Open Access Article
Open Access ArticleCan exosomes transfer the preconditioning effects triggered by (poly)phenol compounds between cells?†
Inês
Figueira
 a,
Paulo
Bastos
a,
Antonio
González-Sarrías
a,
Paulo
Bastos
a,
Antonio
González-Sarrías
 b,
Juan Carlos
Espín
b,
Juan Carlos
Espín
 b,
Bruno
Costa-Silva
c and
Cláudia
Nunes dos Santos
b,
Bruno
Costa-Silva
c and
Cláudia
Nunes dos Santos
 *ad
*ad
aiNOVA4Health, NOVA Medical School| Faculdade de Ciências Médicas, NMS|FCM, Universidade NOVA de Lisboa, Lisboa, Portugal. E-mail: claudia.nunes.santos@nms.unl.pt
bLaboratory of Food & Health, Research Group on Quality, Safety and Bioactivity of Plant Foods, CEBAS-CSIC, Murcia, Spain
cChampalimaud Physiology and Cancer Programme, Champalimaud Foundation, Lisboa, Portugal
diBET, Institute of Experimental and Technological Biology, Oeiras, Portugal
First published on 16th December 2022
Abstract
Effective strategies in prolonging life- and health span are increasingly recognized as acting as mild stressors. Micronutrients and other dietary compounds such as (poly)phenols may act as moderate stressors and confer protective effects via a preconditioning phenomenon. (Poly)phenols and their metabolites may not need to reach their target cells to produce biologically significant responses, so that cells exposed to it at entry points may communicate signals to other cells. One of such “communication” mechanisms could occur through extracellular vesicles, including exosomes. In vitro loading of exosomes with (poly)phenols has been used to achieve targeted exosome homing. However, it is unknown if similar shuttling phenomena occur in vivo upon (poly)phenols consumption. Alternatively, exposure to (poly)phenols might trigger responses in exposed organs, which can subsequently signal to cells distant from exposure sites via exosomes. The currently available studies favor indirect effects of (poly)phenols, tempting to suggest a “billiard-like” or “domino-like” propagating effect mediated by quantitative and qualitative changes in exosomes triggered by (poly)phenols. In this review, we discuss the limited current data available on how (poly)phenols exposure can potentially modify exosomes activity, highlighting major questions regarding how (epi)genetic, physiological, and gut microbiota factors can modulate and be modulated by the putative exosome-(poly)phenolic compound interplay that still remains to be fully understood.
1. Introduction
Effective strategies in prolonging life- and health-span are increasingly recognized as mild stressors capable of triggering a broad spectrum of biological activities in cells to prevent many important chronic diseases. Such mild stressors can lead to the generation of a protective state, termed “preconditioning”, which primes cells to better respond to subsequent and perhaps more aggressive environmental insults. A clear example of this is physical exercise, which has been shown to lead to short-term energy depletion, inhibiting mammalian target of rapamycin (mTOR) while activating AMP-activated protein kinase (AMPK), NAD-dependent deacetylase sirtuin-1 (SIRT1), Forkhead box domain-containing (FOXO), and autophagy. In parallel, the production of reactive oxygen species (ROS) during physical exercise activates Nrf2 and stimulates the expression of endogenous antioxidant enzymes (e.g., superoxide dismutase, catalase, glutathione peroxidase), which in the long run can attenuate ROS levels at the cellular level.1,2 At the same time, caloric restriction and intermittent fasting also increase the levels of NAD+, SIRT1, FOXO, and endogenous antioxidant enzymes, promoting DNA repair, mitochondrial biogenesis, and autophagy.3,4Micronutrients and other dietary compounds [most notably (poly)phenols] can be considered exercise or caloric restriction “mimetics” and thus also act as mild stressors and confer protective effects via preconditioning phenomena similar to hormesis or trained immunity.5,6 Accordingly, certain substances may confer protection when tested within a range of physiologically compatible concentrations, but too much of any given molecule might end up being detrimental. Consequently, the consumption of dietary (poly)phenols has gained increased attention for their putative “hormetic-like” potential.7–9 In parallel, the observation that (poly)phenols act as mild stressors and activate endogenous defense enzymatic machineries10,11 suggests processes akin to those reported for trained immunity12 to be of relevance for their overall protective/beneficial effects.
2. How (poly)phenol compounds and their metabolites may modulate and be modulated by exosome signaling
Among the dietary compounds with preconditioning effects of putative relevance for a plethora of human diseases, (poly)phenolic compounds have been increasingly raised to the spotlight. (Poly)phenolic compounds comprise an important source of bioactive compounds in the human diet as they are present in almost all vegetables, fruits, cereals, beverages (such as tea, coffee, red wine), and other plant-derived foods. (Poly)phenols present multiple hydroxyl groups on aromatic rings and encompass diverse classes of compounds currently represented by >8000 different molecules, such as flavonoids (flavonols, flavan-3-ols, flavones, etc.) and non-flavonoids (hydrolyzable tannins, hydroxycinnamic acids, etc.). Of particular note, an increasing number of such compounds has been associated with increased health benefits in humans, as observed across a growing number of conditions (as reviewed in ref. 13 and 14).Despite the aforementioned benefits, the administration of (poly)phenol compounds may result in unintended consequences. For instance, high doses of (poly)phenols can be toxic to cells and may also provide a fitness advantage to tumor cells.15,16 Here, higher and shorter peak exposures, achieving milder and more prolonged concentrations, could lead to more beneficial biological responses at the organism level.17 Also, it should be noted that, once (poly)phenols are treated as xenobiotics, these are poorly absorbed, rapidly metabolized, and extensively excreted. However, most of the results consist of data on parent (poly)phenol compounds, which, unfortunately, at least most of them, can only reach host cells to a negligible extent.14 Little is known yet whether microbiota- and host-derived metabolic products, which reach multiple host organs in measurable amounts, can trigger comparable effects:14 while parent (poly)phenol compounds are found in circulation only in nanomolar concentration range or even not detected at all, their resulting metabolites (e.g., hydroxy-benzenes and hydroxy hippuric acids) reach the circulation to a significantly greater extent (as recently reviewed in18,19). Studies have shown that (poly)phenols are subjected to a series of metabolic reactions that lead to the production of diverse circulating (poly)phenol metabolites.20 The large majority of such metabolites derive from a group of structurally diverse parent compounds (e.g., chlorogenic acids, flavanols, proanthocyanidins, theaflavins, and thearubigins) that undergo modifications converging to the formation of aromatic/phenolic acids with hydroxyls substituents. In contrast, fewer ones are associated with a unique circulating (poly)phenol metabolite type (e.g., urolithins derived from ellagitannins, S-equol derived from isoflavones). Nevertheless, data are still lacking for some (poly)phenol metabolites (e.g., pyranoanthocyanins, coumarins, and other minor dietary components). But, despite our limited mapping of the complete reactions spectrum leading to circulating (poly)phenol metabolites formation, we do know that their appearance can result from both host- and microbiota-mediated metabolism. Also, (poly)phenols metabolism and interconversion may occur at the level of the small and large intestine, in the liver, and in cells outside the gastrointestinal tract, culminating in a wide variety of derivatives (e.g., sulfated, methylated, and glucuronidated). For instance, (poly)phenols aglycones can be absorbed in the small intestine after undergoing cleavage by epithelial glycosidases; in turn, (poly)phenol conjugates with sugar moieties that are resistant to glycosidases and cannot be absorbed in the small intestine to a significant extent, reach the colon where they can be further metabolized.21 In the colon, such conjugates are cleaved, and the resulting aglycones undergo ring fission by microbiota-encoded enzymes, producing a plethora of (poly)phenols metabolites.13 For instance, ring fission of flavanones produces 3-hydroxy-3-(phenyl)propionic acid, while isoflavones generate 2-(hydroxyphenyl)propanoic acid. In turn, ring fission of flavan-3-ols (i.e., catechin, epicatechin, epigallocatechin) produces 5-(hydroxyphenyl)valeric acid and 5-(hydroxyphenyl)-γ-valerolactones, which are specific and do not derive from any other flavonoid. Another noteworthy class of circulating (poly)phenol metabolites is represented by urolithins resulting from ellagic acid metabolism. The gut microbiota produces these metabolites with significant interindividual variability, leading to individuals’ categorization, according to different “metabotypes”.22,23 Unlike other (poly)phenol metabolites, urolithins (i.e., urolithin A, isourolithin A, and urolithin B) are not further metabolized into smaller compounds. In contrast, urolithins are mainly absorbed and conjugated by phase II enzymes, circulate in the bloodstream, and are excreted as conjugates via urine or unconjugated urolithins via feces.24 All in all, these (poly)phenols metabolites reach the circulation at much higher concentrations than their parent counterparts and are thus of much more biological relevance. However, preclinical research has reported that these circulating metabolites usually show low health effects as conjugated compounds, with lower bioactivity than their deconjugated counterparts.14,25,26 Here, it should be noted that their higher bioavailability is at least partially driven by the fact that multiple structurally different parent compounds converge onto a small but common number of circulating (poly)phenol metabolites. In turn, most circulating (poly)phenol metabolites cannot be specifically mapped to unique parent (poly)phenol compounds.
