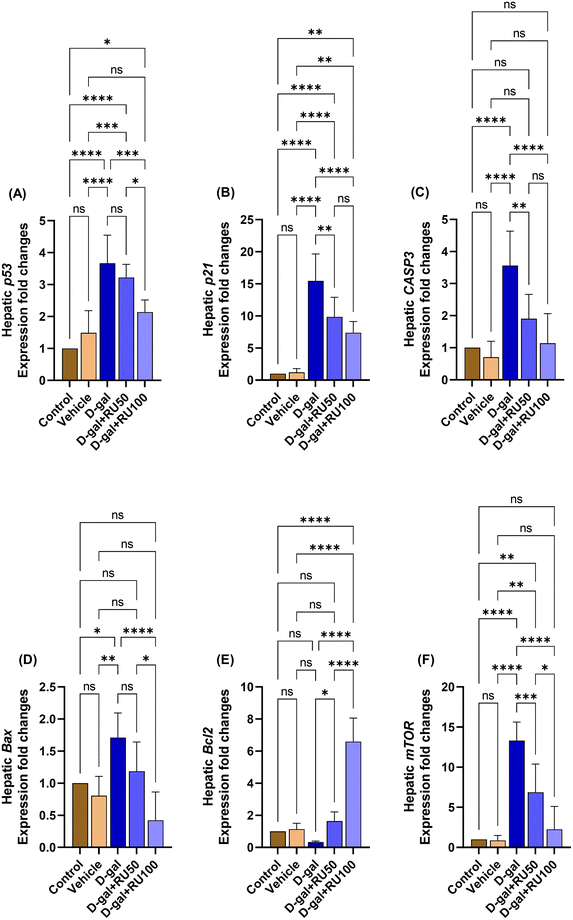
Similarly, hepatic p53 (Fig. 12A), p21 (Fig. 12B), CASP3 (Fig. 12C), Bax (Fig. 12D), and mTOR (Fig. 12F) mRNA expression was significantly increased in the D-gal group compared with the control group, while Bcl2 (Fig. 12E) expression was significantly decreased. In contrast, p53, p21, CASP3, Bax, and mTOR mRNA expression was significantly decreased in the D-gal + RU50 and D-gal + RU100 groups compared with the D-gal group, while Bcl2 expression was significantly increased in a dose dependent manner.
3.5. Molecular docking scores
Data presented in Table 1 and Fig. 13 reveal that rutin exhibited high binding affinity to rat and human PK3CA, PK3CB, AKT1, AKT2, AKT3, mTOR, caspase-8, caspase-9, caspase-3, IL6RA, and IL6RB proteins.| Rats | Human | |||
|---|---|---|---|---|
| Binding free energy (kcal mol−1) | pKi | Binding free energy (kcal mol−1) | pKi | |
| PK3CA | −8.40 | 6.16 | −9.40 | 6.89 |
| PK3CB | −9.40 | 6.89 | −9 | 6.60 |
| AKT1 | −9 | 6.60 | −8.80 | 6.45 |
| AKT2 | −8.20 | 6.01 | −7.50 | 5.50 |
| AKT3 | −7.90 | 5.79 | −7.50 | 5.50 |
| mTOR | −8.30 | 6.09 | −8.90 | 6.53 |
| Caspase-8 | −7.70 | 5.65 | −8.80 | 6.45 |
| Caspase-9 | −6 | 4.40 | −7.10 | 5.21 |
| Caspase-3 | −7.80 | 5.72 | −7.30 | 5.35 |
| IL6RA | −7.3 | 5.35 | −8.1 | 5.94 |
| IL6RB | −7.7 | 5.65 | −8.2 | 6.01 |
4. Discussion
Chronic intraperitoneal and subcutaneous administration of D-gal has been used as a model of aging.42 Aging is caused by the accumulation of senescent cells, which alters the physiological responses in the surrounding microenvironment in an autocrine and paracrine fashion through senescence-associated secretory phenotype.43 Aging is characterized by cellular hallmarks, including the accumulation of β-galactosidase with upregulation of p53 and p21. In the current study, β-galactosidase accumulation was significantly recognized in the D-gal group along with upregulation of p53 and p21 mRNA expression. El-Far et al.44,45 identified significant upregulation of p53 and p21 expression in the rat brain in the D-gal group. Also, Sun et al.46 declared an upregulation of brain p21 in mice injected with D-gal. Also, pancreas and kidney samples of rats injected with D-gal to induce aging exhibited upregulation of p53 and p21.44β-galactosidase, p53, and p21 protein expression has been significantly recognized by western blot analysis in the liver of D-gal-treated rats.47 Furthermore, a high accumulation of β-galactosidase could be recognized in the hippocampus of aged rats using a staining kit.48Oxidative stress is a primary factor in neurodegenerative diseases and the normal aging process.49 The exact mechanism of oxidative stress-induced aging is unknown, but increased ROS levels are likely to cause cellular senescence.50 It was confirmed that hippocampal and hepatic proliferation was reduced after receiving D-gal, as demonstrated by previous studies which confirmed that Ki-67 decreased after D-gal administration.51–53 Besides, D-gal markedly decreased the hepatic glycogen content in the D-gal-treated group, indicating impairment of liver storage ability.54 In the present study, D-gal significantly decreased brain and liver SOD1, GPx-1, and GST-α protein levels. Du