Open Access Article
Open Access ArticleSynthesis and cytotoxicity studies of Cu(I) and Ag(I) complexes based on sterically hindered β-diketonates with different degrees of fluorination†
Maura
Pellei
 *a,
Jo’
Del Gobbo
a,
Miriam
Caviglia
a,
Deepika V.
Karade
b,
Valentina
Gandin
c,
Cristina
Marzano
*c,
Anurag
Noonikara Poyil
*a,
Jo’
Del Gobbo
a,
Miriam
Caviglia
a,
Deepika V.
Karade
b,
Valentina
Gandin
c,
Cristina
Marzano
*c,
Anurag
Noonikara Poyil
 b,
H. V. Rasika
Dias
b,
H. V. Rasika
Dias
 *b and
Carlo
Santini
*b and
Carlo
Santini
 a
a
aSchool of Science and Technology, Chemistry Division, University of Camerino, Via Madonna delle Carceri (ChIP), 62032 Camerino, Macerata, Italy. E-mail: maura.pellei@unicam.it
bDepartment of Chemistry and Biochemistry, The University of Texas at Arlington, Box 19065, Arlington, Texas 76019-0065, USA. E-mail: dias@uta.edu
cDepartment of Pharmaceutical and Pharmacological Sciences, University of Padova, via Marzolo 5, 35131 Padova, Italy. E-mail: cristina.marzano@unipd.it
First published on 3rd August 2023
Abstract
Design, synthesis, and in vitro antitumor properties of Cu(I) and Ag(I) phosphane complexes supported by the anions of sterically hindered β-diketone ligands, 1,3-dimesitylpropane-1,3-dione (HLMes) and 1,3-bis(3,5-bis(trifluoromethyl)phenyl)-3-hydroxyprop-2-en-1-one (HLCF3) featuring trifluoromethyl or methyl groups on the phenyl moieties have been reported. In order to compare the biological effects of substituents on the phenyl moieties, the analogous copper(I) and silver(I) complexes of the anion of the parent 1,3-diphenylpropane-1,3-dione (HLPh) ligand were also synthesized and included in the study. In the syntheses of the Cu(I) and Ag(I) complexes, the phosphane coligands triphenylphosphine (PPh3) and 1,3,5-triaza-7-phosphaadamantane (PTA) were used to stabilize silver and copper in the +1 oxidation state, preventing the metal ion reduction to Ag(0) or oxidation to Cu(II), respectively. X-ray crystal structures of HLCF3 and the metal adducts [Cu(LCF3)(PPh3)2] and [Ag(LPh)(PPh3)2] are also presented. The antitumor properties of both classes of metal complexes were evaluated against a series of human tumor cell lines derived from different solid tumors, by means of both 2D and 3D cell viability studies. They display noteworthy antitumor properties and are more potent than cisplatin in inhibiting cancer cell growth.
Introduction
Medicinal inorganic chemistry allows the design of compounds with therapeutic properties that are not readily available in organic compounds.1,2 Recently, many chelating ligands and delivery systems for metal-based drugs have been developed to obtain more potent, clinically effective, and less toxic metal-based antiproliferative drugs with improved selectivity towards tumour cells.3,4 Group 11 metal complexes showed encouraging perspectives in this regard and several noble metal complexes were investigated for their antitumoral properties.5–7β-Diketone compounds represent a very important class of reagents in the synthesis of heterocyclic compounds.8 The β-diketone scaffold is not very common in nature though it is the main feature of curcumin and its derivatives.9 Although β-diketones represent one of the oldest classes of chelating ligands,10,11 their coordination chemistry continues to attract much interest, due to the ability of related metal complexes to support several unique and important catalytic reactions.12 It is often noted that even modestly sterically hindered β-diketones offer improvements over the parent acetylacetone.13 Truly hindered β-diketones have only recently been made synthetically accessible.14 The presence of steric bulk on β-diketones is of great interest due to their peculiar coordination behavior useful for improving their catalytic activity and selectivity.15,16 β-Diketonates have been used as supporting ligands for Ti(IV),17,18 Ru(II),19–25 Rh(I),26,27 Pd(II)28 and Pt-based anticancer agents.29–33 They have also been reported to induce apoptosis in human tumor cells.34 In an early investigation, several platinum(II) complexes with β-diketonate ligands as leaving groups were studied, revealing that the ligands play an integral role in modulating toxic side effects.35 In particular, the phenyl ring substituents increase the lipophilicity and improve cellular uptake of the resulting complexes, whereas the electron-withdrawing CF3 group increases the hydrolysis rates of the complexes in aqueous solution.