Nevertheless, (poly)phenols and their circulating metabolites may not need to reach their target cells to produce biologically significant responses. Accordingly, it is possible for cells exposed to (poly)phenol compounds at intestinal or hepatic levels, to communicate stress or survival “signals” to other distant cells in the organism, and one of the poorly understood mechanisms via which such “communication” may take place comprises exosomes or other extracellular vesicles (EVs).27
EVs are lipid membrane-enclosed vesicular structures that may shuttle a variety of cellular cargos, and different types and classifications thereof exist based on size, morphology, function, and biogenesis (e.g., apoptotic bodies, macro and microvesicles (MVs), and exosomes). In this manuscript, however, we focus solely on the effects of (poly)phenol-triggered “exosome-mediated” effects, with the caveat that the currently available vesicle characterization methods cannot ensure 100% specificity.
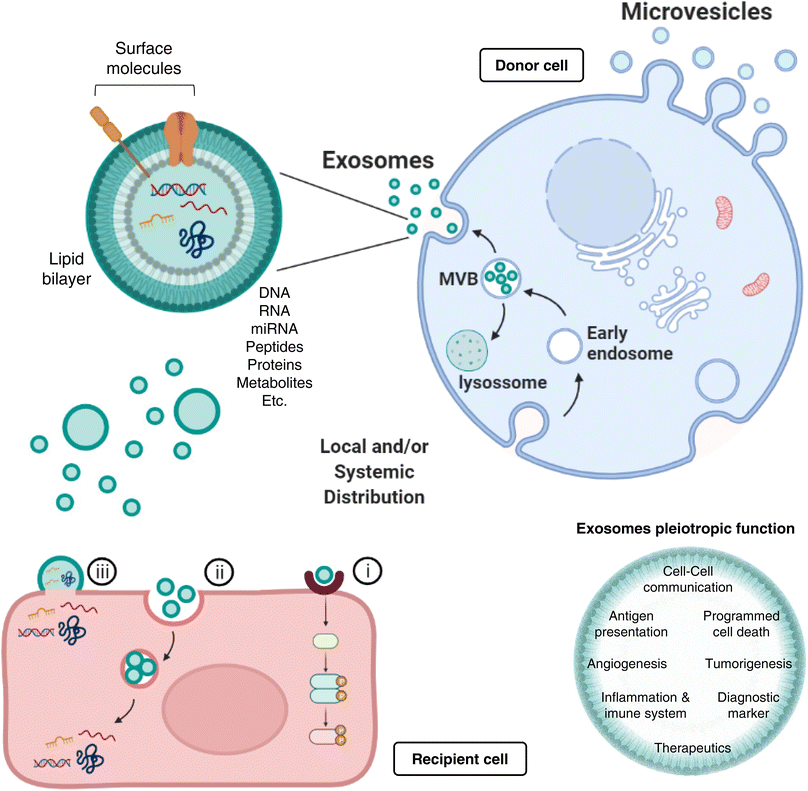
Exosomes represent nanosized (30–150 nm) double-layered lipid membrane EVs, contains every basic cell biomolecule. Exosomes are processed through endosomal compartments and released from cells upon fusion of the endocytic compartment called multivesicular body (MVB) with their surface membrane. MVBs can either fuse with lysosomes for degradation, or with the plasma membrane, resulting in exosomes release to the extracellular milieu. The MVB contains intraluminal vesicles (ILVs) that, upon release, are termed exosomes.28,29 Once released, exosomes can act both locally or systemically by traveling through the circulation. Exosomes communicate with target cells though at least three different mechanisms: (i) By docking at the plasma membrane and activating intracellular signaling by ligand–receptor interaction; (ii) By internalization through phagocytosis, micropinocytosis or receptor-/raft-mediated endocytosis, and subsequent fusion with the delimiting membrane of an endocytic compartment, releasing their content into the cytoplasm of the recipient cells. (iii) Although less frequent than mechanisms i and ii, exosomes may also release their content into the cytoplasm by direct membrane fusion (Fig. 1).
Most protocols for EVs isolation provide enriched (instead of pure) preparations of EVs with a specific origin.30,31 Thus, many studies to date consider small vesicles as exosomes regardless of cellular origin. Exosomes are present in extracellular spaces and all body fluids, allowing the intercellular shuttling of micro and macromolecules (including proteins,32 lipids,33 nucleic acids,33,34 and metabolites35,36) and cell-to-cell communication over distant target cells.37,38 Exosomes pleiotropic function embraces not only intercellular communication, but also antigen presentation, as well as regulation of several mechanisms, such as programmed cell death, angiogenesis, tumorigenesis, or even inflammatory response (Fig. 1). A greater understanding of the role of exosomes as intercellular signaling vesicles in the last decade has resulted in a paradigm shift in our knowledge of how cells communicate, where their cargos play significant role in physiology and pathological processes.39 Accordingly, the autocrine, paracrine, and endocrine signaling mediated by exosomes and the capacity to alter the recipient cell phenotype depends on their cargo, which is dictated by active sorting mechanisms and allow a cell to communicate environmental cues to distant cells.40,41
Such mechanisms of communication enable the horizontal genetic transfer of exosomal cargo to the cytoplasm of the recipient cell, where epigenetic reprogramming through miRNA, lipids and functional proteins can take place.39 Exosomal miRNAs, in particular, play a vital role in inter-cellular and inter-organism signal transduction, since miRNAs are theorized to act as epigenetic regulators on more than half of all mammalian genomes.42,43 Indeed, such exosomal miRNAs have been described to hold multiple functions, like cell differentiation44 or insulin secretion,45 overall highlighting the dramatic regulatory capacity that miRNAs can hold when coupled to the intercellular communication abilities of an exosome.
Importantly, exosome cargo mirrors the physiological status of the secreting tissue.39 In various diseases, exosomes offer a window into altered cellular/tissue states, and their detection in biological fluids potentially offers a multicomponent diagnostic readout, important tools for determining the prognosis of patients. Indeed, exosomes have been associated, for instance, with immune responses, cardiovascular and central nervous system-related diseases, as well as with cancer progression (as reviewed in46–49); consequently, in all these scenarios, the exosome cargo delivered into recipient cells will surely alter their biological response. Such exosome-mediated responses can be either disease-promoting or disease-restraining and, for that reason, constitute invaluable diagnostic or therapeutic tools, respectively (Fig. 1). The intrinsic exosomes nature, entities capable of regulating complex intracellular pathways, have advanced their utility as therapeutics for neurodegenerative conditions and cancer: they can be engineered to deliver diverse therapeutic payloads, as siRNAs, or chemotherapeutics agents, with an ability to direct their delivery to a desired target.50,51 Moreover, exosome lipid and protein composition will affect their pharmacokinetic properties, where their natural constituents may play a role in enhanced bioavailability and in minimizing adverse reactions.
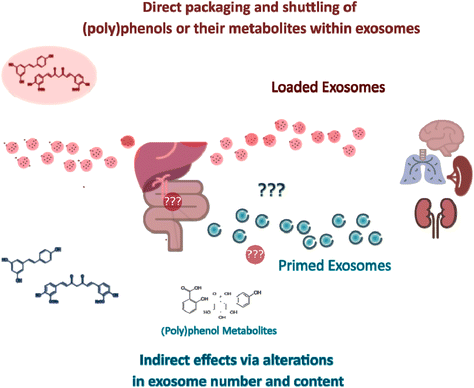
As further detailed below, increased attention has been paid to how the content of exosomes is affected by exposure to different drugs or (poly)phenol compounds. In parallel, many studies have evaluated the efficacy of in vitro (poly)phenol-loaded exosomes as drug delivery systems. As such, there are two different but not necessarily mutually exclusive strategies for studying the exosome-mediated effects of (poly)phenols so far, namely (i) the loading of exosomes with (poly)phenols for shuttling purposes, which effects can be evaluated in vitro (Table 1) or in vivo (Table 2), and (ii) (poly)phenol-mediated alterations in exosomal content (with the isolation of (poly)phenols “primed” exosomes) in cells (Table 3), in animals or humans upon (poly)phenols consumption. Here, it should be noted that the characterization of exosomes from animal or human origin upon (poly)phenols consumption has not been performed to date. A putative explanation for such a gap is that a precise mapping from the administration of (poly)phenol parent compound through the formation of the resulting small phenolic metabolites to their respective targets and exosome-producing cells is technically challenging.