et al.55 stated significant increases in 8-OHdG (the oxidative derivative of guanosine) expression. In rats, they decreased total superoxide dismutase (T-SOD) and GPx activity in the ventral cochlear nucleus of the D-galactose-induced aging model. Similarly, El-Far et al.45 recognized significant decreases in brain GPx and GST activities in D-gal-treated rats. Also, D-gal decreased the protein expression of SOD156 and SOD57 activity in the brain of rats. Similarly, Motevalian et al.51 stated significant decreases in the expression of Sirt1, Bcl2, CAT, and GPx of D-gal-treated mice.
Oxidative stress in aging is accompanied by cellular inflammation monitored by elevated cytokines.58 IL-6 significantly expressed in immunostaining of brain samples of D-gal group has been stated in the present study. Similarly, IL-6 levels were increased in the rat brain.57,59–61 Also, molecular docking study revealed high affinity of rutin to IL6RA and IL6RB indicating the ability of rutin to reduce aging-associated inflammatory process.
Oxygen-derived free radicals exert detrimental effects, including peroxidation of membrane lipids, enzyme inactivation, DNA fragmentation, and activation of apoptosis.62 In the present study, we stated enhancement of apoptosis monitored by the upregulation of CASP3, Bax, and Bax protein along with significant decreases in Bcl2 expression. Atef et al.57 reported an elevation in brain caspase-3 immunostaining expression in rat hippocampus. Also, in D-gal-treated rats, brain and heart levels of caspase-3, Bcl2, Bax, and CASP3.63 In the same context, D-gal induced significant increases in the Bax/Bcl-2 ratio and caspase-3 in mice's64 and rats’65 brains. In the same context, rutin could inhibit caspase-8, caspase-9, and caspase-3 as stated in molecular docking study.
Experimentally induced aging led to significantly elevated IBA1 and GFAP and decreased synaptophysin expression. IBA1 expression was significantly expressed in the brain63,66 and heart63 of D-gal-treated rats. IBA1 is a specific marker of activated microglia, the major factor in the development of neuroinflammation.67 Also, IBA1 was increased in hepatic aging and inflammation with hepatic macrophage abundance.68
GFAP is a protein that is encoded by the GFAP gene in humans that is expressed by astrocytes in the central nervous system69 and stellate cells of the pancreas and liver in rats.70 GFAP levels were increased in neurodegenerative and hepatic oxidative damage and aging.71,72
Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons.73D-gal decreased synaptophysin in the rat brain, reflecting the efficiency of transmission of neurological signals between different neurons and the general weakness of animals.65,74–76 Also, Budni et al.77 observed reductions in synaptophysin and TAU protein levels in the prefrontal cortex in the D-gal-treated rats.
mTOR is known to regulate some aging hallmarks.78 In the present study, we stated significant upregulation of mTOR. Similarly, mTOR expression was increased compared with that in the control group in the D-gal-treated mice79 and rat80,81 models. Besides, rutin possess high affinity to bind and interact with PI3K/AKT/mTOR as indicated by molecular docking assessment.