Despite the enormous amount of work devoted to the synthesis and characterization of copper(II) β-diketonate complexes,36 there are relatively few reports devoted to the corresponding Cu(I) complexes perhaps due to their tendency to undergo disproportionation to copper metal and copper(II) compounds in the absence of stabilizing ligands.37,38 Reports of triorganophosphane adducts, (β-diketonate)Cu(PR3)n, are also relatively scarce.39–52
Several silver(I) β-diketonates have been synthesized and structurally characterized.53–57 In particular, fluorinated β-diketonate ligands were used to construct coordination polymers,58–60 stabilize multinuclear ethynide or thiolate clusters,61,62 and as precursors for chemical vapor deposition processes.63 Moreover, the photosensitivity of silver β-diketonates makes it possible to obtain functional materials even at room temperature.64,65 However, very little attention has been paid to the study of phosphane adducts of silver(I) β-diketonates,40,66–71 although they may also display a rich structural diversity.
To our knowledge, with the exception of Cu(II) derivatives of curcumin,72,73 Casiopeinas®-like compounds (Cas III)74–77 and analogous mixed 1,10-phenanthroline and 4,4,4-trifluoro-1-phenyl-1,3-butanedionate,78 2,2-bipyridine and 4,4,4-trifluoro-1-(2-furyl)-1,3-butanedionate79,80 or benzoylacetonate,81 acetylacetonate82 and 2-thenoyltrifluoroacetonate,83 very few studies on the anticancer activity of group 11 metal complexes of β-diketones have been reported in the literature to date.84 Copper(I)- and silver(I)-based anticancer complexes supported by β-diketonate ligands remain an unexplored research field. Therefore, as part of our continuous investigation on the chemical and biological properties of copper- and silver-containing coordination compounds,85–91 we report here for the first time a study on the syntheses, characterization and biological evaluation of new Ag(I) and Cu(I) complexes containing phosphanes and the anion of the β-diketone ligands, 1,3-dimesitylpropane-1,3-dione (HLMes), 1,3-bis(3,5-bis(trifluoromethyl)phenyl)-3-hydroxyprop-2-en-1-one (HLCF3) and 1,3-diphenylpropane-1,3-dione (HLPh). The ligands were selected to systematically modify the electronic properties and hydrophobicity of the resulting metal complexes using phenyl, mesityl and trifluoromethyl-phenyl groups, respectively. On the other hand, fluorine-containing compounds are of relevant interest in modern medicinal chemistry and in general, they are of special interest for use in drug design because of the good biological activity and low toxicity of molecules containing the trifluoromethyl moieties.92–94 Selective introduction of fluorine into a therapeutic or diagnostic small molecule candidate can enhance/modulate a number of physicochemical and pharmacokinetic properties such as improved metabolic stability and enhanced membrane permeation.95 Indeed, the substitution of a main group by a trifluoromethyl group in a molecule might be expected to induce great changes in molecular properties, in terms of hydrophobicity, solubility and electronegativity affecting not only metabolic stability but also binding affinities toward target proteins.96–99 In designing the novel β-diketonate metal complexes, the lipophilic triphenylphosphine (PPh3) and hydrophilic 1,3,5-triaza-7-phosphaadamantane (PTA) were selected as co-ligands, in order to stabilize copper and silver in their +1 oxidation state and to confer different solubility properties to the corresponding metal complexes.
