Open Access Article
Open Access ArticleAntifungal activity and mechanisms of AgNPs and their combination with azoxystrobin against Magnaporthe oryzae†
Huanbin
Shi
 a,
Hui
Wen
a,
Hui
Wen
 a,
Shuwei
Xie
a,
Shuwei
Xie
 a,
Yuan
Li
a,
Yuan
Li
 b,
Ya
Chen
b,
Ya
Chen
 a,
Zhiquan
Liu
a,
Zhiquan
Liu
 a,
Nan
Jiang
a,
Nan
Jiang
 a,
Jiehua
Qiu
a,
Jiehua
Qiu
 a,
Xueming
Zhu
a,
Xueming
Zhu
 c,
Fucheng
Lin
c,
Fucheng
Lin
 bc and
Yanjun
Kou
bc and
Yanjun
Kou
 *a
*a
aState Key Laboratory of Rice Biology and Breeding, China National Rice Research Institute, Hangzhou 311400, China. E-mail: kouyanjun@caas.cn
bCollege of Agriculture and Biotechnology, Zhejiang University, Hangzhou 310058, China
cState Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
First published on 21st July 2023
Abstract
Silver nanoparticles (AgNPs) exhibit a broad spectrum of antimicrobial activity against various fungal pathogens, including the devastating rice blast fungus Magnaporthe oryzae, which threatens rice production worldwide. However, the antifungal molecular mechanisms of AgNPs against M. oryzae are still poorly understood. Here, it was found that the mycelial growth and virulence of M. oryzae strains were significantly reduced by AgNPs. Two nm-AgNPs at an EC50 concentration of 2.0 μg mL−1 almost impaired 100% virulence of M. oryzae. Further studies revealed that AgNP treatment changes cell wall morphology and suppresses appressorium formation. The defects in appressorium development upon AgNP treatment were caused by the reduced phosphorylation level of the MAPK MoPmk1 and impaired conidial autophagy. Importantly, the combination of AgNPs and azoxystrobin at appropriate ratios enhanced fungitoxicity to azoxystrobin-sensitive/resistant M. oryzae strains. Due to the excellent control effect of AgNPs on rice blast, we established a method for the biosynthesis of AgNPs by using the biocontrol Bacillus strain Tu27 to reduce waste environmental pollution and synthesis costs. Infection assays showed that the biosynthesized AgNPs displayed similar fungal toxicity. Taken together, our results revealed that AgNPs display the antifungal activity by a unique mechanism of damaging cell wall integrity, reducing phosphorylation of the MAPK MoPmk1, and impairing autophagy, and have an additive synergy with azoxystrobin on M. oryzae.
Environmental significanceThe rice blast disease, caused by Magnaporthe oryzae, is the most serious threat to rice production worldwide. Long-term application of the conventional fungicide azoxystrobin to control rice blast results in the emergence of drug-resistant strains in rice paddies. Therefore, it is necessary to develop an environmentally friendly novel fungicide to reduce the application doses of azoxystrobin and delay the occurrence of resistance. In this study, we provide insights into the antifungal activity and mechanism of silver nanoparticle (AgNPs) against M. oryzae. Importantly, we found that AgNPs display enhanced fungitoxicity to M. oryzae when co-applied with lower doses of azoxystrobin. Therefore, our results suggested that AgNPs have great application potential for combating conventional fungicide azoxystrobin resistance and reducing fungicide residues in the environment without decreasing control efficacy of rice blast. In addition, the finding provides strategies for achieving sustainable control of rice blast with different modes of action nano-fungicides and conventional fungicides. |
Introduction
Rice blast disease, caused by the ascomycete Magnaporthe oryzae, is a destructive rice disease that threatens global food security.1,2 The disease causes an approximately 30% loss of rice total production, which is enough to feed 60 million people.3–5 Although blast could be significantly reduced by planting blast-resistant rice cultivars, most rice blast-resistant cultivars have become susceptible after planting for several years due to pathogenic variation in the M. oryzae populations.5 The application of fungicides is an important management of rice blast disease in the field.6,7 However, several studies suggest that the improper and constant use of conventional fungicides over the past decades has resulted in the emergence of many fungicide-resistant populations. The emergence of M. oryzae populations resistant to azoxystrobin, isoprothiolane, phosphorothiolates, kasugamycin and blasticidin S challenged the longevity of their application in rice paddies.8–10 Among them, azoxystrobin is a broad-spectrum fungicide, which has been widely used to control rice blast.11 It suppresses mitochondrial respiration by binding to the Qo site of the cytochrome bc1 complex and thus interrupting electron transfer and ATP synthesis.12 The target of azoxystrobin, the cytochrome b subunit of the cytochrome bc1 complex, is encoded by mitochondrial genomic DNA and prone to mutation resulting in azoxystrobin resistance.13,14 In addition, concerns have also been raised about the potential risks to environmental ecology and human health when fungicides reside in the grains, soil, and waterways.15 Therefore, it is necessary to develop novel fungicides to reduce the use doses of conventional fungicides.Due to their excellent antimicrobial properties, silver nanoparticles (AgNPs) have been successfully applied for countering human and plant pathogens, including bacteria, fungi and viruses.16–18 One of the potential applications of AgNPs is the management of plant diseases.19 Currently, AgNPs are gradually being recognized as a promising alternative to conventional fungicides in rice paddies. With the technological improvement of synthesizing engineered nanomaterials, smaller commercial or biosynthesized AgNPs have been developed, but their antifungal activity remains unknown. In addition, some antimicrobial mechanisms of AgNPs have been proposed, especially in bacteria, including those causing DNA, cell wall and cell membrane damage, preventing electron transport or ATP biosynthesis, disturbing protein synthesis, and activating ROS generation.20 However, the toxicological effect and antifungal molecular mechanisms of AgNPs remain unclear, especially on the intracellular signaling pathways and key biological processes.
The specialized infection structure appressorium, differentiated at the tip of the conidial germ tube, is utilized by M. oryzae to rupture the rice cuticle and colonize host cells.21 Proper appressorium development is required for the virulence of M. oryzae and is tightly regulated by certain signaling pathways and biological processes, including MAPK (mitogen-activated protein kinase) signaling pathways and autophagy.22,23 Pmk1 is a conserved MAPK orchestrating appressorium formation in different fungi.24 Deletion of MoPMK1 results in failure of appressorium formation and plant infection in M. oryzae.25 A group of surface sensors, including MoPth11, is involved in MoPmk1 phosphorylation and sequential activation.26,27 During appressorium formation, phosphorylation of MoPmk1 is directly catalyzed by the kinase Mst7 (MAPKK) and then activates the downstream transcription factor Mst12 to regulate appressorium development and infection.28 In addition to the MoPmk1 signaling pathway, autophagy is another crucial biological process required for appressorium formation and virulence. Autophagy is an evolutionarily conserved process responsible for degrading damaged or obsolete intracellular components, such as organelles and proteins, to maintain energy homeostasis.29 Blocking of autophagy by disrupting ATG8, an essential autophagy factor, abolishes appressorium-mediated penetration in M. oryzae.30 Previous studies showed that either AgNPs 20–30 nm in diameter or biosynthesized AgNPs have the ability to inhibit the hyphal growth and appressorium development of the rice blast fungus M. oryzae.31,32 However, it is unknown how AgNP inhibits the appressorium development in M. oryzae.
In this study, we investigated the toxicological effects and antifungal activity of various sizes of commercial and biosynthesized AgNPs and depicted the molecular mechanism of AgNPs acting against M. oryzae. Our results showed that AgNPs reduce the growth and virulence of M. oryzae by disturbing cell wall morphology, weakening the activation of the MoPmk1-mediated intracellular signaling pathway and conidial autophagy. Further, we found that AgNPs in combination with azoxystrobin enhanced fungitoxic effects on azoxystrobin sensitive/resistant strains. Our findings provide novel insights into the unique antifungal mechanism of AgNPs against M. oryzae and the synergy between AgNPs and azoxystrobin.