Senescent cell killing and/or amelioration of their senescence-associated secretory phenotype-related negative effects are currently considered to be effective gerotherapeutic approaches for the treatment of age-related disorders.82 Natural products have been extensively used for their anti-aging potential in D-gal-inducing models.3,13,27,31,34,44,45,63 In the present study, rutin supplementation to the D-gal-treated rats significantly improved the antioxidant status of the rat brain. Yang et al.83 reported that the activities of SOD and GPx in D-gal-treated mice were significantly decreased compared with those in the control group, and administering rutin significantly attenuated these decreases. These results indicated that rutin prevented D-gal-induced short-term memory retrieval impairment in a passive avoidance paradigm. Rutin has demonstrated the neuroprotective effect on brain ischemia. Administration of rutin caused attenuation of ischemic neural apoptosis due to the embarrassment of p53 expression and lipid peroxidation and increment in endogenous antioxidant enzymes.27 Treatment of rutin to trunk neural crest cells increased their viability without altering cell differentiation and proliferation due to the modulation of ERK2 and PI3K pathways.84 Rutin also possesses anticonvulsant activity and seems safe for patients with epilepsy as it does not alter any of the administered antiepileptic drugs nor demonstrates any adverse effects.85 Rutin suppressed the activity of proinflammatory cytokines by diminishing TNF-α and IL-1β production in microglia. Such an effect seems helpful in the treatment of Alzheimer's disease, as evidenced by the prevention of β-amyloid oligomeric cytotoxicity.86
5. Conclusion
Oxidative stress and inflammation are now widely accepted as the main mechanisms involved in the aging process. Our results suggested that rutin could improve the biochemical indicators of ageing rats by exerting antioxidant effects, regulating apoptosis-related proteins (Bax and CASP3) to inhibit cell apoptosis, and exerting downregulation of aging markers (p53, p21, and β-galactosidase) signalling pathways. In addition, we discovered that rutin played an important role in improving the histopathological features of the brain and liver in rats with D-gal-induced ageing. Nonetheless, because the causes of ageing are complex and assorted, the anti-aging effects of rutin must be investigated further.Data availability
All data generated or analysed during this study are included in this published article and its ESI.†Author contributions
Conceptualization: S. M. S., A. H. E.-F., S. A. M., A. E. N., F. A. E.-T., Y. H. A. E., R. S. F., S. K. A.-J., A. A. A.; formal analysis: S. M. S., A. H. E.-F., S. A. M., A. E. N., F. A. E.-T., Y. H. A. E., R. S. F., S. K. A.-J., A. A. A.; investigation: S. M. S., A. H. E.-F., S. A. M., A. E. N., F. A. E.-T., Y. H. A. E., A. A. A.; supervision: S. M. S., A. H. E.-F., S. A. M., F. A. E.-T., A. A. A.; writing—original draft: S. M. S., A. H. E.-F., S. A. M., A. E. N., A. A. A.; writing—review and editing: S. M. S., A. H. E.-F., S. A. M., A. E. N., F. A. E.-T., Y. H. A. E., R. S. F., S. K. A.-J., A. A. A. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.References
- K. F. Azman, A. Safdar and R. Zakaria, Exp. Gerontol., 2021, 150, 111372 CrossRef CAS PubMed.
- C. A. Juan, J. M. Pérez de la Lastra, F. J. Plou and E. Pérez-Lebeña, Int. J. Mol. Sci., 2021, 22, 4642 CrossRef CAS PubMed.
- E. Birben, U. M. Sahiner, C. Sackesen, S. Erzurum and O. Kalayci, World Allergy Organ. J., 2012, 5, 9–19 CrossRef CAS PubMed.
- R. Rathod, A. Kale and S. Joshi, J. Biomed. Sci., 2016, 23, 17 CrossRef PubMed.
- C. Jiménez-Torres, H. El-Kehdy, L. C. Hernández-Kelly, E. Sokal, A. Ortega and M. Najimi, Front. Neurosci., 2020, 14, 613225 CrossRef PubMed.
- S. Aydin, P. Atukeren, U. Çakatay, H. Uzun and T. Altuǧ, Biogerontology, 2010, 11, 335–346 CrossRef CAS PubMed.
- S. Aydin, K. Yanar, P. Atukeren, E. Dalo, M. E. Sitar, E. Uslu, N. Caf and U. Çkatay, Biogerontology, 2012, 13, 251–260 CrossRef CAS PubMed.
- D. Kumar and S. I. Rizvi, Aging: Clin. Exp. Res., 2014, 26, 261–267 Search PubMed.