A search in the Cambridge Structural Database100 reveals that many of the structurally well-authenticated phosphane adducts of copper(I) and silver(I) involve fluoroalkyl substituted β-diketonates. Among these, copper adducts are relatively more common and often feature two-phosphane ligands bonded to copper producing tetrahedral molecules. A relatively larger number of reported silver complexes are three-coordinate with one phosphane on silver(I). Two representative examples are depicted as A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 and B
52 and B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 71 in Fig. 1.
71 in Fig. 1.
 | ||
| Fig. 1 Several examples of structurally authenticated copper(I) (A, C, D, E) and silver(I) (B) phosphane complexes. | ||
Notably, structural data on copper and silver phosphane complexes supported by diaryl β-diketonates are quite limited. They include (1,3-diferrocenylpropane-1,3-dionato)bis(triphenylphosphine)copper(I) (C, Fig. 1),50 (1,3-diphenyl-1,3-dionato)bis(triphenylphosphine)copper(I) (D, Fig. 1)51 and (1,3-diphenyl-1,3-dionato)(trimethylphosphine)copper(I) (E, Fig. 1).41 In this paper, we report the X-ray crystal structures of [Cu(LCF3)(PPh3)2] and [Ag(LPh)(PPh3)2] as two new additions to this group.
Finally, the in vitro antitumor properties of the new Cu(I) and Ag(I) complexes as well as of the corresponding uncoordinated ligands were evaluated against several human cancer cell lines derived from different solid tumors by means of both 2D and 3D cell viability tests. The cytotoxicity data have been compared with those obtained with cisplatin, the reference metal-based chemotherapeutic drug.
Results and discussion
Synthesis and characterization
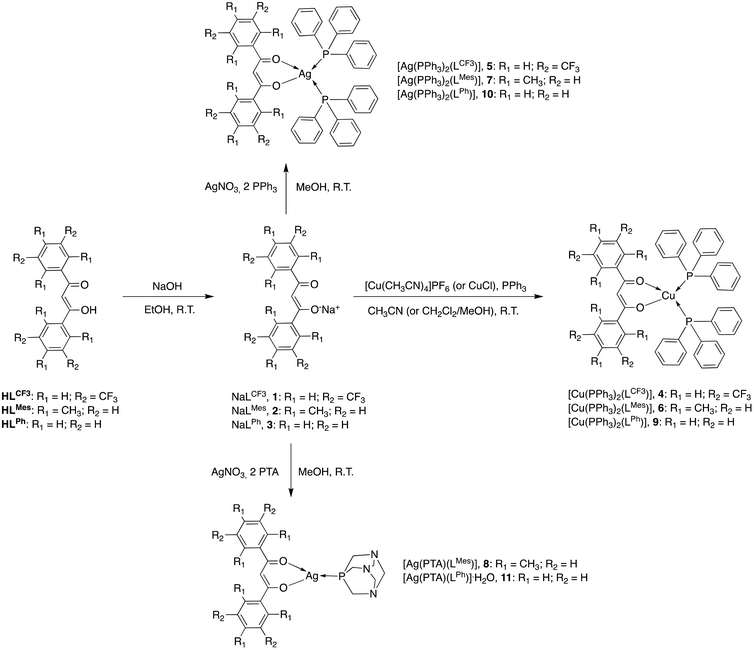
The β-diketones 1,3-bis(3,5-bis(trifluoromethyl)phenyl)propane-1,3-dione (HLCF3)101 and 1,3-dimesityl-propane-1,3-dione (HLMes)102 were prepared according to the literature procedures and fully characterized by several methods. The X-ray crystal structure of HLCF3 is presented in the ESI (Fig. S1†). 1,3-Diphenylpropane-1,3-dione (HLPh) was obtained from commercial sources and used as received. β-Diketones are capable of keto–enol tautomerism, and the tautomers exist in equilibrium with each other in solution; although, in most organic solvents, β-diketones are predominately enolized.103The sodium salts of β-diketonate ligand NaLCF3 (1), NaLMes (2) and NaLPh (3) were prepared, using a modified literature method of analogous sodium β-diketonates,104 from the reaction with HLCF3, HLMes and HLPh respectively, with NaOH in ethanol solution, and isolated as orange whitish solids in 85% yield for NaLCF3, 74% yield for NaLMes and in 80% yield for NaLPh (Scheme 1).