Materials and methods
Strains and culture conditions
The fungal strain M. oryzae wild-type strain (WT) B157 and derivatives ΔMopmk1, ΔMopth11, ΔMoatg8, and ΔMomst12 were used in this study.27 The null mutant ΔMomps1 was derived from the Guy11 background and used to test the sensitivity in response to AgNPs.33 Field isolates of M. oryzae were isolated from diseased panicles. All strains were cultured on complete medium (CM, 6 g L−1 NaNO3, 0.52 g L−1 KCl, 0.52 g L−1 MgSO4, 1.52 g L−1 KH2PO4, 22 mg L−1 ZnSO4, 11 mg L−1 H3BO3, 5 mg L−1 MnCl2, 5 mg L−1 FeSO4, 1.7 mg L−1 CoCl2, 1.6 mg L−1 CuSO4, 1.5 mg L−1 Na2MoO4, 0.05 g L−1 Na4EDTA, 10 g L−1D-glucose, 2 g L−1 peptone, 1 g L−1 yeast extract, 1 g L−1 casamino acid, 0.1 mg L−1 biotin, 0.1 mg L−1 pyridoxine, 0.1 mg L−1 thiamine, 0.1 mg L−1 riboflavin, 0.1 mg L−1p-aminobenzoic acid, 0.1 mg L−1 nicotinic acid, pH 6.5) plates under 16 h light and 8 h dark conditions at 25 °C according to standard procedures.21To isolate the biocontrol strain Bacillus sp. Tu27, one gram of rice seedling rhizosphere soil was resolved in 10 mL of sterile distilled water and homogenized with steel balls. The resultant solution (0.1 mL) was spread on nutrient agar (NA) medium followed by a series of dilutions of 10 and 100-fold, respectively. The single colony grown on the NA plates was purified on another NA plate and then cultured with mycelial plugs of M. oryzae by confrontation assays to screen biocontrol strains that efficiently inhibit the growth of M. oryzae. The potential biocontrol strains were stored at −80 °C in 25% (v/v) glycerol.
To identify the isolated biocontrol strains, 16S rRNA gene sequence analysis was performed. Briefly, bacterial DNA was extracted with a genomic DNA isolation kit (DP302-02, TIANGEN). The 16S rRNA gene was amplified from the extracted genomic DNA with primers 63F and 1387R using high-fidelity polymerase enzyme (Vazyme d105, China) to obtain 16S rRNA gene sequences, which were aligned in the National Center for Biotechnology Information (NCBI) using the nucleotide BLAST tool. The phylogenetic tree was constructed using MEGA 7.0 software via the neighbor-joining method.34,35
Preparation of silver nanoparticles
Silver nanoparticles with particle sizes of 2 nm (N196423) and 15 nm (N186422) were purchased from Shanghai Aladdin Biochemical Technology Corporation, China. The stock solutions had a concentration of 1000 μg mL−1. Biosynthesized AgNPs were synthesized in this study using a cell-free culture filtrate of bacteria Bacillus sp. Tu27. Biosynthesis of AgNPs was performed as described.31 The cell-free supernatant of Tu27 was prepared by culturing Bacillus sp. Tu27 in nutrient broth (NB) medium at 30 °C and 200 rpm for 2 d and filtering twice using a 0.22 μm filter followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 20 min. Cell-free supernatants (10 mL) were mixed with 90 mL of 3 mM silver nitrate solution and incubated at 30 °C with shaking at 200 rpm for 4 d in the dark. A mixture of 10 mL NB medium and 90 mL silver nitrate solution served as a control. The resultant AgNP reaction solutions were tested by UV-visible spectrometry in the wavelength range of 200–800 nm. AgNPs in the reaction solution were collected by centrifugation at 10
000 rpm at 4 °C for 20 min. Cell-free supernatants (10 mL) were mixed with 90 mL of 3 mM silver nitrate solution and incubated at 30 °C with shaking at 200 rpm for 4 d in the dark. A mixture of 10 mL NB medium and 90 mL silver nitrate solution served as a control. The resultant AgNP reaction solutions were tested by UV-visible spectrometry in the wavelength range of 200–800 nm. AgNPs in the reaction solution were collected by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min, washed twice with sterile distilled water and stored at −80 °C.
000 × g for 20 min, washed twice with sterile distilled water and stored at −80 °C.
Characterization of the biosynthesized AgNPs
Characterization of the biosynthesized AgNPs was performed with Fourier transform infrared (FTIR), TEM, SEM, and X-ray diffraction (XRD) as previously reported.31 The diameter of the AgNPs was measured according to TEM images. FTIR was performed to identify the functional groups responsible for reducing Ag ions to AgNPs. The mixture of AgNPs (1 mg) and potassium bromide was compressed and subjected to testing on an AVATAR 370 FTIR spectrometer (Thermo Nicolet, USA) in a spectral range of 500–4000 cm−1. To determine the components of biosynthesized AgNPs, XRD was conducted on a Bruker D8 Advance machine with a detector voltage of 45 kV and a current of 40 mA using CuKo radiation.Sensitivity of M. oryzae to AgNPs and azoxystrobin
To evaluate the sensitivity of M. oryzae strains to a diameter of 2 or 15 nm AgNPs and biosynthesized AgNPs, mycelial plugs were inoculated on CM plates supplemented with 0.5, 1, 1.5, 2, 2.5, 5, and 10 μg mL−1 of AgNPs or 5, 10, 20, and 40 μg ml−1 biosynthesized AgNPs, and cultured in the dark for 6 d. To test the azoxystrobin resistance of M. oryzae strains, mycelial plugs were inoculated on CM plates supplemented with 10, 20, 40, and 100 μg mL−1 of azoxystrobin and cultured in the dark for 6 d. To test the interaction between AgNPs and azoxystrobin, mycelial plugs were inoculated on CM plates supplemented with three different ratios of mixture: azoxystrobin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNPs = 9
AgNPs = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, azoxystrobin
1, azoxystrobin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNPs = 1
AgNPs = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, azoxystrobin
1, azoxystrobin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AgNPs = 1
AgNPs = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9. The final concentrations of azoxystrobin and AgNPs are 1.8 μg mL−1 AgNPs in combination with 1 μg mL−1 of azoxystrobin, 1 μg mL−1 AgNPs in combination with 5 μg mL−1 of azoxystrobin, and 0.2 μg mL−1 AgNPs in combination with 9 μg mL−1 of azoxystrobin, respectively. The treatments of 2 μg mL−1 AgNPs and 10 μg mL−1 azoxystrobin were separately used as individual control treatments. The sensitivity assays were repeated three times with three replicates each time. The relative inhibitory rates and conidiation were calculated based on the colony diameter and conidial numbers as previously described.36
9. The final concentrations of azoxystrobin and AgNPs are 1.8 μg mL−1 AgNPs in combination with 1 μg mL−1 of azoxystrobin, 1 μg mL−1 AgNPs in combination with 5 μg mL−1 of azoxystrobin, and 0.2 μg mL−1 AgNPs in combination with 9 μg mL−1 of azoxystrobin, respectively. The treatments of 2 μg mL−1 AgNPs and 10 μg mL−1 azoxystrobin were separately used as individual control treatments. The sensitivity assays were repeated three times with three replicates each time. The relative inhibitory rates and conidiation were calculated based on the colony diameter and conidial numbers as previously described.36
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) assays
To determine the size and morphology of AgNPs used in this study, purchased and biosynthesized AgNPs solutions were dried on copper mesh and TEM observation was performed using a microscope (JEM-1230, Japan) by placing AgNPs on a copper grid. To observe the effect of AgNPs on the fungal morphology of M. oryzae hyphae, mycelia cultured in CM medium for 2 days were treated with AgNPs with EC50 and EC90 concentrations for 2 hours. Then, the mycelia were collected and fixed with 2.5% (v/v) glutaraldehyde solution, stained with 1% (w/v) osmium tetroxide and dehydrated with a series of ethanol solutions (30–100%) as described.37 The morphology of mycelia and biosynthesized AgNPs was captured with SEM (SU8010, Hitachi, Japan) and TEM (H7650, JEOL, Japan).Observation of appressorium morphogenesis and infection assays
To determine the effect of AgNPs on appressorium formation, the WT strain was cultured on CM plates for 6 d to collect conidia. The resultant conidial suspensions (1 × 105 conidia per mL) amended with AgNPs at final concentrations of 0, 0.05, 0.1, 0.2, 0.4, and 2 μg mL−1 were inoculated on hydrophobic plastic coverslips for 4, 8, and 24 h. The photos were captured using an Olympus BX53 microscope under a bright field.To test the effects of AgNPs and azoxystrobin on virulence, equal volumes (4 mL) of conidial suspensions (1 × 105 conidia per mL) were harvested from 6 d-old cultures of the WT. Conidial suspensions were supplemented with final concentrations of 0, 0.05, 0.1, 0.2, 0.4, and 2 μg mL−1 AgNPs or a mixture of AgNPs and azoxystrobin, and then spray-inoculated on 3 week-old susceptible rice seedlings CO39 (Oryza sativa). The inoculated rice seedlings were placed in the dark for 2 d at 22 °C under humid conditions and transferred to cycles of 16 h light/8 h dark conditions for 5 d. The numbers and lesion area of rice disease lesions on leaves were counted and measured at 5 d post inoculation (dpi). The infection assays were repeated three times with consistent results. Primers are supplemented in Table S1.†
Fluorescence observation of conidial autophagy
Conidial suspensions collected from the fluorescent strain expressing the fusion protein GFP-MoAtg8 were supplemented with final concentrations of 0, 0.05, 0.1, 0.2, 0.4, and 2 μg mL−1 AgNPs. The resultant conidial suspensions were inoculated on hydrophobic surfaces at 25 °C under humid conditions. Fluorescence observation was performed using a confocal fluorescence microscope Zeiss LSM700 at 4 and 24 h post inoculation (hpi). GFP fluorescence was obtained with 488 nm laser excitation. The resultant images were processed with ImageJ software and organized with Adobe Illustrator CS6. The numbers of autophagosomes in conidia were counted upon treatment with different concentrations of AgNPs.Western blotting
To detect the phosphorylation levels of MoPmk1 and MoMps1, mycelia of the WT were cultured in liquid CM for 2 d and then treated with 2 μg mL−1 AgNPs for 2 and 6 h, respectively. Total proteins were extracted from mycelia as previously described.38 Briefly, mycelia were ground into powders with liquid nitrogen and suspended in 10% (m/v) trichloroacetic acid solutions. The resultant pellets were dissolved in a solution of 30% (m/v) sucrose and 2% (m/v) SDS. After centrifugation, the proteins in the supernatant were pelleted with 80% methanol and then dissolved in 8 M urea solution. Samples with equal total proteins were loaded and separated on a 10% SDS-polyacrylamide gel and then transferred to a PVDF membrane. The membrane was blocked and immunoblotted with anti-P-p44/42 MAPK (Cat. 9101S, Cell Signaling Technology, USA), anti-MoPmk1 (Cat. sc-514![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302, Santa Cruz, USA), and anti-MoMps1 antibodies (Cat. 9102S, Cell Signaling Technology, USA). The ECL chemiluminescent kit (Cat. No. 1705060, Bio-Rad, USA) was used for Western blotting detection. GAPDH, which was detected by the anti-GAPDH antibody (Cat. No. R1208-3, HuaBio, China), was used as a loading control.