- A. H. El-Far, Y. H. A. Elewa, E.-Z. A. Abdelfattah, A.-W. A. Alsenosy, M. S. Atta, K. M. Abou-Zeid, S. K. Al Jaouni, S. A. Mousa and A. E. Noreldin, Int. J. Mol. Sci., 2021, 22, 6839 CrossRef CAS PubMed.
- A. H. El-Far, Y. H. A. Elewa, E.-Z. A. Abdelfattah, A.-W. A. Alsenosy, M. S. Atta, K. M. Abou-Zeid, S. K. Al Jaouni, S. A. Mousa and A. E. Noreldin, Int. J. Mol. Sci., 2021, 22, 6839 CrossRef CAS PubMed.
- T. Wang, G. Di, L. Yang, Y. Dun, Z. Sun, J. Wan, B. Peng, C. Liu, G. Xiong, C. Zhang and D. Yuan, J. Pharm. Pharmacol., 2015, 67, 1284–1296 CrossRef CAS PubMed.
- Z. F. Zhang, S. H. Fan, Y. L. Zheng, J. Lu, D. M. Wu, Q. Shan and B. Hu, Food Chem. Toxicol., 2009, 47, 496–501 CrossRef CAS PubMed.
- X. Cui, P. Zuo, Q. Zhang, X. Li, Y. Hu, J. Long, L. Packer and J. Liu, J. Neurosci. Res., 2006, 84, 647–654 CrossRef CAS PubMed.
- F. Fang and G. Liu, Basic Clin. Pharmacol. Toxicol., 2007, 101, 447–454 CrossRef CAS PubMed.
- H. Wei, L. Li, Q. Song, H. Ai, J. Chu and W. Li, Behav. Brain Res., 2005, 157, 245–251 CrossRef CAS PubMed.
- Y. X. Shen, S. Y. Xu, W. Wei, X. X. Sun, J. Yang, L. H. Liu and C. Dong, J. Pineal Res., 2002, 32, 173–178 CrossRef CAS PubMed.
- J. F. Weiss and M. R. Landauer, Toxicology, 2003, 189, 1–20 CrossRef CAS PubMed.
- A. H. El-Far, S. K. Al Jaouni, W. Li and S. A. Mousa, Nutrients, 2018, 10, 1369 CrossRef PubMed.
- M. F. Dora, N. M. Taha, M. A. Lebda, A. E. Hashem, M. S. Elfeky, Y. S. El-Sayed, S. Al Jaouni and A. H. El-Far, Int. J. Mol. Sci., 2021, 22, 3829 CrossRef CAS PubMed.
- A. E.-D. H. Sayed, Y. S. El-Sayed and A. H. El-Far, Ecotoxicol. Environ. Saf., 2017, 143, 344–350 CrossRef CAS PubMed.
- A. H. El-Far, K. Godugu, A. E. Noreldin, A. A. Saddiq, O. A. Almaghrabi, S. K. Al Jaouni and S. A. Mousa, Integr. Cancer Ther., 2021, 20, 1–20 CrossRef.
- K. Godugu, A. H. El-Far, S. Al Jaouni and S. A. Mousa, Molecules, 2020, 25, 2597 CrossRef CAS PubMed.
- T. Sudha, D. S. Mousa, A. H. El-Far and S. A. Mousa, Nutr. Cancer, 2021, 73, 1350–1356 CrossRef CAS PubMed.
- A. H. El-Far, K. Godugu, T. A. Salaheldin, N. H. E. Darwish, A. A. Saddiq and S. A. Mousa, Biomedicines, 2021, 9, 990 CrossRef CAS PubMed.
- M. A. Lebda, W. N. El-Hawarry, R. M. Shourbela, A. H. El-Far, R. S. Shewita and S. A. Mousa, Fish Shellfish Immunol., 2019, 88, 619–626 CrossRef CAS PubMed.
- M. S. Atta, E. A. Almadaly, A. H. El-Far, R. M. Saleh, D. H. Assar, S. K. Al Jaouni and S. A. Mousa, Int. J. Mol. Sci., 2017, 18, 919 CrossRef PubMed.
- M. M. Khan, A. Ahmad, T. Ishrat, G. Khuwaja, P. Srivastawa, M. B. Khan, S. S. Raza, H. Javed, K. Vaibhav, A. Khan and F. Islam, Brain Res., 2009, 1292, 123–135 CrossRef CAS PubMed.
