Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Complementary in vitro and in vivo strategies to assess the biological effects of the nano enabled food additives E171 and E551†
Ana
Peropadre
 ab,
Patricia
Vega-Cuesta
ab,
Patricia
Vega-Cuesta
 c,
Paloma
Fernández Freire
c,
Paloma
Fernández Freire
 b,
Diego
Pulido
b,
Diego
Pulido
 c,
Marie
Carriere
c,
Marie
Carriere
 *a and
Jose F.
de Celis
*a and
Jose F.
de Celis
 *c
*c
aCEA, CNRS, IRIG, SyMMES, CIBEST, Univ. Grenoble-Alpes, 38000 Grenoble, France. E-mail: marie.carriere@cea.fr
bDepartamento de Biología, Facultad de Ciencias, Universidad Autónoma de Madrid, Spain
cCentro de Biología Molecular Severo Ochoa, CSIC-UAM, Nicolás Cabrera 1, Madrid 28049, Spain. E-mail: jfdecelis@cbm.csic.es
First published on 27th March 2023
Abstract
We have analyzed the effects on Drosophila melanogaster intestines and two human cell lines of intestinal origin of two different nano-enabled food additives belonging to the persistent or slowly soluble engineered nanomaterials (NM): silicon dioxide E551 and titanium dioxide E171. These compounds are considered as high priority for toxicological testing. Our analysis combines the advantages of a model organism (Drosophila) with the mechanistic approaches feasible in cellular models. We evaluated general cytotoxicity in both models, and selected 10 and 100 μg mL−1 as exposure concentrations that do not compromise cellular or organism viability. We identified changes in the expression levels of a selected set of genes related to the regulation of oxidative stress and DNA integrity. Interestingly, these changes were in the same direction when comparing Drosophila intestines and human differentiated Caco-2 cells, indicating a conservation in the general cellular response to E551 and E171 exposure in both systems. The use of Drosophila offers unique opportunities in the anatomical, physiological and biochemical analysis of the exposure to engineered nanomaterials, as it complements in vitro approaches in an experimental setting that reproduces many of the biological characteristics of the human intestine. Drosophila is relatively inexpensive, genetically versatile and compliant with 3R principles, and consequently we expect that its use will contribute to other risk assessment strategies aiming to identify the action mechanisms of toxicants.
Environmental significanceLong-term animal experimentation and human epidemiological studies constitute the legislative base regulating the safe use of nanomaterials as alimentary additives. In addition, regulatory agencies recommend the use of alternative methods to reduce animal experimentation and develop less expensive and relevant toxicity testing methodologies. Here we show that Drosophila intestines and differentiated intestinal human cells show similar responses to additives E171 and E551 on oral exposition, including alterations in the expression of genes related to oxidative stress and DNA damage. We suggest that Drosophila could become a powerhouse in the development of in vivo strategies to analyse the biological consequences of nano-enabled materials ingested in the food, and together with human in vitro models promote the standardization of novel risk assessment methodologies. |
Introduction
Nanomaterials (NMs) have significant impact on the food sector, as they are used to produce perishable aliments with better flavoring characteristics or increased shelf life.1,2 For this reason, the oral intake of nanotechnology-derived food products is a relevant component of alimentation and a potential risk to consumers worldwide.3 The use of NMs as alimentary food additives raises safety concerns due to their small size, which allows them to pass through the intestinal barrier.4 Once absorbed into cells, NMs have the potential to disrupt cellular homeostasis and organ physiology, ultimately leading to toxicity. However, and despite considerable efforts devoted to the study of NM effects on mammalian cells in recent years, there is still insufficient evidence to identify health risks associated with NM oral exposure, particularly regarding long-term exposure or chronic risk.5 Synthetic amorphous silica (SAS) and titanium dioxide are among the most predominant nanoengineered compounds used across many industries, with an increased presence in processed food.3,6 Their components are not dissolved during digestion and may be absorbed, retained, and accumulated within the body. In addition, those used as dietary supplements are considered as high priority for testing due to concerns in their safety profiles.4Silicon and titanium dioxide are used as additives in the food industry (E551 and E171 respectively) as well as in other edible consumer goods such as medicines, chewing gums, icings, and toothpaste.3,7,8 E551 has been related to the induction of oxidative stress and genotoxic effects in different cellular types,9–11 though at concentrations not relevant for short-term acute exposures.12 However, other studies examining the toxicity of E551 upon oral exposure report that a health risk of these particles in food could not be excluded.13 E171, in contrast, met the criteria to be classified as a suspected human carcinogen (category 2) at least upon inhalation,14 and cannot be considered as safe when used as a food additive according to the European Food Safety Agency.15 It has been suggested that titanium dioxide might induce or promote colon tumors via inflammation and reactive oxygen species (ROS) production.16 These effects could occur at lower but cumulative doses, but there is insufficient data available on this regard.17 As diet is the main source of exposure to nano-enabled food additives, it is a priority to obtain relevant data on their potential effect upon the gastrointestinal tract. Experimental studies involving oral exposure18 or inhalation19 to nanoparticles revealed that they might cause an excessive ROS production, leading to oxidative stress, inflammatory responses, and ultimately epithelial injury. Chronic exposure can induce persistent injury and DNA damage, leading to tissue and organ effects including regenerative cell proliferation, hyperplasia, and ultimately intestinal tumors.17
The study of possible harmful effects of NM is mostly conducted in vitro, as 2-dimensional cell cultures allow standardized functional assays, high data reproducibility and direct observations concerning the cellular effects and cytotoxicity of nanomaterials present in the cell culture medium.20In vitro approaches have limitations compared to in vivo systems. They include differences in particle aggregation in culture media and variations in uptake, pharmacokinetics and biodistribution of NM in cultured cells. For these reasons, the use of in vivo models is a necessary step in the evaluation of biological risks arisen from NM exposal. A major goal of the in vivo approaches is to promote a better understanding of the biological consequences of NM exposure in the context of the entire organism, preferably in realistic scenarios. Ideally, a combination of in vitro and in vivo experimental approaches should offer a better opportunity to identify relevant biochemical and cellular effects of NM exposure leading to toxicological effects, which combined with human epidemiological studies could improve and accelerate the appraisal of risks to human health.
In this work we have combined experiments carried out in human intestinal cells and in Drosophila melanogaster intestines exposed to different doses of the E551 and E171 additives. Our main goal is to identify to what extent the exposure to these additives elicits common cellular responses in seemly unrelated biological settings. We argue that similar responses are a strong indication of common mechanisms of action, and this should reinforce the use of Drosophila melanogaster as a toxicity testing platform for NMs. Drosophila melanogaster has several strengths to favour its use in general toxicological evaluations. The first is to exploit the readily available possibilities offered by genetic manipulations, which can be targeted to affect its life cycle, physiology, metabolism and multitude of different cellular and biochemical parameters. A second advantage is the overall resemblance existing between Drosophila organs and their vertebrate counterparts. The similarities are evident at the cellular and biochemical levels, and in some instances, they also extend to organs retaining a similar supracellular architecture and physiology. One of such organs is the digestive tract, and in particular the intestine, which in both Drosophila and vertebrates is formed by monolayered epithelia wrapped by longitudinal and transversal visceral muscles.21 The luminal side of the intestinal epithelium is covered by a layer formed by chitin and glycoproteins, the peritrophic matrix, which plays an important role in the defence against enteric pathogens, and is the equivalent of vertebrate intestinal mucus.22 Finally, Drosophila also offers the possibility of detailed in situ gene expression analyses, allowing the use of reporter constructs to visualize the activity of cellular stress response pathways as a first indication of possible harmful effects.
The potential of Drosophila for risk assessment in nanotoxicological research is reflected in the increasing number of experimental works dealing, among others, with silver, gold, zinc, titanium dioxide and silica nanoparticles.23 Many of these works focus in the evaluation of organismal end-points, including developmental timing, fertility, behavioural alterations or survival rates, after dietary ingestion of different NM supplied in the larval or adult food.23 In addition, these approaches also include the study of inflammatory, cytotoxic and genotoxic adverse effects, as well as microscopic examination of nanoparticle subcellular localization, gene expression analyses of cytoprotective genes and a variety of genotoxic assays.23 There is a reasonable agreement in the potential of the Drosophila model, and a general consensus that the main route of putative damage caused by NM ingestion is due to the generation of ROS that may lead to oxidative damage and genotoxic effects in the target cell population. However, in most instances the effects of NM exposure are very weak, and many studies report conflicting results.23–25 This may be due to the focus in organismal end points that might be distant to the actual possible cellular damage caused by ingestion of NM, the difficulty in estimating effective doses, NMs physicochemical characteristics when they are presented in the fly food and the challenges posed to define NMs uptake and distribution once they enter the digestive tract.
In this work we focus on the in vivo analyses of E551 and E171 exposure to Drosophila melanogaster larval intestines. We expect to be closer to the immediate site of action of NMs ingested in the food, allowing a better cause to effect reconstruction of events. In addition, we have used both Drosophila normal larvae and genetically modified larvae displaying an extended period of larval development. These larvae allow the implementation of long-term studies at the main feeding stage of Drosophila development. We first defined exposure conditions in which the basal cytotoxicity is minimal in our two cellular models and have no consequence on Drosophila developmental timing and survival. In these conditions, which are reasonably close to realistic exposure scenarios (Table S1†), we monitored the expression of genes related to cellular stress, oxidative stress and DNA damage in the cells and in the larval intestine. Remarkably, we find a strong similarity in the response of differentiated Caco-2 cells and Drosophila intestines after exposure to E551 and E171. This response consisted in an increase in the expression of oxidative stress markers without associated genotoxicity. The expression of one of these genes, Gst-D1, was also monitored in situ in larval intestinal cells. Changes in gene expression occurred without major alterations to intestine ultrastructure, suggesting that they constitute an early response of intestinal cells to the presence of E551 and E171 in the intestinal tract. We propose that the combination of in vitro and in vivo experimental models in which similar parameters can be measured directly in the target cell population favour the identification of common cellular responses and have the potential to improve the understanding of the consequences of dietary long-term exposures to NMs.
Experimental methods
Experimental design
We used human intestinal cell lines and Drosophila melanogaster larval guts as experimental models (Fig. S1†). We determined viability for human cell lines, and survival and pupariation rates for D. melanogaster. The relative expression of genes belonging to the DNA damage and oxidative stress pathways were evaluated in both model systems by RT-qPCR (Fig. S1†). Undifferentiated (HCT116, ATCC CTL-247) and differentiated (Caco-2, ATCC HTB-37) human intestinal cells were exposed for 24 h (acute exposure) to increasing concentrations (0.5 to 100 μg mL−1) of E171 or E551 diluted in complete cell culture medium at pH 7.4 (Fig. S1A†). Control cells were exposed to the maximum amount of the vehicle used (1% distilled H2O). Drosophila melanogaster with normal and extended development were of GstD1-GFP and GstD1-GFP/+; phm-Gal4/UAS-Smt3-RNAi genotypes, respectively (Fig. S1B†). The phm-Gal4 driver directs the expression of Smt3 RNAi in the prothoracic gland, preventing the synthesis of the molting hormone ecdysone.26,27 Mutant GstD1-GFP/+; phm-Gal4/UAS-Smt3-RNAi larvae are unable to pupariate and remain in an extended third instar stage for up to 35 d after egg laying. Second instar larvae of 24–48 h age were placed in fly media prepared with water (control food) or with water containing different concentrations of E171 or E551 (1–100 μg mL−1). Larval midguts were collected for immunocytochemistry and mRNA extraction after 4 d of culture (GstD1-GFP; normal development) or after 7, 14 and 21 d of culture (GstD1-GFP/+; phm-Gal4/UAS-Smt3-RNAi; extended development, Fig. S1B†).We selected 10 and 100 μg mL−1 of E171 and E551 as experimental doses for immunocytochemistry and mRNA analysis. In order to contextualize these doses, we used the estimated human daily intake (EDI) and their equivalent dose in Caco-2 cell defined by Sohal et al. (2020)28 and Putra et al. (2022).29 We transformed our doses (10 and 100 μg mL−1) to the same units (μg cm−2) for intestinal cells to obtain equivalent EDIs (Table S1†). Following a similar procedure, we calculated the dose range/surface and the equivalent of EDI for Drosophila melanogaster, using estimated values for food intake30 and midgut surface.31 Our low dose lies within the EDI for Caco-2, one order of magnitude higher for HCT116 and three orders of magnitude lower for D. melanogaster (Table S1†). Although these estimations should be taken with caution, especially for the fruit fly, the values obtained support that our selected doses are within the range of realistic exposure scenarios.
Cell cultures
We grew Caco-2 cells (ATCC HTB-37, passages 49 to 60) in DMEM GlutaMAX™ supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids and 1% penicillin–streptomycin. For HTC116 (ATCC CCL-247) cells we used McCoy's 5a medium modified with stable L-glutamine and supplemented with 10% FBS and 1% penicillin–streptomycin. Both cell lines were maintained at 37 °C under a humified atmosphere with 5% CO2. We obtained all cell culture media and supplements from Life Technologies (California, USA).Before initiating the treatments, we seeded Caco-2 cells at a density of 5 × 105 cells per mL and allowed them to differentiate by maintaining the culture for 21 d post-confluence with medium changes every 48–72 h. In the case of the HCT116 cell line, we used a seeding density of 1.5 × 105 cells per mL, and cells grew to confluence (24 h) before treatments.
Drosophila genetics and fly cultures
We use the GstD1 reporters GST-lacZ and GST-GFP to visualize the activity of the GstD1 regulatory region.32 Flies were raised at 25 °C (unless otherwise stated) in fly medium containing glucose (Panreac; 50 g L−1), agar (Condalab; 7.86 g L−1), wheat flour (Gallo; 35.7 g L−1), yeast (LevaReal; 71.4 g L−1), methylparaben (Sigma; 2.8 mL L−1) and propionic acid (Sigma; 4.3 mL L−1).Physicochemical characterization of E171 and E551 nanoparticles
Food-grade TiO2 particles (E171 in Europe) were purchased from a French commercial supplier of food coloring. The stock solution (10 mg mL−1) was suspended in ultrapure sterile water and dispersed using an indirect cup-type sonicator (Vibracell 75041, Bioblock Scientific), operated in continuous mode for 30 min at 80% amplitude and 4 °C. SiO2 particles were obtained from an industrial collaborator producing food grade precipitated silica (E551). The compound, provided as powder, was suspended in ultrapure water at the concentration of 10 mg mL−1 and sterilized by heating at 80 °C overnight. After dispersion, the zeta-potential (ξ) and agglomeration state of suspensions were characterized in water by dynamic light scattering on a ZetaSizer nanoZS (Malvern Instruments Ltd., Worcestershire, UK). The characterization by transmission electron microscopy of these two substances has been previously published.33,34Cytotoxicity assays in HTC116 and Caco-2 cells
We used the WST-1 colorimetric assay (Roche, Mannheim, Germany) for the evaluation of cell metabolic activity. In brief, cells grown in 96-well plates were exposed for 24 h to different concentrations of E171 and E551. After the exposure period, the medium containing nanoparticles was discarded and replaced by 100 μL of WST-1 diluted to the tenth, as indicated by the supplier. After incubation for 1 h at 37 °C, the quantification of metabolic activity was calculated from absorbance measurement at 450 nm in a spectramax M2 (Molecular Devices). The absorbance values of treated cells were referenced to those cultured in control media. We used as positive control cells incubated for 24 h in media containing 10% dimethyl sulfoxide (DMSO). Interference test was performed in control HCT116 cells (not exposed to E171 or E551) incubated with WST-1 for 1 h at 37 °C. After agitation, this solution was mixed with 0, 1, 10 or 100 μg mL−1 of E171 or E551. The plate was then centrifuged for 5 min at 200 rcf, and 50 μL of each well was transferred to a clean plate. Absorbance was measured at 450 nm and corrected via subtraction of background absorbance at 650 nm.Developmental timing and adult viability calculations in Drosophila
We measured the time in days from egg laying to puparium formation in larvae of the same genetic background grown in media prepared with control food or with food containing different concentrations of E171 or E551. We also counted the number of pupae and emerging adults each 24 h. All experiments were conducted at 25 °C in incubation chambers with 70% humidity. To synchronize larvae for survival assays, flies were allowed to lay eggs for 3 h on grape-juice agar plates supplemented with yeast paste. 48 h later newly hatched L2 larvae were transferred into 5 mL vials (20 per vial to prevent crowding) and reared at 25 °C. Dead larvae were scored twice a day. Survival data were plotted using Kaplan–Meier curves (GraphPad Prism 9 software).Immunocytochemistry and confocal microscopy
We dissected guts from larvae of GstD1-GFP and GstD1-lacZ genotype growing in control food or in food containing different concentrations of E171 or E551. Larval guts were extracted in PBS, rinsed in PBS/Triton X100 0.3% (PBT), and fixed in 4% formaldehyde in PBT for 30 min. After two rinses in PBT (15 min), the samples were incubated in PBT containing 4% bovine serum albumin (BSA) for 1 h, and then overnight at 4 °C in PBT-BSA containing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution of rabbit anti phospho-H2AV (ThermoFisher Scientist). The samples were washed in PBT and incubated for 2 h at room temperature with secondary red fluorescent anti-rabbit antibody (ThermoFisher Scientist, 1
200 dilution of rabbit anti phospho-H2AV (ThermoFisher Scientist). The samples were washed in PBT and incubated for 2 h at room temperature with secondary red fluorescent anti-rabbit antibody (ThermoFisher Scientist, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution) in PBT-BSA containing a 1
200 dilution) in PBT-BSA containing a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of To-Pro. After several rinses in PBT the guts were mounted on microscopic slides in Vectashield mounting medium (Vector Laboratories). Images were acquired using a confocal LSM 510 (Zeiss) confocal microscope using the 20× and 40× objectives at 512 × 512–pixel resolution. All images were processed with the program ImageJ 1.48 u (NIH, USA) and Adobe Photoshop CS3 (Adobe Systems Inc.).
1000 dilution of To-Pro. After several rinses in PBT the guts were mounted on microscopic slides in Vectashield mounting medium (Vector Laboratories). Images were acquired using a confocal LSM 510 (Zeiss) confocal microscope using the 20× and 40× objectives at 512 × 512–pixel resolution. All images were processed with the program ImageJ 1.48 u (NIH, USA) and Adobe Photoshop CS3 (Adobe Systems Inc.).
Transmission electron microscopy (TEM)
For electron microscopy, dissected larval guts were rinsed in PBS and fixed in 4% formaldehyde/0.04% glutaraldehyde in PBS. The guts were embedded in epoxy resine, cryosectioned and visualized in a Jem1010 (JEOL) instrument working at 80 kV with a TemCam F416 (TVIPS) camera and EMenu software. Transmission electron microscopy images were processed with Adobe Photoshop CS3.Gene expression analyses
RNA for real-time PCR (RT-qPCR) was extracted from Drosophila larval guts (30 guts per sample in three replicates) dissected in PBS and stored at −80 °C in TRIzol, or from human intestinal cells Caco-2 and HCT116 grown in 12-well plates. Total RNA was extracted using the guanidinium isothiocyanate method (TRIzol reagent; Invitrogen, Carlsbad, CA), followed by purification using RNeasy columns (Qiagen, Valencia, CA). Each RNA preparation was tested for degradation using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA samples were treated with DNAse (DNA-free Dnase Treatment and Removal Reagents, Applied Biosystems). We used 1 mg of total RNA for a first round of reverse transcription employing the Superscript III First Strand Synthesis Supermix kit for qRT-PCR (Invitrogen). Quantitative PCR analysis was performed in a Cfx 384 Real-Time System (BioRad). To normalize the results, we used probes for the genes Act5C and RpL32 (D. melanogaster), RPL32 and 18S (human intestinal cells). The list of oligonucleotides we used is presented in Table S3.† We used the following positive controls:– Oxidative stress response: larvae grown for 8 h previous to dissection in normal media containing 1.5% of H2O2 and cells exposed to 250 μM of H2O2 for 8 h.
– DNA damage response: irradiated larvae with a final dose of 2000 Roentgen 3 h before dissection, and cells (Caco-2/HCT116) grown for 24 h in media containing etoposide 50 μM.
Statistical analysis
Numerical data, including fluorescence intensity values and RT-PCR, were collected from at least three independent experiments and processed in Microsoft Excel (Microsoft Inc.) and Prism 9 (GraphPad Software, LLC). The statistical analyses of survival curves were carried out using log-rank (Mantel–Cox) and Gehan–Breslow–Wilcoxon tests. Quantitative data was assessed for normality (Shapiro–Wilk test) and homoscedasticity (Brown–Forsythe or Bartlett's test) and subsequently analyzed for group comparisons with ordinary one-way ANOVA, Brown–Forsythe and Welch ANOVA or Kruskal–Wallis test, followed by appropriate post hoc analyses (Dunnett, Dunnett T3 or Dunn's respectively). When required, unpaired t test or Mann–Whitney test with two-tails were used for simple comparisons. We consider that there was a significant difference when the p-value is lower than 0.0001 (****), 0.001 (***), 0.01 (**) or 0.05 (*). Benchmark dose response analysis for cytotoxicity assays was performed using the free web-based software PROAST v.70.1 (RIVM National Institute for Public Health and the Environment, Netherlands).Results and discussion
TiO2 E171 and silica E551 characterization
E171 was anatase-dioxide with traces of rutile forms (<5%) and displayed a mean primary diameter of 119 nm.35 Representative images of E171 TEM analysis are provided in Fig. S2.† TiO2 particle solutions were dominated by a mode with a peak-size close to 400 nm in sterile-filtered MilliQ-water, and a tail upwards into the μm-range (Fig. S2†). E551 is a nanostructured material, intentionally composed of aggregated SiO2 nanoparticles linked to one another by covalent bonds to achieve a chaplet shape (Fig. S2†). Our E551 water dispersions showed a peak-size near to 400 nm for 10 μg mL−1 and reaching 800 nm for 100 μg mL−1 (Fig. S2†). Our size analysis indicates that E171 produces the smallest Z-average and polydispersity index (PdI) of the tested materials (Fig. S2†). The water suspensions were stable, and the zeta potential was slightly negative (∼−30 mV), which also suggests a tendency towards agglomeration. A complete characterization in cell culture media of the obtained suspension has been published previously. Thus, in cell culture exposure medium, E551 formed very large agglomerates with diameter >5 μm,35 and E171 showed slight agglomeration (hydrodynamic diameter of 739.3 ± 355.3 nm (ref. 33)).Short-time exposures to E171 or E551 cause minimal cytotoxicity in cultured intestinal cells and do not affect Drosophila developmental time or survival until adult eclosion
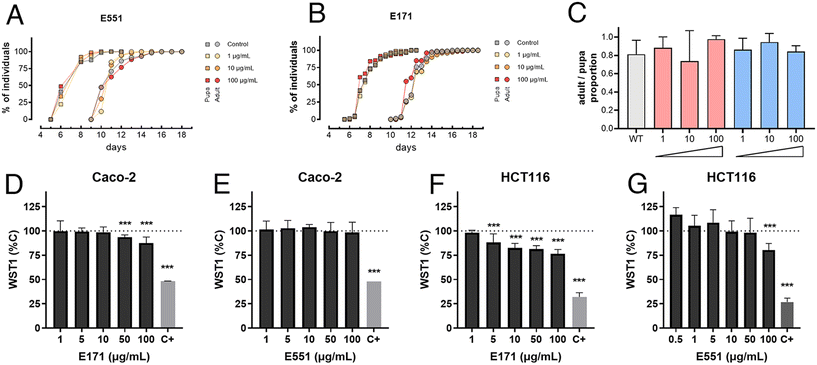
As a preliminary step to characterize the consequences of E171 and E551 exposure, we first measured basic developmental and survival parameters of Drosophila larvae feeding in fly food containing three different concentrations of E171 and E551 (1, 10 and 100 μg mL−1) for 4 d (Fig. 1A–C). We found that the developmental time (measured in time from egg laying to puparium formation) and the rate of survival (percentage of viable pupae) were not affected at these concentrations (Fig. 1A–C). Likewise, the exposure of differentiated Caco-2 cells to a similar range of E171 or E551 (0.5–100 μg mL−1) for 24 h caused mild effects in viability (WST-1; Fig. 1D and E), statistically significant with respect to controls only for the highest E171 concentrations. Undifferentiated intestinal cells HCT116 were more sensitive to increasing concentrations of E171 (Fig. 1F) and E551 particles (Fig. 1G). Regarding benchmark dose response analyses, the lower and upper limits for Caco-2 cells could only be obtained for E171 (14.9–64.2 μg mL−1), while for HCT116 cell line were 2.44 × 10−6–1.11 μg mL−1 for E171 and 1.48–25.8 μg mL−1 for E551. These results are in agreement with other studies on gut models reporting the impact of these NPs, which also failed to identify significant acute toxic effects at low doses.36,37 With these values of organism viability and cellular cytotoxicity, we chose 10 and 100 μg mL−1 as the low and high concentrations for the following set of experiments, assuming that at these concentrations any consequences of E551 or E171 exposure occur without compromising cellular viability.GstD1 expression is induced in Drosophila guts exposed to E171 or E551
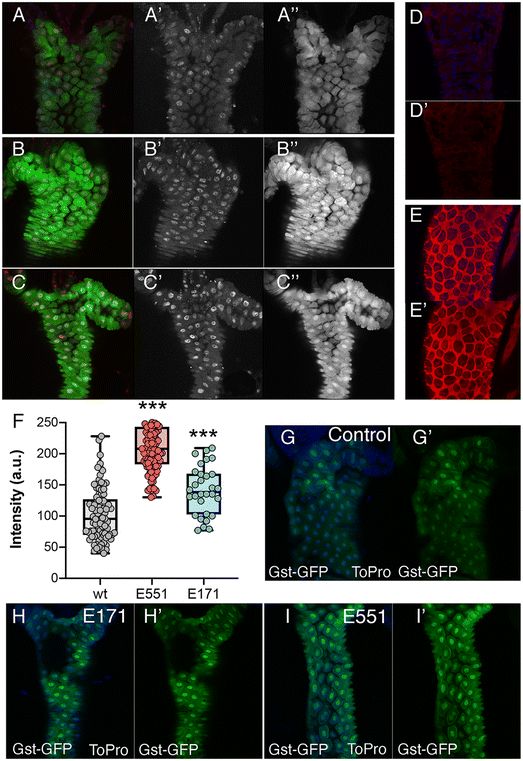
The most prominent consequence of the exposure to silica or titanium particles, observed in a variety of biological settings, is an increased ROS production leading to oxidative stress.38–40 As an indication of oxidative stress, we decided to monitor the expression of genes involved in oxidative stress response pathways, as their expression is triggered by free radicals and serve as an early indication of oxidative stress. We first monitored the expression of reporter constructs containing the regulatory region of the GstD1 gene fused to either lacZ or GFP. In GstD1-GFP or GstD1-lacZ larvae (Fig. 2A–C) and adult flies (Fig. 2D and E) we observed a robust induction of GFP or lacZ expression in response to the presence in the media of H2O2 compared to control flies or larvae growing in normal media (Fig. 2A, B, D and E). We also observed a strong induction of GFP expression, as well as the appearance of DNA damage in intestinal cells of GstD1-GFP larvae irradiated 3 h before dissection (Fig. 2C). The induction of reporter gene expression to increased RedOx misbalance was then used to evaluate the effect of food containing 100 μg mL−1 of E171 or E551. In both cases we found a significant increase in the level of GstD1-GFP expression (Fig. 2F–I). These levels were consistently higher after exposure to E551 than to E171 (Fig. 2F–I). In this manner, exposure to E171 and E551 for 4 d that do not impact on cell or organism viability cause an increase in the expression of GstD1, which is compatible with previous findings in Drosophila40,41 and in human cultured cells.33,38The expression of genes involved in the response to oxidative stress changes in Drosophila guts and human cells exposed to E171 and E551
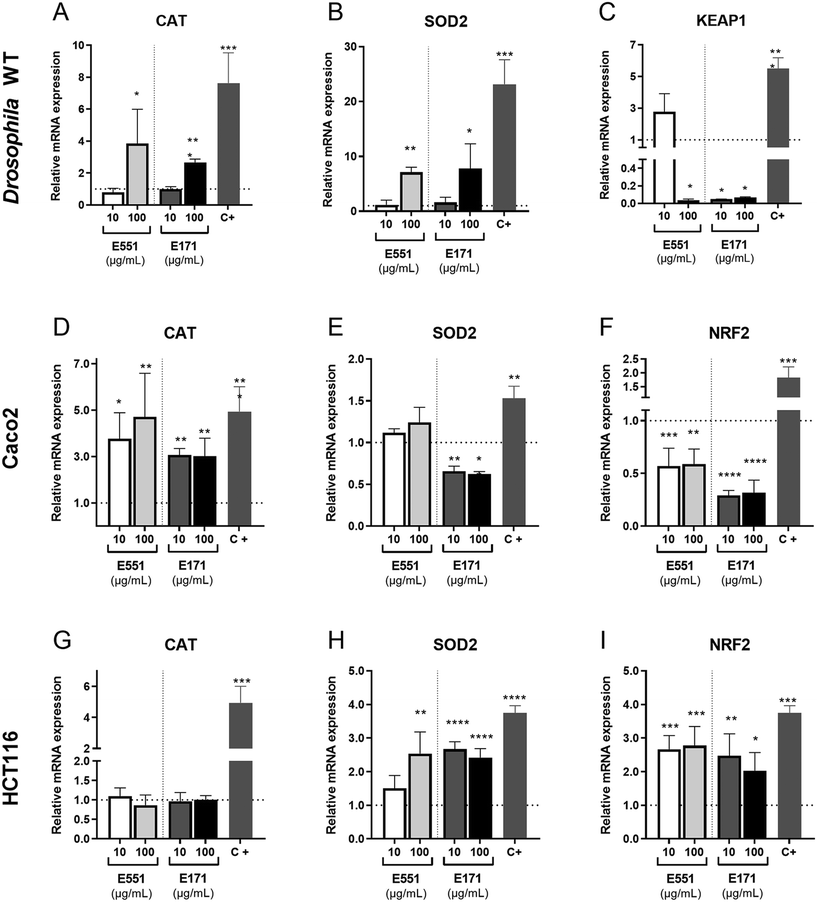
The redox balance is critical for the proliferation and survival of intestinal stem cells.42 The cellular response to oxidative stress involves the activities of several detoxifying enzymes including catalase (Cat) and superoxide dismutase 2 (Sod2), which participate in the elimination of hydrogen peroxide in the cytoplasm (Cat) and superoxide radicals in the mitochondria (Sod2; reviewed in ref. 43). In addition, the conserved transcription factor heterodimer formed by Keap1 and NRF2 (cap-un-collar in Drosophila) participates in the regulation of several genes encoding detoxifying enzymes.44 We found that the expression of both Cat and Sod2 mRNA was increased relative to controls at the highest assayed concentration of E171 and E551 (Fig. 3A and B). In contrast, the transcription of keap-1, which is induced at low E551 concentration, is mostly repressed at high concentration of E551 and at both low and high concentrations of E171 (Fig. 3C).We also found remarkable changes in gene expression associated to E171 and E551 exposure in Caco-2 and HCT116 cells. In these experiments the cells were grow for 24 h in culture media containing identical concentrations of E171 and E551 to those used in the Drosophila food. In the case of Caco-2 cells we found a significant induction of CAT in all treatments (Fig. 3D). In contrast, the expression of SOD2 was reduced upon E171 treatment, but remained unchanged in the E551 treatments (Fig. 3E). The expression of NRF2, the human orthologous of Drosophila cap-n-collar (cnc) and the binding partner of KEAP1, is reduced upon all treatments (Fig. 3F). The changes observed in HCT116 were very different to those found for differentiated Caco-2 cells. Thus, the expression of CAT is not affected by any treatment (Fig. 3G), whereas the expression of SOD2 and NRF2 was consistently increased when the cells were grown in media containing either E171 or E551 (Fig. 3H and I). In this manner, we found similar but cell-specific changes in gene expression associated to both E171 and E551 treatments, and a more similar response comparing differentiated Caco-2 cells and Drosophila intestinal cells. Differentiated Caco-2 cells resemble the transcriptional activity of normal human colon cells,45 and consequently the similarities found in the response of these cells with Drosophila intestinal cells indicates strong consistency in the in vitro and in vivo approaches.
The expression of genes involved in the response to DNA damage changes in Drosophila intestines and human cells exposed to E171 and E551
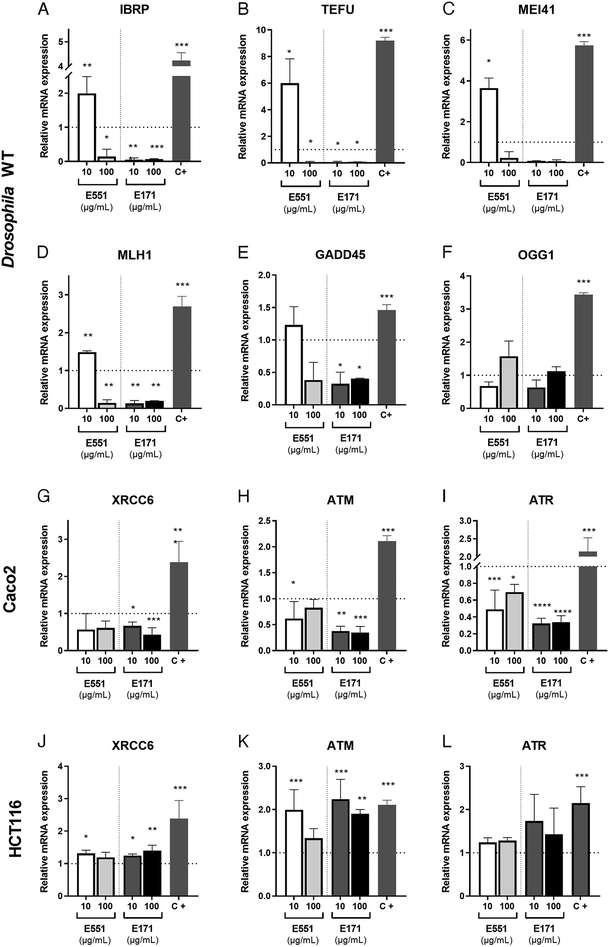
The presence of ROS can lead to DNA lesions and to the activation of DNA damage response pathways.46 DNA lesions induced by external agents (i.e., chemicals or ionizing radiation) or by endogenous factors associated to DNA replication include nucleotide alterations, single-strand breaks (SSBs) and double-strand breaks (DSBs). The DNA damage response (DDR) promotes the elimination of DNA lesions through a variety of mechanisms including non-homologous end joining (NHEJ) and homologous recombination (HR), single-strand break repair, mismatch repair and nucleotide excision repair, among others.47,48 We searched for alterations in the expression levels of genes encoding different components of the DNA repair machinery. In our analysis, carried out in Drosophila guts and human cells exposed to 10 and 100 μg mL−1 of E551 and E171, we included the genes inverted repeat binding protein (Irbp; involved in double-strand break repair via NHEJ), telomere fusion (tefu; ATM in mammals, a serine–threonine kinase involved in sensing DNA damage),49mei11 (ATR in mammals that also senses DNA damage and promotes DDR), Mlh1 (an ATPase involved in mismatch repair), growth arrest and DNA damage-inducible 45 (Gadd45, a modulator of oxidative and genotoxic stress that stimulate DNA scission repair) and 8 oxoguanine DNA glycosylase (Ogg1, a DNA glycosidase required for the removal of oxidized guanines following oxidative stress).50We found strong changes in mRNA levels for all genes monitored in Drosophila intestinal cells (Fig. 4A–F). As a general rule, exposure to the low E551 concentration (10 μg mL−1) causes a significant increase in the expression of Irbp, tefu, mei41 and Mlh1 (Fig. 4A–D). In contrast, exposure to the high dose of E551 (100 μg mL−1) or to both doses of E171 causes a dramatic reduction in the expression of all these genes (Fig. 4A–D), and a moderate reduction in the expression of Gadd45 (Fig. 4E). The expression of Ogg1 was not significantly affected in any media composition (Fig. 4F). Intriguingly, we found similar changes in mRNA expression for the human genes XRCC6 (Fig. 4G), ATM (Fig. 4H) and ATR5 (Fig. 4I) in Caco-2 cells. These changes were always of higher magnitude for Caco-2 cells growing for 24 h in media containing either high or low E171 concentrations (Fig. 4G–I). The expression of these genes was also altered in HTC116 cells (Fig. 4J–L). However, in these cases the tendency we observed was the opposite to that found in Caco-2 cells or Drosophila intestinal cells. Thus, the expression of XRCC6 (Fig. 4J), ATM (Fig. 4K) and to a lower extent ATR5 (Fig. 4L) was increased upon E551 or E171 treatments, without major differences detected between high or low concentrations of exposure. These results are in concordance with the recent EFSA evaluation indicating that TiO2 NPs have the potential to induce DNA damage.15 The molecular mechanism underlying the observed changes in the expression levels of enzymes involved in different DDR genes is unknown, but it is remarkable that Drosophila intestinal cells and human differentiated intestinal cells arising from long-term cell cultures of Caco-2 cells show similar changes in gene expression associated to E171 and E551 exposure. ATM and ATR are PI3K that are recruited to sites of double-strand or single-strand DNA breaks, respectively.51,52 Both kinases share a similar set of substrates involved in DDR and cell cycle arrest, and also substrates encoding ATP-dependent chromatin remodeling proteins that participate in a positive feedback loop to regulate both ATM and ATR transcription.53 Changes in the expression of these genes might underline the genotoxic effects described for E171,25,54 and further indicate that these additives can potentially synergize with other genotoxic chemicals to affect DNA integrity.35,55 Thus, our results show that stress responses are altered by nanosized TiO2 and SiO2 exposure, which may induce cellular susceptibility to oxidative and stress-related damage.
Reporter and gene expression in Drosophila larval intestines with extended development exposed to E171
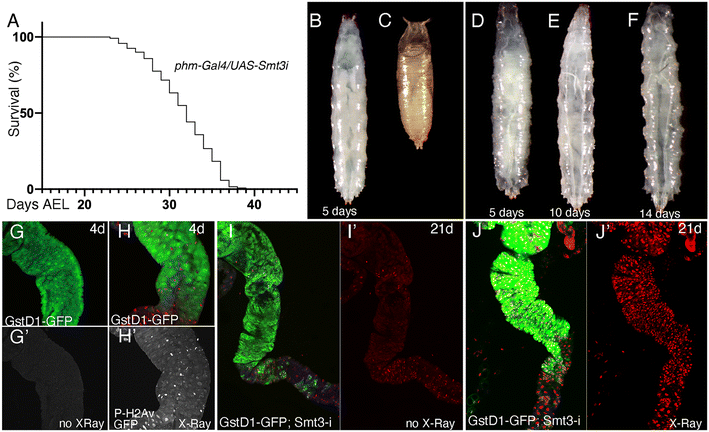
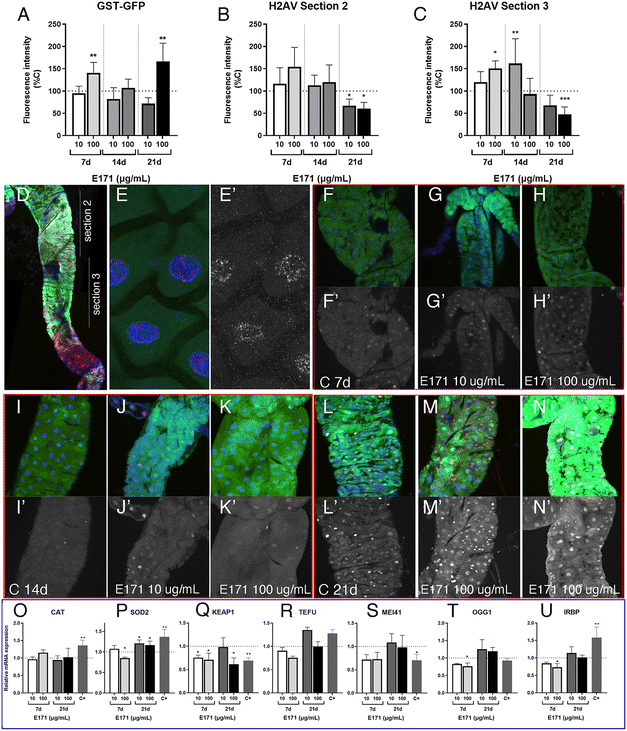
In order to evaluate the consequences of E171 and E551 exposure through oral ingestion, we generated a Drosophila genotype in which the synthesis of ecdysone, the fly hormone regulating developmental transitions,56 is impeded in the ring gland. In these larvae, of phm-Gal4/UAS-Smt3-RNAi genotype, the expression of an RNAi targeting the Drosophila gene encoding the unique fly Sumo protein (UAS-Smt3-RNAi) is directed in the ring gland (phm-Gal4), and as a consequence the synthesis of ecdysone is reduced in this tissue and the larvae remain for up to 35 d in the third larval stage26 (Fig. 5A–F), which we refer to as “extended larval development”. As a reporter for the existence of oxidative stress we used GstD1-GFP, and as a way to measure the existence of DNA damage we studied the accumulation of P-H2Av foci, the phosphorylated form of H2Av that is incorporated upon the presence of double-strand breaks in the DNA.57 The expression of GstD1-GFP and the phosphorylation of H2Av are observed in GstD1-GFP 4 d old larvae upon irradiation (Fig. 5G and H), and also in irradiated larvae with 21 d of extended development (GstD1-GFP/+; phm-Gal4/UAS-Smt3-RNAiFig. 5I and J), suggesting that these larvae exhibit to some extent similar responses compared to normal larvae.The expression of GFP is increased in GstD1-GFP/+; phm-Gal4/UAS-Smt3-RNAi larvae upon exposure to the highest concentration of E171 (100 μg mL−1) after 21 d in the culture (Fig. 6A). In contrast, the accumulation of P-H2Av remains similar in larvae treated with low or high concentrations of E171 (10 μg mL−1 and 100 μg mL−1) for a period of up to 14 d (Fig. 6B and C) in two regions of the anterior midgut (Fig. 6D). After 21 d treatments we observed a moderate reduction in the levels of P-H2Av along the anterior midgut (Fig. 6B and C). The appearance of the P-H2Av staining in the nucleus of intestinal cells was similar at all ages and under all different treatments, and consisted in a weak spotty pattern distributed through the nucleus (Fig. 6E and E′). Some representative examples of the expression of GstD1-GFP and P-H2Av in the anterior midgut (section 2), both in controls and in guts from larvae feeding on E171 media, are shown in Fig. 6F–N.
We also searched for changes in the expression of mRNA from several genes involved in oxidative stress responses (cat, sod2 and keap1) and DNA damage response (tefu, mei41, Ogg1 and Irbp) after 7 and 21 d of extended development in normal food and in food containing 10 and 100 μg mL−1 of E171 (Fig. 6O–U). We found very little differences in mRNA levels for these genes, in particular if we compared these results with those observed in guts from wild type larvae (see Fig. 3 and 4). This result suggests that larvae growing in the absence of ecdysone show more moderate responses to E171 than wild type larvae, even though they have been feeding for up to 21 d in food containing this additive. It is known that ecdysone is required for the activity of the Nrf2–Keap1 pathway during neural remodeling,58 and our results suggest that ecdysone might be also required for the gene expression changes associated to oxidative stress and DNA damage that we observed during normal development in response to oral ingestion of E171.
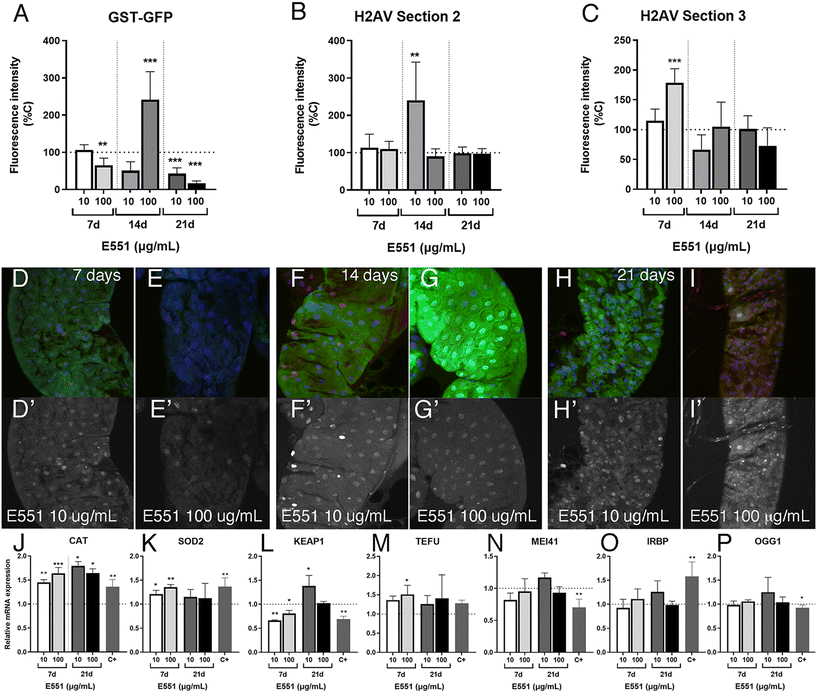
Reporter expression in Drosophila larval guts with extended development exposed to E551
We also analyzed in the same experimental setting (larvae with extended development due to loss of ecdysone production in the ring gland) the effects of low and high concentrations of E551 after 7, 14 and 21 d of extended larval development. In these cases, we found weaker than expected effects on the expression levels of the reporter GstD1-GFP (Fig. 7A) and no indication of genotoxic effects of the additive at any concentration used or in any region of the larval intestine (Fig. 7B and C). In fact, in the case of GstD1-GFP expression we found, contrary to our expectations, reduced expression of the reporter after 21 d of the larvae feeding in media containing E551 (quantified in Fig. 7A, representative examples shown in Fig. 7D–H). As it was the case for E171, we also failed to find strong changes in the expression levels of genes related to oxidative stress, with the exception of Cat expression (Fig. 7J–L), nor significant changes in the expression of genes related to DNA damage responses (Fig. 7M–P) after exposure to E551. We can think of two scenarios to explain these results, both of which grant future research. One possibility is that larval intestinal cells after long-term exposures, from 7 to 21 d beyond their normal developmental time, reach some form of homeostasis in which the presence of the stressor is compensated by the normal mechanisms triggered by oxidative stress and DNA damage. In these scenario cells would have acquired a balance that ensures the reparation of the damages caused by short-term inductions observed in normal flies. In this context, we have to consider that most intestinal cells which we are monitoring are polyploid cells that do not divide and have a low turnover rate, which may grant them enough time to recover from mild toxicological insults. In a second scenario, milder responses could be attributed to the loss of ecdysone signaling that characterizes these larvae with extended development. In this manner, changes in gene expression occurring during larval development might require a co-regulation of transcription by both cellular stress response pathways and ecdysone signaling.58,59 We showed that intestinal cells from larvae with extended development still respond to acute insults by increasing the expression of at least GstD1 reporters, as well as suffer considerable DNA damage in response to irradiation (see Fig. 5G–J). To what extent the way the genes respond to insults or to the loss of ecdysone is universal is entirely unknown, and a further understanding of the molecular mechanism involved is beyond the scope of our analysis.Ultrastructure of the Drosophila intestine after extended development and ingestion of E171 and E551
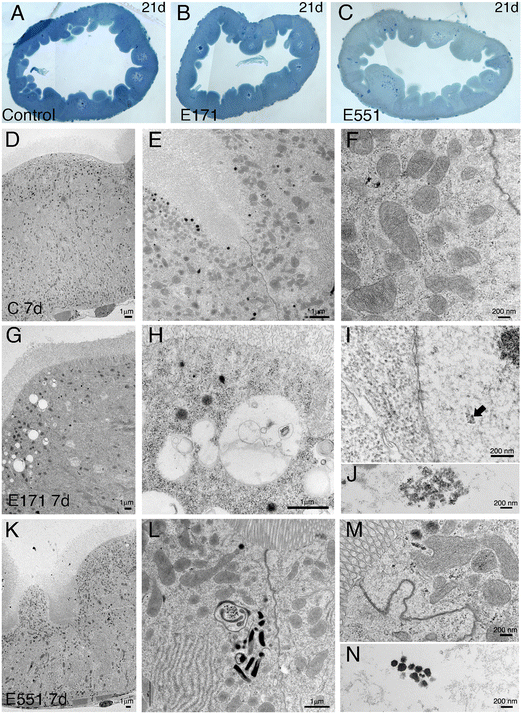
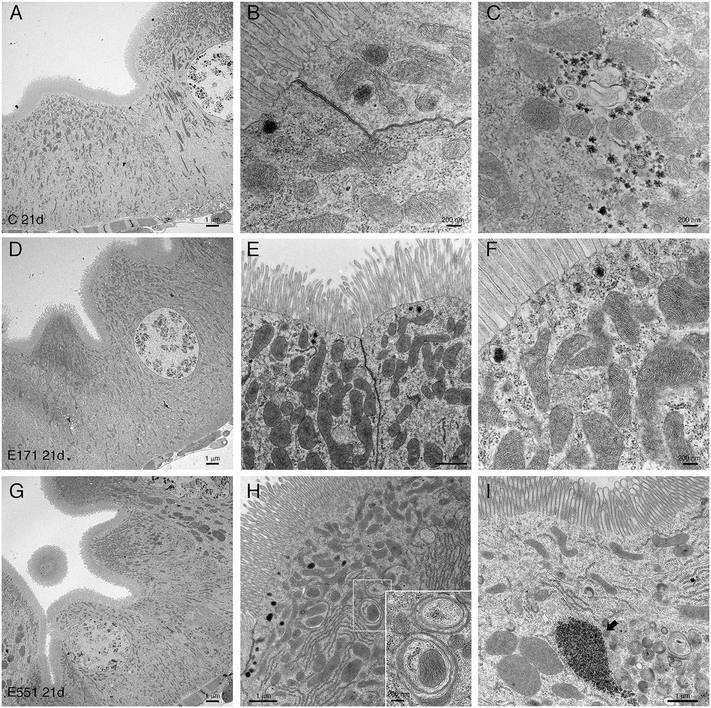
Drosophila intestinal thin sections of the anterior gut from control larvae or from larvae feeding during 21 d in 100 μg mL−1 of E171 or E551 show an overall similar appearance under the light microscope (Fig. 8A–C). We only noticed a larger preponderance to develop larger intestinal crypts when the larvae were fed with E171 or E551 compared to controls (Fig. 8A–C). It has been described that these nanomaterials can disrupt the epithelial barrier in the intestine and spread into internal fluids.39 Furthermore, nanomaterial internalization can reach the nucleus, where it can compromise DNA integrity.39,60 We explored the possibility of subcellular alterations caused by E171 or E551 ingestion using TEM in intestines taken from larvae growing in 100 μg mL−1 E171 or E551 in the food media during 7 and 21 d. At the first time point of extended development control larvae show the characteristic epithelial ultrastructure described for the wild type Drosophila intestine31,61 (Fig. 8D–F). Thus, these intestines show a normal junction between adjacent cells (Fig. 8E and F), suggestive of integrity of the epithelial barrier, a well-developed pattern of microvilli in the apical side of the enterocytes (Fig. 8D and E), and extensive infoldings of the basal plasma membrane, the basal labyrinth (Fig. 8D). These cells also accumulate a large number of normal looking mitochondria in the apical region (Fig. 8E and F). We did not find radical ultrastructural changes in intestines exposed to E171 or E551 during 7 d. Thus, intestinal cells show correct apico-basal differentiation with normally packed apical microvilli, nuclei in central position and a well-defined basal labyrinth (Fig. 8G and K). The presence of cellular junctions (Fig. 8I and L) and normal mitochondria (Fig. 8L) suggests that the integrity of the intestinal barrier is preserved. The only unusual morphological modification we observed was an increase in the presence of apical vacuoles in larvae exposed to E171 (Fig. 8D, compare with H), and of autophagic vesicles along with electrodense material in E551-fed larvae (Fig. 8L). Despite that no major ultrastructural changes were detected, both nanoparticles could penetrate the peritrophic membrane (Fig. 8J and M) and interact directly with the intestinal cellular layer. In fact, E171 could be found inside the enterocytes as has been previously reported by other authors,23 being able to reach the cellular nucleus (ref. 62 and Fig. 8I–J). We found no sign of disruption of the intestinal brush border, as has been reported for TiO2 using the Caco-2BBe1 cell line.63The ultrastructure of the intestine at 21 d of extended development is also similar comparing larvae grown in control medium with larvae growing in E171 or E551 media containing a concentration of 100 μg mL−1. In this manner the overall apico-basal organization of the epithelia is similar (Fig. 9A, D and G), the epithelial barrier appears normal in all cases (Fig. 9B and E), the microvilli were densely packed in the apical side of the cell, and the mitochondria organized mostly in the apical side of the cell (Fig. 9B, E, F and H). We did find some differences regarding mitochondrial size, consisting in the appearance of enlarged mitochondria in larvae feeding in E171 (Fig. 9B, E and F). The dynamic nature of the mitochondrial network is central for their proper functioning. An imbalance towards increased fusion, with the appearance of enlarged mitochondria, has been related to protection from apoptotic stimuli, and could be related to cancer progression.64 However, there is no clear consensus regarding the direct or indirect influence of fission/fusion mitochondrial status and cellular toxicity on the development of diseases.65 We also found clear indications of mitophagy in larvae feeding with E551 (Fig. 9H). Thus, the presence of mitochondria enclosed by double membranes in autophagosomes was recurrently observed in sections from intestines grown in this food additive (see for example Fig. 9H), and never observed in control or E171 intestines. Mitophagy is a conserved cellular homeostatic process that ensures the appropriate function of the mitochondrial network by eliminating damaged or malfunctioning mitochondria.66 Acute mitochondrial clearance has been related to various stresses, such as mitochondrial depolarization and proteotoxic stress,67 as well as with different pathological conditions.66,68 Finally, we also observed large accumules of electrodense granules in intestines from 21 d larvae growing in E551, compatible with glycogene accumulation (Fig. 9I). Abnormal glycogen accumulation has been related to redox imbalance through alterations in gluconeogenesis metabolic pathways.69
Conclusions
We have evaluated two alimentary additives, E551 and E171, in exposure concentrations that do not compromise cellular or organism viability and that may represent a realistic exposure scenario. Our work in human intestinal cells and in Drosophila melanogaster intestines identified changes in the expression levels of genes related to the regulation of oxidative stress and DNA integrity. These changes were remarkably similar in differentiated intestinal Caco-2 cells and in Drosophila intestines, indicating that the cellular response to E551 and E171 exposure are very similar in differentiated intestinal cells. We searched for ultrastructural differences in intestinal cells exposed to these alimentary additives, and could find differences related to autophagy and mitochondrial biology upon prolonged exposure to E551 and E171, respectively. The use of in vivo models likely reproduce accurately the complex interactions that occur between ingested NMs and the different intestine microenvironments. How our data translate into human risk is not known, as the determination of relevant dose responses for mammals from Drosophila data is not yet well-established. In any case the similarities observed between Drosophila and human cells suggest that flies could be useful to determine safety concerns for different chemicals at realistic exposure doses. In this context, Drosophila melanogaster offer a variety of genetic tools to modelate different genetic susceptibility scenarios that could impact in the biological consequences of ingested NMs.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We would like to thank the Genomic, Confocal and Electronic microscopy scientific support services of the CBMSO for their skillfull help. We also thank Milagros Guerra for the preparation of TEM microscopy samples shown in Fig. 8 and 9. We also will like to thank Dr Jacques-Aurélien Sergent and two annonimous reviewers for criticism that greatly improved this manuscript. Our work was supported by grants Labex Serenade (No. ARN-II-LABX-0064) funded by the Investiments d'Avenir (French government program of the French National Research Agency), Secretaría de Estado de Investigación, Desarrollo e Innovación (grant number PGC2018-094476-B-I00) and Department of Biology, UAM (grant BIOUAM03-2021). The CBMSO enjoys institutional support from the Ramón Areces Foundation.References
- M. Eleftheriadou, G. Pyrgiotakis and P. Demokritou, Nanotechnology to the rescue: using nano-enabled approaches in microbiological food safety and quality, Curr. Opin. Biotechnol., 2017, 44, 87–93 CrossRef CAS.
- M. Mattarozzi, M. Suman, C. Cascio, D. Calestani, S. Weigel, A. Undas and R. Peters, Analytical approaches for the characterization and quantification of nanoparticles in food and beverages, Anal. Bioanal. Chem., 2016, 409(1), 63–80 CrossRef.
- A. Weir, P. Westerhoff, L. Fabricius, K. Hristovski and N. Von Goetz, Titanium Dioxide nanoparticles in food and personal care products, Environ. Sci. Technol., 2012, 46, 2242 CrossRef CAS PubMed.
- I. Sohal, K. O'Fallon, P. Gaines, P. Demokritou and D. Bello, Ingested engineered nanomaterials: state of science in nanotoxicity testing and future research needs, Part. Fibre Toxicol., 2018, 15, 29 CrossRef PubMed.
- C. McCracken, P. K. Dutta and W. J. Waldman, Critical assessment of toxicological effects of ingested nanoparticles, Environ. Sci.: Nano, 2016, 3, 256–282 RSC.
- S. Dekkers, A. G. Oomen, E. A. J. Bleeker, R. J. Vandebriel, C. Micheletti, J. Cabellos, G. Janer, N. Fuentes, S. Vázquez-Campos, T. Borges, M. J. Silva, A. Prina-Mello, D. Movia, F. Nesslany, A. R. Ribeiro, P. E. Leite, M. Groenewold, F. R. Cassee, A. J. A. M. Sips, A. Dijkzeul, T. van Teunenbroek and S. W. P. Wijnhoven, Towards a nanospecific approach for risk assessment, Regul. Toxicol. Pharmacol., 2016, 80, 46–59 CrossRef PubMed.
- H. Chen, M. C. Roco, J. Son, S. Jiang, C. A. Larson and Q. Gao, Global nanotechnology development from 1991 to 2012: Patents, scientific publications, and effect of NSF funding, J. Nanopart. Res., 2013, 15, 1–21 Search PubMed.
- C. Rompelberg, M. Heringa, G. van Donkersgoed, J. Drijvers, A. Roos, S. Westenbrink, R. Peters, G. van Bemmel, W. Brand and A. Oomen, Oral intake of added titanium dioxide and its nanofraction from food products, food supplements and toothpaste by the Dutch population, Nanotoxicology, 2016, 10, 1404–1414 CrossRef CAS PubMed.
- S. J. So, I. S. Jang and C. S. Han, Effect of micro/nano Silica particle feeding for mice, J. Nanosci. Nanotechnol., 2008, 8, 5367–5371 CrossRef CAS PubMed.
- C. Fruijtier-Pölloth, The safety of nanostructured synthetic amorphous silica (SAS) as a food additive (E 551), Arch. Toxicol., 2016, 90, 2885–2916 CrossRef PubMed.
- E. Demir, S. Aksakal, F. Turna, B. Kaya and R. Marcos, In vivo genotoxic effects of four different nano-sizes forms of silica nanoparticles in Drosophila melanogaster, J. Hazard. Mater., 2015, 283, 260–266 CrossRef CAS PubMed.
- M. Younes, P. Aggett, F. Aguilar, R. Crebelli, B. Dusemund, M. Filipič, M. J. Frutos, P. Galtier, D. Gott, U. Gundert-Remy, G. G. Kuhnle, J. C. Leblanc, I. T. Lillegaard, P. Moldeus, A. Mortensen, A. Oskarsson, I. Stankovic, I. Waalkens-Berendsen, R. A. Woutersen, M. Wright, P. Boon, D. Chrysafidis, R. Gürtler, P. Mosesso, D. Parent-Massin, P. Tobback, N. Kovalkovicova, A. M. Rincon, A. Tard and C. Lambré, Re-evaluation of silicon dioxide (E 551) as a food additive, EFSA J., 2018, 16, e05088 Search PubMed.
- W. Brand, P. C. E. van Kesteren, R. J. B. Peters and A. G. Oomen, Issues currently complicating the risk assessment of synthetic amorphous silica (SAS) nanoparticles after oral exposure, Nanotoxicology, 2021, 15, 905–933 CAS.
- European Commission, Directorate-General for Health and Food Safety, Guidance on the safety assessment of nanomaterials in cosmetics, SCCS/1611/19, 2019.
- M. Younes, G. Aquilina, L. Castle, K. H. Engel, P. Fowler, M. J. Frutos Fernandez, P. Fürst, U. Gundert-Remy, R. Gürtler, T. Husøy, M. Manco, W. Mennes, P. Moldeus, S. Passamonti, R. Shah, I. Waalkens-Berendsen, D. Wölfle, E. Corsini, F. Cubadda, D. de Groot, R. FitzGerald, S. Gunnare, A. C. Gutleb, J. Mast, A. Mortensen, A. Oomen, A. Piersma, V. Plichta, B. Ulbrich, H. van Loveren, D. Benford, M. Bignami, C. Bolognesi, R. Crebelli, M. Dusinska, F. Marcon, E. Nielsen, J. Schlatter, C. Vleminckx, S. Barmaz, M. Carfí, C. Civitella, A. Giarola, A. M. Rincon, R. Serafimova, C. Smeraldi, J. Tarazona, A. Tard and M. Wright, Safety assessment of titanium dioxide (E171) as a food additive, EFSA J., 2021, 19, e06585 CAS.
- S. Bettini, E. Boutet-Robinet, C. Cartier, C. Coméra, E. Gaultier, J. Dupuy, N. Naud, S. Taché, P. Grysan, S. Reguer, N. Thieriet, M. Réfrégiers, D. Thiaudière, J. Cravedi, M. Carrière, J. Audinot, F. Pierre, L. Guzylack-Piriou and E. Houdeau, Food-grade TiO 2 impairs intestinal and systemic immune homeostasis, initiates preneoplastic lesions and promotes aberrant crypt development in the rat colon, Sci. Rep., 2017, 7, 40373 CrossRef CAS PubMed.
- D. Kirkland, M. J. Aardema, R. v. Battersby, C. Beevers, K. Burnett, A. Burzlaff, A. Czich, E. M. Donner, P. Fowler, H. J. Johnston, H. F. Krug, S. Pfuhler and L. F. Stankowski, A weight of evidence review of the genotoxicity of titanium dioxide (TiO2), Regul. Toxicol. Pharmacol., 2022, 136, 105263 CrossRef CAS PubMed.
- I. L. Bergin and F. A. Witzmann, Nanoparticle toxicity by the gastrointestinal route: evidence and knowledge gaps, Int. J. Biomed. Nanosci. Nanotechnol., 2013, 3, 163 CrossRef CAS PubMed.
- P. H. Hoet, I. Brüske-Hohlfeld and O. v. Salata, Nanoparticles – known and unknown health risks, J. Nanobiotechnol., 2004, 2, 1–15 CrossRef PubMed.
- B. Drasler, P. Sayre, K. G. Steinhäuser, A. Petri-Fink and B. Rothen-Rutishauser, In vitro approaches to assess the hazard of nanomaterials, NanoImpact, 2017, 8, 99–116 CrossRef.
- I. Miguel-Aliaga, H. Jasper and B. Lemaitre, Anatomy and physiology of the digestive tract of Drosophila melanogaster, Genetics, 2018, 210, 357–396 CrossRef CAS PubMed.
- T. Kuraishi, O. Binggeli, O. Opota, N. Buchon and B. Lemaitre, Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15966–15971 CrossRef CAS PubMed.
- M. Alaraby, B. Annangi, R. Marcos and A. Hernández, Drosophila melanogaster as a suitable in vivo model to determine potential side effects of nanomaterials: A review, J. Toxicol. Environ. Health, Part B, 2016, 19, 65–104 CAS.
- B. Jovanović, V. J. Cvetković and T. L. Mitrović, Effects of human food grade titanium dioxide nanoparticle dietary exposure on Drosophila melanogaster survival, fecundity, pupation and expression of antioxidant genes, Chemosphere, 2016, 144, 43–49 CrossRef PubMed.
- B. Jovanović, N. Jovanović, V. J. Cvetković, S. Matić, S. Stanić, E. M. Whitley and T. Lj Mitrović, The effects of a human food additive, titanium dioxide nanoparticles E171, on Drosophila melanogaster-a 20 generation dietary exposure experiment, Sci. Rep., 2018, 8, 17922 CrossRef PubMed.
- A. Talamillo, L. Herboso, L. Pirone, C. Pérez, M. González, J. Sánchez, U. Mayor, F. Lopitz-Otsoa, M. S. Rodriguez, J. D. Sutherland and R. Barrio, Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila Steroidogenesis, PLoS Genet., 2013, 9, e1003473 CrossRef CAS PubMed.
- L. Herboso, M. M. Oliveira, A. Talamillo, C. Pérez, M. González, D. Martín, J. D. Sutherland, A. W. Shingleton, C. K. Mirth and R. Barrio, Ecdysone promotes growth of imaginal discs through the regulation of Thor in D. melanogaster, Sci. Rep., 2015, 5, 1–14 Search PubMed.
- I. S. Sohal, G. M. DeLoid, K. S. O'Fallon, P. Gaines, P. Demokritou and D. Bello, Effects of ingested food-grade titanium dioxide, silicon dioxide, iron (III) oxide and zinc oxide nanoparticles on an in vitro model of intestinal epithelium: Comparison between monoculture vs. a mucus-secreting coculture model, NanoImpact, 2020, 17, 100209 CrossRef.
- C. Putra, D. Bello, K. L. Tucker, S. L. Kelleher and K. M. Mangano, Estimation of Titanium Dioxide intake by diet and stool assessment among US healthy adults, J. Nutr., 2022, 152, 1525–1537 CrossRef PubMed.
- O. V. Lushchak, B. M. Rovenko, D. V. Gospodaryov and V. I. Lushchak, Drosophila melanogaster larvae fed by glucose and fructose demonstrate difference in oxidative stress markers and antioxidant enzymes of adult flies, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2011, 160, 27–34 CrossRef CAS PubMed.
- S. Shanbhag and S. Tripathi, Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut, J. Exp. Biol., 2009, 212, 1731–1744 CrossRef CAS PubMed.
- G. P. Sykiotis and D. Bohmann, Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila, Dev. Cell, 2008, 14, 76 CrossRef CAS PubMed.
- M. Dorier, D. Béal, C. Marie-Desvergne, M. Dubosson, F. Barreau, E. Houdeau, N. Herlin-Boime and M. Carriere, Continuous in vitro exposure of intestinal epithelial cells to E171 food additive causes oxidative stress, inducing oxidation of DNA bases but no endoplasmic reticulum stress, Nanotoxicology, 2017, 11, 751–761 CAS.
- M. Dorier, D. Béal, C. Tisseyre, C. Marie-Desvergne, M. Dubosson, F. Barreau, E. Houdeau, N. Herlin-Boime, T. Rabilloud and M. Carriere, The food additive E171 and titanium dioxide nanoparticles indirectly alter the homeostasis of human intestinal epithelial cells in vitro, Environ. Sci.: Nano, 2019, 6, 1549–1561 RSC.
- F. Dussert, P. A. Arthaud, M. E. Arnal, B. Dalzon, A. Torres, T. Douki, N. Herlin, T. Rabilloud and M. Carriere, Toxicity to RAW264.7 macrophages of Silica nanoparticles and the E551 food additive, in combination with genotoxic agents, Nanomaterials, 2020, 10, 1418 CrossRef CAS PubMed.
- J. Athinarayanan, A. A. Alshatwi, V. S. Periasamy and A. A. Al-Warthan, Identification of nanoscale ingredients in commercial food products and their induction of mitochondrially mediated cytotoxic effects on human mesenchymal stem cells, J. Food Sci., 2015, 80, N459–N464 CrossRef CAS PubMed.
- H. Proquin, C. Rodríguez-Ibarra, C. G. J. Moonen, I. M. Urrutia Ortega, J. J. Briedé, T. M. de Kok, H. van Loveren and Y. I. Chirino, Titanium dioxide food additive (E171) induces ROS formation and genotoxicity: contribution of micro and nano-sized fractions, Mutagenesis, 2017, 32, 139–149 CrossRef CAS PubMed.
- J. Athinarayanan, V. S. Periasamy, M. A. Alsaif, A. A. Al-Warthan and A. A. Alshatwi, Presence of nanosilica (E551) in commercial food products: TNF-mediated oxidative stress and altered cell cycle progression in human lung fibroblast cells, Cell Biol. Toxicol., 2014, 30, 89–100 CrossRef CAS PubMed.
- E. Demir, An in vivo study of nanorod, nanosphere, and nanowire forms of titanium dioxide using Drosophila melanogaster: toxicity, cellular uptake, oxidative stress, and DNA damage, J. Toxicol. Environ. Health, Part A, 2020, 83, 456–469 CrossRef CAS PubMed.
- E. Demir, F. T. Demir and R. Marcos, Drosophila as a suitable in vivo model in the safety assessment of nanomaterials, Adv. Exp. Med. Biol., 2022, 1357, 275–301 CrossRef PubMed.
- A. Pandey, S. Chandra, L. K. S. Chauhan, G. Narayan and D. K. Chowdhuri, Cellular internalization and stress response of ingested amorphous silica nanoparticles in the midgut of Drosophila melanogaster, Biochim. Biophys. Acta, 2013, 1830, 2256–2266 CrossRef CAS PubMed.
- C. Xu, J. Luo, L. He, C. Montell and N. Perrimon, Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca2+ signaling in the Drosophila midgut, eLife, 2017, 6, e22441 CrossRef PubMed.
- C. M. C. Andrés, J. M. P. de la Lastra, C. A. Juan, F. J. Plou and E. Pérez-Lebeña, Chemistry of Hydrogen Peroxide formation and elimination in mammalian cells, and Its role in various pathologies, Stresses, 2022, 2, 256–274 CrossRef.
- P. Jennings, A. Limonciel, L. Felice and M. O. Leonard, An overview of transcriptional regulation in response to toxicological insult, Arch. Toxicol., 2013, 87, 49–72 CrossRef CAS PubMed.
- A. M. Sääf, J. M. Halbleib, X. Chen, T. Y. Siu, Y. L. Suet, W. J. Nelson and P. O. Brown, Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer, Mol. Biol. Cell, 2007, 18, 4245–4260 CrossRef PubMed.
- M. Valko, C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur, Free radicals, metals and antioxidants in oxidative stress-induced cancer, Chem.-Biol. Interact., 2006, 160, 1–40 CrossRef CAS PubMed.
- F. Vernì, DNA Damage Response (DDR) and DNA Repair, Int. J. Mol. Sci., 2022, 23, 7204 CrossRef PubMed.
- A. Sancar, L. A. Lindsey-Boltz, K. Ünsal-Kaçmaz and S. Linn, Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints, Annu. Rev. Biochem., 2004, 73, 39–85 CrossRef CAS PubMed.
- A. Maréchal and L. Zou, DNA Damage Sensing by the ATM and ATR Kinases, Cold Spring Harbor Perspect. Biol., 2013, 5, a012716 CrossRef PubMed.
- K. Okumura, S. Nishihara and Y. H. Inoue, Genetic identification and characterization of three genes that prevent accumulation of oxidative DNA damage in Drosophila adult tissues, DNA Repair, 2019, 78, 7–19 CrossRef CAS PubMed.
- J. H. Lee and T. T. Paull, ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex, Science, 2005, 308, 551–554 CrossRef CAS PubMed.
- D. A. Mordes and D. Cortez, Activation of ATR and related PIKKs, Cell Cycle, 2008, 7, 2809 CrossRef CAS PubMed.
- R. Sethy, R. Rakesh, K. Patne, V. Arya, T. Sharma, D. T. Haokip, R. Kumari and R. Muthuswami, Regulation of ATM and ATR by SMARCAL1 and BRG1, Biochim. Biophys. Acta, Gene Regul. Mech., 2018, 1861, 1076–1092 CrossRef CAS PubMed.
- D. Kirkland, M. J. Aardema, R. v. Battersby, C. Beevers, K. Burnett, A. Burzlaff, A. Czich, E. M. Donner, P. Fowler, H. J. Johnston, H. F. Krug, S. Pfuhler and L. F. Stankowski, A weight of evidence review of the genotoxicity of titanium dioxide (TiO2), Regul. Toxicol. Pharmacol., 2022, 136, 105263 CrossRef CAS PubMed.
- X. Cao, G. M. Deloid, D. Bitounis, R. De La Torre-Roche, J. C. White, Z. Zhang, C. G. Ho, K. W. Ng, B. D. Eitzer and P. Demokritou, Co-exposure to the food additives SiO2 (E551) or TiO2 (E171) and the pesticide boscalid increases cytotoxicity and bioavailability of the pesticide in a tri-culture small intestinal epithelium model: potential health implications, Environ. Sci.: Nano, 2019, 6, 2786–2800 RSC.
- E. T. Danielsen, M. E. Moeller, N. Yamanaka, Q. Ou, J. M. Laursen, C. Soenderholm, R. Zhuo, B. Phelps, K. Tang, J. Zeng, S. Kondo, C. H. Nielsen, E. B. Harvald, N. J. Faergeman, M. J. Haley, K. A. O'Connor, K. King-Jones, M. B. O'Connor and K. F. Rewitz, A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing, Dev. Cell, 2016, 37, 558–570 CrossRef CAS PubMed.
- S. Baldi and P. B. Becker, The variant histone H2A.V of Drosophila - Three roles, two guises, Chromosoma, 2013, 122, 245–258 CrossRef CAS PubMed.
- A. Liang Yuh Chew, H. Zhang, J. He and F. Yu, Correspondence, The Nrf2-Keap1 pathway is activated by steroid hormone signaling to govern neuronal remodeling, Cell Rep., 2021, 36, 109466 CrossRef PubMed.
- H. Deng, Multiple roles of Nrf2-Keap1 signaling: Regulation of development and xenobiotic response using distinct mechanisms, Fly, 2014, 8, 7 CrossRef PubMed.
- N. El Yamani, L. Rubio, A. García-Rodríguez, A. Kažimírová, E. Rundén-Pran, B. Magdalena, R. Marcos and M. Dusinska, Lack of mutagenicity of TiO2 nanoparticles in vitro despite cellular and nuclear uptake, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2022, 882, 503545 CrossRef CAS PubMed.
- O. Baumann, Posterior midgut epithelial cells differ in their organization of the membrane skeleton from other Drosophila epithelia, Exp. Cell Res., 2001, 270, 176–187 CrossRef CAS PubMed.
- N. el Yamani, L. Rubio, A. García-Rodríguez, A. Kažimírová, E. Rundén-Pran, B. Magdalena, R. Marcos and M. Dusinska, Lack of mutagenicity of TiO2 nanoparticles in vitro despite cellular and nuclear uptake, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2022, 882, 503545 CrossRef CAS PubMed.
- J. J. Faust, K. Doudrick, Y. Yang, P. Westerhoff and D. G. Capco, Food grade titanium dioxide disrupts intestinal brush border microvilli in vitro independent of sedimentation, Cell Biol. Toxicol., 2014, 30, 169–188 CrossRef CAS PubMed.
- J. Nunnari and A. Suomalainen, Mitochondria: In sickness and in health, Cell, 2012, 148, 1145–1159 CrossRef CAS PubMed.
- K. Kim and E. Y. Lee, Excessively enlarged mitochondria in the kidneys of diabetic nephropathy, Antioxidants, 2021, 10, 741 CrossRef CAS PubMed.
- K. Palikaras, E. Lionaki and N. Tavernarakis, Mechanisms of mitophagy in cellular homeostasis, physiology and pathology, Nat. Cell Biol., 2018, 20, 1013–1022 CrossRef CAS PubMed.
- S. Sekine and R. J. Youle, PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol, BMC Biol., 2018, 16, 1–12 CrossRef PubMed.
- L. Liu, X. Liao, H. Wu, Y. Li, Y. Zhu and Q. Chen, Mitophagy and its contribution to metabolic and aging-associated disorders, Antioxid. Redox Signaling, 2020, 32, 906–927 CrossRef CAS PubMed.
- B. della Noce, R. Martins Da Silva, M. Vianna De Carvalho Uhl, S. Konnai, K. Ohashi, C. Calixto, A. Arcanjo, L. Araujo De Abreu, S. Serafim De Carvalho, I. Da, S. Vaz and C. Logullo, Redox imbalance induces remodeling of glucose metabolism in Rhipicephalus microplus embryonic cell line, J. Biol. Chem., 2022, 298(3), 101599 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00009e |
| This journal is © The Royal Society of Chemistry 2023 |