Compounds 1, 2 and 3 were fully characterized by multinuclear NMR spectroscopy, FT-IR, ESI-MS and elemental analysis. The two (CF3)2Ph, mesityl and phenyl groups are magnetically equivalent in 1H and 13C NMR. The 1H-NMR spectrum of 1, recorded in DMSO-d6 solution, shows a single set of resonances for the two CH protons in ortho- and para-positions of aromatic rings at 8.48 and 8.08 ppm, respectively, while the signal at δ 6.57 ppm is assignable to the COCHCO proton of the diketone. The 19F NMR spectrum of 1 displayed a singlet at δ −61.21 ppm. Analogously, the 1H NMR spectrum of compound 2 in CDCl3 includes single peaks assignable to (CH3)3Ph protons (δ 2.23, 2.24 and 6.72 ppm) and to the COCHCO proton of the diketone (δ 5.31 ppm), while in the spectrum of 3 recorded in acetone-d6 the aromatic protons are at 7.46–8.07 ppm and the COCHCO proton of the diketone is at δ 6.87 ppm. Deprotonation of β-diketonate ligands leads to a slight shift of the COCHCO group resonance with respect the protonated species (δ 6.89 ppm for HLCF3, 5.77 ppm for HLMes and 6.89 for HLPh, in CDCl3). The infrared (FT-IR) spectra of β-diketones generally exhibit very strong bands in the 1200–1650 cm−1 region.105–108 For compounds 1–3, the bands in the range of 1565–1643 cm−1 are assigned to the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) stretching modes. For compound 1 the bands at 1163–1164 and 1110–1121 cm−1 can be assigned to the C–F stretching and CF3 deformation, respectively.
O) stretching modes. For compound 1 the bands at 1163–1164 and 1110–1121 cm−1 can be assigned to the C–F stretching and CF3 deformation, respectively.
For derivatives of the HLCF3 ligand, the Cu(I) complex [Cu(LCF3)(PPh3)2] (4) was prepared from the reaction of PPh3, Cu(CH3CN)4PF6 and the sodium salt NaLCF3, while the Ag(I) complex [Ag(LCF3)(PPh3)2] (5) was prepared from the reaction of PPh3, AgNO3 and the sodium salt NaLCF3 (Scheme 1). The IR spectra recorded for a solid sample of 4 and 5 show all the expected bands for the β-diketone ligand and the triphenylphosphine co-ligands. The absorptions due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching are at 1582–1626 cm−1, while bands due to C–F stretching and CF3 deformation are at 1169–1171 and 1125–1126 cm−1, respectively. They don't significantly vary with respect to the same absorptions of the carbonyl group detectable in the spectrum of the free ligand salt 1. The 1H-NMR spectra of complexes 4 and 5, recorded in CDCl3 solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalent, with a slight shift due to the coordination to the metal centre. The PPh3 co-ligands show a characteristic series of peaks in the range of 7.22–7.44 ppm. 19F NMR spectra of 4 and 5 in CDCl3 displayed singlets at δ −62.64 and −62.66 ppm, respectively. The ESI-MS study was performed by dissolving 4 and 5 in CH3CN and recording the spectra in positive- and negative-ion modes. The structure of 4 and 5 was confirmed by the presence of peaks attributable to the [Cu(PPh3) + CH3CN]+, [Cu(PPh3)2]+, [Ag(PPh3) + CH3CN]+ and [Ag(PPh3)2]+ species, being positive fragments of the dissociation of the ligand from the complex. In addition, in the negative-ion mode spectra we observe peaks at 495 due to the [LCF3]− fragment.