- A. K. Ratty and N. P. Das, Biochem. Med. Metab. Biol., 1988, 39, 69–79 CrossRef CAS PubMed.
- M. Nassiri-Asl, S. R. Mortazavi, F. Samiee-Rad, A. A. Zangivand, F. Safdari, S. Saroukhani and E. Abbasi, Epilepsy Behav., 2010, 18, 50–53 CrossRef PubMed.
- S. K. Richetti, M. Blank, K. M. Capiotti, A. L. Piato, M. R. Bogo, M. R. Vianna and C. D. Bonan, Behav. Brain Res., 2011, 217, 10–15 CrossRef CAS PubMed.
- H. Javed, M. M. Khan, A. Ahmad, K. Vaibhav, M. E. Ahmad, A. Khan, M. Ashafaq, F. Islam, M. S. Siddiqui, M. M. Safhi and F. Islam, Neuroscience, 2012, 210, 340–352 CrossRef CAS PubMed.
- J. Fan, X. Yang, J. Li, Z. Shu, J. Dai, X. Liu, B. Li, S. Jia, X. Kou, Y. Y. Yang and N. Chen, Oncotarget, 2017, 8, 17475–17490 CrossRef PubMed.
- R. A. Khan, M. R. Khan and S. Sahreen, BMC Complementary Altern. Med., 2012, 12, 204 CrossRef CAS PubMed.
- J. Arowoogun, O. O. Akanni, A. O. Adefisan, S. E. Owumi, A. S. Tijani and O. A. Adaramoye, J. Biochem. Mol. Toxicol., 2021, 35, e22623 CrossRef CAS PubMed.
- J. D. Bancroft and C. Layton, The hematoxylin and eosin, in Theory Practice of histological techniques, ed. S. K. Suvarna, C. Layton and J. D. Bancroft, Churchill Livingstone of Elsevier, Philadelphia, 7th edn, 2013 Search PubMed.
- C. Layton and J. D. Bancroft, Carbohydrates, in Bancroft's theory and practice of histological techniques, ed. S. K. Suvarna, C. Layton and J. D. Bancroft, Churchill Livingstone, Philadelphia, 2013 Search PubMed.
- K. N. Gibson-Corley, A. K. Olivier and D. K. Meyerholz, Vet. Pathol., 2013, 50, 1007–1015 CrossRef CAS PubMed.
- A. E. Noreldin, Y. H. A. Elewa, Y. Kon, K. Warita and Y. Z. Hosaka, Acta Histochem., 2018, 120, 323–328 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- N. M. Hassan, A. A. Alhossary, Y. Mu and C.-K. K. Kwoh, Sci. Rep., 2017, 7, 15451 CrossRef PubMed.
- J. Budni, M. L. Garcez, F. Mina, T. Bellettini-Santos, S. da Silva, A. P. da Luz, G. L. Schiavo, H. Batista-Silva, G. Scaini, E. L. Streck and J. Quevedo, Metab. Brain Dis., 2017, 32, 811–817 CrossRef CAS PubMed.
- B. G. Childs, M. Durik, D. J. Baker and J. M. van Deursen, Nat. Med., 2015, 21, 1424–1435 CrossRef CAS PubMed.
- A. H. El-Far, M. A. Lebda, A. E. Noreldin, M. S. Atta, Y. H. A. Elewa, M. Elfeky and S. A. Mousa, Int. J. Mol. Sci., 2020, 21, 4348 CrossRef CAS PubMed.
- A. H. El-Far, H. H. Mohamed, D. A. Elsabagh, S. A. Mohamed, A. E. Noreldin, S. K. Al Jaouni and A. A. Alsenosy, Environ. Sci. Pollut. Res., 2022, 29, 47436–47447 CrossRef CAS PubMed.
- K. Sun, P. Yang, R. Zhao, Y. Bai and Z. Guo, Oxid. Med. Cell. Longevity, 2018, 2018, 1–12 Search PubMed.
- C.-C. Huang, W.-D. Chiang, W.-C. Huang, C.-Y. Huang, M.-C. Hsu and W.-T. Lin, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 1–9 Search PubMed.
- X. Li, Y. Chen, S. Shao, Q. Tang, W. Chen, Y. Chen and X. Xu, Exp. Gerontol., 2016, 83, 89–93 CrossRef CAS PubMed.
