Open Access Article
Open Access ArticleThe anti-hepatocellular carcinoma effects of polysaccharides from Ganoderma lucidum by regulating macrophage polarization via the MAPK/NF-κB signaling pathway
Guo-li
Li
 ac,
Jia-feng
Tang
ac,
Wen-li
Tan
c,
Tao
Zhang
bc,
Di
Zeng
a,
Shuang
Zhao
a,
Jian-hua
Ran
b,
Jing
Li
a,
Ya-ping
Wang
*a and
Di-long
Chen
*ac
ac,
Jia-feng
Tang
ac,
Wen-li
Tan
c,
Tao
Zhang
bc,
Di
Zeng
a,
Shuang
Zhao
a,
Jian-hua
Ran
b,
Jing
Li
a,
Ya-ping
Wang
*a and
Di-long
Chen
*ac
aLab of Stem Cell and Tissue Engineering, Department of Histology and Embryology, Chongqing Medical University, Chongqing, PR China. E-mail: 100474@cqmu.edu.cn; xinmengyuandlc@163.com
bNeuroscience Research Center, College of Basic Medicine, Chongqing Medical University, Chongqing, PR China
cChongqing Key Laboratory of Development and Utilization of Genuine Medicinal Materials in Three Gorges Reservoir Area, Chongqing Three Gorges Medical College, Chongqing, PR China
First published on 24th February 2023
Abstract
The response of macrophages to environmental signals demonstrates its heterogeneity and plasticity. After different forms of polarized activation, macrophages reach the M1 or M2 activation state according to their respective environment. Ganoderma lucidum polysaccharide (GLPS) is a major bioactive component of Ganoderma lucidum, a well-known medicinal mushroom. Although the immunomodulatory and anti-tumor effects of GLPS have been proven, GLPS's effect on inhibiting hepatocellular carcinoma (HCC) by regulating macrophage polarization is little known. Our data showed that GLPS notably inhibited the growth of a Hepa1-6 allograft. The expression of M1 marker CD86 was higher in the tumor tissue of the GLPS treatment group than in the control group in vivo. In vitro, the phagocytic activity and NO production of macrophages were increased by GLPS treatment. Moreover, it was discovered that GLPS was able to increase the expression of the M1 phenotype marker CD86, iNOS, and pro-inflammatory cytokines comprising IL-12a, IL-23a, IL-27 and TNF-α, but inhibited macrophage polarization towards the M2 phenotype by decreasing the expression of CD206, Arg-1, and inflammation-related cytokines comprising IL-6 and IL-10. The data suggest that GLPS may regulate macrophage polarization. Mechanistically, GLPS increased the phosphorylation of MEK and ERK. In addition, the phosphorylation of IκBα and P65 was increased by GLPS treatment. These data showed that GLPS can regulate the MAPK/NF-κB signaling pathway responsible for M1 polarization. In a nutshell, our research puts forward a new application of GLPS in anti-HCC treatment by regulating macrophage polarization through activating MAPK/NF-κB signaling.
1. Introduction
Macrophages have diverse functions in a variety of diseases.1,2 They play a key role in anti-tumor immune response and make up a major part of the leukocyte infiltrate in the tumor microenvironment (TME). The heterogeneity of macrophages may result in the plasticity and diversity of their responses to signals from the microenvironment.3 Macrophage's function and subtypes are profoundly affected by various cytokines and microbial products.4 Macrophages have been categorized into a classically activated (pro-inflammatory) M1 state or an alternatively activated (anti-inflammatory) M2 state according to different environments and stimuli.5 M1 macrophages are polarized by microbial lipopolysaccharide (LPS) and interferon-γ (IFN-γ). Various functional phenotypes of macrophages were induced by different M2 polarization molecules, which included IL-10, IL-4 or IL-13, glucocorticoid hormones, and vitamin D3.6 Based on the information available, M1 macrophages can inhibit tumor cell growth and produce pro-inflammatory cytokines. Contrastingly, M2 cells regulate inflammatory responses and adaptive Th1 immunity, scavenge debris, and boost angiogenesis, tissue remodeling and repair.7 The macrophages in tumor tissues are called tumor-associated macrophages (TAMs). They are mainly the M2 phenotype. The abnormal function of macrophages facilitates cancer progression. In the tumor microenvironment, TAMs can boost tumor cell proliferation, immunosuppression, and angiogenesis, thus supporting tumor growth and metastasis.8 There is considerable evidence showing that TAM abundance in tumors is related to poor disease prognosis and therapy resistance.9 In HCC, the central function of TAMs is to promote tumor development by the production of chemokines, growth factors, cytokines, and matrix metalloproteases. These products facilitate angiogenesis, metastasis, tumor cell proliferation and protection from apoptosis of tumor cells.10 Hepatic macrophages offer an inflammatory microenvironment susceptible to tumors. Meanwhile, macrophages actively facilitate the development and progression of HCC, responding to tumor and other signals derived from stromal cells.11 Therefore, the development of cancer immunotherapies that target these TAMs by regulating macrophage polarization deserves greater attention.Polysaccharides extracted from Ganoderma lucidum have shown extensive pharmacological activities, such as anti-tumor, immunomodulating and strong antioxidant activities.12–14Ganoderma lucidum, a basidiomycete, has been extensively utilized as an appealing source of functional food and drug precursors in physiology for many centuries. Several bioactive substances, polysaccharides in particular, were extracted and identified from Ganoderma lucidum. Ganoderma lucidum polysaccharide (GLPS) has been broadly investigated recently and it presented various biological activities such as immunomodulatory and anti-tumor effects.15 It has been reported that GLPS exerts anti-tumor effects by immunomodulatory effects, but it cannot directly kill tumor cells in vitro. GLPS can regulate the activity of various immune cells. The activation of murine peritoneal macrophages by GLPS has been investigated in vitro.16 Antagonism by GLPS against the inhibition by tumor cell culture supernatants of macrophages has been shown.17 GLPS's antagonistic effect on the prohibition of IL-2, IFN-γ and TNF-α production in macrophages by culture supernatants of tumor cells might help to control cancer.18
Although it has been reported that GLPS can exert anti-tumor effects by enhancing the activity of macrophages, it is still unclear whether GLPS can induce the polarization of macrophages to achieve anti-tumor effects. Herein, the influences of GLPS on macrophage polarization were evaluated in vivo and in vitro. The objective of this study was to further clarify the influence of GLPS on the polarization of macrophages in terms of anti-tumor immunity, which is of great significance to offer theoretical evidence for developing GLPS with anti-tumor activity as clinical anti-tumor adjuvant drugs.
2. Materials and methods
2.1. Drugs and reagents
GLPS (batch no.: DST201119-142) was provided by Chendu Esite Biology Technology in China. It was extracted from Ganoderma lucidum (Leyss. et Fr.) Karst. The appearance of this extract was a water-soluble powder of sandy beige color. The purity of GLPS was analyzed to be more than 98% by high-performance liquid chromatography (HPLC). GLPS contained D-glucose, D-galactose, D-mannose, D-xylose, L-arabinose, and L-rhamnose, and their molar ratio was 5.82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.23
2.23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.00
1.00![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.35
1.35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.72
0.72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.51 as determined by gas chromatography analysis. For in vivo experiments, GLPS was dissolved in normal saline; for in vitro experiments, it was dissolved in serum-free DMEM (Gibco, USA). The solution was sterilized through a 0.22 μm filter and stored at 4 degrees Celsius for further use. GLPS had an endotoxin concentration of below 0.01 EU mg−1, showing negligible endotoxin contamination.