302, Santa Cruz, USA), and anti-MoMps1 antibodies (Cat. 9102S, Cell Signaling Technology, USA). The ECL chemiluminescent kit (Cat. No. 1705060, Bio-Rad, USA) was used for Western blotting detection. GAPDH, which was detected by the anti-GAPDH antibody (Cat. No. R1208-3, HuaBio, China), was used as a loading control.
Statistical analysis
All values presented in the column are the mean ± standard deviation (SD). Significant differences among different columns were calculated by Duncan's test at P < 0.01 and indicated with different letters.Results
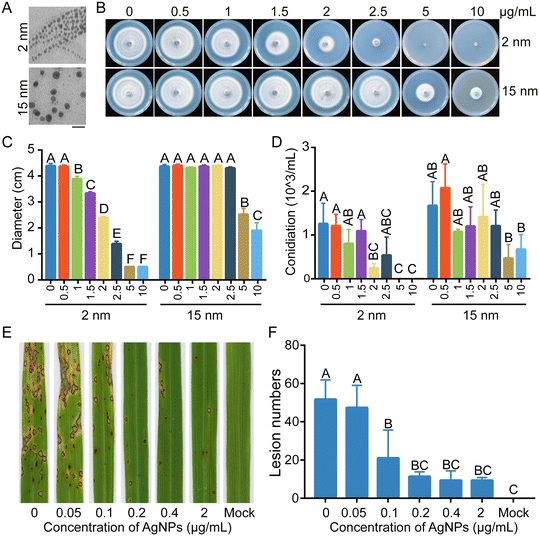
AgNPs suppress mycelial growth and virulence of M. oryzae dependent on size and dosage
The morphology and UV-vis absorption spectrum of the AgNPs used in this study were first determined. Uniform spherical silver particles were present, and the diameters of each AgNP particle were consistent with theoretical values of 2 nm and 15 nm (Fig. 1A and S1†). To determine the sensitivity of M. oryzae to AgNPs, a series of concentrations of 0.5, 1, 1.5, 2, 2.5, 5, and 10 μg mL−1 AgNPs with 2 nm or 15 nm diameters were used to obtain fungitoxicity curves and calculate the effective concentration of AgNPs inhibiting colony growth by 50% (EC50) values. Mycelial plugs of the WT strain B157 were inoculated on CM plates supplemented with a series of concentrations of AgNPs. As shown in Fig. 1B, the growth of the B157 strain was significantly inhibited with increasing concentrations of supplemented AgNPs. In addition, 2 nm in diameter AgNPs displayed higher inhibition efficacy than the same concentrations of 15 nm AgNPs. Based on the fungitoxicity-curves of M. oryzae to AgNPs, the EC50 of 2 nm AgNPs was 2.0 μg mL−1. Mycelial growth was completely inhibited by 2 nm AgNPs at concentrations of 5 and 10 μg mL−1. For 15 nm AgNPs, significant growth inhibition (50%) was observed until the concentration reached 5 μg mL−1 (Fig. 1C). Thus, 2 nm AgNPs were used to further elucidate the antifungal mechanism against M. oryzae. In contrast to vegetative growth, conidiation showed a slightly decreasing tendency when treated with different concentrations of AgNPs (Fig. 1D). Further, the sensitivities of M. oryzae to AgNPs of 2 nm in diameter were determined using 14 single-spore isolates from diseased rice paddies. The 50% effective inhibition concentrations of these 14 isolates against AgNPs ranged from 2.7 to 3.3 μg mL−1 (Table S2†), which is comparable to the model strain B157.To evaluate the effect of AgNP treatment on the virulence of M. oryzae, a conidial suspension of M. oryzae amended with 2.0 μg mL−1 (EC50 of 2 nm AgNPs) AgNPs was spray-inoculated onto rice seedlings. The conidial suspension without AgNPs was used as a control. In contrast to the large amount of rice blast lesions formed on the rice leaves, few lesions were observed on the rice leaves inoculated conidia with 2.0 μg mL−1 AgNPs, indicating that AgNPs severely impaired the virulence of M. oryzae (Fig. 1E). To screen a lower concentration of AgNPs sufficient for the inhibition of M. oryzae virulence, the AgNPs were further diluted and added into conidial suspensions to inoculate rice seedlings. As shown in Fig. 1E and F, the number of lesions increased by 50% at a concentration of 0.1 μg mL−1, in contrast to that at a concentration of 2.0 μg mL−1 of AgNPs. These data indicated that AgNPs suppress the virulence of M. oryzae in a dosage-dependent manner.
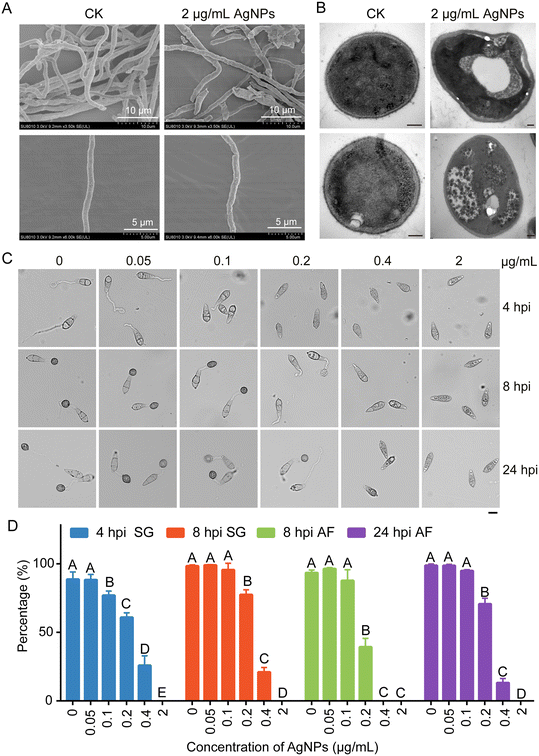
AgNPs cause damaged fungal cell walls and suppress appressorium development of M. oryzae
To explain why AgNPs in combination with azoxystrobin enhanced the reduction in virulence of M. oryzae, the antifungal mechanism of AgNPs was explored. Firstly, we conducted SEM and TEM to monitor the fungal morphologies treated with 2 μg mL−1 AgNPs or not. The results of SEM showed that hyphae were collapsed and aberrant after treatment with AgNPs in comparison with the control (Fig. 2A), implicating damage to fungal cell wall integrity. TEM observation further revealed the changes of hyphal cell walls when treated with AgNPs (Fig. 2B). These data suggested that the inhibition of mycelial growth may be associated with the cell wall damage caused by AgNPs.In addition to growth, the appressorium, which is the most important infection structure of M. oryzae, keeps a tight relationship with virulence.21 Therefore, appressorium development processes, including conidial germination and appressorium formation, were monitored. Conidial suspensions amended with 0, 0.05, 0.1, 0.2, 0.4, and 2 μg mL−1 of AgNPs were dropped on the hydrophobic surface and incubated for 4, 8, and 24 h. With the increase of AgNP concentrations, the rates of conidial germination and appressorium formation decreased gradually (Fig. 2C). At 0.05 μg mL−1 AgNPs, the ratios of conidial germination (88.6 ± 5.4%) and appressorium formation (98.7 ± 1.2%) were comparable to the ratios of conidial germination (88.2 ± 4.0%) and appressorium formation (98.6 ± 0.9%) of the control at 0 μg mL−1 at 4 hpi and 24 hpi, respectively. In contrast to the mock treatment, when treated with 0.1 μg mL−1 AgNPs, conidial germination was suppressed. The rates of conidial germination were decreased to 76.8 ± 3.2% and 95.5 ± 4.8% at 4 hpi and 8 hpi, respectively. However, at 24 hpi, the rate of appressorium formation at 0.1 μg mL−1 (94.8 ± 0.8%) was comparable to that of the control (98.6 ± 0.9%). Significant inhibition effects of conidial germination and appressorium formation were found at 0.2 μg mL−1, which was consistent with the conidial inoculation assays (Fig. 2D). Moreover, conidial germination was completely blocked at an EC50 concentration of AgNPs, suggesting that the same concentration of AgNPs is more fungitoxic to spores than hyphae. These results indicated that AgNPs play an inhibitory role in appressorium formation.
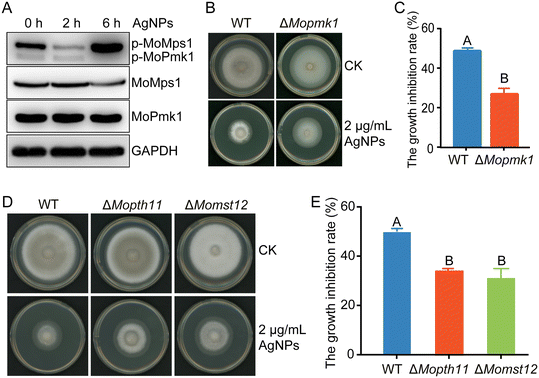
AgNPs reduce appressorium formation by weakening the phosphorylation of MAPK MoPmk1
The MAPK MoPmk1 is an essential regulator of appressorium development and invasive growth in M. oryzae. MoPmk1 null mutants of M. oryzae cannot form appressoria to infect rice plants.25 We speculated that defects in appressorium development and virulence caused by AgNP treatment may be associated with MoPmk1 activity. Thus, the MoPmk1 activity, indicated by the phosphorylation levels of MoPmk1, was detected by Western blotting. With AgNP treatment at 2 and 6 hpi, the phosphorylation level of MoPmk1 was significantly suppressed, while the protein level of MoPmk1 did not change obviously (Fig. 3A). We also found that compared with the WT, the deletion mutant ΔMopmk1 was less sensitive to AgNPs at 2 μg mL−1 (Fig. 3B and C), indicating that MoPmk1 is involved in the tolerance to AgNP stress in M. oryzae. In addition, the phosphorylation level of MAPK MoMps1, which is involved in regulating cell wall integrity, was decreased at 2 hpi but apparently increased at 6 hpi (Fig. 3A). In contrast to ΔMopmk1, the ΔMomps1 became more sensitive to AgNPs than the WT (Fig. S2†), which is consistent with cell wall damage during TEM observation.In the MoPmk1-mediated signaling cascade, phosphorylated MoPmk1 could be activated by the upstream surface sensor MoPth11, which then activates the downstream transcription factor Mst12 to control appressorium formation and infection.22 To further analyze the molecular mechanism underlying the response of MoPmk1 to AgNP stress, the roles of MoPth11 and MoMst12 in response to AgNP stress were investigated. When treated with an EC50 concentration of 2 nm AgNPs, ΔMopth11 and ΔMomst12 displayed lower growth inhibition rates than the WT (Fig. 3D and E), suggesting that the MoPmk1-mediated appressorium formation signaling pathway is involved in responding to AgNP stress.
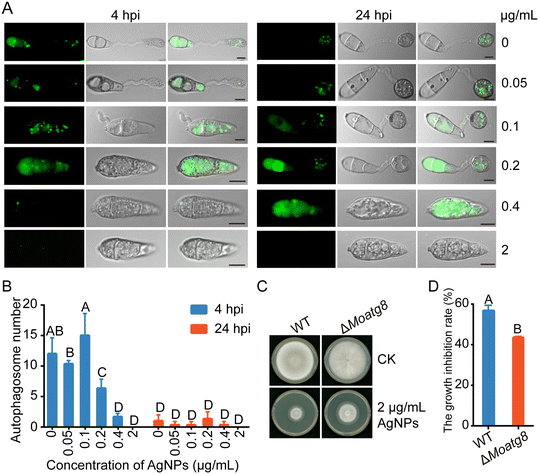
AgNPs impair conidial autophagy during appressorium development
Autophagy is required for appressorium development and infection in M. oryzae.39 Thus, we determined whether conidial autophagy is further affected when treated with AgNPs. Conidial autophagy was observed using the green fluorescent protein GFP-labeled protein Atg8, which is a common marker used to detect autophagy levels.40 When conidial drops were inoculated on the hydrophobic surface, conidial autophagy was indicated by the presence of GFP-MoAtg8 labeled fluorescent dots in conidia at 4 hpi, and ended with no obvious green fluorescence observed in conidia at 24 hpi. In contrast, at 4 hpi, the green fluorescent dots in conidia expressing the fusion protein GFP-Atg8 decreased when conidia were treated with increased concentrations of AgNPs (Fig. 4A), indicating the suppression of autophagy by AgNPs. In addition, green fluorescence was still detected at 24 hpi in conidia treated with 0.2 μg mL−1 AgNPs, further implicating a suppressive role of AgNPs in conidial autophagy. Consistent with fluorescence observation, the numbers of autophagosomes in conidia were reduced with increasing concentrations of AgNPs (Fig. 4B). These data suggested that AgNPs play an inhibitory role in autophagy during appressorium formation in M. oryzae. Furthermore, the deletion mutant ΔMoatg8 was less sensitive to AgNPs than the WT when incubated on CM plates amended with 2 μg mL−1 AgNPs (Fig. 4C and D), implicating a role of MoAtg8-mediated autophagy in responding to AgNP stress.AgNPs display antifungal activity against azoxystrobin-resistant strains and their combination with azoxystrobin shows a significantly enhanced antifungal activity
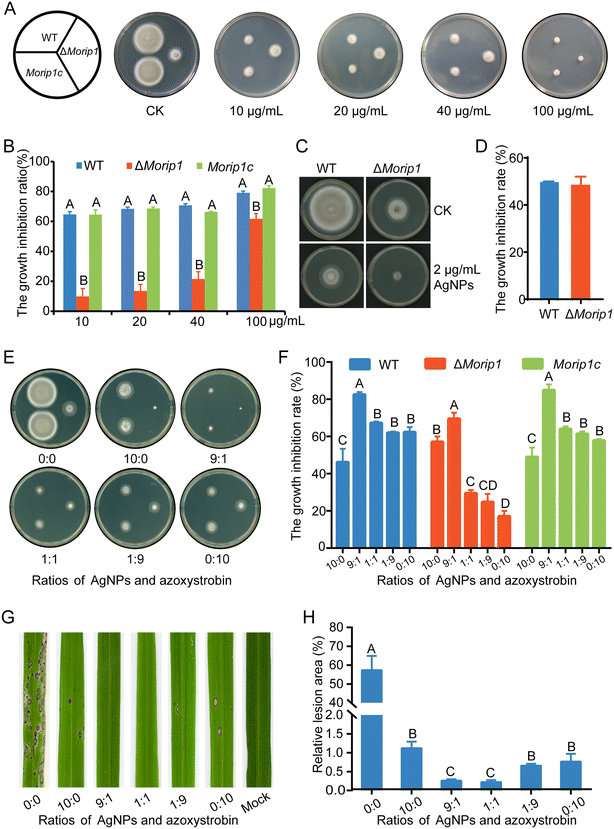
MoRIP1 is a putative subunit of the cytochrome bc1 complex and encodes a Rieske iron–sulfur protein. Its gene deletion mutant ΔMorip1 was resistant to azoxystrobin and showed lower sensitivity to azoxystrobin compared to those of the WT and complementary strain Morip1c (Fig. 5A and B). When inoculated on AgNP-containing plates, AgNPs displayed a similar inhibitory rate to the ΔMorip1 mutant in sensitivity to EC50 of AgNPs when compared with the WT (Fig. 5C and D), suggesting that AgNPs are effective to azoxystrobin-resistant strains.To determine the control efficacy of AgNPs in combination with azoxystrobin, inhibitory effects of AgNPs mixed with azoxystrobin at various ratios on azoxystrobin-sensitive/resistant strains were measured in vitro. As shown in Fig. 5E, the mixture of AgNPs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) azoxystrobin (9
azoxystrobin (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) significantly increased the inhibitory rate on azoxystrobin-sensitive (increased by 78.6% and 32.4%) and resistant strains (increased by 21.7% and 306.4%) compared to the individual treatments of 2 μg mL−1 AgNPs (10
1) significantly increased the inhibitory rate on azoxystrobin-sensitive (increased by 78.6% and 32.4%) and resistant strains (increased by 21.7% and 306.4%) compared to the individual treatments of 2 μg mL−1 AgNPs (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and 10 μg mL−1 of azoxystrobin (0
0) and 10 μg mL−1 of azoxystrobin (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). In addition, the mixture of AgNPs
10). In addition, the mixture of AgNPs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) azoxystrobin (1
azoxystrobin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) displayed a slightly enhanced inhibitory rate on azoxystrobin-sensitive/resistant strains when compared with individual treatments of AgNPs (10
1) displayed a slightly enhanced inhibitory rate on azoxystrobin-sensitive/resistant strains when compared with individual treatments of AgNPs (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) or azoxystrobin (0
0) or azoxystrobin (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) at the P < 0.05 level. The inhibitory effects of the AgNPs and azoxystrobin mixture on the complementary strain Morip1c were similar to the WT strain (Fig. 5E and F). These data indicated that AgNPs are more effective on counting M. oryzae when AgNPs are used in combination with azoxystrobin.
10) at the P < 0.05 level. The inhibitory effects of the AgNPs and azoxystrobin mixture on the complementary strain Morip1c were similar to the WT strain (Fig. 5E and F). These data indicated that AgNPs are more effective on counting M. oryzae when AgNPs are used in combination with azoxystrobin.
To further confirm the synergy between AgNPs and azoxystrobin, the fungitoxic effects were tested on rice seedlings inoculated with M. oryzae conidial suspensions supplemented with mixtures of AgNPs and azoxystrobin. As shown in the results, large numbers of lesions were formed on the leaves inoculated with conidial suspensions without fungicide AgNPs or azoxystrobin. In contrast, the combination of AgNPs and azoxystrobin at the ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 1
1 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 caused a significant decrease in the lesion area than individual treatments with AgNPs or azoxystrobin (Fig. 5G and H). All these results suggested that AgNPs have an additive synergy with azoxystrobin when co-applied on rice seedlings.
1 caused a significant decrease in the lesion area than individual treatments with AgNPs or azoxystrobin (Fig. 5G and H). All these results suggested that AgNPs have an additive synergy with azoxystrobin when co-applied on rice seedlings.
Biosynthesis and characterization of AgNPs
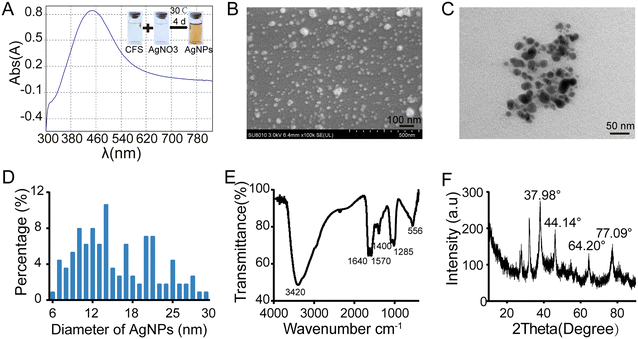
The aforementioned results confirmed that AgNPs show high antifungal efficacy against M. oryzae. In addition, the biosynthesis of AgNPs has green, rapid, low-cost, and eco-friendly advantages.41 Next, we biosynthesized AgNPs using a bacteria-mediated pathway and compared their antifungal efficacy. By screening the potential biocontrol strains isolated from the rice rhizosphere, the strain named Tu27 showed antagonistic effects on M. oryzae in the plate confrontation assays and had the ability to catalyze the synthesis of AgNPs. Through 16S rRNA sequence analysis and phylogenetic tree analysis, the Tu27 strain was identified as Bacillus sp. (Fig. S3†).Using the cell-free supernatant (CFS) of cultured Tu27, AgNPs were synthesized by incubating CFSs with silver nitrate solution. As shown in Fig. 6A, the color of the mixture converted from light yellow to dark brown following 4 d incubation with shaking at 30 °C. The formation of nanoparticles was demonstrated by showing surface plasmon resonance at 440 nm using a UV spectrophotometer. SEM and TEM observation showed that spherical particles were present with sizes ranging from 6 nm to 30 nm (Fig. 6B–D). The functional groups of the biosynthesized AgNPs were confirmed by FTIR (Fourier transform infrared) analysis. Six peaks at 3420, 1640, 1570, 1400, 1265, and 556 cm−1 were found in the FTIR spectrum, corresponding to the stretching vibrations of –NH, –OH, and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (Fig. 6E). These groups indicated a role of peptides or proteins in reducing Ag+ to Ag0. The components of biosynthesized AgNPs were determined by XRD analysis. Four emission peaks at 37.98°, 44.14°, 64.20°, and 77.09° were suitable for crystalline silver planes (1 1 1), (2 0 0), (2 2 0), and (3 1 1) (Fig. 6F), respectively, suggesting the presence of crystalline AgNPs. The above characterization assays confirmed that AgNPs were biosynthesized successfully.
O (Fig. 6E). These groups indicated a role of peptides or proteins in reducing Ag+ to Ag0. The components of biosynthesized AgNPs were determined by XRD analysis. Four emission peaks at 37.98°, 44.14°, 64.20°, and 77.09° were suitable for crystalline silver planes (1 1 1), (2 0 0), (2 2 0), and (3 1 1) (Fig. 6F), respectively, suggesting the presence of crystalline AgNPs. The above characterization assays confirmed that AgNPs were biosynthesized successfully.
Biosynthesized AgNPs display similar antifungal activity to commercial AgNPs
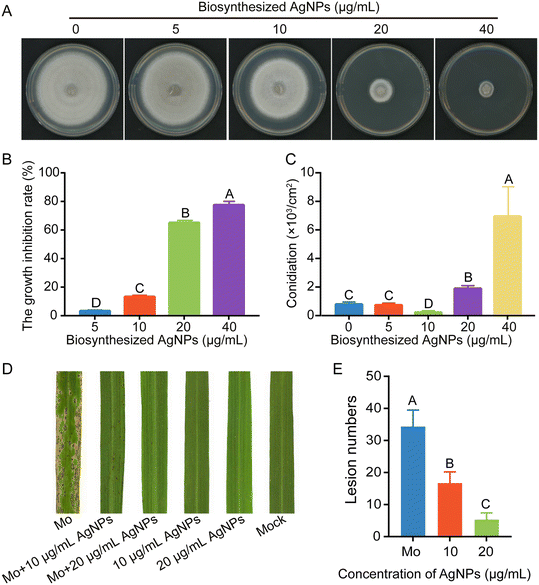
The inhibitory effects of biosynthesized AgNPs on mycelial growth, conidiation, and virulence were evaluated. Biosynthesized AgNPs at 20 μg mL−1 significantly reduced the mycelial growth of M. oryzae by more than 60%. Consistent with commercialized AgNPs of 2 nm or 15 nm in diameter, the higher the concentrations of biosynthesized AgNPs, the more significant the inhibitions (Fig. 7A and B). Interestingly, the inhibitory effects of AgNPs at 20 and 40 μg mL−1 on the conidiation of M. oryzae were significantly increased compared to those at lower concentrations (Fig. 7C). When conidial suspensions were amended with 10 and 20 μg mL−1 biosynthesized AgNPs, few lesions were formed on the rice leaves (Fig. 7D and E), indicating a high efficacy of biosynthesized AgNPs in reducing the virulence of M. oryzae. These data indicated that AgNPs biosynthesized by the strain Tu27 display inhibitory effects on the development and virulence of M. oryzae as commercialized AgNPs.Discussion
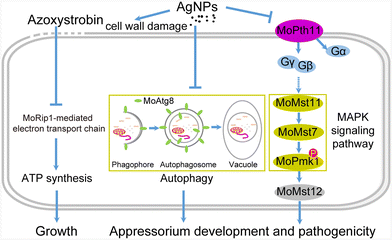
Recently, there have been more applications and toxicological analysis of AgNPs on various organisms, due to their outstanding antimicrobial activity.42–46 To date, the detailed intracellular signal transduction mediated by AgNPs against microbes has not been clearly defined. In this study, we systematically evaluated the fungitoxicity of AgNPs against the rice blast fungus M. oryzae. Then, it is revealed that AgNPs inhibit growth, appressorium development, and virulence in M. oryzae by impairing cell wall integrity and suppressing MoPmk1 signaling and autophagy (Fig. 8). And further we found that AgNPs have an additive synergy when co-applied with the conventional fungicide azoxystrobin. Previous studies revealed that the proposed mechanisms of AgNPs functioning in antimicrobials mainly involve three aspects: first, AgNPs attach to the cell membrane and disturb phospholipid bilayers resulting in increased cell membrane permeability; second, AgNPs enter cells, causing damage to intracellular organelles or large biomolecules; and third, AgNPs modulate intracellular biological processes by affecting signal transduction. To our knowledge, this is the first report that AgNPs inhibit fungal development and virulence by novel mechanisms of down-regulating the MAPK MoPmk1 signaling pathway and autophagy in fungi.AgNPs show different degrees of antimicrobial activity based on various factors such as shape, size and concentration.20 The sphere-shaped AgNPs were confirmed to be more effective against Klebsiella sp. and Escherichia coli than rod-shaped AgNPs.47 The AgNPs used in this study are all spherical, as observed by TEM. In addition to shape, the particle size of AgNPs is one of the most important factors influencing inhibitory efficacy. The antifungal activity of AgNPs against Fusarium graminearum was shown to be dependent on the particle size.44 The smaller size of AgNPs means a higher surface![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) volume ratio, which facilitates the release of silver ions. In our study, excluding the effects of shape, 2 nm AgNPs displayed better inhibitory activities on M. oryzae than 15 nm AgNPs at the same concentrations, further demonstrating that the size of AgNPs is a key characteristic of antimicrobial activity. In addition, mycelial growth was inhibited more with increasing concentration of AgNPs supplemented in the medium, suggesting that the efficacy of AgNPs is directly associated with the concentration of AgNP application.
volume ratio, which facilitates the release of silver ions. In our study, excluding the effects of shape, 2 nm AgNPs displayed better inhibitory activities on M. oryzae than 15 nm AgNPs at the same concentrations, further demonstrating that the size of AgNPs is a key characteristic of antimicrobial activity. In addition, mycelial growth was inhibited more with increasing concentration of AgNPs supplemented in the medium, suggesting that the efficacy of AgNPs is directly associated with the concentration of AgNP application.
According to our results, the antifungal activity of AgNPs is associated with cell wall damage and negative regulation of Pmk1 signaling and autophagy. The integrity of the cell wall is important for hyphal growth and virulence.48 Cell wall damage caused by AgNP treatment could be confirmed by the distorted morphology of direct SEM and TEM observation and the indirect increased sensitivity of ΔMomps1 to AgNPs. In addition to cell wall damage, our further observation showed that AgNPs reduce the virulence of M. oryzae by affecting conidial germination. Transcriptome sequencing of F. solani species mycelia upon AgNP treatment revealed that energy and substance metabolism-related genes were down-regulated, implicating an interruption of the cell cycle and ATP synthesis.49 Repression of energy metabolism may be one reason why conidial germination was blocked at the EC50 concentration of 2 μg mL−1 AgNPs. In addition, AgNPs inhibit the activation of the MAPK MoPmk1 to disrupt the appressorium formation. The MoPmk1 signaling pathway plays important roles in regulating appressorium formation. Disruption of key components of the MoPmk1 signaling pathway resulted in failure of appressorium development and virulence in M. oryzae.23,25 In the nematode Caenorhabditis elegans, the MAPK p38-based oxidative stress signaling network is involved in defense against AgNP exposure.50 Moreover, the expression of MAPK was induced by AgNPs in F. solani mycelia.49 Consistent with these studies, we found that phosphorylation of MoPmk1 was suppressed upon AgNP treatment. Furthermore, the disruptions of the MoPmk1 signaling pathway genes, including MoPTH11, MoPMK1, and MoMST12, resulted in the lower sensitivities to AgNPs, suggesting that AgNPs reduce appressorium formation by interfering with the MoPmk1 signaling pathway. Furthermore, AgNPs cause defects in appressorium formation by suppressing conidial autophagy. AgNPs impair mitochondrial antiviral immunity by blocking autophagic flux in lung epithelial cells.51 In our study, the autophagy marker protein GFP-MoAtg8 decreased upon treatment with increasing concentrations of AgNPs, indicating that the autophagy was suppressed by AgNPs. Blocking autophagy by deletion of MoATG8 decreased the sensitivity of AgNPs to M. oryzae. In addition, several studies have indicated that MoPmk1 signaling is associated with autophagy. In M. oryzae, MoPmk1 is responsible for conidial autophagy during appressorium development.52 A recent report revealed that MoPmk1 signaling and autophagy could be activated by one common activator MoMka1.53 Taken together, we postulated that AgNPs show antifungal activity by disturbing cell wall integrity, as well as MoPmk1- and Atg8-mediated pathways during appressorium development, resulting in reduced virulence of M. oryzae.
In recent years, the widespread use of azoxystrobin in disease control has resulted in the occurrence of azoxystrobin-resistant strains. Azoxystrobin resistance has been reported to be related to mutations at the M. oryzae target protein Cytb subunit of the cytochrome bc1 complex.54 Previous reports suggested that there exists conformationally linked interaction between inhibitors of the Qo site and the Rieske iron–sulfur protein Rip1 in the cytochrome bc1 complex.55 This conclusion was further confirmed by the evidence that deletion of MoRIP1 resulted in resistance to azoxystrobin, which is the first report in filamentous fungi. AgNPs are reported to count fruit brown rot pathogen Monilinia fructicola isolates resistant to benzimidazole fungicides.43 Further, we found that AgNPs are not only effective to azoxystrobin-resistant strains, but also their combination with azoxystrobin at appropriate ratios enhances the fungitoxic effect on M. oryzae both in vitro and when applied on preventing rice seedling blast. According to our results, we assumed that cell wall damage caused by AgNP treatment may prompt the entry of azoxystrobin into fungal cells to disrupt ATP synthesis and inhibit growth. Together, our results provide some important references for resistance management and sustainable application of azoxystrobin in the field.
Biosynthesis of AgNPs is an environmentally friendly, low-cost, and easy method to synthesize AgNPs using extracts from plants, fungi, and bacteria.41,56 Some bacteria such as Bacillus licheniformis, Bifidobacterium bifidum, and Lactobacillus sp. are reported to synthesize silver nanoparticles for antimicrobial, antioxidant, and anticancer applications.57,58 The endophytic strain Pseudomonas poae has the ability to synthesize AgNPs to protect wheat from F. graminearum infection.59 An endophytic bacterium Bacillus endophyticus strain H3 isolated from onion is able to synthesize AgNPs to inhibit M. oryzae.31 A recent report showed that biosynthesized AgNPs by using cucumber leaves and rice husk extracts exhibit long-term antimicrobial properties in comparison with chemically synthesized AgNPs.60 In this study, the biocontrol strain Bacillus sp. Tu27 was identified by screening strains isolated from the rice rhizosphere and plate confrontation assays. Using the cell-free culture filtrate of Tu27, AgNPs were biosynthesized and showed good antifungal activity against M. oryzae as commercial AgNPs. The biosynthesized AgNPs are spherical with sizes ranging from 6 to 30 nm. Therefore, the EC50 concentration of biosynthesized AgNPs against M. oryzae is higher than that of 2 nm AgNPs. In contrast to chemically synthesized AgNPs, biosynthesized AgNPs increased conidiation at concentrations higher than the EC50, which may be caused by functional groups in the biosynthesized AgNPs. In the future, it is crucial to further optimize the biosynthesis reaction conditions of AgNPs to improve the antimicrobial ability of biosynthesized AgNPs.
Overall, we revealed that the MoPmk1 signaling pathway and conidial autophagy were novel pathways suppressed by AgNPs in addition to cell wall damage. These contributed to elucidating the underlying antifungal mechanism of AgNP-mediated management of rice blast. Co-application with azoxystrobin will improve the antifungal activity of AgNPs and environmental impact. Our finding provided fundamental theoretical support for foliar application of AgNPs and azoxystrobin in future rice blast disease control.
Accession number
The M. oryzae genes from this article can be found in the GenBank database under the following accession numbers: MoRIP1 (MGG_16866), MoPMK1 (MGG_09565), MoMPS1 (MGG_04943), MoPTH11 (MGG_05871), MoMST12 (MGG_12958), and MoATG8 (MGG_01062).Data availability
All data generated or analyzed during the present study could be found within the manuscript and the supplemental files.Author contributions
Conceptualization, H. S. and Y. K.; methodology, H. W., Y. C., and Z. L.; validation, H. S.; formal analysis, H. S. and H. W.; investigation, H. S., H. W., S. X.; resources, Y. K., F. L., and X. Z.; data curation, Y. C. and Y. K.; writing – original draft preparation, H. S. and Y. K.; writing – review & editing, H. S., N. J., J. Q. and Y. K.; visualization, H. S. and Y. K.; supervision, Y. K.; project administration, Y. K.; funding acquisition, H. S, Y. K. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that there are no conflicts-of-interest related to this article. The authors declare no competing financial interests.Acknowledgements
This research was supported by the National Natural Science Foundation of China (grant no. 31900127 and 32100161), National Key Research and Development Program (2022YFD1401500), Zhejiang Provincial Natural Science Foundation of China (grant no. LY22C140006), Zhejiang Science and Technology Major Program on Rice New Variety Breeding (grant no. 2021C02063), Key R&D project of China National Rice Research Institute (grant no. CNRRI-2020-04), and the Open Project Program of State Key Laboratory of Rice Biology (grant no. 20210106). This project was also supported by the Chinese Academy of Agricultural Sciences under the Agricultural Sciences and Technologies Innovation Program. We thank Mrs. Lijuan Mao of Zhejiang University for technique assistance in FITR detection and the Rice-Pathogen Interaction Group at China National Rice Research Institute for useful discussion and suggestions.References
- A. B. Eseola, L. S. Ryder, M. Oses-Ruiz, K. Findlay, X. Yan and N. Cruz-Mireles, et al., Investigating the cell and developmental biology of plant infection by the rice blast fungus Magnaporthe oryzae, Fungal Genet. Biol., 2021, 154, 103562 CrossRef CAS PubMed.
- M. C. Fisher, D. A. Henk, C. J. Briggs, J. S. Brownstein, L. C. Madoff and S. L. McCraw, et al., Emerging fungal threats to animal, plant and ecosystem health, Nature, 2012, 484(7393), 186–194 CrossRef CAS PubMed.
- R. Dean, J. A. Van Kan, Z. A. Pretorius, K. E. Hammond-Kosack, A. Di Pietro and P. D. Spanu, et al., The Top 10 fungal pathogens in molecular plant pathology, Mol. Plant Pathol., 2012, 13(4), 414–430 CrossRef PubMed.
- X. Yan and N. J. Talbot, Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae, Curr. Opin. Microbiol., 2016, 34, 147–153 CrossRef CAS PubMed.
- L. Nalley, F. Tsiboe, A. Durand-Morat, A. Shew and G. Thoma, Economic and environmental impact of rice blast pathogen (Magnaporthe oryzae) alleviation in the United States, PLoS One, 2016, 11(12), e0167295 CrossRef PubMed.
- N. Kongcharoen, N. Kaewsalong and T. Dethoup, Efficacy of fungicides in controlling rice blast and dirty panicle diseases in Thailand, Sci. Rep., 2020, 10(1), 16233 CrossRef CAS PubMed.
- G. A. Bezerra, A. A. Chaibub, M. I. S. Oliveira, E. S. G. Mizubuti and M. C. C. Filippi, Evidence of Pyricularia oryzae adaptability to tricyclazole, J. Environ. Sci. Health, Part B, 2021, 56(10), 869–876 CrossRef CAS PubMed.
- A. Kunova, C. Pizzatti, M. Bonaldi and P. Cortesi, Sensitivity of nonexposed and exposed populations of Magnaporthe oryzae from rice to tricyclazole and azoxystrobin, Plant Dis., 2014, 98(4), 512–518 CrossRef CAS PubMed.
- Z. Q. Wang, F. Z. Meng, M. M. Zhang, L. F. Yin, W. X. Yin and Y. Lin, et al., A putative Zn2Cys6 transcription factor is associated with isoprothiolane resistance in Magnaporthe oryzae, Front. Microbiol., 2018, 9, 2608 CrossRef PubMed.
- C. Avila-Adame and W. Koller, Characterization of spontaneous mutants of Magnaporthe grisea expressing stable resistance to the Qo-inhibiting fungicide azoxystrobin, Curr. Genet., 2003, 42(6), 332–338 CrossRef CAS PubMed.
- N. Kongcharoen, N. Kaewsalong and T. Dethoup, Efficacy of fungicides in controlling rice blast and dirty panicle diseases in Thailand, Sci. Rep., 2020, 10, 16233 CrossRef CAS PubMed.
- N. Fisher, B. Meunier and G. A. Biagini, The cytochrome bc(1) complex as an antipathogenic target, FEBS Lett., 2020, 594(18), 2935–2952 CrossRef CAS PubMed.
- C. Z. Wei, H. Katoh, K. Nishimura and H. Ishii, Site-directed mutagenesis of the cytochrome b gene and development of diagnostic methods for identifying QoI resistance of rice blast fungus, Pest Manage. Sci., 2009, 65(12), 1344–1351 CrossRef CAS PubMed.
- V. L. Castroagudin, P. C. Ceresini, S. C. de Oliveira, J. T. A. Reges, J. L. N. Maciel and A. L. V. Bonato, et al., Resistance to QoI fungicides is widespread in Brazilian populations of the wheat blast pathogen Magnaporthe oryzae, Phytopathology, 2015, 105(3), 284–294 CrossRef CAS PubMed.
- T. M. Mac Loughlin, M. L. Peluso and D. J. G. Marino, Multiple pesticides occurrence, fate, and environmental risk assessment in a small horticultural stream of Argentina, Sci. Total Environ., 2022, 802, 149893 CrossRef CAS PubMed.
- S. Gharpure, A. Akash and B. Ankamwar, A review on antimicrobial properties of metal nanoparticles, J. Nanosci. Nanotechnol., 2020, 20(6), 3303–3339 CrossRef PubMed.
- Q. Liu, Y. Zhang, J. Huang, Z. Xu, X. Li and J. Yang, et al., Mesoporous silica-coated silver nanoparticles as ciprofloxacin/siRNA carriers for accelerated infected wound healing, J. Nanobiotechnol., 2022, 20(1), 386 CrossRef CAS PubMed.
- T. Mocan, C. T. Matea, T. Pop, O. Mosteanu, A. D. Buzoianu and C. Puia, et al., Development of nanoparticle-based optical sensors for pathogenic bacterial detection, J. Nanobiotechnol., 2017, 15(1), 25 CrossRef PubMed.
- A. L. M. Terra, R. D. C. Kosinski, J. B. Moreira, J. A. V. Costa and M. G. Morais, Microalgae biosynthesis of silver nanoparticles for application in the control of agricultural pathogens, J. Environ. Sci. Health, Part B, 2019, 54(8), 709–716 CrossRef CAS PubMed.
- M. Akter, M. T. Sikder, M. M. Rahman, A. Ullah, K. F. B. Hossain and S. Banik, et al., A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives, J. Adv. Res., 2018, 9, 1–16 CrossRef CAS PubMed.
- N. J. Talbot, Appressoria, Curr. Biol., 2019, 29(5), R144–R146 CrossRef CAS PubMed.
- G. Li, X. Zhou and J. R. Xu, Genetic control of infection-related development in Magnaporthe oryzae, Curr. Opin. Microbiol., 2012, 15(6), 678–684 CrossRef CAS PubMed.
- X. H. Liu, H. M. Gao, F. Xu, J. P. Lu, R. J. Devenish and F. C. Lin, Autophagy vitalizes the pathogenicity of pathogenic fungi, Autophagy, 2012, 8(10), 1415–1425 CrossRef CAS PubMed.
- C. Jiang, X. Zhang, H. Liu and J. R. Xu, Mitogen-activated protein kinase signaling in plant pathogenic fungi, PLoS Pathog., 2018, 14(3), e1006875 CrossRef PubMed.
- K. S. Bruno, F. Tenjo, L. Li, J. E. Hamer and J. R. Xu, Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea, Eukaryotic Cell, 2004, 3(6), 1525–1532 CrossRef CAS PubMed.
- T. M. DeZwaan, A. M. Carroll, B. Valent and J. A. Sweigard, Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues, Plant Cell, 1999, 11(10), 2013–2030 CrossRef CAS PubMed.
- Y. Kou, Y. H. Tan, R. Ramanujam and N. I. Naqvi, Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast, New Phytol., 2017, 214(1), 330–342 CrossRef CAS PubMed.
- X. Zhao and J. R. Xu, A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea, Mol. Microbiol., 2007, 63(3), 881–894 CrossRef CAS PubMed.
- N. Mizushima, T. Yoshimori and Y. Ohsumi, The role of Atg proteins in autophagosome formation, Annu. Rev. Cell Dev. Biol., 2011, 27, 107–132 CrossRef CAS PubMed.
- C. Veneault-Fourrey, M. Barooah, M. Egan, G. Wakley and N. J. Talbot, Autophagic fungal cell death is necessary for infection by the rice blast fungus, Science, 2006, 312(5773), 580–583 CrossRef CAS PubMed.
- E. Ibrahim, J. Luo, T. Ahmed, W. Wu, C. Yan and B. Li, Biosynthesis of silver nanoparticles using onion endophytic bacterium and its antifungal activity against rice pathogen Magnaporthe oryzae, J. Fungi, 2020, 6(294), 1–13 Search PubMed.
- Y. K. Jo, B. H. Kim and G. Jung, Antifungal activity of silver ions and nanoparticles on phytopathogenic fungi, Plant Dis., 2009, 93(10), 1037–1043 CrossRef CAS PubMed.
- X. H. Liu, S. Liang, Y. Y. Wei, X. M. Zhu, L. Li and P. P. Liu, et al., Metabolomics analysis identifies sphingolipids as key signaling moieties in appressorium morphogenesis and function in Magnaporthe oryzae, MBio, 2019, 10, e01467-19 CrossRef PubMed.
- K. Tamura, M. Nei and S. Kumar, Prospects for inferring very large phylogenies by using the neighbor-joining method, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(30), 11030–11035 CrossRef CAS PubMed.
- S. Kumar, G. Stecher and K. Tamura, MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, 33(7), 1870–1874 CrossRef CAS PubMed.
- H. Shi, S. Meng, J. Qiu, C. Wang, Y. Shu and C. Luo, et al., MoWhi2 regulates appressorium formation and pathogenicity via the MoTor signalling pathway in Magnaporthe oryzae, Mol. Plant Pathol., 2021, 22(8), 969–983 CrossRef CAS PubMed.
- H. Wen, H. Shi, N. Jiang, J. Qiu, F. Lin and Y. Kou, Antifungal mechanisms of silver nanoparticles on mycotoxin producing rice false smut fungus, iScience, 2023, 26(1), 105763 CrossRef CAS PubMed.
- X. M. Zhu, S. Liang, H. B. Shi, J. P. Lu, B. Dong and Q. S. Liao, et al., VPS9 domain-containing proteins are essential for autophagy and endocytosis in Pyricularia oryzae, Environ. Microbiol., 2018, 20(4), 1516–1530 CrossRef CAS PubMed.
- M. Oses-Ruiz, N. Cruz-Mireles, M. Martin-Urdiroz, D. M. Soanes, A. B. Eseola and B. Tang, et al., Appressorium-mediated plant infection by Magnaporthe oryzae is regulated by a Pmk1-dependent hierarchical transcriptional network, Nat. Microbiol., 2021, 6(11), 1383–1397 CrossRef CAS PubMed.
- H. Cheong and D. J. Klionsky, Biochemical methods to monitor autophagy-related processes in yeast, Methods Enzymol., 2008, 451, 1–26 CAS.
- K. S. Siddiqi, A. Husen and R. A. K. Rao, A review on biosynthesis of silver nanoparticles and their biocidal properties, J. Nanobiotechnol., 2018, 16(1), 14 CrossRef PubMed.
- K. Lamsal, S. W. Kim, J. H. Jung, Y. S. Kim, K. S. Kim and Y. S. Lee, Application of silver nanoparticles for the control of Colletotrichum species in vitro and pepper anthracnose disease in field, Mycobiology, 2011, 39(3), 194–199 CrossRef CAS PubMed.
- A. A. Malandrakis, N. Kavroulakis and C. V. Chrysikopoulos, Use of silver nanoparticles to counter fungicide-resistance in Monilinia fructicola, Sci. Total Environ., 2020, 747, 141287 CrossRef CAS PubMed.
- Y. Jian, X. Chen, T. Ahmed, Q. Shang, S. Zhang and Z. Ma, et al., Toxicity and action mechanisms of silver nanoparticles against the mycotoxin-producing fungus Fusarium graminearum, J. Adv. Res., 2022, 38, 1–12 CrossRef CAS PubMed.
- H. Fouad, G. Yang, A. A. El-Sayed, G. Mao, D. Khalafallah and M. Saad, et al., Green synthesis of AgNP-ligand complexes and their toxicological effects on Nilaparvata lugens, J. Nanobiotechnol., 2021, 19(1), 318 CrossRef CAS PubMed.
- H. H. Lara, D. G. Romero-Urbina, C. Pierce, J. L. Lopez-Ribot, M. J. Arellano-Jimenez and M. Jose-Yacaman, Effect of silver nanoparticles on Candida albicans biofilms: an ultrastructural study, J. Nanobiotechnol., 2015, 13, 91 CrossRef PubMed.
- M. A. Raza, Z. Kanwal, A. Rauf, A. N. Sabri, S. Riaz and S. Naseem, Size- and shape-dependent antibacterial studies of silver nanoparticles synthesized by wet chemical routes, Nanomaterials, 2016, 6(74), 1–15 Search PubMed.
- I. Geoghegan, G. Steinberg and S. Gurr, The role of the fungal cell wall in the infection of plants, Trends Microbiol., 2017, 25(12), 957–967 CrossRef CAS PubMed.
- T. Shen, Q. Wang, C. Li, B. Zhou, Y. Li and Y. Liu, Transcriptome sequencing analysis reveals silver nanoparticles antifungal molecular mechanism of the soil fungi Fusarium solani species complex, J. Hazard. Mater., 2020, 388, 122063 CrossRef CAS PubMed.
- J. Y. Roh, H. J. Eom and J. Choi, Involvement of Caenohabditis elegans MAPK signaling pathways in oxidative stress response induced by silver nanoparticles exposure, Toxicol. Res., 2012, 28(1), 19–24 CrossRef CAS PubMed.
- B. Villeret, A. Dieu, M. Straube, B. Solhonne, P. Miklavc and S. Hamadi, et al., Silver nanoparticles impair retinoic acid-inducible gene I-mediated mitochondrial antiviral immunity by blocking the autophagic flux in lung epithelial cells, ACS Nano, 2018, 12(2), 1188–1202 CrossRef CAS PubMed.
- E. Thines, R. W. Weber and N. J. Talbot, MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea, Plant Cell, 2000, 12(9), 1703–1718 CAS.
- W. Lv, Y. Xiao, Z. Xu, H. Jiang, Q. Tong and Z. Wang, The paxillin MoPax1 activates mitogen-activated protein(MAP) kinase signaling pathways and autophagy through MAP kinase activator MoMka1 during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae, MBio, 2022, e0221822 CrossRef PubMed.
- Y. S. Kim, E. W. Dixon, P. Vincelli and M. L. Farman, Field resistance to strobilurin (Q(o)I) fungicides in Pyricularia grisea caused by mutations in the mitochondrial cytochrome b gene, Phytopathology, 2003, 93(7), 891–900 CrossRef CAS PubMed.
- E. A. Berry and L. S. Huang, Conformationally linked interaction in the cytochrome bc(1) complex between inhibitors of the Q(o) site and the Rieske iron-sulfur protein, Biochim. Biophys. Acta, 2011, 1807(10), 1349–1363 CrossRef CAS PubMed.
- S. Qamer, M. H. Romli, F. Che-Hamzah, N. Misni, N. M. S. Joseph and N. A. Al-Haj, et al., Systematic review on biosynthesis of silver nanoparticles and antibacterial activities: application and theoretical perspectives, Molecules, 2021, 26(5057), 1–21 Search PubMed.
- S. Shanthi, B. D. Jayaseelan, P. Velusamy, S. Vijayakumar, C. T. Chih and B. Vaseeharan, Biosynthesis of silver nanoparticles using a probiotic Bacillus licheniformis Dahb1 and their antibiofilm activity and toxicity effects in Ceriodaphnia cornuta, Microb. Pathog., 2016, 93, 70–77 CrossRef CAS PubMed.
- Y. Tang, C. Chen, B. Jiang, L. Wang, F. Jiang and D. Wang, et al., Bifidobacterium bifidum-mediated specific delivery of nanoparticles for tumor therapy, Int. J. Nanomed., 2021, 16, 4643–4659 CrossRef PubMed.
- E. Ibrahim, M. Zhang, Y. Zhang, A. Hossain, W. Qiu and Y. Chen, et al., Green-synthesization of silver nanoparticles using endophytic bacteria isolated from garlic and its antifungal activity against wheat Fusarium head blight pathogen Fusarium graminearum, Nanomaterials, 2020, 10(2), 219 CrossRef CAS PubMed.
- H. Zhang, S. Chen, X. Jia, Y. Huang, R. Ji and L. Zhao, Comparation of the phytotoxicity between chemically and green synthesized silver nanoparticles, Sci. Total Environ., 2021, 752, 142264 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00168g |
| This journal is © The Royal Society of Chemistry 2023 |