| (Poly)phenol | Exosomes’ source/loading approach | Exosomes characterization | Goal | Model | Dose | Post-exposure duration until analysis | Main effects | Ref. |
|---|---|---|---|---|---|---|---|---|
| Abbreviations: ABC: ATP-binding cassette, AChE: acetylcholinesterase activity assay, AFM: atomic force microscopy, Bax: Bcl-2-associated X protein, Bcl-2: B-cell lymphoma 2, CD: cluster of differentiation, DLS: dynamic light scattering, EM: electron microscopy, LC3: microtubule-associated protein 1A/1B-light chain 3, NTA: nanoparticle tracking analysis, NF-κB: nuclear factor kappa-light-chain-enhancer of activated B cells, PINK1: PTEN induced putative kinase 1, RhoB: ras homolog family member B, ROS: reactive oxygen species, TNF-α: tumor necrosis factor-alpha, TSG101: tumor susceptibility gene 101, VCAM1: vascular cell adhesion molecule 1, WB: western blotting, ZO: zonula occludens. | ||||||||
| Epicatechin gallate (ECG) | Bovine milk/sonication 6×, on ice 2 min each cycle | WB (CD63 and CD9) | To enhance the activity and bioavailability of ECG | Rotenone-induced SHSY5Y cell model | 10 μM ECG-Exo | 21 h | ↑ Viability | Luo et al. (2021)63 |
| DLS | ↓ ROS | |||||||
| EM | ↓ LC3 | |||||||
| ↓ PINK1 | ||||||||
| To evaluate the neuroprotective effects of ECG-Exo against rotenone insult | ↓Parkin | |||||||
| ↓ Caspase-3 | ||||||||
| ↓ Bax | ||||||||
| ↑ Bcl-2 | ||||||||
| Curcumin | Human chronic myelogenous leukemia K562 or LAMA84 cells/coincubation at 37 °C for 24 h at 10, 20 or 40 μM | NA | To study how curcumin exposure modulates exosome tumor cell composition | Endothelial permeability (HUVEC cells) | ∼50–∼100 μM | 6 h | ↑ miR-21 | Taverna et al. (2016)64 |
| ↓ RhoB | ||||||||
| ↓ Motility | ||||||||
| ↓ IL-8 | ||||||||
| ↓ VCAM1 | ||||||||
| To evaluate how curcumin-primed exosomes modulated endothelium monolayer integrity | ↓ Angiogenesis | |||||||
| ↑ ZO-1 | ||||||||
| ↑ VE-Cadherin | ||||||||
| ↓ Endothelial permeability | ||||||||
| Curcumin | Murrah buffaloes’ milk/5 mg incubated with 1.5 mL milk whey overnight at 4 °C | DLS | Stability in solution | NA | 3 h | ↑ Solubility | Vashisht et al. (2017)65 | |
| To evaluate free curcumin vs exosomal curcumin stability | Resistance to in vitro enzymatic digestion | ↑ Stability | ||||||
| To study the uptake and trans-epithelial transport of curcumin upon encapsulation | ↑ Resistance to digestion | |||||||
| Intestinal permeability-(Caco-2 cells) | ↑ trans-Epithelial crossing | |||||||
| Curcumin | Cow milk/coincubation at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 concentration ratio to exosomes at room temperature 5 concentration ratio to exosomes at room temperature |
DLS | To study the loading efficiency of exosomes with curcumin, their stability, and cancer cells’ antiproliferative activity | Human lung cancer (H1299 and A549), cervical cancer (HeLa), and breast cancer (MDA-MB-231 and T47D) cells | 0.25–50 μM | 72 h | ↓ Proliferation | Aqil et al. (2017)66 |
| AFM | ↓ TNF-α | |||||||
| ↓ NF-κB | ||||||||
| Curcumin | Cow milk or Caco-2 cell-derived/coincubation at2 mg mL−1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio to exosomes overnight at room temperature 5 ratio to exosomes overnight at room temperature |
NTA | To compare the intestinal translocation of cow milk exosomes with that of epithelial cell-derived exosomes | Intestinal permeability and uptake (Caco-2 cells) | NA | 4 h or 72 h | ↑ Uptake of cell-derived exosomes, particularly in differentiated cells | Carobolante et al. (2020)67 |
| To compare the uptake and antiproliferative activity of exosomes-incorporated curcumin to that of free curcumin | ↑ trans-Epithelial translocation | |||||||
| ↓ Cell proliferation | ||||||||
| Curcumin | PANC-1 or MIA PaCa-2 cells/24 h coincubation | AChE | To determine the “functional alterations” of exosomes released by pancreatic cancer cells upon exposure to curcumin | Pancreatic adenocarcinoma PANC-1 or MIA PaCa-2 cells | 50 μM | 24 h, 48 h, or 72 h | ↑ Uptake | Osterman et al. (2015)68 |
| DLS | ↑ Apoptosis | |||||||
| ↓ Cell viability | ||||||||
| Curcumin or Resveratrol | Bovine milk/passive diffusion and further purified by size exclusion chromatography (SEC) | WB (presence of CD63 and TSG101) (absence of calnexin and β-casein) | To compare the uptake and anticancer activity of exosomes-incorporated curcumin or resveratrol to that of free curcumin or resveratrol on breast cancer cells models | Human MCF-7 and MDA-MB-231 breast cancer and MCF-10A non-tumorigenic cells | Curcumin: 48 and 96 nM | 4 and 72 h | ↓ Cell proliferation on cancer cells by curcumin-loaded exosomes (arrest at G0/G1 phase) with no effect on normal cells | González-Sarrías et al. (2022)56 |
| Resveratrol: 375 or 750 nM | ↑ Apoptosis (via mitochondrial pathway) ↑ Uptake of curcumin-loaded exosomes via clathrin-mediated endocytosis, avoiding ABC transporters | |||||||
| (Poly)phenol | Exosomes’ source/loading approach | Exosomes characterization | Model target | Dose | Post-administration duration until analysis | Main effects | Ref. |
|---|---|---|---|---|---|---|---|
| Abbreviations: AChE: acetylcholinesterase activity assay, AFM: atomic force microscopy, BBB: blood–brain barrier, CD: cluster of differentiation, c(RGDyK): cyclic arginine-glycine-aspartic acid-D-tyrosine-lysine, DLS: dynamic light scattering, EM: electron microscopy, GFAP: glial fibrillary acidic protein, GRP94: glucose-regulated protein 94, ICAM: intercellular adhesion molecule, IL-1β: interleukin 1 beta, LPS: lipopolysaccharide, NeuN: neuronal marker, NTA: nanoparticle tracking analysis, NMDR1: N-methyl-D-aspartate (NMDA) receptor 1, ROS: reactive oxygen species, TEM: transmission electron microscopy, TNF-α: tumor necrosis factor-alpha, TSG101: tumor susceptibility gene 101, WB: western blotting. | |||||||
| Curcumin | Mouse lymphoma | Intraperitoneal administration in C57BL/6J mice | 100 mg kg−1 | 48 h | 5–10× ↑ blood concentration | Sun et al. (2010)55 | |
| EL-4 cells, 22 °C, 5 min co-incubation | EM | 4 mg kg−1 | 4 days | ↓ Mortality to LPS challenge | |||
| ↓ IL-6 in serum | |||||||
| WB (TSG101 and CD81) | ↓ TNF-α in serum | ||||||
| 100 μg | 1 day | ↓ CD11b+Gr-1+ cells in the lungs | |||||
| Curcumin | Mouse lymphoma | NA | LPS-induced inflammation model, C57BL/6J mice (intranasal) | 1.5 nM | 2 h | ↓ IL-1β expression in CD45.2 brain microglia | Zhuang et al. (2011)57 |
| EL-4 cells, 22 °C, 5 min co-incubation | Experimental autoimmune encephalomyelitis model, C57BL/6J mice (intranasal) | 1.5 nM | 31 days | ↓ IL-1β expression in CD45.2 brain microglia | |||
| Curcumin | Mouse embryonic stem cells, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 coincubation at RT, 15 min + 3× freeze-thawing 4 coincubation at RT, 15 min + 3× freeze-thawing |
WB (TSG101) | Ischemia-reperfusion injury model in C57BL/6J mice (intranasal) | 10 μL 2× per day | 7 days | ↓ Neurological deficits | Kalani et al. (2016)58 |
| NTa | ↓ Infarct volume | ||||||
| AChE | ↓ Edema | ||||||
| ↓ Inflammation | |||||||
| ↓ Astrogliosis | |||||||
| ↓ GFAP expression | |||||||
| ↑ NeuN | |||||||
| ↓ NMDR1 mRNA | |||||||
| ↓ TNF-α mRNA | |||||||
| ↓ ROS in the brain | |||||||
| ↓ ICAM | |||||||
| ↑ Claudin-5 (BBB) | |||||||
| ↑ Occludin (BBB) | |||||||
| ↑ VE-cadherin (BBB) | |||||||
| Curcumin | Cow milk/coincubation at RT at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 concentration ratio 5 concentration ratio |
DLS | Oral administration to Sprague-Dawley rats | 1.25 or 2.5 mg kg−1 daily | 14 days | ↑ 3–5× higher homing to different organs | Aqil et al. (2017)66 |
