Open Access Article
Open Access ArticleRaw milk kefir: microbiota, bioactive peptides, and immune modulation†
Ton
Baars‡
*a,
Betty
van Esch‡
 a,
Luuk
van Ooijen
ab,
Zuomin
Zhang
c,
Pieter
Dekker
a,
Luuk
van Ooijen
ab,
Zuomin
Zhang
c,
Pieter
Dekker
 cd,
Sjef
Boeren
d,
Mara
Diks
a,
Johan
Garssen
cd,
Sjef
Boeren
d,
Mara
Diks
a,
Johan
Garssen
 a,
Kasper
Hettinga
a,
Kasper
Hettinga
 c and
Remco
Kort
c and
Remco
Kort
 *be
*be
aDivision of Pharmacology, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science, Utrecht University, Utrecht, The Netherlands. E-mail: a.baars@uu.nl
bAmsterdam Institute for Life and Environment, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands. E-mail: r.kort@vu.nl
cFood Quality and Design Group, Wageningen University & Research, Wageningen, The Netherlands
dLaboratory of Biochemistry, Wageningen University & Research, Wageningen, The Netherlands
eARTIS-Micropia, Amsterdam, The Netherlands
First published on 24th January 2023
Abstract
This study aims to characterize the microbiota and peptidomic composition of raw milk kefir, and to address the potential anti-allergic effects of raw milk kefir using validated research models for food allergy. Raw milk kefir was produced after incubation with a defined freeze-dried starter culture. Kefir was sampled during fermentation at seven time intervals. For comparison, kefir was also prepared from heat-treated milk. Peptide compositions were determined for the raw and heated milk, and kefir end products made from these milks. In a murine food allergy model, the two kefir end products were investigated for their allergy modulating effects. In both kefirs, we identified amplicon sequence variants identical to those in the starter culture, matching the bacteria Lactococcus lactis, Streptococcus thermophilus, Leuconostoc and the yeast Debaryomyces. In raw milk kefir, additional sequence variants of Lactococcus lactis and the yeasts Pichia and Galactomyces could be identified, which were absent in heated milk kefir. Analysis of peptide compositions in both kefirs indicated that the number and intensity of peptides drastically increased after fermentation. Heating of the milk negatively affected the diversity of the peptide composition in kefir. Only raw milk kefir suppressed the acute allergic skin response to the food allergen ovalbumin in sensitised mice. These effects coincided with differences in the T-cell compartment, with lower percentages of activated Th1 cells and IFNg production after treatment with kefir made from heated milk. The results of this study indicate specific properties of raw milk kefir that may contribute to its additional health benefits.
1. Introduction
Fermentation is the oldest way of milk conservation, transforming milk into kefir and yoghurt, as well as fresh or ripened cheese. Defined starter cultures (SC) based on selected microorganisms were not used until the early 1900s, implicating that fermentation was initiated by microorganisms originating from the environment and equipment.1 Over the last few decades, there has been increasing interest in kefir, a drinkable milk product that originates from Eastern Europe, Turkey, or the Caucasus region. In contrast to yoghurt fermentation, which is based on thermophilic bacteria, kefir is fermented by mesophilic bacteria as well as yeasts. Wild starters of kefir are based on kefir grains or SCOBY, a symbiotic community of bacteria and yeast containing a matrix of the exopolysaccharide kefiran and proteins. The microbial species in these kefir grains include lactic acid bacteria, acetic acid bacteria and yeasts. The most common method to produce kefir on an industrial scale is based on fermentation with defined, often freeze-dried SCs.2 These SCs are based on a selection of microbial species, including the bacteria Streptococcus, Lactobacillus, and Lactococcus as well as yeasts Kluyveromyces and Debaryomyces.3 These bacteria and yeasts multiply during the anaerobic fermentation process. According to the Codex Alimentarius (2003), kefir should contain at least 107 cfu mL−1 bacteria, and at least 104 cfu mL−1 yeast.Fermented dairy belongs to the group of ‘functional foods’ or ‘nutraceuticals’.4 The health impact of fermented food consumption is widely accepted, and benefits may be based on multiple mechanisms.5 For kefir, lactose fermenting micro-organisms are responsible for the transformation of lactose into lactate and other organic acids, causing a drop in pH. During fermentation, peptides are produced because of proteolytic degradation of predominantly caseins,6 along with the generation of short chain fatty acids, ethanol, and vitamins.4 Some kefir peptides show biological activity, including angiotensin-converting enzyme (ACE)-inhibitory, antimicrobial, immunomodulating, opioid, mineral binding, antioxidant, and antithrombotic effects.6,7 The impact of kefir on gastrointestinal health is partly based on the changes in the gut microbiome, and the improvement of gut dysbiosis and gut barrier.4 Altered gut colonization and gut dysbiosis are associated with increased risk of immune related diseases, like asthma and allergies.8 Overall health claims associated with kefir consumption were reviewed from different perspectives,9–13 but studies on the impact on food allergic diseases are limited. In healthy volunteers, two weeks of kefir consumption changed the level of several cytokines, indicating an increase of the Th1 immune response and a decrease of the Th2 immune response.14 In a study utilizing an ovalbumin sensitisation murine asthma model, it was found that mice receiving intra-gastric kefir had lower levels of airway hyper-responsiveness (AHR) than control mice, and, impressively, had lower levels of AHR than the positive control group receiving an anti-asthma drug.15 In young rats, kefir consumption improved the intestinal mucosal and systemic immune responses after the ingestion of cholera toxin.16 Although pre-clinical studies on kefir affected allergy related molecules like IgE after kefir consumption as well as reduced inflammation and airway hyperresponsiveness in allergic asthma, it is currently unknown whether kefir has the potential to prevent food allergy symptoms.
Kefir is usually made from heat-treated milk, using skimmed milk, or homogenized full fat milk. Knowledge about the impact of kefir made from unheated and non-homogenized full fat milk is missing, and there is an increasing amount of literature on the impact of milk processing on its composition in relation to human health.17–19 Various epidemiological studies have shown reduced incidences of asthma and allergies in children who consumed raw bovine milk.20 After the raw milk was heated, the protection was lost, even in farm children.21 These epidemiologic observations were confirmed in recent studies where we showed anti-allergic effects of raw milk in murine food allergy models and a clinical pilot study with eleven allergic children.22–24 It could be shown that the protection is related to the whey fraction of the milk, containing a wide range of heat-sensitive proteins.23
The aim of this study was to evaluate the impact of kefir either made from raw or heated whole milk. Hereto, raw milk kefir (RMK) and heated milk kefir (HMK) were characterized by the analysis of the microbiota and peptide compositions. Both RMK and HMK were investigated for their allergy protective effects in a murine food allergy model. Microbiota and peptide composition were used to generate hypotheses about potential mechanisms of allergy protection.