0.51 as determined by gas chromatography analysis. For in vivo experiments, GLPS was dissolved in normal saline; for in vitro experiments, it was dissolved in serum-free DMEM (Gibco, USA). The solution was sterilized through a 0.22 μm filter and stored at 4 degrees Celsius for further use. GLPS had an endotoxin concentration of below 0.01 EU mg−1, showing negligible endotoxin contamination.
2.2. Cell culture
Mouse RAW264.7 macrophages and hepatocellular carcinoma cell line Hepa1-6 were obtained from ATCC (Manassas, VA, USA). We cultured them in DMEM (Gibco, USA) with high glucose containing 10% fetal bovine serum (FBS, Gibco, USA), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin at a body temperature of 37 degrees Celsius under a 5% CO2 atmosphere. In addition, RAW 264.7 cells were incorporated into the Hepa1-6 culture using 0.4 μm Transwell hanging cell culture inserts, and the co-culture was incubated for 24 h thereafter.2.3. Cell proliferation assay
The influence of GLPS on the proliferation of cells was detected by a cell counting Kit-8 (CCK-8, Dojindo Laboratories, Japan) assay. We seeded Hepa1-6 or RAW264.7 cells in 96-well plates at 6 × 103 cells per well and cultured them in DMEM containing 10% FBS. After incubation for 24 h, we cultured cells in a maintenance medium with 0–200 μg mL−1 GLPS for 24 h, respectively. Briefly, 100 μL of fresh DMEM containing 10 μL of CCK-8 solution was supplemented in each well. Later, the plate was subjected to incubation for 2 h at 37 degrees Celsius. A microplate reader (Varioskan LUX, Thermo scientific, USA) was used to measure the value of absorbance at a wavelength of 450 nm.2.4. TUNEL staining assay
Cell apoptosis was examined using a one-step TUNEL apoptosis assay kit (Beyotime, China) to verify GLPS-induced apoptosis in Hepa1-6 cells based on the manufacturer's instructions. We seeded cells at 1 × 104 cells per well in a 24-well plate with sterile coverslips per well. After incubation for 24 h, 200 μg mL−1 GLPS was used for cell treatment for another 24 h. Subsequently, the single-layer adherent cells were rinsed two times with cool PBS and fixed with 4% paraformaldehyde for half an hour. Next, the cells were incubated in TUNEL test solution at 37 degrees Celsius for an hour in the dark. Finally, 5 μg mL−1 DAPI was adopted to counterstain cell nuclei for 5 min. After rinsing three times with PBS, images of the treated cells were randomly obtained under a fluorescence microscope (Olympus, Japan), and at least 5 regions in each well were randomly selected in three independent experiments.2.5. Neutral red phagocytosis assay
We seeded Raw264.7 cells at 1 × 105 cells per well in 96-well microplates. They were incubated for 2 h at 37 degrees Celsius under 5% CO2. Thereafter, we discarded the supernatant. For 24 h, the samples were treated with or without 200 μg mL−1 GLPS in DMEM. Then, each well was supplemented with 100 μL neutral red solution (0.1%, w/w) and incubated for another 30 min. After discarding the supernatant, the cells were rinsed three times with PBS. Then, they were photographed using a light microscope or lysed for half an hour at RT by supplementing 200 μL of cell lysis buffer (Vethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vacetic acid = 1
Vacetic acid = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A microplate reader was utilized to detect the absorbance of each well at 540 nm.
1). A microplate reader was utilized to detect the absorbance of each well at 540 nm.
2.6. Animal experiments
Hepatocellular carcinoma cell lines Hepa1-6, derived from a BW7756 tumor in a C57 mouse, were cultured and separated from the medium. The suspension contained 1 × 107 cells per mL in normal saline, and then 0.2 mL of suspension (2 × 106 cells) was inoculated subcutaneously into the axilla of the right foreleg of 4–6 week-old male C57 BL/6 mice bought from Chongqing Medical University Experimental Animal Center. Once the tumors grew to the size of 100 mm3 on average, the mice were stochastically divided into control and GLPS treatment groups. The 40 mice were stochastically separated into four groups (10 mice per group) and treated for 14 days consecutively with GLPS (50, 100, and 200 mg kg−1) once every day by gavage and sterile physiological saline once every day by gavage as negative controls. All mice were sacrificed on the 14th day after the completion of treatment. We removed and weighed the tumors. This study was performed in strict accordance with the Chinese guidelines for the ethical review of animal welfare of laboratory animals (GB/T35892-2018) and was approved by the Institutional Biomedical Ethics Committee of Chongqing Three Gorges Medical College (Chongqing, China).2.7. Flow cytometry assay
We stained RAW264.7 cells with anti-mouse antibodies phycoerythrin (PE)-conjugated CD86 (M1) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Biolegend, USA) and fluorescein isothiocyanate (FITC)-conjugated CD206 (M2) (1
200, Biolegend, USA) and fluorescein isothiocyanate (FITC)-conjugated CD206 (M2) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Biolegend, USA), which were used to examine macrophage polarization. Cells were treated with 50, 100, and 200 μg mL−1 GLPS for 48 h in treatment groups, 100 ng mL−1 lipopolysaccharide (LPS) and 20 ng mL−1 recombinant mouse IFN-γ for 48 h in the M1 positive control group, and 20 ng mL−1 IL-4 for 48 h in the M2 positive control group. Based on the manufacturer's guidelines, we fixed cells with 4% paraformaldehyde in PBS for 20 min at RT, rinsed them thrice with 3% FBS/PBS and permeabilized them with 0.1% Triton X-100 in PBS. Thereafter, protected from light, the antibodies were stained for 20 min at RT in PBS. After rinsing with 3% FBS/PBS, briefly, flow cytometry was used to measure the cell samples (FACSCalibur, Becton Dickinson, USA).
200, Biolegend, USA), which were used to examine macrophage polarization. Cells were treated with 50, 100, and 200 μg mL−1 GLPS for 48 h in treatment groups, 100 ng mL−1 lipopolysaccharide (LPS) and 20 ng mL−1 recombinant mouse IFN-γ for 48 h in the M1 positive control group, and 20 ng mL−1 IL-4 for 48 h in the M2 positive control group. Based on the manufacturer's guidelines, we fixed cells with 4% paraformaldehyde in PBS for 20 min at RT, rinsed them thrice with 3% FBS/PBS and permeabilized them with 0.1% Triton X-100 in PBS. Thereafter, protected from light, the antibodies were stained for 20 min at RT in PBS. After rinsing with 3% FBS/PBS, briefly, flow cytometry was used to measure the cell samples (FACSCalibur, Becton Dickinson, USA).