| AFM | Oral gavage, cervical CaSki tumor xenograft model, athymic nude mice | 20 mg kg−1 and 80 mg kg−1 curcumin and exosomes | 7-8 weeks | ↓ Tumor growth | |||
| Curcumin | Murine bone marrow-derived MSCs/c(RGDyK)-conjugated exosomes via click-chemistry | EM | Cerebral ischemia (middle cerebral artery occlusion) model, C57BL/6J mice (intravenous) | 100 μg, 12 h after reperfusion | 12 h (mRNA) or 24 h (protein) | ↓ IL-1β mRNA | Tian et al. (2018)62 |
| WB (TSG101 and Alix) | ↓ TNF-α mRNA | ||||||
| AFM | ↓ IL-6 mRNA | ||||||
| NTA | ↓ Caspase-3 | ||||||
| ↓ p-p65 | |||||||
| ↓ Microgliosis (within the lesioned brain region) | |||||||
| Curcumin | Mouse Raw264.7 cells with a RGERPPR peptide (RGE)-tagged exosomes via click-chemistry, loading by electroporation | WB (CD63) | Orthotopic glioma-bearing model, 5-week-old BALB/c nude female mice (intravenous) | 800 μg curcumin/200 μg exosome | Every 2 days (7×) | ↑ Targeting to glioma site | Jia et al. (2018)69 |
| EM | ↑ Survival | ||||||
| NTA | ↓ Tumor volume | ||||||
| Resveratrol | Primary microglia from spinal cords of fetal rats, co-incubation for 24 h (40 μM) | TEM | Spinal cord injury model, 16-week-old Sprague-Dawley rats (intraperitoneal) | 100 mg kg−1 resveratrol | 14 days | ↑ BBB resveratrol permeability | Fan et al. (2020)70 |
| DLS | 0.2 mL of resveratrol-loaded exosomes after injury | ↑ Functional motor recovery | |||||
| WB (CD63, CD81, TSG101, and GRP94) | ↑ Autophagy | ||||||
| ↓ Apoptosis | |||||||
| (Poly)phenol | Model Target | Exosomes Characterization | Dose | Exposure Duration | Upregulated in exosomes | Downregulated in exosomes | Ref. |
|---|---|---|---|---|---|---|---|
| Abbreviations: ACTB: actin beta, ACTN4: alpha-actinin-4, C3: complement component 3, CD: cluster of differentiation, CLIC1: chloride intracellular channel protein 1, EEF1G. elongation factor 1-gamma, EM: electron microscopy, ENO1: alpha-enolase, FACS: fluorescence-activated cell sorting, FTL: ferritin light chain, GAPDHS: glyceraldehyde-3-phosphate dehydrogenase, GNAI3: guanine nucleotide-binding protein G(i) subunit alpha-3, GNAS: guanine nucleotide-binding protein G(s) subunit alpha isoforms short, GNB2: guanine nucleotide-binding protein subunit beta-2-like 1, H2AC18: histone H2A type 2-A, H2AX: histone H2AX, H4C1: histone H4, HIST2H2BE: histone H2B type 2-E, HSP90AA1: heat shock protein HSP 90-alpha, HSP90AB1: heat shock protein HSP 90-beta, HSP90AB2P: putative heat shock protein HSP 90-beta 2, IF: immunofluorescence, IFITM1: interferon induced transmembrane protein 1, KRT14: keratin, type I cytoskeletal 14, KRT18: keratin, type I cytoskeletal 18, LTF: lactotransferrin, MARCKSL1: MARCKS-related protein, MDK: Midkine, NPM1: nucleophosmin, NTA: nanoparticle tracking analysis, PLTP: phospholipid transfer protein, RAP1B: ras-related protein Rap-1b precursor, REEP6: receptor expression-enhancing protein 6, RPL12: 60S ribosomal protein L12, RPL15: ribosomal protein L15, RPL27: 60S ribosomal protein L27, RPS4X: 40S ribosomal protein S4, X isoform, RPSA: 40S ribosomal protein SA, SLC3A2: 4F2 cell-surface antigen heavy chain, SLC7A5: large neutral amino acids transporter small subunit 1, TCF21: transcription Factor 21, TRAP1: TNF receptor associated protein 1, TRPS: tunable resistive pulse sensing, TSG101: tumor susceptibility gene 101, TUBA1B: tubulin alpha-1B chain, WB: western blotting. | |||||||
| Epigallocatechin gallate | Murine breast cancer cell line 4T1 | WB (TSG101 and CD63) | 100 μM | 24 h | ↑ let-7 | ↓ miR-10a | Jang et al. (2013)80 |
| EM | ↑ miR-16 | ↓ miR-18a | |||||
| ↑ miR-18b | ↓ miR-19a | ||||||
| ↑ miR-20a | ↓ miR-26b | ||||||
| ↑ miR-25 | ↓ miR-29b | ||||||
| ↑ miR-92 | ↓ miR-34b | ||||||
| ↑ miR-93 | ↓ miR-98 | ||||||
| ↑ miR-221 | ↓ miR-129 | ||||||
| ↑ miR-320 | ↓ miR-181d | ||||||
| Curcumin | Human non-small cell lung carcinoma | TRPS | 10 μM | 48 h | ↑ TCF21 mRNA | Wu et al. (2016)81 | |
| H1299 cells | |||||||
| Curcumin | Human chronic myelogenous leukemia K562 cells | NA | 20 μM | 24 h | ↑ miR-21 | ↓ H4C1 | Taverna et al. (2016)64 |
| ↑ PLTP (B3KUE5) | ↓ CLIC1 | ||||||
| ↑ MDK (E9PPJ5) | ↓ TUBA1B | ||||||
| ↑ Pleckstrin (P08567) | ↓ NPM1 | ||||||
| ↑ KRT14 (P02533) | ↓ ENO1 | ||||||
| ↓ HSP90AB1 | |||||||
| ↓ H2AC18 | |||||||
| ↓ HIST2H2BE | |||||||
| ↓ RPS4X | |||||||
| ↓ EEF1G | |||||||
| ↓ RPL15 | |||||||
| ↓ SLC7A5 | |||||||
| ↓ RPL27 | |||||||
| ↓ MARCKSL1 | |||||||
| ↓ HSP90AA1 | |||||||
| ↓FTL | |||||||
| ↓ GAPDHS | |||||||
| ↓ RPSA | |||||||
| ↓ GNB2 | |||||||
| ↓ Basigin | |||||||
| ↓ REEP6 | |||||||
| ↓ SLC3A2 | |||||||
| ↓ CD81 antigen | |||||||
| ↓ ACTN4 | |||||||
| ↓ RPL12 | |||||||
| ↓ IFITM1 | |||||||
| Resveratrol | Human glioblastoma U251 cells | EM | 100 μM | 48 h | ↑Exosome release | ↓ KRT18 | Nie et al. (2019)74 |
| NTA | ↑ RAP1B | ↓ H2AX | |||||
| WB (CD63) | ↑ ACTB | ↓ LTF | |||||
| ↑ GNAS | |||||||
| ↑ GNAI3 | |||||||
| Human glioblastoma LN428 cells | 100 μM | 48 h | ↑ Exosome release | ↓ C3 | |||
| ↑ KRT18 | ↓ RAP1B | ||||||
| ↑ H2AX | ↓ ACTB | ||||||
| ↓ TRAP1 | |||||||
| ↓ HSP90AB2P | |||||||
| Curcumin | HepG2 hepatocarcinoma cells treated with antipsychotics and loaded with LDL | EM | 30 μM | 2 h | ↑ Exosome release | Canfrán-Duque et al. (2015)82 | |
| WB (CD63) | ↑ CD63 expression in Exos | ||||||
| FACS (CD63) | |||||||
| Curcumin | Rat C6 glial cells | NTA | 30 μM | 4 h | ↑ Exosome release | García-Seisdedos et al. (2020)75 | |
| WB (CD63) | ↑ Ceramide | ||||||
| FACS (CD63) | ↑ Hexosylceramide | ||||||
| ↑ Sphingomyelin | |||||||
| ↑ Free cholesterol | |||||||
| ↑ Cholesterol ester | |||||||
| Magniferin | Perivascular adipose tissue cells | EM | 0.1, 1, 10 μM | 2 h | ↑ Exosome release | Zhao et al. (2019)83 | |
| IF (CD63) | |||||||
| Hydroxytyrosol | Human Simpson-Golabi-Behmel syndrome (SGBS) preadipocytes | NA | 10 μM | 1 h | ↑let-7c | ↓ miR-34a | Scoditti et al. (2019)77 |
| ↓ miR-155 | |||||||
| Oleocanthal | Human SGBS preadipocytes | NA | 25 μM | 6 h | ↑let-7c | ↓ miR-34a | Carpi et al. (2019)76 |
| ↓ miR-155 | |||||||
| Oleacein | Human SGBS preadipocytes | NA | 25 μM | 6 h | ↑let-7c | ↓ miR-34a | Carpi et al. (2019)76 |
| ↓ miR-155 | |||||||
3. Exosomes as direct dietary (poly)phenols nanocarriers
Exosomes have been suggested as delivery systems of dietary (poly)phenols for several reasons, including their endogenous nature and biocompatibility, their nanoscale size and capacity to cross host barriers, as the blood–brain barrier (BBB),52 their negative zeta potential, and relatively long circulation time,53 as well as their hydrophobicity.54 Most of these studies have focused on the incremental effects of (poly)phenol compound encapsulation within exosomes followed by in vitro exposure (Table 1) or in vivo administration (Table 2).Except for two studies with epicatechin gallate (ECG) and resveratrol, most of the work published so far regarding in vitro evaluation of the putative modulating effects of (poly)phenol compound encapsulation within exosomes have used curcumin (Table 1). Likewise, most in vivo studies reported so far were conducted with curcumin as well (Table 2). As a general trend, encapsulation of these parent compounds does seem to increase their stability and the overall amount taken up by exposed cells. Furthermore, curcumin encapsulation seems to increase apoptosis and decrease proliferation and cell viability, while opposite effects have been reported for encapsulated ECG in a rotenone-induced Parkinson's disease model in vitro. However, different cell models have been employed as readouts, which precludes any valid comparison (Table 1).