2. Materials and methods
2.1. Kefir production and sampling
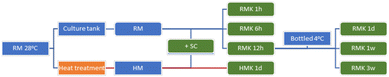
Kefir was produced at a commercial dairy plant, the Raw Milk Company (De Lutte, The Netherlands). The raw milk came from a mix of 80 dairy cows in one morning milking. Raw bulk milk was cooled at the farm to approximately 25 °C. The time between the end of milking and the start of fermentation was less than two hours. The farm was organic certified, according to SKAL/EU regulations. The kefir produced this way is referred to as RMK. RMK was produced by 2%-addition to raw milk of a freeze-dried defined starter EXACT® kefir culture, containing the bacterial species Leuconostoc sp., Streptococcus thermophilus, Lactococcus lactis subsp. cremoris, L. lactis subsp. lactis, L. lactis biovar diacetylactis, and the yeast Debaryomyces hansenii (Chr. Hansen, Hoersholm, Denmark). Fermentation was carried out at 28 °C for 24 hours in tanks of 1000 L. Before bottling, the kefir was gently mixed. Bottles of 1 L were stored at 4 °C.At seven different time points, samples were taken both during the fermentation process of the raw milk as well as during the storage of the final kefir. To control the fermentation process, the pH was measured every 1 to 3 hours. To evaluate the impact of heating, raw milk was heated to produce HM kefir (HMK). After thermisation, 3 L of milk was brought to 100 °C (microbiota profiling and peptidomics) or heated for 30 min at 72 °C (murine allergy model), followed by immediate cooling to 24 °C. In contrast to RMK, HMK was only sampled at two timepoints: milk after heating before inoculation and at the end of kefir processing after 24 hours. A schematic overview of the fermentation process and the samples collected and analysed is displayed in Fig. 1.
2.2. Enumeration of total bacteria, lactic acid bacteria and yeasts
Three different types of nutrient agar plates were used to determine the number of total bacteria, lactic acid bacteria and yeasts. Petri dishes with culture media and the NaCl solutions (0.9%, pH = 7.0) for the dilution series were provided by Biotrading Benelux BV (Mijdrecht, The Netherlands). Total bacterial counts were determined on Tryptic Soy Agar (TSA), lactic acid bacteria on de Man, Rogosa and Sharpe (MRS) agar, and yeasts on Sabouraud Dextrose Agar (SDA) with chloramphenicol to prevent bacterial growth. For each sample (Fig. 1), serial dilutions were made and colony forming units were determined after 24 to 30 hours of incubation at 30 °C.2.3. Amplicon sequencing of 16S rRNA and ITS
A specific milk bacterial DNA isolation kit (Norgen Biotek, Thorold, Canada) was used with slight modifications of the manufacturer's protocol. The following amount of the sample was used: for unfermented milk 1 mL per sample, for fermented samples 0.2 mL with the addition of 0.8 mL phosphate buffer. For the defined SC, 20 mg was added to 1 mL phosphate buffered saline solution. The manufacturer's recommendation of addition of 75 μL mock microbial community suspension was used, equivalent to a total of 220 ng of genomic DNA. All fermented samples were passed through a 21-gauge syringe (Sigma-Aldrich, St Louis, MS, Unites States) to reduce the viscosity. After centrifuging for 2 min at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, the supernatant was removed. The cell pellet was resuspended in 100 μL resuspension solution A from the DNA isolation kit and transferred to a 2 mL screw cap tube preloaded with 0.5 g autoclaved glass beads. The samples were chilled on ice for 1 minute, followed by 10 seconds of bead shaking treatment at the speed set to 6 meters per second on a Fastprep®-24 5G Instrument (MP Biomedicals, Solon, OH, United States). Cycles of chilling and shaking were performed eight times. After mechanical cell lysis, the DNA isolation kit manufacturer's protocol was applied as specified for Gram-positive and unknown bacterial species.
000g, the supernatant was removed. The cell pellet was resuspended in 100 μL resuspension solution A from the DNA isolation kit and transferred to a 2 mL screw cap tube preloaded with 0.5 g autoclaved glass beads. The samples were chilled on ice for 1 minute, followed by 10 seconds of bead shaking treatment at the speed set to 6 meters per second on a Fastprep®-24 5G Instrument (MP Biomedicals, Solon, OH, United States). Cycles of chilling and shaking were performed eight times. After mechanical cell lysis, the DNA isolation kit manufacturer's protocol was applied as specified for Gram-positive and unknown bacterial species.
2.4. Kefir peptidomics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 10 min to remove caseins and fat.7 Remaining proteins were precipitated by the addition of an equal volume of 200 g L−1 trichloroacetic acid in Milli-Q water, followed by centrifugation at 3000g and 4 °C for 10 min. The peptide fraction in the supernatant was cleaned-up with a solid phase extraction (SPE) on C18+ Stage tip columns (prepared in-house).30 SPE was carried out as described before,31 using 100 μL supernatant of each sample. Finally, peptides were reconstituted to 50 μL with 1 mL L−1 formic acid in water.
000g and 4 °C for 10 min to remove caseins and fat.7 Remaining proteins were precipitated by the addition of an equal volume of 200 g L−1 trichloroacetic acid in Milli-Q water, followed by centrifugation at 3000g and 4 °C for 10 min. The peptide fraction in the supernatant was cleaned-up with a solid phase extraction (SPE) on C18+ Stage tip columns (prepared in-house).30 SPE was carried out as described before,31 using 100 μL supernatant of each sample. Finally, peptides were reconstituted to 50 μL with 1 mL L−1 formic acid in water.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The 25 most abundant positively charged peaks (2–5+) in the MS scan were isolated and fragmented (HCD) with an isolation width of 1.2 m/z and 24% normalized collision energy. MSMS scans were recorded at a resolution of 15
000. The 25 most abundant positively charged peaks (2–5+) in the MS scan were isolated and fragmented (HCD) with an isolation width of 1.2 m/z and 24% normalized collision energy. MSMS scans were recorded at a resolution of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in the data-dependent mode with a threshold of 1.2 × 105 and 15 s exclusion duration for the selected m/z ± 10 ppm.
000 in the data-dependent mode with a threshold of 1.2 × 105 and 15 s exclusion duration for the selected m/z ± 10 ppm.
In a second search, a new database was created comprising only the protein sequences of which peptides were identified in the first, initial search. This new database comprised 39 protein sequences and was used for a new search in which the minimum peptide length was set to 4 and the maximum peptide length to 45 amino acids. The rationale behind this search strategy was to comprehensively map the complete kefir peptidome with short processing times. In both searches, oxidation of methionine, n-terminal acetylation, deamidation of asparagine and glutamine and phosphorylation of serine and threonine were set as variable modifications. Peptides were filtered on identification score (>80) and only peptides from proteins with >5 unique peptides were retained. Raw, relative peptide intensities were used for data analysis and precursor proteins were determined from the MaxQuant proteinGroups result file as described before.35 Bioactive properties of identified peptide sequences were searched using the Milk Bioactive Peptide Database (MBPDB).36 The search type used in the database was ‘precursor’ with a similarity threshold of 100%. Results were filtered on the requirement that the length of the bioactive sequence > 4 amino acids and that the length of the identified sequence <2 × length bioactive sequence.