2.8. Immunofluorescence assay
Immunofluorescence staining of RAW264.7 cells was carried out to evaluate the level of protein expression and the distribution of NF-κB p65. We seeded 1 × 103 cells per well in 24-well plates containing glass coverslips. The test cells were subjected to treatment with GLPS for 24 h. After the treatment and removal of medium, we fixed the cells with 4% paraformaldehyde for 15 min and then permeabilized them in 0.1% Triton X-100 (Solarbio, China) for 10 min. Subsequently, nonspecific binding sites were blocked at RT for an hour in 5% bovine serum albumin (BSA) buffer (Solarbio, China). After rinsing three times with PBST (PBS containing 0.1% Tween-20), the cells were incubated with mouse anti-CD86 (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Santa Cruz, USA), goat anti-CD206 (diluted 1
200, Santa Cruz, USA), goat anti-CD206 (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, RD, USA), and rabbit anti-P65 (diluted 1
200, RD, USA), and rabbit anti-P65 (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Abcam, UK) overnight at 4 degrees Celsius. Then, we stained the cells with the corresponding secondary FITC-conjugated antibodies (diluted 1
200, Abcam, UK) overnight at 4 degrees Celsius. Then, we stained the cells with the corresponding secondary FITC-conjugated antibodies (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Proteintech, USA) or PE-conjugated antibodies (diluted 1
200, Proteintech, USA) or PE-conjugated antibodies (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Proteintech, USA) at RT for an hour in the dark. Finally, an antifade mounting medium containing DAPI (Solarbio, China) to visualize the nucleus was dripped on the glass slides. The slides were stained for 5 minutes at RT. A fluorescence microscope (IX81, Olympus, Japan) was used to observe and photograph the labeled cells.
200, Proteintech, USA) at RT for an hour in the dark. Finally, an antifade mounting medium containing DAPI (Solarbio, China) to visualize the nucleus was dripped on the glass slides. The slides were stained for 5 minutes at RT. A fluorescence microscope (IX81, Olympus, Japan) was used to observe and photograph the labeled cells.
2.9. Real-time PCR
We treated RAW264.7 cells with 200 μg mL−1 GLPS for 0 and 24 h. Briefly, RNA was extracted from RAW264.7 by the Trizol method (Invitrogen, CA). To synthesize cDNA, total RNA was reverse transcribed with MMLV and oligo primers (Invitrogen, CA) in 20 μL reactions. A Bio-Rad system utilizing an SYBR Green I PCR Kit (Takara, Japan) was adopted to perform the real-time quantitative polymerase chain reaction (RT-qPCR). The expression level of mRNA was quantified using the −ΔΔCt method, and β-actin was taken as the control. Table 1 shows the corresponding primers.| Gene name | Primer 5′–3′ | |
|---|---|---|
| TNF-α | F: CCCTCACACTCAGATCATCTTCT | R: GCTACGACGTGGGCTACAG |
| IL-10 | F: GCTCTTACTGACTGGCATGAG | R: CGCAGCTCTAGGAGCATGTG |
| IL-12a | F: AGACATCACACGGGACCAAAC | R: CAGGCAACTCTCGTTCTTGT |
| IL-23a | F: ATGCTGGATTGCAGAGCAGTA | R: ACGGGGCACATTATTTTTAGTCT |
| IL-27 | F: GCTCGTTTGCCTTAGACCAG | R: GCATGGTGGCCTAACTCAAG |
| iNOS | F: GTTCTCAGCCCAACAATACAAGA | R: GTGGACGGGTCGATGTCAC |
| Arg-1 | F: CTCCAAGCCAAAGTCCTTAGAG | R: AGGAGCTGTCATTAGGGACATC |
| β-Actin | F: TAGGCGGACTGTTACTGAGC | R: GCCTTCACCGTTCCAGTTTT |
2.10. Immunohistochemistry
The test tissues were fixed with formalin and paraffin-embedded. Then, they were analyzed by IHC experiments. We cut paraffin blocks into sections with a thickness of 5 μm. The tissue sections were placed on slides that were coated with poly-L-lysine, and dried overnight. After the dried slides were de-paraffinized for 20 min by xylene treatment, the slides were subsequently subjected to antigen retrieval for 20 min by applying an automated antigen retrieval machine under ethylenediaminetetraacetic acid conditions (pH 9.0). The sections were incubated in 10% normal goat serum (Gibco, USA) for an hour to block non-specific binding before being incubated with CD86 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Santa Cruz, USA) primary antibodies at 4 degrees Celsius overnight. After rinsing with PBS thrice, the sections were subjected to incubation with secondary antibodies with streptavidin-tagged horse-radish peroxidase. Signaling was induced by diaminobenzidine (Sigma-Aldrich, USA). The tissue sections of IHC experiments were observed with an optical microscope (Leica) and analyzed with Leica software.
500, Santa Cruz, USA) primary antibodies at 4 degrees Celsius overnight. After rinsing with PBS thrice, the sections were subjected to incubation with secondary antibodies with streptavidin-tagged horse-radish peroxidase. Signaling was induced by diaminobenzidine (Sigma-Aldrich, USA). The tissue sections of IHC experiments were observed with an optical microscope (Leica) and analyzed with Leica software.
2.11. ELISA assay
RAW264.7 (1 × 105 cells) were incubated with/without GLPS (200 μg mL−1) for 24, 48, and 72 h. The supernatants of the conditioned culture media were collected at the indicated times to evaluate the expression levels of mouse TNF-α and IL-10 in fluids free of cells using an ELISA Kit (Boshide, China) based on the manufacturer's guidance.2.12. Western blotting analysis
The resulting extracts of protein from RAW264.7 macrophages were quantified and resolved by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Afterwards, we moved them to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Skimmed milk powder (5%) was used to block the membranes, which were immunoblotted with either rabbit anti-iNOS, anti-Arg-1 or anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Cell Signaling, USA) at 4 degrees Celsius overnight. For signaling analysis, cell lysates from RAW264.7 were transferred to PVDF membranes and incubated with rabbit antibodies to phospho-ERK1/2, ERK1/2, phospho-MEK1, MEK1/2 (1
1000, Cell Signaling, USA) at 4 degrees Celsius overnight. For signaling analysis, cell lysates from RAW264.7 were transferred to PVDF membranes and incubated with rabbit antibodies to phospho-ERK1/2, ERK1/2, phospho-MEK1, MEK1/2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1,000, Bimake, USA), phospho-IκBα, IκBα, phospho-P65, P65, GAPDH, and β-actin (1
1,000, Bimake, USA), phospho-IκBα, IκBα, phospho-P65, P65, GAPDH, and β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1,000, Cell Signaling, USA). The membranes were rinsed thrice in Tris-buffered saline with 0.1% Tween 20 (TBST), and subjected to incubation with HRP-conjugated anti-rabbit secondary antibodies for an hour at RT and developed using a hypersensitive enhanced chemiluminescence (ECL) reagent (Biosharp, China). National Institutes of Health ImageJ software was employed to analyze the band relative density of various proteins.
1,000, Cell Signaling, USA). The membranes were rinsed thrice in Tris-buffered saline with 0.1% Tween 20 (TBST), and subjected to incubation with HRP-conjugated anti-rabbit secondary antibodies for an hour at RT and developed using a hypersensitive enhanced chemiluminescence (ECL) reagent (Biosharp, China). National Institutes of Health ImageJ software was employed to analyze the band relative density of various proteins.