O stretching are at 1582–1626 cm−1, while bands due to C–F stretching and CF3 deformation are at 1169–1171 and 1125–1126 cm−1, respectively. They don't significantly vary with respect to the same absorptions of the carbonyl group detectable in the spectrum of the free ligand salt 1. The 1H-NMR spectra of complexes 4 and 5, recorded in CDCl3 solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalent, with a slight shift due to the coordination to the metal centre. The PPh3 co-ligands show a characteristic series of peaks in the range of 7.22–7.44 ppm. 19F NMR spectra of 4 and 5 in CDCl3 displayed singlets at δ −62.64 and −62.66 ppm, respectively. The ESI-MS study was performed by dissolving 4 and 5 in CH3CN and recording the spectra in positive- and negative-ion modes. The structure of 4 and 5 was confirmed by the presence of peaks attributable to the [Cu(PPh3) + CH3CN]+, [Cu(PPh3)2]+, [Ag(PPh3) + CH3CN]+ and [Ag(PPh3)2]+ species, being positive fragments of the dissociation of the ligand from the complex. In addition, in the negative-ion mode spectra we observe peaks at 495 due to the [LCF3]− fragment.
The Cu(I) complexes of HLMes and HLPh ligands, [Cu(LMes)(PPh3)2] (6) and [Cu(LPh)(PPh3)2] (9) were prepared from the reaction of PPh3, Cu(CH3CN)4PF6 and NaLMes (2) and NaLPh (3), respectively (Scheme 1). Analogously, the Ag(I) complexes [Ag(LMes)(PPh3)2] (7) and [Ag(LPh)(PPh3)2] (10) were prepared from the reaction of PPh3, AgNO3 and the sodium salts 2 and 3, respectively (Scheme 1). The IR spectra recorded of solid samples of 6, 7, 9 and 10 show all the expected bands for the β-diketone ligand and the triphenylphosphine co-ligands. The absorptions due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching are at 1548–1651 cm−1 and they don't significantly vary with respect to the same absorptions in the spectra of the free ligand salts 2 and 3. The 1H-NMR spectra of complexes 6, 7, 9 and 10, recorded in CDCl3 or CD3OD solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalent, with a slight shift due to coordination with the metal center. The PPh3 co-ligands show a characteristic series of peaks in the range of 7.21–7.50 ppm. The ESI-MS study was performed by dissolving 6, 7, 9 and 10 in CH2Cl2/CH3CN, CH3OH or CH3CN. In the positive-ion mode spectra, we observed the presence of peaks attributable to the [Cu(PPh3)2]+ and [Ag(PPh3)2]+ species, which are positive fragments of the dissociation of the ligand from the complexes. In addition, in the negative-ion mode spectra we observed peaks at m/z 307 and 223, due to the [LMes]− and [LPh]− fragments, respectively.
O stretching are at 1548–1651 cm−1 and they don't significantly vary with respect to the same absorptions in the spectra of the free ligand salts 2 and 3. The 1H-NMR spectra of complexes 6, 7, 9 and 10, recorded in CDCl3 or CD3OD solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalent, with a slight shift due to coordination with the metal center. The PPh3 co-ligands show a characteristic series of peaks in the range of 7.21–7.50 ppm. The ESI-MS study was performed by dissolving 6, 7, 9 and 10 in CH2Cl2/CH3CN, CH3OH or CH3CN. In the positive-ion mode spectra, we observed the presence of peaks attributable to the [Cu(PPh3)2]+ and [Ag(PPh3)2]+ species, which are positive fragments of the dissociation of the ligand from the complexes. In addition, in the negative-ion mode spectra we observed peaks at m/z 307 and 223, due to the [LMes]− and [LPh]− fragments, respectively.