Sequence logos were created for the P1 and P1′ positions of the identified peptides. N- and C-terminals of the peptides were summed, and intensities of the amino acids were plotted using the R package ggseqlogo, version 0.1.37
2.5. Murine food allergy model
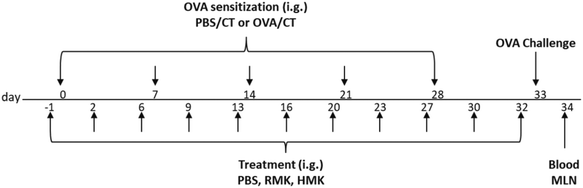
On experimental days 0, 7, 14, 21 and 28, mice (n = 8 per group) were orally sensitised, by using a blunt needle, to the hen's egg protein ovalbumin (OVA) (20 mg/0.5 mL PBS; grade V; Sigma-Aldrich) using cholera toxin (CT; 15 μg/0.5 mL; List Biological Laboratories, Campbell, CA, United States) as an adjuvant. Sham-sensitised mice (PBS/PBS) received CT alone (15 μg/0.5 mL PBS). The following experimental groups (treatment/sensitisation and challenge): PBS/PBS, PBS/OVA (allergic control), RMK/OVA and HMK/OVA. HMK was prepared from heated milk with the same starter culture. After 24 hours of fermentation, both RMK and HMK kefir samples were kept at −18 °C and thawed on the day of challenge. For the treatment with the different kefirs 0.5 ml of PBS, RMK or HMK were given 3×/week by oral gavage from day −1 to 32. On day 33, mice were challenged intradermally (i.d.) in both ear pinnae with OVA (10 μg/20 μL PBS) to evaluate the acute allergic symptoms after i.d. challenge. Allergen-specific IgE in blood and percentages of activated Th1, Th2, Foxp3+ regulatory cells and IFNg production were measured on day 34 and 16 hours after oral OVA challenge (50 mg OVA/0.5 mL PBS) in MLN or spleen. A schematic overview of the experimental timeline is shown in Fig. 2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Serum was subsequently obtained and stored at −20 °C until analysis of OVA-specific IgE and IgG levels by means of ELISA as described elsewhere.24
000 rpm for 10 min. Serum was subsequently obtained and stored at −20 °C until analysis of OVA-specific IgE and IgG levels by means of ELISA as described elsewhere.24
3. Results
3.1. Microbial growth and acidification during fermentation and storage
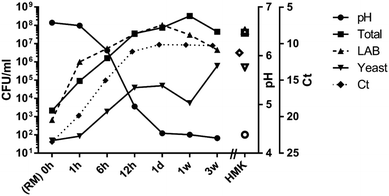
The growth of lactic acid bacteria at 28 °C and concomitant production of lactic acid showed a typical pattern: a relatively slow reduction of pH during the first hour, followed by a rapid decrease to pH 4.3 at 24 hours. A short lag phase was evident for yeast, but lactic acid bacteria show exponential growth from the first hour of incubation for starter cultures grown on lactose. The storage period from one day to three weeks at 4 °C was characterized by a further slow decline of the pH (Fig. 3). The total increase in total bacterial load during the 24 hours of fermentation at 28 °C for RMK was about five orders of magnitude, from approximately 103 to 108 CFU mL−1. An increase between four and five orders of magnitude in bacterial load can also be derived from the 16S rRNA gene-based cycle threshold values (Ct) of the quantitative PCR, as the Ct values decreased from 24 to 10 in the first 24 hours during the fermentation process. RMK and HMK do not significantly differ in Ct values or CFU counts of total or lactic acid bacteria after 1 day of fermentation. However, HMK showed about 40-fold higher levels of yeasts (0.8 × 106 CFU mL−1) than RMK (0.2 × 105 CFU mL−1) after one day of fermentation at 28 °C. The significance of differences in CFU counts between RMK and HMK was confirmed by including enumeration results of a kefir batch fermented from different raw milk, leading to p-values of 0.5 (total bacteria), 0.4 (lactic acid bacteria) and 0.03 (yeasts). During storage of bottled RMK at 4 °C from 24 hours to three weeks, a reduction of CFU count of lactic acid bacteria was observed, while the CFU count of yeasts further increased during this period.3.2. Composition of microbiota during fermentation and storage of raw milk kefir
3.3. Composition of peptides in milk and kefir end products
3.4. Murine food allergy model
4. Discussion
The safety of the RMK is based on two principles, a high hygienic standard during milking and processing as well as the immediate acidification of raw milk. In several countries, legal raw milk has been produced under very strict hygienic conditions. The zoonotic risk is almost reduced to zero, and if any zoonotic bacteria were detected, there were no adverse health effects found by the authorities.39 As a result of the high hygienic standards applied at the dairy farm delivering raw milk for this study, the bacterial contamination of the raw milk is kept around and below 103 CFU mL−1. Furthermore, there was a very short period between the end of milking and the start of culturing for less than two hours, leading to the rapid production of lactic acid, resulting in a lowering of the pH to 4.3, limiting the risk for microbial contamination and foodborne diseases,40,41 also in raw fermented milk.42After 24 h of fermentation with the added SC, kefir contains approximately 108 bacteria and 105 yeasts per mL. This agrees with previous reports, indicating ranges of lactic acid bacteria between 108 and 109 CFU mL−1 and for the yeast of 105 and 106 CFU mL−1 in the final kefir products.43 With respect to storage, we observed a further increase of yeast counts over the period of three weeks, while the lactic acid bacteria reached their highest levels after one day of fermentation and the numbers started to decline during storage, possibly due to further acidification. Indeed, other studies confirmed a slight decrease in lactic acid bacteria and an increase of yeasts in kefir produced from a starter culture stored over a period of three weeks.44 Strikingly, HMK showed higher levels of fungi than RMK after one day of fermentation. This could result from the inactivation of the antifungal lactobacilli known to be present in RM.45
Most of the detected microbial amplicons are identical to or are derived from the added SC and could be considered as safe or even beneficial for consumption. Although human infections with food derived yeasts harbouring virulence factors such as those seen in some food derived from Kluyveromyces marxianus are potentially possible.46Streptococcus dysgalactiae was detected in RM. It is most frequently encountered as a human commensal of the alimentary tract or genital tracts, and exposure to it could contribute to trained immunity.47