2.13. Statistics
All values are presented as means plus or minus mean standard error. The two-tailed unpaired Student's t-test was employed to perform statistical analyses between two groups. When more than two groups needed to be compared, one or two-way analysis of variance (ANOVA) with Bonferroni post-hoc analysis was adopted. We repeated all experiments three times or more. For all the cases studied, a P-value <0.05 was considered statistically significant: *P < 0.05, **P < 0.01, and ***P < 0.001. All statistical analyses were performed with GraphPad Prism 8.0 software (GraphPad, San Diego, CA, USA).3. Results
3.1. GLPS has anti-HCC activity related to macrophage polarization
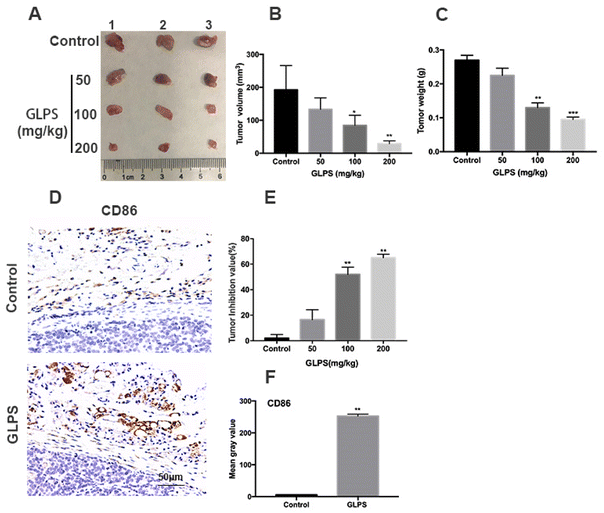
A hepatic carcinoma Hepa1-6 allograft mouse model was adopted to measure the in vivo anti-tumor activity of GLPS. Compared with the control group, tumor development was markedly inhibited in the GLPS group, after 14 days of drug administration (Fig. 1A). The tumor weight of treatment group mice gradually decreased with increasing drug dose, and the tumor volume was obviously smaller than that of the control group (Fig. 1B and C). Furthermore, as the GLPS concentration increased, tumor inhibition values were, respectively, 16.5%, 52% and 65% of tumor weight for the drug treatment group. In 100 and 200 mg kg−1 GLPS treatment groups, GLPS inhibited tumor growth effectively (Fig. 1E). Moreover, CD86 expression was higher in the mouse Hepa1-6 allograft tumor of the 200 mg kg−1 GLPS treatment group than that in the control group (Fig. 1D and F). These above results indicated that GLPS may exert its anti-tumor effect related to macrophage polarization in vivo.3.2. GLPS exerts its anti-tumor effect by increasing macrophage phagocytic activity
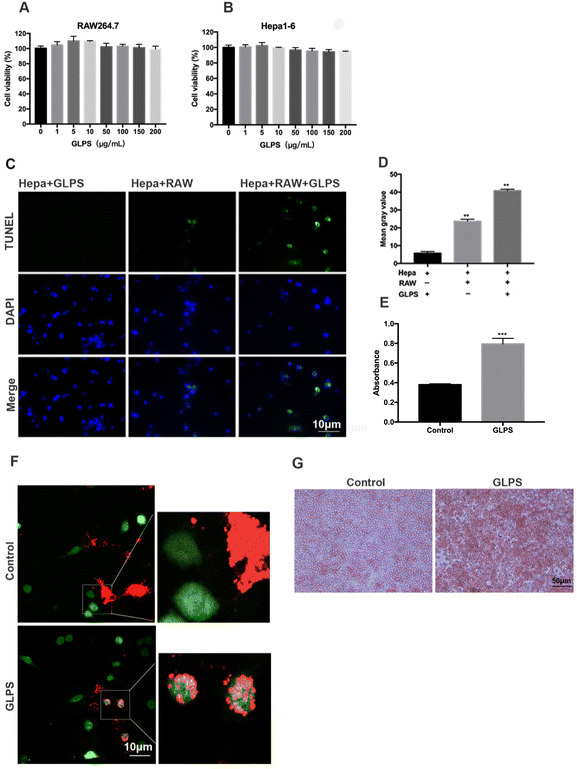
To evaluate whether GLPS can directly inhibit cell viability, Hepa1-6 and RAW264.7 were, respectively, measured using CCK8. The results showed that their cell viability was not significantly suppressed by GLPS (Fig. 2A and B). In order to analyze whether GLPS kills tumor cells directly or indirectly, Hepa1-6 and RAW264.7 macrophages were co-cultured using Transwell inserts with or without GLPS treatment. The representative images of TUNEL analysis are shown. In the GLPS-treated tumor group, apoptotic cells were not obviously observed. We observed a few apoptotic cells in the group of tumor cells with macrophages. More apoptotic cells were seen in the group of GLPS-treated tumor cells with macrophages (Fig. 2C and D). It is known that macrophages are able to phagocytose cancer cells to promote antitumor immunity. Enhancing the phagocytosis of tumor cells by macrophages can increase the frequency of TAMs and improve their survival. To further access the effect of GLPS on macrophage phagocytic activity, CMFDA, a green fluorescent dye, was used to label RAW264.7 macrophages. Also, CM-dil, a red fluorescent dye, was used to label Hepa1-6 tumor cells. Then, the two kinds of cells were co-cultured and treated with 200 μg mL−1 GLPS for 2 h. It was found that GLPS can enhance the phagocytosis of tumor cell debris by macrophages (Fig. 2F). Moreover, GLPS enhanced macrophage RAW264.7 phagocytosis of the neutral red dye (Fig. 2G and E). The above results indicated that GLPS can exert its anti-tumor effect by enhancing the phagocytic activity of macrophages.3.3. GLPS polarizes RAW264.7 macrophages to the M1-like phenotype
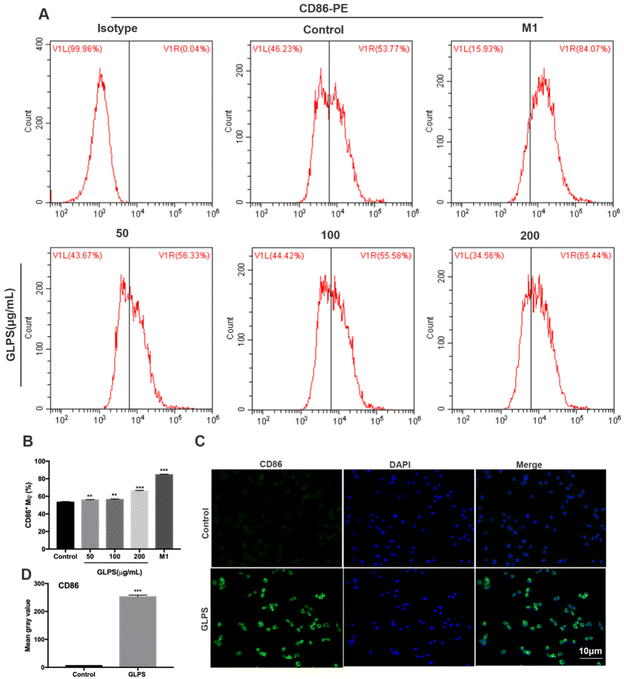
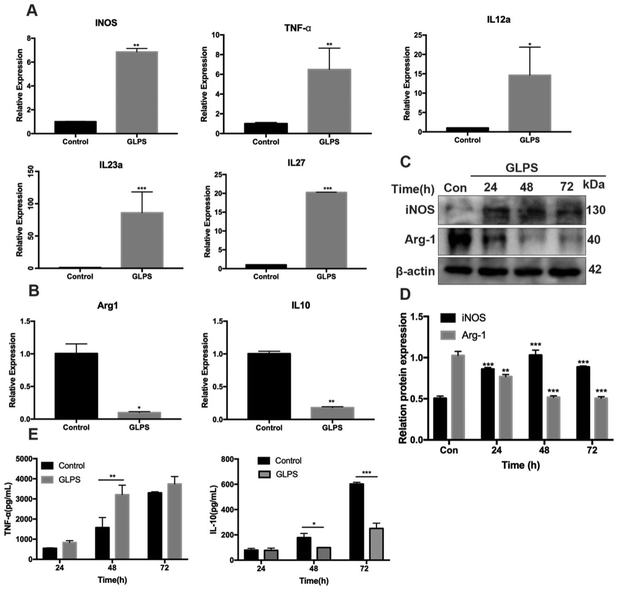
To determine the phenotype of RAW264.7 macrophages after GLPS treatment, the expression of the M1 marker (CD86) was detected by flow cytometric analysis. RAW264.7 macrophages were administered, respectively, with 50, 100, and 200 μg mL−1 GLPS for 24 h. LPS and IFN-γ were used as the positive control for M1. Compared with the control group, GLPS promoted a significant increase in CD86 expression. In particular, 200 μg mL−1 GLPS substantially increased CD86 expression on RAW264.7 macrophages (Fig. 3A and B). In addition, immunofluorescence assay was performed to analyze CD86 expression. The results also indicated that GLPS significantly increased CD86 expression in the treatment group treated with 200 μg mL−1 GLPS (Fig. 3C and D). In addition, RT-PCR illustrated that the representative M1 genes, comprising iNOS, TNF-α, IL-12a, IL-23a and IL-27, were increased in the GLPS treatment group (Fig. 5A). In comparison with the control, the iNOS protein expression level (Fig. 5C) and TNF-α production (Fig. 5E) were significantly increased after GLPS treatment. These results indicated that GLPS can promote macrophage polarization to M1.3.4. GLPS inhibits macrophage polarization to the M2-like phenotype
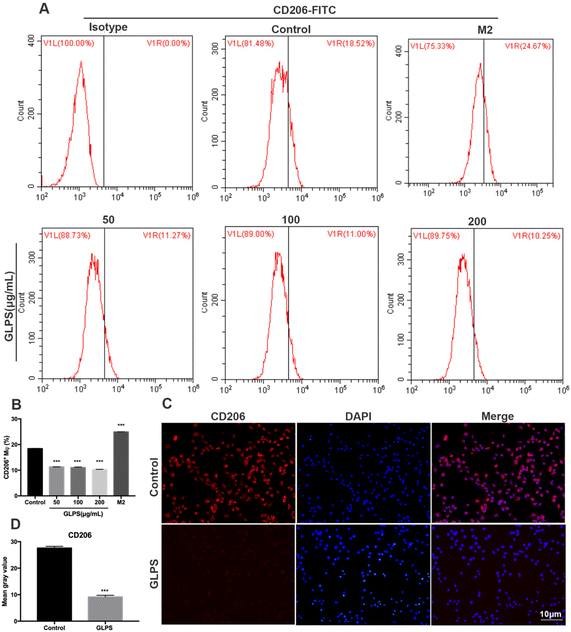