The Ag(I) complexes of HLMes and HLPh ligands, [Ag(LMes)(PTA)] (8) and [Ag(LPh)(PTA)]·H2O (11), were prepared from the reaction of PTA, AgNO3 and the sodium salts 2 and 3, respectively (Scheme 1). Several attempts to synthesize Ag(I) complexes with two PTA as coligands have been unsuccessful, even modifying the reaction conditions and stoichiometric ratio between the reagents. Analytical and spectroscopic data suggest the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry for complexes 8 and 11, with PTA coordinated via the phosphorus atom. On the other hand, PTA acts as a monodentate P-donor ligand in a vast majority of known complexes,109 although in the absence of crystallography data we cannot exclude that the PTA binds the metal in the bridging N,P-coordination mode.110
1 stoichiometry for complexes 8 and 11, with PTA coordinated via the phosphorus atom. On the other hand, PTA acts as a monodentate P-donor ligand in a vast majority of known complexes,109 although in the absence of crystallography data we cannot exclude that the PTA binds the metal in the bridging N,P-coordination mode.110
The IR spectra recorded for the solid samples of 8 and 11 show all the expected bands for the β-diketone ligand and the 1,3,5-triazaphosphaadamantane co-ligands. The absorptions due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching are at 1513–1610 cm−1 and they don't significantly vary with respect to the same absorptions in the spectra of the free ligand salts. The 1H-NMR spectra of complexes 8 and 11, recorded in CD3OD solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalents, with a slight shift due to the coordination to the metal center. The PTA co-ligands show a characteristic series of peaks in the range of 4.15–4.67 ppm, with an integration that confirms the 1
O stretching are at 1513–1610 cm−1 and they don't significantly vary with respect to the same absorptions in the spectra of the free ligand salts. The 1H-NMR spectra of complexes 8 and 11, recorded in CD3OD solution at room temperature, show a single set of resonances for the β-diketone moiety, indicating that the protons of the aromatic rings are equivalents, with a slight shift due to the coordination to the metal center. The PTA co-ligands show a characteristic series of peaks in the range of 4.15–4.67 ppm, with an integration that confirms the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The ESI-MS study was performed by dissolving 8 and 11 in CH3OH. In the positive-ion mode spectra we observed at m/z 158 and 420 the presence of peaks attributable to the [PTA + H]+ and [Ag(PTA)2]+ species, respectively, due to the dissociation of the ligand from the complexes.
1 stoichiometry. The ESI-MS study was performed by dissolving 8 and 11 in CH3OH. In the positive-ion mode spectra we observed at m/z 158 and 420 the presence of peaks attributable to the [PTA + H]+ and [Ag(PTA)2]+ species, respectively, due to the dissociation of the ligand from the complexes.
It's interesting to note that the diagnostic COCHCO signal is at 6.17 and 6.21 ppm in the spectra recorded in CDCl3 of [Cu(LCF3)(PPh3)2] (4) and [Ag(LCF3)(PPh3)2] (5), respectively, and the related peak is present at 6.57 ppm in the spectrum of sodium salt NaLCF3 (1) in DMSO-d6 solution and at 6.91 in the spectrum of HLCF3 in CDCl3 solution. In the 13C{1H}-NMR spectra of 4 and 5 the COCHCO signals are at 92.02 and 92.35 ppm, respectively, and the related peaks are at 90.91 and 93.82 ppm in the spectra of the free ligands 1 and HLCF3. In the 1H-NMR spectra of [Cu(LMes)(PPh3)2] (6), [Ag(LMes)(PPh3)2] (7) and [Ag(LMes)(PTA)] (8), recorded in CDCl3 or DMSO-d6, the COCHCO signals are at 4.81–5.34, and the related peaks are present at 5.29 and 5.77 ppm in the spectra of NaLMes (2) and HLMes, respectively, in CDCl3 solution. In the 13C{1H}-NMR spectra of 6–8 the COCHCO signals are at 101.66–103.75, and the related peak is at 103.85 and 105.48 ppm, in the spectra of the free ligands 2 and HLMes. Finally, the COCHCO signals are at 6.40–6.78 ppm in the spectra recorded in CDCl3 or CD3OD for [Cu(LPh)(PPh3)2] (9), [Ag(LPh)(PPh3)2] (10) and [Ag(LPh)(PTA)]·H2O (11), respectively, and the related peaks are present at 6.87 and 6.89 ppm in the spectra of the sodium salt NaLPh (3) and HLPh in acetone-d6 and CDCl3 solution, respectively. In the 13C{1H}-NMR spectra of 9–11 the COCHCO signals are at 90.35–93.38 ppm and the related peak is at 93.21 ppm in the spectrum of the free ligand 3. In the 13C{1H}-NMR spectra of 9–11 the COCHCO signals are at 90.35–93.38 ppm, and the related peaks are at 93.21 and 93.18 ppm, in the spectra of the free ligands 3 and HLPh.