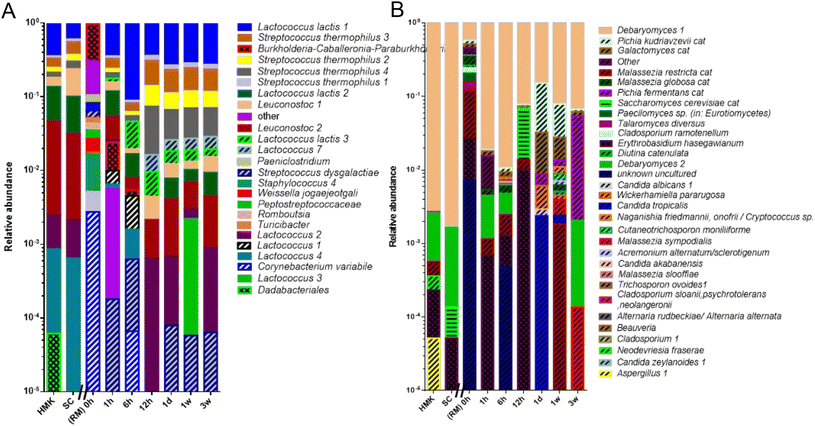
Both RM and HM showed a very high richness of over one hundred microbial species. As these species have been identified based on their DNA in RM, this number may overrepresent the number of viable microbial species in the milk. Previous studies also led to the detection of hundreds of bacterial species in the RM microbiota. In agreement with the present study, members of the phyla Firmicutes and Proteobacteria account for most of these species.48 Their abundance depends on specific environmental conditions. In our fresh raw milk microbiota, we found a rather even distribution of microbial species, in contrast to other studies where the milk-fermenting species Streptococcus thermophilus and Lactococcus lactis dominated the raw milk microbiota.49 The numbers of detected fungi were limited in our raw milk to about 30 ASVs, including yeasts Debaryomyces, Mallasezia, and Candida and the mold Cladosporium, all previously detected members of the raw milk mycobiota with a potential role in milk fermentation as some of them are efficient lactose degraders and show proteolytic and lipolytic activity.48 During the process of fermentation, the richness in microbial species drastically reduced. The most prominent members include the bacteria Streptococcus thermophilus, Lactococcus lactis, Lactococcus, Leuconostoc and the yeast Debaryomyces showing a perfect ASV match to those of the microbial species present in the defined kefir starter culture. It should be noted that the kefir fermentation with a defined starter culture reported here is rather different from the kefir fermentation with natural grains, which includes a bacterial core community consisting of the kefir matrix former Lactobacillus kefiranofaciens and Lactobacillus kefiri, Leuconostoc sp., Lactococcus lactis and Acetobacter sp.3 In the latter case a niche has been created by early fermenters by the production of amino acids and lactate, which is converted to acetic acid by Acetobacter sp. Although the last process appears to be absent in our raw milk kefir production, as we did not identify any acetic acid bacteria, we did identify bacteria and yeasts that were not detectable or at low levels in early kefir fermentation and were more predominantly present after 6 to 12 hours of fermentation. These late fermenting microbial species included a specific variant of L. lactis and species of the fungi Galactomyces and Pichia. The former possibly uses galactose produced from lactose,50 while the latter species may be involved in proteolytic activity.51
The production of kefir from raw milk produced in this study raises the question of whether the raw milk microbiota participates in the kefir fermentation process. A comparative analysis of the microbiota from heated milk kefir indicates that several ASVs identified in raw milk kefir were absent in heated milk kefir. These ASVs match specific variants of the L. lactis and the fungus Galactomyces geotrichum and yeast Pichia kudriavzevii. It should be noted that the latter two species also have been identified in raw milk which has been used for fermentation, whereas the variants of L. lactis were possibly below the detection limit. Interestingly, co-cultures of Galactomyces geotrichum or Pichia kudriavzevii with Lactobacillus sp. showed a strong stimulating effect on the production of peptides.51 These peptides include bioactive anti-hypertensive peptides52 with a strong activity against Angiotensin I-Converting Enzyme (ACE), a dipeptidyl carboxypeptidase that plays a role in the regulation of blood pressure.53 In addition, the presence of wild L. lactis strains in raw milk kefir could increase bioactive peptide levels in the final product.54
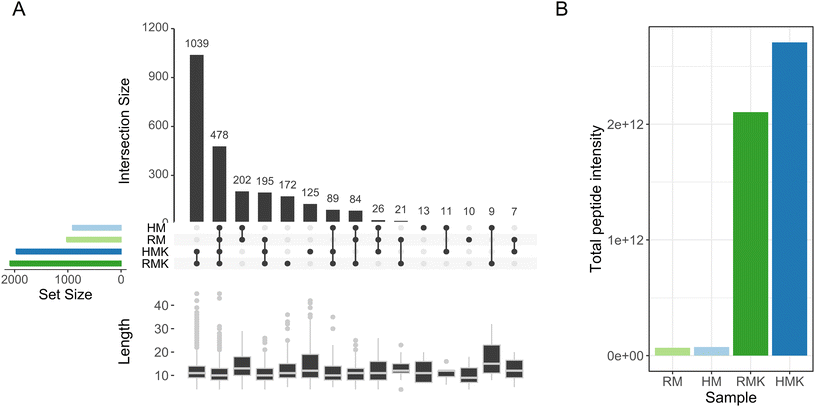
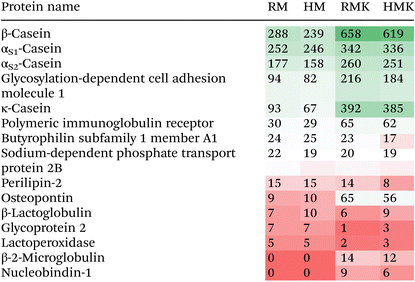
The main result emerging from the peptide analysis is that upon fermentation, the number of unique peptides and the total peptide abundance increases by several orders of magnitude (Fig. 5). This increase results from the proteolytic degradation either by endogenous or microbial proteolytic enzymes. The microbial proteases may originate from both the defined starter culture for both types of kefirs and the raw milk specifically for the RMK samples.7 Both before and after fermentation, most of the peptides were derived from the hydrolysis of caseins, especially β-casein (Table 1). The majority of peptides has a length of less than 20 amino acids, as observed in previous studies of kefir peptides.55,56 For endogenous milk proteases like plasmin, it is known that caseins are their major target.57 Also, for lactic acid bacteria, it is known that their proteases preferentially hydrolyse caseins.58 Interestingly, additional bioactive peptides may be formed by the unique co-cultures of Galactomyces geotrichum or Pichia kudriavzevii with Lactobacillus sp. originating from raw milk, as discussed above.51 Many peptides present in milk were not identified after fermentation to kefir (Fig. 5A), as 202 peptides were only found in the milk samples. The most probable explanation would be that these peptides are further degraded upon fermentation, leading to the formation of shorter peptides or free amino acids. This is underpinned by the peptide length data in Fig. 5A, which shows that these 202 peptides present only in unfermented milk were on average longer than the peptides present only in kefir, or in both kefir and milk. When looking at the amino acids in the P1 and P1′ positions of the peptides (ESI 5†), there is a difference in cleavage specificity between RMK and HMK. However, as the proteases produced by the unique cultures in RMK are not characterized, it is not possible to determine whether there is a direct relation.