In order to determine the phenotype of macrophages induced by GLPS treatment, flow cytometry analysis was conducted to measure the level of M2 marker (CD206) expression. IL-4 was used as the positive control. CD206 expression was significantly increased in the IL-4 treatment group. Upon treatment with GLPS, GLPS substantially inhibited CD206 expression on RAW264.7 macrophages (Fig. 4A and B). In addition, immunofluorescence analysis was performed to analyze CD206 expression. The results also indicated that GLPS significantly decreased CD206 expression in the treatment group (Fig. 4C and D). Thereafter, RT-qPCR analysis indicated that the representative M2 marker genes, comprising Arg-1 and IL-10, were significantly reduced in the treatment group (Fig. 5B). The expression level of Arg-1 protein was also decreased after GLPS treatment (Fig. 5C). In addition, GLPS-treated macrophages secreted lower IL-10 levels than the control. GLPS also acted time-dependently to inhibit the generation of IL-10 (Fig. 5E). All the data indicated that GLPS can inhibit macrophage polarization to M2.3.5. GLPS induces RAW264.7 macrophage polarization to the M1 phenotype by MAPK signaling
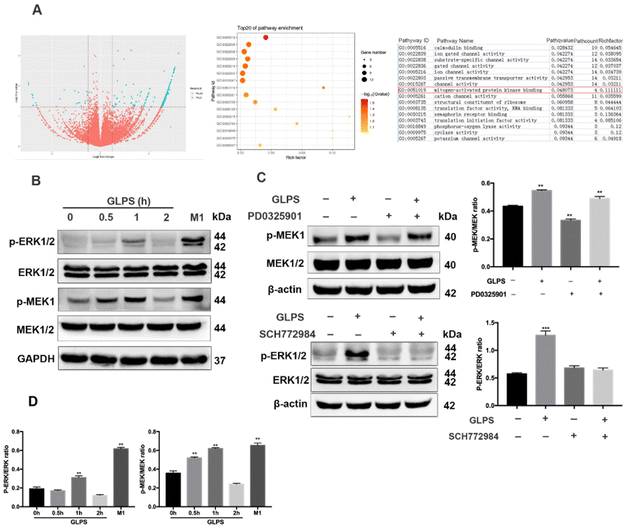
To further determine GLPS-induced signals in macrophage polarization, transcriptome sequencing of RAW264.7 macrophages was performed in the control group and 200 μg mL−1 GLPS treatment group. Bioinformatics analysis showed that MAPK signaling was a potential signal pathway when GLPS polarized macrophages (Fig. 6A). MAPK signaling was measured by western blot analysis. After the stimulation of RAW264.7 with 200 μg mL−1 GLPS, p-MEK1 and p-ERK1/2 expression increased, but decreased rapidly at 2 h. When macrophages were treated with PD0325901, a MEK inhibitor, for 1 h, GLPS induced-MEK phosphorylation was inhibited. When they were treated with ERK inhibitor SCH772984 (100 nM) for 4 h, GLPS induced-ERK phosphorylation was inhibited (Fig. 6C). These data indicated that GLPS can activate MAPK signaling.3.6. GLPS activated the NF-κB signaling pathway
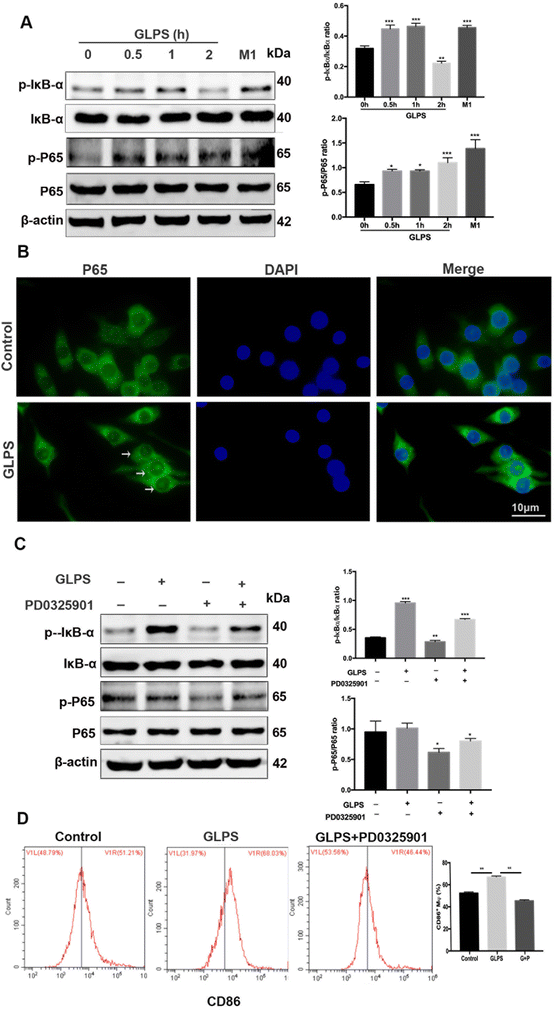
To further determine other GLPS-induced signals in macrophage polarization, western blotting was used to measure NF-κB signaling. After treatment of RAW264.7 with 200 μg mL−1 GLPS at indicated time points, it was observed that GLPS induced IκBα and P65 phosphorylation (Fig. 7A). After stimulation of RAW264.7 with 200 μg mL−1 GLPS for 1 h, IκBα and P65 phosphorylation increased compared with those in the control group. The representative images show obvious p-P65 nuclear translocation in the GLPS treatment group (Fig. 7B). When macrophages were treated with PD0325901, a MEK inhibitor, for 1 h, GLPS induced-IκBα and -P65 phosphorylation was inhibited (Fig. 7C). These data indicated that GLPS can activate NF-κB signaling. Moreover, NF-κB signaling might be a downstream signaling of MEK. However, it is unclear whether a MEK inhibitor can inhibit the expression of CD86, an M1 marker, in the GLPS-induced macrophages. CD86 expression was analysed by flow cytometry after the treatment with a MEK inhibitor (Fig. 7D). It showed that MEK inhibitor PD0325901 can inhibit the expression of M1 marker CD86. Therefore, GLPS can induce the M1 polarization of macrophages through the MAPK-mediated NF-κB signaling pathway.4. Discussion
This paper demonstrates the inhibiting role of GLPS in HCC progression by regulating macrophage polarization. In addition, a pathway is proposed in which GLPS regulated the activation of the MAPK/NF-κB signaling pathway, skewing macrophage function to the M1 phenotype. Macrophages have two different phenotypes, including M1 activation and M2 responses, which can be modulated by the surrounding microenvironment. M1/M2 imbalance is closely related to the progression of malignant tumors. The aggregation of M2 macrophages facilitates the reduction of immune response and the development of tumors, whereas the infiltration of M1 macrophages can elicit a protective immune response against tumors. Therefore, the conversion of macrophages from M2 to M1 is a promising strategy for finding new potential therapeutic strategies.GLPS is the key bioactive component of Ganoderma lucidum. Ganoderma lucidum (Fr.) Krast, a famous medicinal fungus, has been broadly utilized as medicine for hundreds of years in Asia to boost long life and to improve health with no significant side effects. Recently, GLPS has been extensively studied and it has been proved that GLPS has many biological activities such as immunomodulatory and anti-tumor effects.