- S. T. Yin, M. L. Tang, H. M. Deng, T. R. Xing, J. T. Chen, H. L. Wang and D. Y. Ruan, Naunyn-Schmiedeberg's Arch. Pharmacol., 2009, 379, 551–564 CrossRef CAS PubMed.
- I. Liguori, G. Russo, F. Curcio, G. Bulli, L. Aran, D. Della-Morte, G. Gargiulo, G. Testa, F. Cacciatore, D. Bonaduce and P. Abete, Clin. Interventions Aging, 2018, 13, 757–772 CrossRef CAS PubMed.
- M. Motevalian, N. T. Maroof, M. H. Nematollahi, F. Khajehasani and I. Fatemi, Iran. J. Basic Med. Sci., 2021, 24, 1388–1394 Search PubMed.
- R. Prajit, N. Sritawan, K. Suwannakot, S. Naewla, A. Aranarochana, A. Sirichoat, W. Pannangrong, P. Wigmore and J. U. Welbat, Nutrients, 2020, 12, 1100 CrossRef CAS PubMed.
- E. B. Kalaz, J. Çoban, A. F. Aydin, I. Doǧan-Ekici, S. Doǧru-Abbasoǧlu, S. Öztezcan and M. Uysal, J. Physiol. Biochem., 2014, 70, 15–25 CrossRef CAS PubMed.
- J. Y. Xia, Y. L. Fan, D. Y. Jia, M. S. Zhang, Y. Y. Zhang, J. Li, P. W. Jing, L. Wang and Y. P. Wang, Zhonghua Ganzangbing Zazhi, 2016, 24, 214–219 CAS.
- Z. D. Du, S. Yu, Y. Qi, T. F. Qu, L. He, W. Wei, K. Liu and S. S. Gong, Neurochem. Int., 2019, 124, 31–40 CrossRef CAS PubMed.
- H.-C. Kuo, S.-Y. Tong, M.-W. Chao and C.-Y. Tseng, PLoS One, 2022, 17, e0266331 CrossRef CAS PubMed.
- M. M. Atef, M. N. Emam, R. E. Abo El Gheit, E. M. Elbeltagi, H. A. Alshenawy, D. A. Radwan, R. L. Younis and R. N. Abd-Ellatif, Neurochem. Res., 2022, 47, 1664–1678 CrossRef CAS PubMed.
- A. Garrido, J. Cruces, N. Ceprián, E. Vara and M. de la Fuente, Int. J. Mol. Sci., 2019, 20, 769 CrossRef CAS PubMed.
- X. Ren, Q.-Q. Wang, X. Zhang, G. Wang, T. Liu, N. Deng and D. Yan, Exp. Ther. Med., 2020, 20, 3228–3236 CAS.
- S. Singh, R. Kumar, G. Garg, A. K. Singh, A. K. Verma, A. Bissoyi and S. I. Rizvi, Biogerontology, 2021, 22, 35–47 CrossRef CAS PubMed.
- B. Zhang, W. Lian, J. Zhao, Z. Wang, A. Liu and G. Du, Oxid. Med. Cell. Longevity, 2021, 2021, 1–31 Search PubMed.
- M. Valko, D. Leibfritz, J. Moncol, M. T. D. D. Cronin, M. Mazur and J. Telser, Int. J. Biochem. Cell Biol., 2007, 39, 44–84 CrossRef CAS PubMed.
- A. H. El-Far, Y. H. A. Elewa, E.-Z. A. Abdelfattah, A.-W. A. Alsenosy, M. S. Atta, K. M. Abou-Zeid, S. K. Al Jaouni, S. A. Mousa and A. E. Noreldin, Int. J. Mol. Sci., 2021, 22, 6839 CrossRef CAS PubMed.
- X. Zhang, J. Z. Wu, Z. X. Lin, Q. J. Yuan, Y. C. Li, J. L. Liang, J. Y. X. Zhan, Y. L. Xie, Z. R. Su and Y. H. Liu, J. Ethnopharmacol., 2019, 234, 44–56 CrossRef CAS PubMed.
- F. Ullah, T. Ali, N. Ullah and M. O. Kim, Neurochem. Int., 2015, 90, 114–124 CrossRef CAS PubMed.
- C. He, Z. S. Huang, C. C. Yu, X. S. Wang, T. Jiang, M. Wu and L. H. Kong, Iran. J. Basic Med. Sci., 2021, 24, 341–348 Search PubMed.