A comparative analysis of RMK and HMK showed that the number of unique peptides was higher in RMK than in HMK (172 vs. 125; Fig. 5A). This may be explained by the larger diversity in the microbiota of RMK (Fig. 4), which could have led to proteolysis by a larger set of different proteases with different cleavage site specificities. In particular, the presence of additional Lactococcus lactis or fungal strains in RMK may play a role, as it is known that these species produce a wide range of proteases and peptidases that will cleave casein at many different cleavage sites, leading amongst others to the formation of so-called peptide ladders,59 which were also found in our peptidomics dataset.
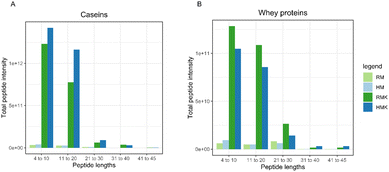
As studies on the digestion of whey proteins have shown that denatured whey proteins are generally digested faster,60 we expected the HMK to contain a larger amount of whey protein-derived peptides. However, our data (Table 1 and Fig. 6) do not confirm this hypothesis. As the sample underwent a short boiling step, which is a rather intense heat treatment, more extensive whey protein aggregation than during low pasteurisation may have occurred. It has previously been shown that the effect of whey protein digestion depends on the specific aggregation structure61 and that intense heating may reduce whey protein proteolysis during fermentation.62 The specific, rather intense, heat treatment used in this study, may thus be the reason why no increase in whey protein-derived peptides was found in the HMK sample.
Nevertheless, although HMK showed to have less unique peptides than RMK, the total peptide abundance was higher. This is in line with the finding that proteolysis in HMK is carried out by a less diverse set of proteases, with less variation in cleavage specificity, leading to a less diverse peptide profile.
A search for known bioactivities showed that HMK has a higher bioactive potential than RMK when peptide abundances are considered (ESI 4†). Nevertheless, for peptides that were identified only in HMK or RMK, most bioactive sequences were found in RMK. Although this could indicate a difference in the effect that HMK and RMK can have on health, caution is needed in interpretation since the extent of bioactivity is different for each peptide. Therefore, it is not possible to speculate on the role of specific peptides in the allergy-modulating effect of RMK.
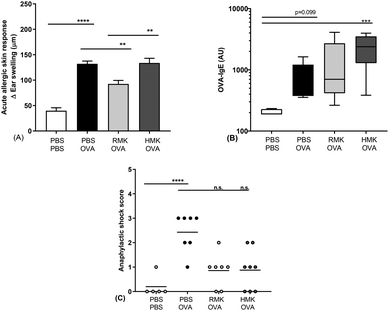
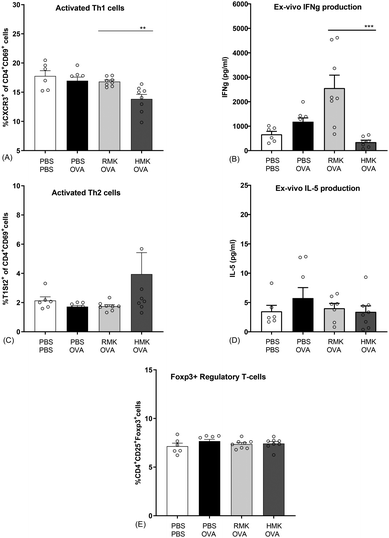
In this study we show for the first time the protective effect of kefir on the food allergic response towards a food allergen. The outcomes are in accordance with the experiences in adults, consuming raw milk kefir. In a retrospective study, people experienced an improved score for their health, immunity, bowel, and mood after regular consumption of raw milk kefir, and health improvements were the largest among people mentioning a poor health.63 In the current pre-clinical study, beneficial anti-allergic effects are only observed in kefir produced from raw, unprocessed milk. A reduced acute allergic skin response is observed in RMK, but not in the kefir prepared from heated milk (HMK). The acute allergic skin response resembles the skin prick test in humans used to identify sensitisation to specific allergens. Previous studies showed health benefits in relation to reduced inflammation and airway hyperresponsiveness in murine allergic asthma models.13,64 In those studies, kefir from pasteurised milk was used. The current study shows that only RMK possesses the putative capacity to redirect the food allergic response to the unrelated food allergen ovalbumin. Allergen-specific IgE is one of the key biomarkers responsible for the induction of allergic symptoms. Specific IgE is produced after sensitisation, i.e., the first encounter with the food allergen. Then IgE specifically binds to a high-affinity Fc receptor on the surface of mast cells or basophils that binds to allergen epitopes and triggers the release of inflammatory mediators and as a result the induction of food allergic symptoms. Although the effects of milk fermentation have been studied,65–67 only one study describes kefir related effects on food sensitisation. Hong et al. showed suppression of IgE production and modulation of the T-cell compartment in LAB, Lactobacillus kefiranofaciens M1 treated mice.68 However, they did not investigated kefir as a matrix and the measurement of clinically related symptoms were not included in the study. Although no difference was observed in OVA-IgE between RMK and HMK in the current study, higher OVA-specific IgE levels in HMK treated allergic mice compared to control mice might underly the protective effect of RMK. One of the main mechanisms leading to a heightened IgE response in food allergy is an imbalance in the Th1/Th2 cells.69–71 The allergy protective effect of RMK was related to a changed Th1/Th2 ratio as shown as an increase in activated Th1-cell percentages in mesenteric lymph nodes and splenic IFNg production in RMK treated mice (Fig. 8). Differences were limited to the Th1 cells, no effect of kefir was observed on the percentage of activated Th2 cells nor their cytokine IL-5. The change in the Th1/Th2 ratio may be in part due to the increased number of regulatory T-cells. However, regulatory T-cell percentages are not changed in the current study. It should be noted that the applied milk heat treatment was at a higher temperature for the HMK used for the microbiota analysis and peptidomics than the HMK sample used for the murine allergy model in this study. However, this will not affect our conclusions on differences between RMK and HMK, because HMK served as a ‘negative control’ in our allergy model experiments.