In vivo, exosomes were first exploited as delivery systems for (poly)phenolic compounds approximately one decade ago using curcumin, whereby both increased plasma concentration and higher bioavailability were observed. In the first study, intraperitoneal administration of curcumin at 100 mg per kg body weight achieved a plasma concentration of 1.25 μg mL−1 at 30 min post-administration, a concentration 5 to 10-fold higher than that achieved with intraperitoneal administration of curcumin alone.55 Since then, the increased bioavailability and stability of curcumin upon in vitro exosomal encapsulation has been confirmed either using in vitro permeability studies and tumor cells antiproliferative potential (Table 1) or in vivo studies upon oral administration (Table 2). In this regard, recently, the encapsulation of curcumin and resveratrol in milk-derived exosomes was reported to enhance their in vivo bioavailability and breast tissue disposition in rats, compared to the administration of similar concentrations of both free forms. Besides, a high antiproliferative activity mediated by cell cycle arrest and apoptosis induction against breast cancer models of representative concentrations (nanomolar) reached in breast tissue was observed when they were loaded into exosomes but not with the equivalent free polyphenol concentrations.56
Regarding their efficacy in delivering curcumin to the brain and putative anti-inflammatory effects, a single treatment consisting of repeated intranasal administration of curcumin-containing exosomes (1.5 nmol) for 10 min, was effective at reducing the number of activated inflammatory microglial cells (CD45.2+, IL-1β+) via apoptosis induction in the brain of LPS-challenged mice (single IP injection of 2.5 mg kg−1).57 Likewise, the same study revealed that curcumin-containing exosomes intraperitoneally administered daily for 26 days (1.5 nmol) significantly reduced disease severity in a mouse model of encephalomyelitis compared to exosomes alone or curcumin alone, an effect accompanied by decreased IL-1β expression in microglial cells after the one-month-long treatment period.57 Intranasal administration allowed exosomes to reach the brain within 1 h post-administration, suggesting perineuronal and(or) perivascular channels as transportation routes. Furthermore, repeated administration of curcumin-containing exosomes every 12 h maintained a constant curcumin concentration at ∼2.6 ± 0.4 nmol g−1 of brain tissue. In the brain, the exosomes used (EL-4 exosomes from T cell lines) were taken up by both microglial (∼60%) and non-microglial cells (∼40%), and both resting and activated microglia were observed to take exosomes up.57 No apparent toxicity or behavioral deficits could be observed after one month of treatment.
Exosomes from mouse embryonic stem cells loaded with curcumin significantly reduced neurological deficits, infarct volume, edema, inflammation, astrogliosis, and N-methyl-D-aspartate receptor 1 expression upon intranasal administration (2× per day) for 7 days in a mouse model of ischemia-reperfusion injury. These curcumin-containing exosomes were located In astrocytes, neurons, and blood vessels.58 In the blood vessels, the expression of vascular endothelial tight (claudin-5 and occludin) and adherens junction (VE-cadherin) proteins could also be rescued by the treatment.58 However, as highlighted by the authors, the simple administration of exosomes from embryonic stem cells had profound neuroprotective effects in similar settings,59–61 but no exosomes group without curcumin was provided as a control in the aforementioned study.
In turn, in a mouse model of cerebral ischemia, curcumin-containing engineered c(RGDyK)-conjugated exosomes (cRGD-Exo, 100 μg) targeting the brain ischemic brain regions (high in αvβ3 integrin) administered 12 h after reperfusion reduced the levels of TNF-α, IL-1β and IL-6 compared to intravenously administered curcumin alone, and those of caspase-3 and phosphorylated p65 compared to either exosomes or curcumin alone.62
These results show that exosomes can be engineered to target specific organs as shuttling vehicles/nanocarriers predominantly. Most studies performed to date have been focused on delivery to the brain, which reflects both the recent hype associated with nutrition-based attenuation of neuroinflammation and the inherent challenges associated with drug delivery across the blood–brain barrier. Overall, even though the above-discussed results do support the efficacy of exosomes as protective shuttles, these fail to mimic active sorting and delivery mechanisms. Moreover, if exosomes actually have any shuttling function for (poly)phenolic compounds under (patho)physiological conditions remains poorly elucidated so far. Nevertheless, in head and neck cancer patients’ serum-derived EVs, particular small metabolites were found to be markedly downregulated compared to healthy controls, like citric acid and 4-hydroxybenzoic acid,71 important metabolomic alterations in exosome cargo observed in pathological conditions. This observation highlights that small compounds, despite chemically different, can comprise valid exosome payloads in addition to proteins, nucleic acids or other metabolites like glucose, glutamate, etc.72,73 Furthermore, how much of the benefit results from the administration of exosomes (as opposed to polyphenol-containing exosomes) is frequently not discriminated. Notably, to the best of our knowledge, no studies have been performed to date using circulating (poly)phenol metabolites in exosome-loading experiments, but this ought to be addressed as circulating (poly)phenol metabolites are the molecules found in vivo at relevant circulating concentrations.
4. Indirect (poly)phenol-triggered exosome-mediated responses
(Poly)phenol compounds may also trigger exosome-mediated cellular responses that do not necessarily require the presence of the parent (poly)phenols (or their metabolites) inside exosomes (Table 3). On the one hand, (poly)phenols may enhance the release of exosomes, which can thus boost their effects. Conversely, (poly)phenols may alter the content of exosomes at multiple different levels (e.g., proteome, transcriptome, lipidome). This question has only been addressed in cell-based systems to the best of our knowledge.Exosomes from drug-sensitive tumour cells exposed to resveratrol (100 μM) for 48 h (but not exosomes from resveratrol-naïve cells) have their protein content altered, which allows for enhanced resveratrol sensitivity of otherwise resistant cells upon transfer of these primed exosomes.74 In turn, the mechanism by which curcumin stimulates exosome release has been shown in rat C6 glial cells to comprise alterations in ceramide synthesis and may thus be associated with alterations in their lipid profile. Accordingly, curcumin (30 μM) exposure for 4 h increased the content of ceramide, hexosylceramide, sphingomyelin, free cholesterol, and cholesterol ester in exosomes,75 which in and of itself may lead to significant biological responses.
Alterations at the level of anti-inflammatory and pro-inflammatory mediators triggered by (poly)phenols at the cellular level are mirrored and propagated in the exosomes released by the cells directly exposed to (poly)phenols.76 For instance, the deregulation of miRNAs (e.g., miR-155-5p, miR-34a-5p, let-7c-5p), NF-κB, and accompanying inflammatory mediators induced by TNF-α (10 ng mL−1 for 18 h) in adipocytes could be observed at the cellular and exosomal levels. Likewise, the reversal brought about by the phenolic secoiridoids oleocanthal and oleacein (25 μM for 6 h) was reflected on both cellular and exosomal compartments.76
In addition to the studies summarized in Table 3, exosomes derived from microglia treated with resveratrol (40 μM) for 24 h have been shown to significantly protect resveratrol from degradation in PBS and plasma (with 40% vs. 70% intact molecules at 2 h post-incubation, respectively).70 In a spinal cord injury model, exosomes secreted by macrophage-like cells were assumed to maximize cargo uptake due to the presence of lymphocyte function-associated antigen 1 (which biases the homing towards the brain vasculature) and used as vehicles for resveratrol. Animals received either resveratrol alone (100 mg kg−1) or resveratrol-primed exosomes (40 μM) for 14 days.70 Compared to free resveratrol alone, intraperitoneal injection of resveratrol-primed exosomes significantly increased hind limb muscle tension and “foot functional movements” in rats after spinal cord injury at days 7, 14, and 28 post-injury, a functional recovery mirrored by increased numbers of spinal anterior horn motor neurons.70 In this model, neuronal viability was attributed to increased autophagy (LC3B and Beclin-1) and diminished apoptosis (Caspase-3 and TUNEL), with the former being inhibited by 3-ethyladenine and thus mediated by the PI3K signaling pathway.70 Notably, to the best of our knowledge, only one single study has analyzed the direct effects of a circulating (poly)phenol metabolite on exosomes in vitro, whereby 10 μM hydroxytyrosol attenuated the inflammatory response mediated by miR-155-5p, miR-34a-5p, and let-7c-5p expression in both cells and exosomes in an obesity model.77
Regarding humans, in elderly subjects, it was shown that the consumption of walnuts, a food naturally rich in (poly)phenolic compounds, despite did not influence the size and concentration of exosomes, hsa-miR-32-5p and hsa-miR-29b-3p exosomal levels were consistently induced by walnut consumption.78
Overall, the aforementioned beneficial effects of primed exosomes’ administration favor indirect (over direct) effects of (poly)phenols. Nevertheless, it is still not clear if the effects observed result from the ingested phenolics and(or) their microbial metabolites, and in both cases, if the phase-II conjugates participate actively, or if the effects are mediated by indirect signaling cascades where it is not necessary the direct interaction of the molecule with the systemic target.14 It is thus tempting to suggest the anti-inflammatory effects of (poly)phenolic compounds to be (at least for the most part) mediated indirectly over long distances in a “billiard-like” or “domino-like” propagating effect.14,79
5. (Lack of) evidence for the encapsulation of (poly)phenols within exosomes upon oral administration
Even though available evidence does support significant modulation of the effects of (poly)phenol compounds upon encapsulation within exosomes, no data supporting such encapsulation to occur in vivo under physiological conditions has been put forward to date. Nevertheless, exosomes are carriers of all the main biomolecules, including lipids, proteins, metabolites, DNAs, messenger RNAs, and microRNA. On the other hand, it is widely acknowledged that (poly)phenols can specifically interact with protein targets and modulate signaling and metabolic pathways relevant to several disorders.84,85 Indeed, a computationally-driven analysis of (poly)phenol-binding proteins has predicted 369 (poly)phenols to interact with 5699 unique human proteins using publicly available data.84 Across all (poly)phenol-protein interactions, flavones, hydroxybenzoic acids, and alkylphenols were the sub-classes with the largest number of protein interactions.84 More than half of these were mediated by quercetin (2500 interactions), coumestrol (1802 interactions), genistein (916 interactions), trans-resveratrol (738 interactions), and acetylsalicylic acid (510 interactions), with 65% of the (poly)phenols having more than 10 reported (poly)phenol-protein interactions. Furthermore, while most (poly)phenol-interacting proteins (95%) interacted with less than 5 (poly)phenols, certain proteins (e.g., ABC-transporters, lipoxygenases, and estrogen receptors) were reported to interact with at least 50 (poly)phenols.84We compared the aforementioned dataset of (poly)phenol-binding proteins with known human plasma exosomal proteins listed in the ExoCarta repository at the end of September 2021.86 The aim was to gather some cues regarding a putative overlap between (poly)phenol-binding proteins and exosomal proteins. We observed an overlap of approximately 1500 proteins (Fig. 2a), suggesting that the specificity of (poly)phenol-binding proteins in humans seems to be rather low, which complicates the task of pinpointing which exosome proteins (if any) might sequester or otherwise bind to (poly)phenolic compounds within exosomes. In turn, it should be highlighted that very little is known regarding the targets of the circulating (poly)phenol metabolites produced upon host- and microbiota-mediated (poly)phenol compound metabolism. Here, once again, specificity seems low, and redundancy high (with circa 60 circulating (poly)phenol metabolites predicted to bind more than 300 human exosomal proteins) when querying about the exosomal “targets” or putative protein shuttles of circulating (poly)phenol metabolites (Fig. 2b). Notwithstanding this limitation, when probing whether exosomal circulating (poly)phenol metabolites-binding proteins are significantly biased towards any given biological phenomenon, one observes that compared to all human metabolite-binding proteins, those within exosomes are significantly enriched in proliferation and survival regulating processes, protein complexes mediating apoptosis, inflammation, and tissue remodeling signaling pathways (Fig. 2c, d, and e). Despite the interesting results obtained in our analysis, the amount of protein interactions of (poly)phenol and of (poly)phenol metabolites obtained are, in our opinion, still insufficient and not fully representative of the complexity of exosomal proteins and (poly)phenols/(poly)phenol metabolites known nowadays to draw robust conclusions. It will be very important to pinpoint, for instance, if particular chemical structures like sulfate, glucuronide, methoxy, or even if the number of free hydroxyl groups in the phenolic ring could influence the binding interactions with plasma exosomal proteins. We believe that a similar analysis to the one here presented using updated and more comprehensive data in the future should be pursued to fully elucidate such paramount questions.