Plenty of studies have shown that GLPS inhibits the growth of various tumors in vivo.20,21 In this work, we have investigated that GLPS inhibited transplanted tumors from HCC cell lines Hepa1-6 dose-dependently. Interestingly, although GLPS suppressed the development of tumors in vivo, the drug did not directly inhibit Hepa1-6 cell proliferation in vitro. Therefore, GLPS might play a role in anti-tumor activity by immunomodulatory effects. This result is consistent with previous reports.22 Macrophages, an innate immune population, are essential to maintain body homeostasis. Macrophages also exhibit high heterogeneity and plasticity and perform various critical functions in different tissue compartments. Therefore, aberrant macrophage function contributes substantially to the growth of numerous diseases including cancer.23 Our study demonstrated that GLPS treatment promoted tumor cell apoptosis in Hepa1-6 tumor cells co-cultured with RAW264.7 macrophage cells. Moreover, GLPS enhanced macrophage phagocytosis and NO production. These data showed that GLPS activated macrophage functions.
Macrophages are a type of highly plastic cell capable of rapidly changing their polarization depending on microenvironmental cues. The polarized subtype of macrophages varies in terms of receptor expression, effector function, cytokine production and chemokine repertoires. For example, arginine metabolism features a high iNOS level in M1 macrophages, whereas high levels of arginase1 are observed in M2-polarized macrophages.24 M1 macrophages overexpressed cell-surface markers CD86, CD80 and CD16/32, whereas M2 macrophages expressed high levels of markers Arg-1, CD163 and CD206. Our study demonstrated that GLPS increased CD86 expression in transplanted tumor tissue. GLPS inhibited the growth of tumors by regulating M1 macrophage polarization. In this research, only CD86 was analyzed for M1 macrophages, and not other cell markers in tumor tissues, for example, CD206.
In addition, the key characteristic of polarized macrophages is differential cytokine production. The M1 phenotype includes IL-12a, IL-23a, IL-27 and TNF-α, whereas M2 macrophages generally produce IL-6 and IL-10.25,26 However, the role of GLPS in regulating macrophage polarization is unclear. This study showed that GLPS can boost the expression of CD86, iNOS and pro-inflammatory cytokines, which suggests that GLPS can boost M1 macrophage polarization. The M1 phenotype increases the production of pro-inflammatory cytokines within the TME and promotes cancer cell damage via recruiting pro-immunostimulating leukocytes and phagocytosis of tumor cells. Our results also illustrate that GLPS inhibits macrophage polarization to the M2 phenotype by the inhibition of CD206, Arg-1, and anti-inflammatory cytokines. Previous studies have revealed that M2-polarized macrophages had a significant role in enhancing the propagation of tumors. The M2 cells facilitate tumor growth in primary and metastatic sites by their effects on the breakdown and deposition of basement membranes, angiogenesis, recruitment of leukocytes, and overall immune inhibition.27 In conclusion, GLPS can regulate macrophage polarization to inhibit tumor growth.
Macrophage polarization is regulated by a complex network, comprising signaling pathways, transcription factors, epigenetic mechanisms, and post-transcriptional regulators.28 The imprinting of long-term macrophage function by environmental signals, including polarizing molecules, has been inconstantly regarded as memory, adaptive-innate or trained immunity.29 The past decade has revealed some important macrophage polarization regulators, such as the interferon regulatory factor (IRF), signal transducer and activator of transcription (STAT), and suppressor of cytokine signaling (SOCS) families.30 The activation of the IRF–STAT-signaling pathway by IFNs and TLR ligands can switch macrophage function to the M1 phenotype via STAT1, whereas IL-4 and IL-13 polarize macrophages to the M2 phenotype through STAT6. IL-10 skews macrophage function to an M2-like phenotype by STAT3-mediated expression of genes (Tgfb1 and Mrc1), and IL-3-mediated STAT5 activation is demonstrated to enhance the M2 polarization of macrophages.31 Notch activation mediated by IRF5 and IRF8 is part of the transcriptional network associated with M1.32,33
This study showed that GLPS can activate MAPK and NF-κB signaling pathways to regulate macrophage polarization. As a crucial regulator of cellular pathology and physiology, MAPKs are composed of MEK, ERK, p38, and JNK MAPK subfamilies.19 Existing research has revealed that MAPK is important in macrophage polarization. The improvement of M1 macrophage polarization was uplifted by MEK and ERK signaling activation.34–36 Consistent with these reports, our research results revealed that p-MEK and p-ERK activities were elevated in macrophages with GLPS treatment. Moreover, we further confirmed this hypothesis by suppressing the activation of these signaling molecules with their specific inhibitors.