5. Conclusions
The raw milk kefir made in this study was assumed to be safe based on high hygienic standards and immediate milk acidification. The overall microbial composition of raw milk kefir resembled that of the defined starter culture. In addition, raw milk kefir contained specific bacterial strains and yeasts which could not be identified in heated milk kefir. Their proteolytic activity may be responsible for the wider range of peptides identified and associated bioactivities in raw milk compared to heated milk kefir. In line with these results, raw milk kefir reduced the acute allergic skin response and modulated T-cell responses in a murine food allergy model.Abbreviations
| ASV | Amplicon sequence variant |
| cat | Concatenated |
| CFU | Colony forming unit |
| Ct | Cycle threshold |
| SC | Defined starter culture eXact® 2 kefir, Chr. Hansen |
| EV | Extracellular vesicles |
| ITS | Internal transcribed spacer |
| NCBI | National Centre for Biotechnology Information |
| PBS | Phosphate buffered saline solution |
| HM | Heated milk (shortly boiled) |
| HMK | Heated milk kefir based on SC |
| qPCR | Quantitative real-time polymerase chain reaction |
| RA | Relative abundance |
| RM | Raw milk |
| RMK | Raw milk kefir based on SC |
Author contributions
TB brought the research partners together. TB, BvE, KH, JG and RK designed the research. TB, BvE, ZZ, PD, LvO, SB and MD conducted the research. BvE, ZZ, LvO and PD analysed the data. TB, BvE, KH, PD and RK wrote the paper. RK carried out the final editing of the manuscript. All authors read and approved the final manuscript.Conflicts of interest
TB is partly sponsored for his research activity by the Raw Milk Company. JG and BvE are partly employed at Danone Nutricia Research. All other authors report no conflict of interest.Acknowledgements
Software AG Stiftung, Darmstadt, Germany is acknowledged for funding the murine allergy study. Raw Milk Company, De Lutte, The Netherlands is acknowledged for providing the milk and kefir samples in this study as well as funding for the microbial enumeration experiments. Funding for microbiota profiling was obtained by R. K. from his NPN James Lind Award 2018.References
- J. P. Tamang, P. D. Cotter, A. Endo, N. S. Han, R. Kort and S. Q. Liu, et al., Fermented foods in a global age: East meets West, Compr. Rev. Food Sci. Food Saf., 2020, 19(1), 184–217 CrossRef PubMed.
- W. Gao, L. Zhang, Z. Feng, H. Liu, N. Shigwedha and X. Han, et al., Microbial diversity and stability during primary cultivation and subcultivation processes of Tibetan kefir, Int. J. Food Sci. Technol., 2015, 50(6), 1468–1476 CrossRef CAS.
- S. Blasche, Y. Kim, R. A. T. Mars, D. Machado, M. Maansson and E. Kafkia, et al., Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community, Nat. Microbiol., 2021, 6(2), 196–208 CrossRef CAS PubMed.
- D. E. Kiousi, N. Chorianopoulos, C. C. Tassou and A. Galanis, The clash of microbiomes: from the food matrix to the host gut, Microorganisms, 2022, 10(1), 116 CrossRef PubMed.
- M. L. Marco, D. Heeney, S. Binda, C. J. Cifelli, P. D. Cotter and B. Foligné, et al., Health benefits of fermented foods: microbiota and beyond, Curr. Opin. Biotechnol., 2017, 44, 94–102 CrossRef CAS PubMed.
- J. Ebner, A. Aşçi Arslan, M. Fedorova, R. Hoffmann, A. Küçükçetin and M. Pischetsrieder, Peptide profiling of bovine kefir reveals 236 unique peptides released from caseins during its production by starter culture or kefir grains, J. Proteomics, 2015, 117, 41–57 CrossRef CAS PubMed.
- D. C. Dallas, F. Citerne, T. Tian, V. L. M. Silva, K. M. Kalanetra and S. A. Frese, et al., Peptidomic analysis reveals proteolytic activity of kefir microorganisms on bovine milk proteins, Food Chem., 2016, 197, 273–284 CrossRef CAS PubMed.
- B. C. T. Bourrie, B. P. Willing and P. D. Cotter, The microbiota and health promoting characteristics of the fermented beverage kefir, Front. Microb., 2016, 7, 647 Search PubMed.
- D. H. Kim, D. Jeong, H. Kim and K. H. Seo, Modern perspectives on the health benefits of kefir in next generation sequencing era: Improvement of the host gut microbiota, Crit. Rev. Food Sci. Nutr., 2019, 559, 1782–1793 CrossRef PubMed.
- C. Slattery, P. D. Cotter and P. W. O’Toole, Analysis of health benefits conferred by Lactobacillus species from kefir, Nutrients, 2019, 11(6), 1252 CrossRef CAS PubMed.
- M. A. Farag, S. A. Jomaa, A. A. El-wahed and H. R. El-seedi, The many faces of kefir fermented dairy products: Quality characteristics, flavour chemistry, nutritional value, health benefits, and safety, Nutrients, 2020, 12(2), 346 CrossRef CAS PubMed.
- N. F. Azizi, M. R. Kumar, S. K. Yeap, J. O. Abdullah, M. Khalid and A. R. Omar, et al., Kefir and its biological activities, Foods, 2021, 10(6), 1210 CrossRef CAS PubMed.
- M. Y. Lee, K. S. Ahn, O. K. Kwon, M. J. Kim, M. K. Kim and I. Y. Lee, et al., Anti-inflammatory and anti-allergic effects of kefir in a mouse asthma model, Immunobiology, 2007, 212(8), 647–654 CrossRef CAS PubMed.
- M. Tanaka and J. Nakayama, Development of the gut microbiota in infancy and its impact on health in later life, Allergol. Int., 2017, 66(4), 515–522 CrossRef CAS PubMed.
- A. K. Adiloğlu, N. Gönülateş, M. Işler and A. Şenol, The effect of kefir consumption on human immune system: a cytokine study, Mikrobiyol. Bul., 2013, 47(2), 273–281 CrossRef PubMed.
- K. Thoreux and D. L. Schmucker, Kefir milk enhances intestinal immunity in young but not old rats, J. Nutr., 2001, 131(3), 807–812 CrossRef CAS PubMed.
- T. Brick, M. Ege, S. Boeren, A. Böck, E. von Mutius and J. Vervoort, et al., Effect of processing intensity on immunologically active bovine milk serum proteins, Nutrients, 2017, 9(9), 1–14 CrossRef PubMed.
- E. Kontopodi, S. Boeren, B. Stahl, J. B. van Goudoever, R. M. van Elburg and K. Hettinga, High-temperature short-time preserves human milk’s bioactive proteins and their function better than pasteurization techniques with long processing times, Front. Pediatr., 2022, 9, 798609 CrossRef PubMed.
- M. C. Michalski and C. Januel, Does homogenization affect the human health properties of cow's milk?, Trends Food Sci. Technol., 2006, 17(8), 423–437 CrossRef CAS.
- C. Braun-Fahrländer and E. von Mutius, Can farm milk consumption prevent allergic diseases?, Clin. Exp. Allergy, 2011, 41(1), 29–35 CrossRef PubMed.
- G. Loss, S. Apprich, M. Waser, W. Kneifel, J. Genuneit and G. Büchele, et al., The protective effect of farm milk consumption on childhood asthma and atopy: The GABRIELA study, J. Allergy Clin. Immunol., 2011, 128(4), 766–773 CrossRef PubMed.