- S. Hickman, S. Izzy, P. Sen, L. Morsett and J. El Khoury, Nat. Neurosci., 2018, 21, 1359–1369 CrossRef CAS PubMed.
- S. A. Bloomer, Int. J. Mol. Sci., 2022, 23, 6502 CrossRef CAS PubMed.
- C. M. Jacque, C. Vinner, M. Kujas, M. Raoul, J. Racadot and N. A. Baumann, J. Neurol. Sci., 1978, 35, 147–155 CrossRef CAS PubMed.
- M. V. Apte, P. S. Haber, T. L. Applegate, I. D. Norton, G. W. McCaughan, M. A. Korsten, R. C. Pirola and J. S. Wilson, Gut, 1998, 43, 128–133 CrossRef CAS PubMed.
- S. Hassan, S. Syed and S. I. Kehar, Pak. J. Med. Sci., 2014, 30, 1027–1032 Search PubMed.
- H. Phatnani and T. Maniatis, Cold Spring Harbor Perspect. Biol., 2015, 7, 1–18 Search PubMed.
- S. E. Kwon and E. R. Chapman, Neuron, 2011, 70, 847–854 CrossRef CAS PubMed.
- M. L. Garcez, R. C. S. Cassoma, F. Mina, T. Bellettini-Santos, A. P. da Luz, G. L. Schiavo, E. B. Medeiros, A. C. B. F. Campos, S. da Silva, L. C. T. Rempel, A. V. Steckert, T. Barichello and J. Budni, Metab. Brain Dis., 2021, 36, 213–224 CrossRef CAS PubMed.
- S. U. Rehman, S. A. Shah, T. Ali, J. Il Chung and M. O. Kim, Mol. Neurobiol., 2017, 54, 255–271 CrossRef PubMed.
- C. Prisila Dulcy, H. K. Singh, J. Preethi and K. Emmanuvel Rajan, J. Neurosci. Res., 2012, 90, 2053–2064 CrossRef CAS PubMed.
- J. Budni, R. Pacheco, S. da Silva, M. L. Garcez, F. Mina, T. Bellettini-Santos, J. de Medeiros, B. C. Voss, A. V. Steckert, S. da Silva Valvassori and J. Quevedo, Behav. Brain Res., 2016, 302, 35–43 CrossRef CAS PubMed.
- T. Weichhart, Gerontology, 2018, 64, 127–134 CrossRef CAS PubMed.
- P. Chen, F. Chen, J. Lei, Q. Li and B. Zhou, Neurotherapeutics, 2019, 16, 1269–1282 CrossRef CAS PubMed.
- J. Yuan, X. Zhao, Y. Hu, H. Sun, G. Gong, X. Huang, X. Chen, M. Xia, C. Sun, Q. Huang, Y. Sun, W. Kong and W. Kong, Int. J. Mol. Med., 2018, 41, 2086–2098 CAS.
- L. Wang, J. Du, F. Zhao, Z. Chen, J. Chang, F. Qin, Z. Wang, F. Wang, X. Chen and N. Chen, Biomed. Pharmacother., 2018, 98, 516–522 CrossRef CAS PubMed.
- M. C. Atayik and U. Çakatay, Biogerontology, 2022, 23, 401–423 CrossRef PubMed.
- Y. C. Yang, H. Y. Lin, K. Y. Su, C. H. Chen, Y. L. Yu, C. C. Lin, S. L. Yu, H. Y. Yan, K. J. Su and Y. L. S. Chen, J. Evidence-Based Complementary Altern. Med., 2012, 2012, 980276 Search PubMed.
- J. Nones, A. P. Costa, R. B. Leal, F. C. A. Gomes and A. G. Trentin, Cell Tissue Res., 2012, 350, 305–315 CrossRef CAS PubMed.
- D. Nieoczym, K. Socała, G. Raszewski and P. Wlaź, Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 2014, 54, 50–58 CrossRef CAS PubMed.
- S. W. Wang, Y. J. Wang, Y. J. Su, W. W. Zhou, S. G. Yang, R. Zhang, M. Zhao, Y. N. Li, Z. P. Zhang, D. W. Zhan and R. T. Liu, NeuroToxicology, 2012, 33, 482–490 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo03301a |
| This journal is © The Royal Society of Chemistry 2023 |