Sustained phosphorylation of MAPK is required for NF-κB RELA degradation.37 NF-κB, an important transcription factor, regulates inflammation and tumor development. NF-κB activation in macrophages is concerned with inflammation-associated tumor generation. The plasticity change of macrophages from M1 to M2 might be connected with the dynamics of NF-κB activity. NF-κB signaling is usually activated in M1 macrophages to secrete high levels of pro-inflammatory cytokines. However, M2 macrophages are regarded to have less NF-κB activity.38 Whether GLPS is able to upregulate NF-κB activity to control macrophage polarization has not been defined before. We have demonstrated that GLPS can activate NF-κB signaling to trigger macrophage polarization to the M1 phenotype. It has been suggested that such macrophage phenotype switching might be induced by the TME. Interestingly, previous studies revealed that most extracted TAMs from solid tumor tissues exhibit the M2 phenotype with less NF-κB activation and present a strong immuno-suppressive activity. The signaling pathway of NF-κB is key for inducing many cytokines and mediators related to inflammation, which include inducible iNOS and TNF-α.39 Thus, GLPS may activate NF-κB to change the macrophage phenotype for anti-tumor activity.
5. Conclusion
In summary, our data show that GLPS modulates macrophage polarization via the MAPK/NF-κB signaling pathway, leading to the growth inhibition of hepatocellular carcinoma. This work demonstrates that GLPS can regulate the MAPK/NF-κB signaling pathway to change macrophage polarization. Our study has provided evidence supporting the link between GLPS and macrophage polarization, laying the groundwork for subsequent GLPS studies. These results offer a rationale for the future research of GLPS to treat patients with tumors characterized by targeting macrophage polarization.Abbreviations
| GLPS | Ganoderma lucidum polysaccharide |
| HCC | Hepatocellular carcinoma |
| TME | Tumor microenvironment |
| LPS | Lipopolysaccharide |
| IFN-γ | Interferon-γ |
| TAMs | Tumor-associated macrophages |
Author contributions
Guo-li Li: methodology, investigation, formal analysis, visualization, and writing – original draft. Jia-feng Tang: methodology, investigation, resources, and visualization. Wen-li Tan: writing – review and editing. Tao Zhang: methodology, formal analysis, and investigation. Di Zeng: methodology and investigation. Shuang Zhao: methodology and investigation. Jian-hua Ran: conceptualization and writing – review and editing. Jing Li: conceptualization and writing – review and editing. Ya-ping Wang: conceptualization, supervision, and writing – review and editing. Di-long Chen: conceptualization, methodology, funding acquisition, project administration, and writing – review and editing.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by a Key Project and a Youth Project of the Science and Technology Research Program of the Chongqing Education Commission of China (No. KJZD-K201802701 and KJQN202002710), a Key Project and a Lab Project of Chongqing Three Gorges Medical College of China (No. 2019XZZ002 and SYS20210005).References
- H. Zeng, B. Zhao, D. Zhang, X. Rui, X. Hou, X. Chen, B. Zhang, Y. Yuan, H. Deng and G. Ge, Viola yedoensis Makino formula alleviates DNCB-induced atopic dermatitis by activating JAK2/STAT3 signaling pathway and promoting M2 macrophages polarization, Phytomedicine, 2022, 103, 154228 CrossRef CAS PubMed.
- S. Prabhu, H. Deng, T. L. Cross, S. H. Shahoei, C. J. Konopka, N. G. Medina, C. C. Applegate, M. A. Wallig, L. W. Dobrucki, E. R. Nelson, A. M. Smith and K. S. Swanson, Nanocarriers targeting adipose macrophages increase glucocorticoid anti-inflammatory potency to ameliorate metabolic dysfunction, Biomater. Sci., 2021, 9, 506–518 RSC.
- N. Dehne, J. Mora, D. Namgaladze, A. Weigert and B. Brune, Cancer cell and macrophage cross-talk in the tumor microenvironment, Curr. Opin. Pharmacol., 2017, 35, 12–19 CrossRef CAS PubMed.
- H. W. Lee, H. J. Choi, S. J. Ha, K. T. Lee and Y. G. Kwon, Recruitment of monocytes/macrophages in different tumor microenvironments, Biochim. Biophys. Acta, 2013, 1835, 170–179 CAS.
- H. Degroote, A. Van Dierendonck, A. Geerts, H. Van Vlierberghe and L. Devisscher, Preclinical and Clinical Therapeutic Strategies Affecting Tumor-Associated Macrophages in Hepatocellular Carcinoma, J. Immunol. Res., 2018, 7819520 CAS.
- C. Ngambenjawong, H. H. Gustafson and S. H. Pun, Progress in tumor-associated macrophage (TAM)-targeted therapeutics, Adv. Drug Delivery Rev., 2017, 114, 206–221 CrossRef CAS PubMed.
- A. Mantovani, S. Sozzani, M. Locati, P. Allavena and A. Sica, Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes, Trends Immunol., 2002, 23, 549–555 CrossRef CAS PubMed.
- C. Anfray, A. Ummarino, F. T. Andon and P. Allavena, Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses, Cells, 2019, 9, 46–70 CrossRef.
- S. Lee, S. Kivimae, A. Dolor and F. C. Szoka, Macrophage-based cell therapies: The long and winding road, J. Controlled Release, 2016, 240, 527–540 CrossRef CAS.
- D. Capece, M. Fischietti, D. Verzella, A. Gaggiano, G. Cicciarelli, A. Tessitore, F. Zazzeroni and E. Alesse, The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages, BioMed Res. Int., 2013, 187204 Search PubMed.
- F. Heindryckx and P. Gerwins, Targeting the tumor stroma in hepatocellular carcinoma, World J. Hepatol., 2015, 7, 165–176 CrossRef PubMed.
- S. Joseph, B. Sabulal, V. George, K. R. Antony and K. K. Janardhanan, Antitumor and anti-inflammatory activities of polysaccharides isolated from Ganoderma lucidum, Acta Pharm., 2011, 61, 335–342 CAS.
- S. Zhang, G. Pang, C. Chen, J. Qin, H. Yu, Y. Liu, X. Zhang, Z. Song, J. Zhao, F. Wang, Y. Wang and L. W. Zhang, Effective cancer immunotherapy by Ganoderma lucidum polysaccharide-gold nanocomposites through dendritic cell activation and memory T cell response, Carbohydr. Polym., 2019, 205, 192–202 CrossRef CAS PubMed.
- A. Li, X. Shuai, Z. Jia, H. Li, X. Liang, D. Su and W. Guo, Ganoderma lucidum polysaccharide extract inhibits hepatocellular carcinoma growth by downregulating regulatory T cells accumulation and function by inducing microRNA-125b, J. Transl. Med., 2015, 13, 100 CrossRef PubMed.
- L. X. Sun, Z. B. Lin, X. S. Duan, J. Lu, Z. H. Ge, X. F. Li, X. J. Li, M. Li, E. H. Xing, Y. X. Song, J. Jia and W. D. Li, Enhanced MHC class I and costimulatory molecules on B16F10 cells by Ganoderma lucidum polysaccharides, J. Drug Targeting, 2012, 20, 582–592 CrossRef CAS PubMed.