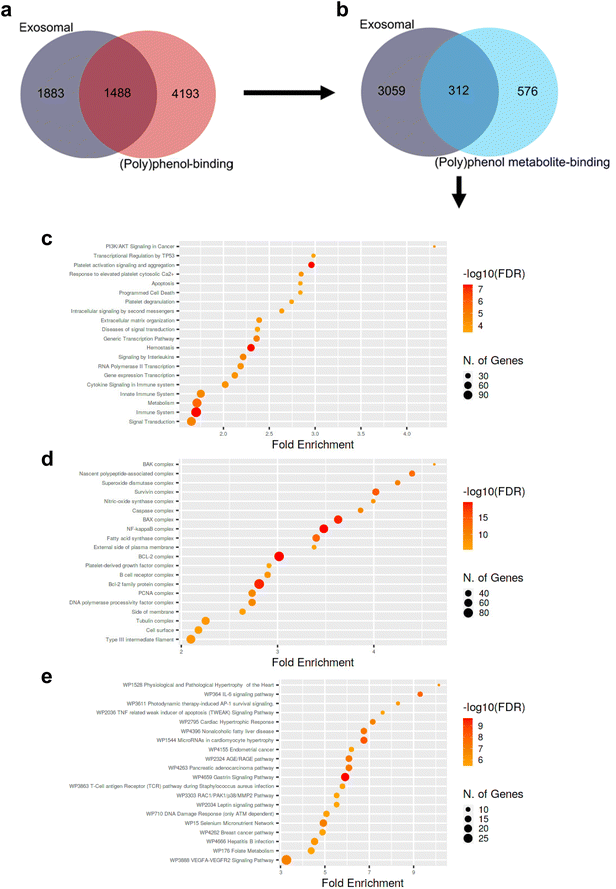 | ||
| Fig. 2 Venn diagrams depicting the overlap between human exosomal proteins cataloged in ExoCarta and human polyphenol-binding (a) or circulating (poly)phenol metabolite-binding proteins (b) put forward by Lacroix et al. 2018.84 Dot plots depicting significantly enriched biological processes (c), protein complexes and subcellular localization (d), and signaling pathways (e) involving human exosomal circulating (poly)phenol metabolites-binding proteins (i.e., 312) compared to all human circulating (poly)phenol metabolites-binding proteins (i.e., 576 + 312). Circle areas are proportional to the number of genes involved. Redder circles represent smaller p-values than the remaining/parallel processes/pathways. Fold enrichment corresponds to the percentage of exosomal circulating (poly)phenol metabolites-binding proteins belonging to each pathway/process/location divided by the corresponding percentage of human circulating (poly)phenol metabolites-binding proteins belonging to that same pathway/process/location, indicating to which extent proteins in the numerator are overrepresented compared to the denominator. Data analyzed according to Ge et al. 202089 and as detailed in ESI.† All p-values < 0.05. FDR: false discovery rate; WP: wikipathway. | ||
Alternatively, to try to pinpoint differences in nature between exosomal and non-exosomal circulating (poly)phenol metabolites protein “targets” (i.e., qualitative differences), an attempt has also been made to measure the exosomal quantitative load in (poly)phenol metabolites. One study has recently investigated whether proanthocyanidins, their gut microbiota-derived and(or) the respective phase II enzymes-derived metabolites could be found within exosomes from rat plasma 3 h or 7 h after oral gavage with 250 mg kg−1 of grape seed proanthocyanidin extract: only residual amounts of (poly)phenolic metabolites derived from both host and microbiota metabolism could be detected by LC-MS/MS, even though sensitivity was not a limiting factor.87 Indeed, these minor amounts found in the exosomes fraction could be simply due to passive diffusion. One could argue that the relative amount of (poly)phenolic compounds and their derivates found in the exosomes fraction (∼6% of phase II metabolites, ∼7% of non-metabolized compounds, ∼8% of all compounds, 12% of microbial-derived metabolites, and 15% of the mixed origin compounds in the exosomes fraction) is considerably high when taking into account the much smaller percentage of circulating volume occupied by exosomes under normal conditions (<1%). Such observation could theoretically point against simple passive diffusion phenomena. However, the grape seed proanthocyanidin extract used was shown to increase the number of extracellular vesicles released.87 Furthermore, exosomes precipitation induces protein precipitation and thus leads to artifactual findings in the otherwise “exosomal” fractions.87 Accordingly, plasma proteins, lipoproteins, and red blood cell constituents have been previously shown to bind to (poly)phenolic compounds88 When ultracentrifugation was employed as the isolation method in the aforementioned study, no (poly)phenols could be found in the exosomal compartment. Overall, there is still insufficient evidence to support the physiological role of exosomes as nanocarriers of polyphenols in vivo,87 an observation that, for the time being, seems to hold for both parent compounds and respective metabolites.
Conclusions
While the currently available literature fails to support a hypothetical scenario where exosomes could function as (poly)phenol nanocarriers under physiological (or pathological) conditions, it is possible for the formation and loading of exosomes in vivo to be responsive to exogenous (poly)phenol administration and to have substantial effects across a plethora of biological phenomena. As such, it is plausible to assume that some of the effects triggered by (poly)phenols in vivo are exosome-mediated. Accordingly, because in vitro studies suggest that (poly)phenols can modulate exosomes at both quantitative and qualitative (i.e., lipidome, proteome, microtranscriptome) levels, exosomes may allow for the propagation of (poly)phenol-triggered responses over longer distances and for more extended periods. For the time being, the most likely conciliating explanations for the presented studies encompass quantitative and qualitative changes in exosomes triggered by (poly)phenols administration (Fig. 3). Nonetheless, the most pressing issues to be addressed in the near future should be highlighted:- How does the content of exosomes derived from (poly)phenol metabolite-exposed cells differ at the protein, lipid, nucleic acid, and metabolite levels across different tissues and cell types?
- Is this indirect communication and transportation route of relevance enough to justify in vivo manipulation/hijacking when aiming at modulating host (patho)physiology?
- If so, which genetic, epigenetic, microbiota, and health status background is likely to benefit from different exosome-polyphenols combinations?
- How do the hormetic effects of (poly)phenols fit into the (poly)phenol-exosome interplay?
- Which stress, survival, or otherwise biological cues are being transferred by exosomes upon (poly)phenols exposure?
- How do these cues differ across the host organs, and how are they altered across human diseases?
The breath of these questions brings to the spotlight how far-reaching the biological impact resulting from the putative interplay between exosomes, (poly)phenols, and their circulating metabolites may be.
Author contributions
Conceptualization: PB, IF and CNS; Investigation PB and IF; Formal analysis: PB and IF; Writing – original draft: PB and IF; Review & editing: PB; IF, AGS, JCE, BCS and CNS; Visualization: PB; Supervision: CNS; Project administration: CNS; Funding acquisition: CNS.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme under grant agreement No 804229. This work was also supported by “iNOVA4Health – UIDB/04462/2020 and UIDP/04462/2020, and by the Associated Laboratory LS4FUTURE (LA/P/0087/2020), two programs financially supported by Fundação para a Ciência e Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior. This work has also been funded by the project PID2019-103914RB-I00 from the Ministry of Science and Innovation (MICINN, Spain). We also acknowledge EMBO Installation Grant 3921, the grant 2017NovPCC1058 from Breast Cancer Now's Catalyst Programme, which is supported by funding from Pfizer, the grant 765492 from H2020-MSCA-ITN-2017, the Champalimaud Foundation and the grant LCF/PR/HR19/52160014 from “La Caixa” Foundation.Notes and references
- C. Handschin, Pharmacol. Res., 2016, 103, 158–166 CrossRef PubMed.