- S. Abbring, J. T. Ryan, M. A. P. Diks, G. Hols, J. Garssen and B. C. A. M. van Esch, Suppression of food allergic symptoms by raw cow’s milk in mice is retained after skimming but abolished after heating the milk—A promising contribution of alkaline phosphatase, Nutrients, 2019, 11(7), 1499 CrossRef CAS PubMed.
- S. Abbring, L. Xiong, M. A. P. Diks, T. Baars, J. Garssen and K. Hettinga, et al., Loss of allergy-protective capacity of raw cow's milk after heat treatment coincides with loss of immunologically active whey proteins, Food Funct., 2020, 11(6), 4982–4993 RSC.
- S. Abbring, D. Kusche, T. C. Roos, M. A. P. Diks, G. Hols and J. Garssen, et al., Milk processing increases the allergenicity of cow's milk—Preclinical evidence supported by a human proof-of-concept provocation pilot, Clin. Exp. Allergy, 2019, 49(7), 1013–1025 CrossRef CAS PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet and G. A. Al-Ghalith, et al., Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37(8), 852–857 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13(7), 581–583 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer and P. Yarza, et al., The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2012, 41(D1), D590–D596 CrossRef PubMed.
- R. H. Nilsson, K. H. Larsson, A. F. S. Taylor, J. Bengtsson-Palme, T. S. Jeppesen and D. Schigel, et al., The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications, Nucleic Acids Res., 2019, 47(D1), D259–D264 CrossRef CAS PubMed.
- https://www.ncbi.nlm.nih.gov/bioproject/PRJNA716278 .
- J. Lu, S. Boeren, S. C. de Vries, H. J. F. van Valenberg, J. Vervoort and K. Hettinga, Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins, J. Proteomics, 2011, 75(1), 34–43 CrossRef CAS PubMed.
- K. A. Dingess, M. de Waard, S. Boeren, J. Vervoort, T. T. Lambers and J. B. van Goudoever, et al., Human milk peptides differentiate between the preterm and term infant and across varying lactational stages, Food Funct., 2017, 8(10), 3769–3782 RSC.
- Y. Liu, A. de Groot, S. Boeren, T. Abee and E. J. Smid, Lactococcus lactis mutants obtained from laboratory evolution showed elevated vitamin K2 content and enhanced resistance to oxidative stress, Front. Microbiol., 2021, 12, 746770 CrossRef PubMed.
- J. Cox and M. Mann, MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol., 2008, 26(12), 1367–1372 CrossRef CAS PubMed.
- A. Bateman, M. J. Martin, S. Orchard, M. Magrane, R. Agivetova and S. Ahmad, et al., UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res., 2021, 49(D1), D480–D489 CrossRef PubMed.
- P. M. Dekker, S. Boeren, A. H. Wijga, G. H. Koppelman, J. J. M. Vervoort and K. A. Hettinga, Maternal allergy and the presence of nonhuman proteinaceous molecules in human milk, Nutrients, 2020, 12, 1169 CrossRef CAS PubMed.
- S. D. Nielsen, R. L. Beverly, Y. Qu and D. C. Dallas, Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization, Food Chem., 2017, 232, 673–682 CrossRef CAS PubMed.
- O. Wagih, ggseqlogo: a versatile R package for drawing sequence logos, Bioinformatics, 2017, 33(22), 3645–3647 CrossRef CAS PubMed.
- X. M. Li, B. H. Schofield, C. K. Huang, G. I. Kleiner and H. A. Sampson, A murine model of IgE-mediated cow's milk hypersensitivity, J. Allergy Clin. Immunol., 1999, 103(2 Pt 1), 206–214 CrossRef CAS PubMed.
- A. C. Berge and T. Baars, Raw milk producers with high levels of hygiene and safety, Epidemiol. Infect., 2020, 148, e14 CrossRef CAS PubMed.
- L. O'Sullivan, R. P. Ross and C. Hill, Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality, Biochimie, 2002, 84(5–6), 593–604 CrossRef PubMed.
- M. J. R. Nout, Fermented foods and food safety, Food Res. Int., 1994, 27(3), 291–298 CrossRef CAS.
- S. Schoustra, C. van der Zon, A. Groenenboom, H. B. Moonga, J. Shindano and E. J. Smid, et al., Microbiological safety of traditionally processed fermented foods based on raw milk, the case of Mabisi from Zambia, LWT, 2022, 113997 CrossRef CAS.
- C. Hecer, B. Ulusoy and D. Kaynarca, Effect of different fermentation conditions on composition of kefir microbiota, Int. Food Res. J., 2019, 26, 401–409 CAS.
- T. Kök-Taş, A. C. Seydim, B. Özer and Z. B. Guzel-Seydim, Effects of different fermentation parameters on quality characteristics of kefir, J. Dairy Sci., 2013, 96(2), 780–789 CrossRef PubMed.
- E. Delavenne, J. Mounier, F. Déniel, G. Barbier and G. le Blay, Biodiversity of antifungal lactic acid bacteria isolated from raw milk samples from cow, ewe and goat over one-year period, Int. J. Food Microbiol., 2012, 155(3), 185–190 CrossRef CAS PubMed.
- L. Peréz-Través, R. de Llanos, A. Flockhart, L. García-Domingo, M. Groenewald and R. Pérez-Torrado, et al., Virulence related traits in yeast species associated with food; Debaryomyces hansenii, Kluyveromyces marxianus, and Wickerhamomyces anomalus, Food Control, 2021, 124, 107901 CrossRef.
- M. G. Netea, J. Domínguez-Andrés, L. B. Barreiro, T. Chavakis, M. Divangahi and E. Fuchs, et al., Defining trained immunity and its role in health and disease, Nat. Rev. Immunol., 2020, 20, 375–388 CrossRef CAS PubMed.
- L. Quigley, O. O'Sullivan, C. Stanton, T. P. Beresford, R. P. Ross and G. F. Fitzgerald, et al., The complex microbiota of raw milk, FEMS Microbiol. Rev., 2013, 37(5), 664–698 CrossRef CAS PubMed.
- W. Masoud, F. K. Vogensen, S. Lillevang, W. Abu Al-Soud, S. J. Sørensen and M. Jakobsen, The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR, Int. J. Food Microbiol., 2012, 153(1–2), 192–202 CrossRef CAS PubMed.
- A. R. Neves, W. A. Pool, A. Solopova, J. Kok, H. Santos and O. P. Kuipers, Towards enhanced galactose utilization by Lactococcus lactis, Appl. Environ. Microbiol., 2010, 76(21), 7048–7060 CrossRef CAS PubMed.
- C. Chaves-López, A. Serio, A. Paparella, M. Martuscelli, A. Corsetti and R. Tofalo, et al., Impact of microbial cultures on proteolysis and release of bioactive peptides in fermented milk, Food Microbiol., 2014, 42, 117–121 CrossRef PubMed.
- B. Hernández-Ledesma, B. Miralles, L. Amigo, M. Ramos and I. Recio, Identification of antioxidant and ACE-inhibitory peptides in fermented milk, J. Sci. Food Agric., 2005, 85(6), 1041–1048 CrossRef.
- J. F. Riordan, Angiotensin-I-converting enzyme and its relatives [Internet], Genome Biol., 2003, 4(8), 225 CrossRef PubMed.
- J. C. Rodríguez-Figueroa, A. F. González-Córdova, M. J. Torres-Llanez, H. S. Garcia and B. Vallejo-Cordoba, Novel angiotensin I-converting enzyme inhibitory peptides produced in fermented milk by specific wild Lactococcus lactis strains, J. Dairy Sci., 2012, 95(10), 5536–5543 CrossRef PubMed.
- Y. Liu and M. Pischetsrieder, Identification and relative quantification of bioactive peptides sequentially released during simulated gastrointestinal digestion of commercial kefir, J. Agric. Food Chem., 2017, 65(9), 1865–1873 CrossRef CAS PubMed.
- M. de Lima, R. A. da Silva and M. F. da Silva, et al., Brazilian kefir-fermented sheep's milk, a source of antimicrobial and antioxidant peptides, Probiotics Antimicrob. Proteins, 2018, 10, 446–455 CrossRef PubMed.
- B. Ismail and S. S. Nielsen, Invited review: Plasmin protease in milk: Current knowledge and relevance to dairy industry, J. Dairy Sci., 2010, 93, 4999–5009 CrossRef CAS PubMed.
- J. Law and A. Haandrikman, Proteolytic enzymes of lactic acid bacteria, Int. Dairy J., 1997, 7(1), 1–11 CrossRef CAS.
- P. S. Tan, B. Poolman and W. N. Konings, Proteolytic enzymes of Lactococcus lactis, J. Dairy Res., 1993, 60(2), 269–286 CrossRef PubMed.
- B. Miralles, R. del Barrio, C. Cueva, I. Recio and L. Amigo, Dynamic gastric digestion of a commercial whey protein concentrate, J. Sci. Food Agric., 2018, 98(5), 1873–1879 CrossRef CAS PubMed.
- T. K. Singh, S. K. Øiseth, L. Lundin and L. Day, Influence of heat and shear induced protein aggregation on the in vitro digestion rate of whey proteins, Food Funct., 2014, 5(11), 2686–2698 RSC.
- F. Guyomarc'h, Formation of heat-induced protein aggregates in milk as a means to recover the whey protein fraction in cheese manufacture, and potential of heat-treating milk at alkaline pH values in order to keep its rennet coagulation properties, A review, Laitiere, 2006, 86(1), 1–20 CrossRef.
- T. Baars, C. Berge, J. Garssen and J. Verster, The impact of raw fermented milk products on perceived health and mood among Dutch adults, Nutr. Food Sci., 2019, 49(6), 1195–1206 CrossRef.
- E. Mendes, M. B. Casaro, C. Fukumori, W. R. Ribeiro, A. L. dos Santos and P. Sartorelli, et al., Preventive oral kefir supplementation protects mice from ovariectomy-induced exacerbated allergic airway inflammation, Benefic. Microbes, 2021, 12(2), 187–197 CrossRef CAS PubMed.
- E. Velez, I. Novotny-Nuñez, S. Correa, G. Perdigón and C. Maldonado-Galdeano, Modulation of gut immune response by probiotic fermented milk consumption to control IgE in a respiratory allergy model, Benefic. Microbes, 2021, 12(2), 175–186 CrossRef CAS PubMed.
- L. Zhao, F. Shi, Q. Xie, Y. Zhang, S. E. Evivie and X. Li, et al., Co-fermented cow milk protein by Lactobacillus helveticus KLDS 1.8701 and Lactobacillus plantarum KLDS 1.0386 attenuates its allergic immune response in Balb/c mice, J. Dairy Sci., 2022, 105(9), 7190–7202 CrossRef CAS PubMed.
- X. Pi, Y. Yang, Y. Sun, Q. Cui, Y. Wan and G. Fu, et al., Recent advances in alleviating food allergenicity through fermentation, Crit. Rev. Food Sci. Nutr., 2022, 62(26), 7255–7268 CrossRef CAS PubMed.
- W. S. Hong, Y. P. Chen and M. J. Chen, The antiallergic effect of kefir Lactobacilli, J. Food Sci., 2010, 75(8), H244–H253 CrossRef CAS PubMed.
- B. Alashkar Alhamwe, L. A. P. M. Meulenbroek, D. H. Veening-Griffioen, T. M. D. Wehkamp, F. Alhamdan and S. Miethe, et al., Decreased histone acetylation levels at Th1 and regulatory loci after induction of food allergy, Nutrients, 2020, 12(10), 3193 CrossRef PubMed.
- N. Acevedo, B. Alashkar Alhamwe, L. Caraballo, M. Ding, A. Ferrante and H. Garn, et al., Perinatal and early-life nutrition, epigenetics, and allergy, Nutrients, 2021, 13(3), 724 CrossRef CAS PubMed.
- Q. S. Li, Y. Q. Wang, Y. R. Liang and J. L. Lu, The anti-allergic potential of tea: a review of its components, mechanisms and risks, Food Funct., 2021, 12(1), 57–69 RSC.
Footnotes |
† Electronic supplementary information (ESI) available: Supplementary file 1: Overview of the sample cycle threshold values, amplicon sequence variants, and metadata. Supplementary file 2: Relative abundance of microbial species in raw milk and heated milk. Dynamics of the microbial population after the first day of fermentation at 28 °C and during three weeks of storage at 4 °C. Supplementary file 3: Precursor proteins of kefir peptides with the total peptide intensity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) in raw milk (RM), heated milk (HM), raw milk kefir (RMK), and heated milk kefir (HMK). Supplementary file 4: Overview of bioactivities found in the Milk Bioactive Peptide Database (MBPDB),35 which match with identified peptide sequences. Supplementary file 5: Relative intensities of amino acids in the P1 and P1′ position for all peptides identified in (A) heated milk kefir (HMK) and (B) raw milk kefir (RMK). Relative intensities of amino acids in the P1 and P1′ position for uniquely identified peptides in (C) HMK and (D) RMK. See DOI: https://doi.org/10.1039/d2fo03248a 10) in raw milk (RM), heated milk (HM), raw milk kefir (RMK), and heated milk kefir (HMK). Supplementary file 4: Overview of bioactivities found in the Milk Bioactive Peptide Database (MBPDB),35 which match with identified peptide sequences. Supplementary file 5: Relative intensities of amino acids in the P1 and P1′ position for all peptides identified in (A) heated milk kefir (HMK) and (B) raw milk kefir (RMK). Relative intensities of amino acids in the P1 and P1′ position for uniquely identified peptides in (C) HMK and (D) RMK. See DOI: https://doi.org/10.1039/d2fo03248a |
| ‡ Shared first authors. |
| This journal is © The Royal Society of Chemistry 2023 |