The room temperature 31P{H}-NMR spectra of the Cu(I) complexes 4, 6 and 9, recorded in CDCl3 or CD2Cl2 solution, exhibited single signals at −3.68, −5.41 and −3.96 ppm, respectively, downfield shifted with respect to the value of the free triphenylphosphine PPh3 (δ = −4.85 ppm in CDCl3 and −5.55 ppm in CD2Cl2). The room temperature 31P{H}-NMR spectra of the Ag(I) complexes, recorded in CDCl3 solution (compounds 5, 7, and 10) or CD3OD solution (compounds 8 and 11), gave singlet signals downfield shifted with respect to the value of the free phosphanes PPh3 and PTA (δ = −4.85 and −102.07 ppm, respectively). At 223 K, the spectra of 5 and 7 (in the CDCl3 solvent) show one pair of doublets in which the coupling of 31P to the 107Ag and 109Ag is resolved, in accordance with a stopped or slow triphenylphosphane exchange process: the 1J(107Ag–31P) and 1J(109Ag–31P) coupling constants are respectively in the range of 408–430 and 472–496 Hz for compounds 5 and 7, being of the same order of magnitude as those reported for analogous silver(I) bis(triphenylphosphine) species.111–113 At 223 K, compound 10 exhibits a broad doublet with a 1J(Ag–31P) coupling constant of 439 Hz. The ratio of 1J(109Ag–31P)/1J(107Ag–31P) is in good agreement with the 107Ag/109Ag gyromagnetic ratio of 1.15. The room temperature 31P{H}-NMR spectra of complexes 8 and 11 with PTA coligands, recorded in CD3OD solution, gave singlets centered at δ −83.73 and −83.82 ppm, while at 243 K they exhibited broad signals, centered at about −81 ppm, in which the coupling of 31P to 107/109Ag is not resolved.
Compounds [Cu(LCF3)(PPh3)2] (4) and [Ag(LPh)(PPh3)2] (10) produced a crystalline material suitable for X-ray crystallography. They crystallize as discrete molecules in the space groups P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and P21/n, respectively, and the molecular structures are illustrated in Fig. 2 and 3. There are two chemically similar molecules in the asymmetric unit of [Cu(LCF3)(PPh3)2]. Both the copper and silver complexes adopt a distorted tetrahedral geometry. Complete details of the bond distances and angles are given in the ESI.† Although the diketonate based CuO2C3 metallacycle is essentially planar in [Cu(LCF3)(PPh3)2] (Fig. 2), the AgO2C3 core of the silver complex [Ag(LPh)(PPh3)2] (Fig. 3) adopts a half-boat conformation with the silver atom residing out of the O2C3 plane. The average Cu–P distance and Cu–O distances of [Cu(LCF3)(PPh3)2] (2.2256 and 2.071 Å, respectively) are shorter than the Ag–P and Ag–O distances in [Ag(LPh)(PPh3)2] (average 2.424 and 2.343 Å, respectively), which is expected due to the larger covalent radius of silver relative to copper. The P–M–P angles (M = Cu, Ag) are very similar between the two systems (128.50° and 129.20° for 4 and 10, respectively). A comparison of Cu–O distances of [Cu(LCF3)(PPh3)2] (av. 2.071 Å) and [Cu(LPh)(PPh3)2] (av. 2.058 Å) indicates that the latter featuring more electron donating β-diketonate has slightly shorter Cu–O contacts.51 The Cu–P distances of these two molecules (av. 2.2256 Å of [Cu(LCF3)(PPh3)2] and av. 2.250 Å of [Cu(LPh)(PPh3)2]) show an opposite trend. We have also confirmed the identity of [Cu(LMes)(PPh3)2] (6) using X-ray crystallography. It is also a four-coordinate, pseudo-tetrahedral complex (see the ESI, Fig. S2†). Unfortunately, the weakly diffracting sample and crystal twinning of [Cu(LMes)(PPh3)2] prevented us from obtaining high quality data suitable for detailed analysis of structural features.