- Z. Liu, J. Xing, Y. Huang, R. Bo, S. Zheng, L. Luo, Y. Niu, Y. Zhang, Y. Hu, J. Liu, Y. Wu and D. Wang, Activation effect of Ganoderma lucidum polysaccharides liposomes on murine peritoneal macrophages, Int. J. Biol. Macromol., 2016, 82, 973–978 CrossRef CAS PubMed.
- J. Lu, L. X. Sun, Z. B. Lin, X. S. Duan, Z. H. Ge, E. H. Xing, T. F. Lan, N. Yang, X. J. Li, M. Li and W. D. Li, Antagonism by Ganoderma lucidum polysaccharides against the suppression by culture supernatants of B16F10 melanoma cells on macrophage, Phytother. Res., 2014, 28, 200–206 CrossRef CAS.
- L. X. Sun, W. D. Li, Z. B. Lin, X. S. Duan, E. H. Xing, M. M. Jiang, N. Yang, H. H. Qi, Y. Sun, M. Li, Y. D. Niu and J. Lu, Cytokine production suppression by culture supernatant of B16F10 cells and amelioration by Ganoderma lucidum polysaccharides in activated lymphocytes, Cell Tissue Res., 2015, 360, 379–389 CrossRef CAS PubMed.
- S. Francisco, A. Arranz, J. Merino, C. Punzon, R. Perona and M. Fresno, Early p38 Activation Regulated by MKP-1 Is Determinant for High Levels of IL-10 Expression Through TLR2 Activation, Front. Immunol., 2021, 12, 660065 CrossRef CAS PubMed.
- J. Zhu, J. Xu, L. L. Jiang, J. Q. Huang, J. Y. Yan, Y. W. Chen and Q. Yang, Improved antitumor activity of cisplatin combined with Ganoderma lucidum polysaccharides in U14 cervical carcinoma-bearing mice, Kaohsiung J. Med. Sci., 2019, 35, 222–229 CrossRef CAS PubMed.
- C. Wang, S. Shi, Q. Chen, S. Lin, R. Wang, S. Wang and C. Chen, Antitumor and Immunomodulatory Activities of Ganoderma lucidum Polysaccharides in Glioma-Bearing Rats, Integr. Cancer Ther., 2018, 17, 674–683 CrossRef CAS PubMed.
- D. Cor, Z. Knez and M. K. Hrncic, Antitumour, Antimicrobial, Antioxidant and Antiacetylcholinesterase Effect of Ganoderma Lucidum Terpenoids and Polysaccharides: A Review, Molecules, 2018, 23, 649–670 CrossRef PubMed.
- C. Ngambenjawong, H. H. Gustafson and S. H. Pun, Progress in tumor-associated macrophage (TAM)-targeted therapeutics, Adv. Drug Delivery Rev., 2017, 114, 206–221 CrossRef CAS PubMed.
- Y. Zhuang, X. Zhao, B. Yuan, Z. Zeng and Y. Chen, Blocking the CCL5-CCR5 Axis Using Maraviroc Promotes M1 Polarization of Macrophages Cocultured with Irradiated Hepatoma Cells, J. Hepatocell. Carcinoma, 2021, 8, 599–611 CrossRef PubMed.
- U. Avila-Ponce de Leon, A. Vazquez-Jimenez, M. Matadamas-Guzman, R. Pelayo and O. Resendis-Antonio, Transcriptional and Microenvironmental Landscape of Macrophage Transition in Cancer: A Boolean Analysis, Front. Immunol., 2021, 12, 642842 CrossRef CAS PubMed.
- D. Artemova, P. Vishnyakova, E. Khashchenko, A. Elchaninov, G. Sukhikh and T. Fatkhudinov, Endometriosis and Cancer: Exploring the Role of Macrophages, Int. J. Mol. Sci., 2021, 22, 5196–5212 CrossRef CAS PubMed.
- D. F. Quail and J. A. Joyce, Microenvironmental regulation of tumor progression and metastasis, Nat. Med., 2013, 19, 1423–1437 CrossRef CAS PubMed.
- A. Sica, P. Invernizzi and A. Mantovani, Macrophage plasticity and polarization in liver homeostasis and pathology, Hepatology, 2014, 59, 2034–2042 CrossRef PubMed.
- M. Locati, A. Mantovani and A. Sica, Macrophage activation and polarization as an adaptive component of innate immunity, Adv. Immunol., 2013, 120, 163–184 CAS.
- A. Mantovani, S. Sozzani, M. Locati, P. Allavena and A. Sica, Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes, Trends Immunol., 2002, 23, 549–555 CrossRef CAS PubMed.
- R. Lang, D. Patel, J. J. Morris, R. L. Rutschman and P. J. Murray, Shaping gene expression in activated and resting primary macrophages by IL-10, J. Immunol., 2002, 169, 2253–2263 CrossRef CAS PubMed.
- T. Krausgruber, K. Blazek, T. Smallie, S. Alzabin, H. Lockstone, N. Sahgal, T. Hussell, M. Feldmann and I. A. Udalova, IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses, Nat. Immunol., 2011, 12, 231–238 CrossRef CAS PubMed.
- A. Sica and A. Mantovani, Macrophage plasticity and polarization: in vivo veritas, J. Clin. Invest., 2012, 122, 787–795 CrossRef CAS PubMed.
- H. Pan, W. Huang, Z. Wang, F. Ren, L. Luo, J. Zhou, M. Tian and L. Tang, The ACE2-Ang-(17)-Mas Axis Modulates M1/M2 Macrophage Polarization to Relieve CLP-Induced Inflammation via TLR4-Mediated NF-small ka, Cyrillicb and MAPK Pathways, J. Inflammation Res., 2021, 14, 2045–2060 CrossRef PubMed.
- J. Lu, H. Zhang, J. Pan, Z. Hu, L. Liu, Y. Liu, X. Yu, X. Bai, D. Cai and H. Zhang, Fargesin ameliorates osteoarthritis via macrophage reprogramming by downregulating MAPK and NF-kappaB pathways, Arthritis Res. Ther., 2021, 23, 142 CrossRef CAS PubMed.
- C. He, S. Sun, Y. Zhang, F. Xie and S. Li, The role of irreversible electroporation in promoting M1 macrophage polarization via regulating the HMGB1-RAGE-MAPK axis in pancreatic cancer, OncoImmunology, 2021, 10, 1897295 CrossRef PubMed.
- M. T. He, H. S. Park, Y. S. Kim, A. Y. Lee and E. J. Cho, Protective Effect of Membrane-Free Stem Cells against Lipopolysaccharide and Interferon-Gamma-Stimulated Inflammatory Responses in RAW 264.7 Macrophages, Int. J. Mol. Sci., 2021, 22, 6894–6904 CrossRef CAS PubMed.
- C. P. Chang, Y. C. Su, P. H. Lee and H. Y. Lei, Targeting NFKB by autophagy to polarize hepatoma-associated macrophage differentiation, Autophagy, 2013, 9, 619–621 CrossRef CAS PubMed.
- R. Han, M. Hu, Q. Zhong, C. Wan, L. Liu, F. Li, F. Zhang and W. Ding, Perfluorooctane sulphonate induces oxidative hepatic damage via mitochondria-dependent and NF-kappaB/TNF-alpha-mediated pathway, Chemosphere, 2018, 191, 1056–1064 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |