Emerging investigator series: differential effects of carbon nanotubes and graphene on the tomato rhizosphere microbiome†
Abstract
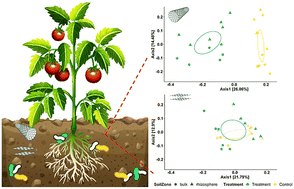
Application of carbonaceous nanomaterials (CNMs) to the soil–plant system can affect plant physiology, with positive results ranging from enhanced seed germination and root system development to improved stress tolerance. The underlying mechanisms are not fully understood. Plant rhizosphere microbiomes at the soil–root interface are strongly influenced by the host plant and play a key role in the plant host's development and health. Yet few studies have characterized changes in plant rhizosphere microbiomes following application of CNMs to the soil–plant system. Here we investigated the effects of multi-walled carbon nanotubes (CNTs) and graphene on microbial communities in the ectorhizosphere of tomato plants versus the surrounding bulk soil. Pot experiments were conducted where tomato plants were exposed to CNTs or graphene at 200 mg kg−1 soil for four weeks. The ectorhizosphere and bulk soils were then collected and analyzed for physicochemical properties and the microbiome structure and function. While graphene had a limited impact on the tomato rhizosphere microbiome, CNTs significantly increased microbial alpha diversity, induced greater divergence of beta diversity, enhanced microbial interactions, and potentially impacted community functions such as aromatic compound degradation, antioxidant synthesis, and redox cofactor biosynthesis. Furthermore, CNTs induced stronger and/or unique microbiome alterations in the tomato rhizosphere compared to the bulk soil. Our findings reveal the differential modulating effects of the two widely-used CNMs on plant rhizosphere microbiomes and highlight an imminent need to understand complex plant root–microbe interplays in the CNM-impacted rhizosphere. These results have implications for realizing the full potential of phytoapplication of CNMs toward improved and sustainable plant production.
- This article is part of the themed collections: Nano-bio interactions, Outstanding Papers 2023 – Environmental Science: Nano and Emerging Investigators Series


 Please wait while we load your content...
Please wait while we load your content...