- Z. Radak, H. Y. Chung and S. Goto, Free Radicals Biol. Med., 2008, 44, 153–159 CrossRef CAS PubMed.
- E. Verdin, Science, 2015, 350, 1208–1213 CrossRef CAS PubMed.
- J. C. Newman and E. Verdin, Trends Endocrinol. Metab., 2014, 25, 42–52 CrossRef CAS PubMed.
- E. J. Calabrese, Crit. Rev. Toxicol., 2001, 31, 425–470 CrossRef CAS PubMed.
- T. G. Son, S. Camandola and M. P. Mattson, NeuroMol. Med., 2008, 10, 236–246 CrossRef CAS PubMed.
- K. Takahashi, S. Yanai, K. Shimokado and A. Ishigami, Nutrition, 2017, 38, 1–8 CrossRef CAS PubMed.
- M. Concetta Scuto, C. Mancuso, B. Tomasello, M. Laura Ontario, A. Cavallaro, F. Frasca, L. Maiolino, A. Trovato Salinaro, E. J. Calabrese and V. Calabrese, Nutrients, 2019, 11(10), 2417 CrossRef PubMed.
- C. Zhang, C. Li, S. Chen, Z. Li, L. Ma, X. Jia, K. Wang, J. Bao, Y. Liang, M. Chen, P. Li, H. Su, S. M. Y. Lee, K. Liu, J.-B. Wan and C. He, Sci. Rep., 2017, 7, 41082 CrossRef CAS PubMed.
- J. Kanner, Antioxidants, 2020, 9, 797 CrossRef CAS PubMed.
- A. Singh, Y. F. Yau, K. S. Leung, H. El-Nezami and J. C.-Y. Lee, Antioxidants, 2020, 9(8), 669 CrossRef CAS PubMed.
- I. Santecchia, F. Vernel-Pauillac, O. Rasid, J. Quintin, M. Gomes-Solecki, I. G. Boneca and C. Werts, PLoS Pathog., 2019, 15, e1007811 CrossRef CAS PubMed.
- D. Del Rio, A. Rodriguez-Mateos, J. P. E. Spencer, M. Tognolini, G. Borges and A. Crozier, Antioxid. Redox Signal., 2013, 18, 1818–1892 CrossRef CAS PubMed.
- C. E. Iglesias-Aguirre, A. Cortés-Martín, M. Á. Ávila-Gálvez, J. A. Giménez-Bastida, M. V. Selma, A. González-Sarrías and J. C. Espín, Food Funct., 2021, 12, 10324–10355 RSC.
- A. Dey, P. Guha, S. Chattopadhyay and S. K. Bandyopadhyay, Biochem. Biophys. Res. Commun., 2009, 381, 90–95 CrossRef CAS PubMed.
- M. C. Maiuri and G. Kroemer, Cell Death Differ., 2019, 26, 680–689 CrossRef PubMed.
- E. J. Calabrese, E. Agathokleous, W. J. Kozumbo, E. J. Stanek and D. Leonard, Environ. Res., 2019, 170, 337–343 CrossRef CAS PubMed.
- D. Carregosa, R. Carecho, I. Figueira and C. N. Santos, J. Agric. Food Chem., 2020, 68, 1790–1807 CrossRef CAS PubMed.
- D. Carregosa, C. Pinto, M. Á. Ávila-Gálvez, P. Bastos, P. Berry and C. Nunes dos Santos, Compr. Rev. Food Sci. Food Saf., 2022 DOI:10.1111/1541-4337.13006.
- C. D. Kay, G. Pereira-Caro, I. A. Ludwig, M. N. Clifford and A. Crozier, Annu. Rev. Food Sci. Technol., 2017, 8, 155–180 CrossRef CAS PubMed.
- A. J. Day, F. J. Cañada, J. C. Díaz, P. A. Kroon, R. Mclauchlan, C. B. Faulds, G. W. Plumb, M. R. Morgan and G. Williamson, FEBS Lett., 2000, 468, 166–170 CrossRef CAS PubMed.
- A. González-Sarrías, R. García-Villalba, M. Romo-Vaquero, C. Alasalvar, A. Örem, P. Zafrilla, F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, Mol. Nutr. Food Res., 2017, 61(5), 1–14 CrossRef PubMed.
- A. Cortés-Martín, M. V. Selma, F. A. Tomás-Barberán, A. González-Sarrías and J. C. Espín, Mol. Nutr. Food Res., 2020, 64, e1900952 CrossRef PubMed.
- J. C. Espín, M. Larrosa, M. T. García-Conesa and F. Tomás-Barberán, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 270418 Search PubMed.
- J. C. Espín, A. González-Sarrías and F. A. Tomás-Barberán, Biochem. Pharmacol., 2017, 139, 82–93 CrossRef PubMed.
- A. González-Sarrías, J. C. Espín and F. A. Tomás-Barberán, Trends Food Sci. Technol., 2017, 69, 281–288 CrossRef.
- R. Soleti, R. Andriantsitohaina and M. C. Martinez, Arch. Biochem. Biophys., 2018, 644, 57–63 CrossRef CAS PubMed.
- D. Wei, W. Zhan, Y. Gao, L. Huang, R. Gong, W. Wang, R. Zhang, Y. Wu, S. Gao and T. Kang, Cell Res., 2021, 31, 157–177 CrossRef CAS PubMed.
- T. Matsui, F. Osaki, S. Hiragi, Y. Sakamaki and M. Fukuda, EMBO Rep., 2021, 22, e51475 CrossRef CAS PubMed.
- C. Théry, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer and G. K. Atkin-Smith, et al. , J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Théry, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E968–E977 CrossRef CAS PubMed.
- W. Li, C. Li, T. Zhou, X. Liu, X. Liu, X. Li and D. Chen, Mol. Cancer, 2017, 16, 145 CrossRef PubMed.
- Z. Sun, K. Shi, S. Yang, J. Liu, Q. Zhou, G. Wang, J. Song, Z. Li, Z. Zhang and W. Yuan, Mol. Cancer, 2018, 17, 147 CrossRef PubMed.
- J. Elzanowska, C. Semira and B. Costa-Silva, Mol. Oncol., 2021, 15, 1701–1714 CrossRef CAS PubMed.
- C. Williams, M. Palviainen, N.-C. Reichardt, P. R.-M. Siljander and J. M. Falcón-Pérez, Metabolites, 2019, 9, 276 CrossRef CAS PubMed.
- A. Zebrowska, A. Skowronek, A. Wojakowska, P. Widlak and M. Pietrowska, Int. J. Mol. Sci., 2019, 20(14), 3461 CrossRef CAS PubMed.
- M. Z. Ratajczak and J. Ratajczak, Leukemia, 2020, 34, 3126–3135 CrossRef PubMed.
- J. Maia, S. Caja, M. C. Strano Moraes, N. Couto and B. Costa-Silva, Front. Cell Dev. Biol., 2018, 6, 18 CrossRef PubMed.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS PubMed.
- S. H. Saber, H. E. A. Ali, R. Gaballa, M. Gaballah, H. I. Ali, M. Zerfaoui and Z. Y. Abd Elmageed, Cells, 2020, 9(3), 564 CrossRef CAS PubMed.
- H. Wei, Q. Chen, L. Lin, C. Sha, T. Li, Y. Liu, X. Yin, Y. Xu, L. Chen, W. Gao, Y. Li and X. Zhu, Int. J. Biol. Sci., 2021, 17, 163–177 CrossRef CAS PubMed.
- L. He and G. J. Hannon, Nat. Rev. Genet., 2004, 5, 522–531 CrossRef CAS PubMed.
- V. Ambros, Nature, 2004, 431, 350–355 CrossRef CAS PubMed.
- A. Forterre, A. Jalabert, K. Chikh, S. Pesenti, V. Euthine, A. Granjon, E. Errazuriz, E. Lefai, H. Vidal and S. ROME, Cell Cycle, 2014, 13, 78–89 CrossRef CAS PubMed.
- M. N. Poy, L. Eliasson, J. Krutzfeldt, S. Kuwajima, X. Ma, P. E. MacDonald, S. Pfeffer, T. Tuschl, N. Rajewsky, P. Rorsman and M. Stoffel, Nature, 2004, 432, 226–230 CrossRef CAS PubMed.
- P. Qiu, J. Zhou, J. Zhang, Y. Dong and Y. Liu, Front. Pharmacol., 2021, 12, 671164 CrossRef CAS PubMed.
- R. Xue, W. Tan, Y. Wu, B. Dong, Z. Xie, P. Huang, J. He, Y. Dong and C. Liu, Front. Cardiovasc. Med., 2020, 7, 592412 CrossRef CAS PubMed.
- W. Liu, X. Bai, A. Zhang, J. Huang, S. Xu and J. Zhang, Front. Mol. Neurosci., 2019, 12, 240 CrossRef CAS PubMed.
- N. Couto, S. Caja, J. Maia, M. C. Strano Moraes and B. Costa-Silva, Biochimie, 2018, 155, 2–10 CrossRef CAS PubMed.
- Z. Zhang, J. A. Dombroski and M. R. King, Cell. Mol. Bioeng., 2020, 13, 1–16 CrossRef CAS PubMed.
- C. Wang, L. Chen, Y. Huang, K. Li, A. Jinye, T. Fan, R. Zhao, X. Xia, B. Shen, J. Du and Y. Liu, Oncol. Lett., 2019, 17, 1953–1961 CAS.
- P. Vader, E. A. Mol, G. Pasterkamp and R. M. Schiffelers, Adv. Drug Delivery Rev., 2016, 106, 148–156 CrossRef CAS PubMed.
- G. Midekessa, K. Godakumara, J. Ord, J. Viil, F. Lättekivi, K. Dissanayake, S. Kopanchuk, A. Rinken, A. Andronowska, S. Bhattacharjee, T. Rinken and A. Fazeli, ACS Omega, 2020, 5, 16701–16710 CrossRef CAS PubMed.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed.
- D. Sun, X. Zhuang, X. Xiang, Y. Liu, S. Zhang, C. Liu, S. Barnes, W. Grizzle, D. Miller and H.-G. Zhang, Mol. Ther., 2010, 18, 1606–1614 CrossRef CAS PubMed.
- A. González-Sarrías, C. E. Iglesias-Aguirre, A. Cortés-Martín, F. Vallejo, A. Cattivelli, L. Del Pozo-Acebo, A. Del Saz, M. C. López de Las Hazas, A. Dávalos and J. C. Espín, Int. J. Mol. Sci., 2022, 23(5), 2860 CrossRef PubMed.
- X. Zhuang, X. Xiang, W. Grizzle, D. Sun, S. Zhang, R. C. Axtell, S. Ju, J. Mu, L. Zhang, L. Steinman, D. Miller and H.-G. Zhang, Mol. Ther., 2011, 19, 1769–1779 CrossRef CAS PubMed.
- A. Kalani, P. Chaturvedi, P. K. Kamat, C. Maldonado, P. Bauer, I. G. Joshua, S. C. Tyagi and N. Tyagi, Int. J. Biochem. Cell Biol., 2016, 79, 360–369 CrossRef CAS PubMed.
- T. R. Doeppner, J. Herz, A. Görgens, J. Schlechter, A.-K. Ludwig, S. Radtke, K. de Miroschedji, P. A. Horn, B. Giebel and D. M. Hermann, Stem Cells Transl. Med., 2015, 4, 1131–1143 CrossRef CAS PubMed.
- M. Chopp and Z. G. Zhang, Expert Opin. Emerging Drugs, 2015, 20, 523–526 CrossRef CAS PubMed.
- H. Xin, Y. Li, Y. Cui, J. J. Yang, Z. G. Zhang and M. Chopp, J. Cereb. Blood Flow Metab., 2013, 33, 1711–1715 CrossRef CAS PubMed.
- T. Tian, H.-X. Zhang, C.-P. He, S. Fan, Y.-L. Zhu, C. Qi, N.-P. Huang, Z.-D. Xiao, Z.-H. Lu, B. A. Tannous and J. Gao, Biomaterials, 2018, 150, 137–149 CrossRef CAS PubMed.
- S. Luo, X. Sun, M. Huang, Q. Ma, L. Du and Y. Cui, J. Agric. Food Chem., 2021, 69, 5134–5143 CrossRef CAS PubMed.
- S. Taverna, S. Fontana, F. Monteleone, M. Pucci, L. Saieva, V. De Caro, V. G. Cardinale, M. Giallombardo, E. Vicario, C. Rolfo, G. De Leo and R. Alessandro, Oncotarget, 2016, 7, 30420–30439 CrossRef PubMed.
- M. Vashisht, P. Rani, S. K. Onteru and D. Singh, Appl. Biochem. Biotechnol., 2017, 183, 993–1007 CrossRef CAS PubMed.
- F. Aqil, R. Munagala, J. Jeyabalan, A. K. Agrawal and R. Gupta, AAPS J., 2017, 19, 1691–1702 CrossRef CAS PubMed.
- G. Carobolante, J. Mantaj, E. Ferrari and D. Vllasaliu, Pharmaceutics, 2020, 12(3), 226 CrossRef CAS PubMed.
- C. J. D. Osterman, J. C. Lynch, P. Leaf, A. Gonda, H. R. Ferguson Bennit, D. Griffiths and N. R. Wall, PLoS One, 2015, 10, e0132845 CrossRef PubMed.
- G. Jia, Y. Han, Y. An, Y. Ding, C. He, X. Wang and Q. Tang, Biomaterials, 2018, 178, 302–316 CrossRef CAS PubMed.
- Y. Fan, Y. Li, S. Huang, H. Xu, H. Li and B. Liu, Neurosci. Lett., 2020, 736, 135262 CrossRef CAS PubMed.
- A. Wojakowska, A. Zebrowska, A. Skowronek, T. Rutkowski, K. Polanski, P. Widlak, L. Marczak and M. Pietrowska, J. Pers. Med., 2020, 10, 229 CrossRef PubMed.
- K. C. Vallabhaneni, P. Penfornis, S. Dhule, F. Guillonneau, K. v. Adams, Y. Y. Mo, R. Xu, Y. Liu, K. Watabe, M. C. Vemuri and R. Pochampally, Oncotarget, 2015, 6, 4953–4967 CrossRef PubMed.
- A. Zebrowska, K. Jelonek, S. Mondal, M. Gawin, K. Mrowiec, P. Widłak, T. Whiteside and M. Pietrowska, Cells, 2022, 11, 1965 CrossRef CAS PubMed.
- J.-H. Nie, H. Li, M.-L. Wu, X.-M. Lin, L. Xiong and J. Liu, Int. J. Mol. Sci., 2019, 20(1), 191 CrossRef PubMed.
- D. García-Seisdedos, B. Babiy, M. Lerma, M. E. Casado, J. Martínez-Botas, M. A. Lasunción, Ó. Pastor and R. Busto, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2020, 1865, 158638 CrossRef PubMed.
- S. Carpi, E. Scoditti, M. Massaro, B. Polini, C. Manera, M. Digiacomo, J. Esposito Salsano, G. Poli, T. Tuccinardi, S. Doccini, F. M. Santorelli, M. A. Carluccio, M. Macchia, M. Wabitsch, R. De Caterina and P. Nieri, Nutrients, 2019, 11(12), 2855 CrossRef PubMed.
- E. Scoditti, S. Carpi, M. Massaro, M. Pellegrino, B. Polini, M. A. Carluccio, M. Wabitsch, T. Verri, P. Nieri and R. De Caterina, Nutrients, 2019, 11(10), 2493 CrossRef CAS PubMed.
- M.-C. López de Las Hazas, J. Gil-Zamorano, M. Cofán, D. C. Mantilla-Escalante, A. Garcia-Ruiz, L. Del Pozo-Acebo, O. Pastor, M. Yañez-Mo, C. Mazzeo, M. Serra-Mir, M. Doménech, C. Valls-Pedret, S. Rajaram, J. Sabaté, E. Ros, A. Sala-Vila and A. Dávalos, Eur. J. Nutr., 2021, 60, 1999–2011 CrossRef PubMed.
- J. Tomé-Carneiro, M. Larrosa, A. González-Sarrías, F. A. Tomás-Barberán, M. T. García-Conesa and J. C. Espín, Curr. Pharm. Des., 2013, 19, 6064–6093 CrossRef PubMed.
- J.-Y. Jang, J.-K. Lee, Y.-K. Jeon and C.-W. Kim, BMC Cancer, 2013, 13, 421 CrossRef PubMed.
- H. Wu, J. Zhou, C. Zeng, D. Wu, Z. Mu, B. Chen, Y. Xie, Y. Ye, J. Liu, H. Wu, J. Zhou, C. Zeng, D. Wu, Z. Mu, B. Chen, Y. Xie, Y. Ye and J. Liu, Oncotarget, 2016, 7, 87081–87090 CrossRef PubMed.
- A. Canfrán-Duque, O. Pastor, M. Reina, M. Lerma, A. J. Cruz-Jentoft, M. A. Lasunción and R. Busto, PLoS One, 2015, 10, e0141829 CrossRef PubMed.
- Q. Zhao, J. Yang, B. Liu, F. Huang and Y. Li, Mol. Med. Rep., 2019, 19, 4797–4805 CAS.
- S. Lacroix, J. Klicic Badoux, M.-P. Scott-Boyer, S. Parolo, A. Matone, C. Priami, M. J. Morine, J. Kaput and S. Moco, Sci. Rep., 2018, 8, 2232 CrossRef PubMed.
- N. Couto, S. Caja, J. Maia, M. C. Strano Moraes and B. Costa-Silva, Biochimie, 2018, 155, 2–10 CrossRef CAS PubMed.
- S. Keerthikumar, D. Chisanga, D. Ariyaratne, H. Al Saffar, S. Anand, K. Zhao, M. Samuel, M. Pathan, M. Jois, N. Chilamkurti, L. Gangoda and S. Mathivanan, J. Mol. Biol., 2016, 428, 688–692 CrossRef CAS PubMed.
- A. Arola-Arnal, M.-C. López de las Hazas, L. Iglesias-Carres, D. C. Mantilla-Escalante, M. Suárez, R. Busto, F. Visioli, C. Bladé and A. Dávalos, Food Funct., 2020, 11, 7784–7792 RSC.
- M. Kaiser, L. Müller-Ehl, M. Passon and A. Schieber, Molecules, 2020, 25(3), 518 CrossRef CAS PubMed.
- S. X. Ge, D. Jung and R. Yao, Bioinformatics, 2020, 36, 2628–2629 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo00876a |
| This journal is © The Royal Society of Chemistry 2023 |