and P21/n, respectively, and the molecular structures are illustrated in Fig. 2 and 3. There are two chemically similar molecules in the asymmetric unit of [Cu(LCF3)(PPh3)2]. Both the copper and silver complexes adopt a distorted tetrahedral geometry. Complete details of the bond distances and angles are given in the ESI.† Although the diketonate based CuO2C3 metallacycle is essentially planar in [Cu(LCF3)(PPh3)2] (Fig. 2), the AgO2C3 core of the silver complex [Ag(LPh)(PPh3)2] (Fig. 3) adopts a half-boat conformation with the silver atom residing out of the O2C3 plane. The average Cu–P distance and Cu–O distances of [Cu(LCF3)(PPh3)2] (2.2256 and 2.071 Å, respectively) are shorter than the Ag–P and Ag–O distances in [Ag(LPh)(PPh3)2] (average 2.424 and 2.343 Å, respectively), which is expected due to the larger covalent radius of silver relative to copper. The P–M–P angles (M = Cu, Ag) are very similar between the two systems (128.50° and 129.20° for 4 and 10, respectively). A comparison of Cu–O distances of [Cu(LCF3)(PPh3)2] (av. 2.071 Å) and [Cu(LPh)(PPh3)2] (av. 2.058 Å) indicates that the latter featuring more electron donating β-diketonate has slightly shorter Cu–O contacts.51 The Cu–P distances of these two molecules (av. 2.2256 Å of [Cu(LCF3)(PPh3)2] and av. 2.250 Å of [Cu(LPh)(PPh3)2]) show an opposite trend. We have also confirmed the identity of [Cu(LMes)(PPh3)2] (6) using X-ray crystallography. It is also a four-coordinate, pseudo-tetrahedral complex (see the ESI, Fig. S2†). Unfortunately, the weakly diffracting sample and crystal twinning of [Cu(LMes)(PPh3)2] prevented us from obtaining high quality data suitable for detailed analysis of structural features.
 | ||
| Fig. 2 Molecular structure of [Cu(LCF3)(PPh3)2] (4) (only one of the two molecules present in the asymmetric unit is shown). | ||
Biological studies
The stability of the new complexes in 0.5% DMSO/physiological solution was evaluated by UV-Vis spectroscopy. Spectra were recorded every 24 h in the range of 240–640 nm over 72 h. The collected spectra for all complexes are reported in the ESI, Fig. S3† showing that all compounds were sufficiently stable under physiological conditions. All the spectra, and in particular the spectra registered for complexes 4 and 10 showed curves with a progressive increase in the baseline adsorption, probably due to solubility issues.The Ag(I) and Cu(I) complexes and the corresponding uncoordinated ligands and their salts were evaluated for their cytotoxic activity towards various human cancer cell lines representative of different solid tumors. Cytotoxicity of PPh3 and PTA ligands have already been published.114 In particular, the in-house cancer cell panel contained examples of human colon (HCT-15), pancreatic (PSN-1 and BxPC3), testicular (NTERA-2), and breast (MDA-MB-231), as well as SCLC (U1285) and NSCLC (A549). The cytotoxicity parameters, expressed in terms of IC50 values obtained after 72 h of exposure to the MTT assay, are reported in Table 1. For comparison, the cytotoxicity of the reference metal-based chemotherapeutic drug cisplatin was assessed under the same experimental conditions.
| IC50 (μM) ± S.D. | |||||||
|---|---|---|---|---|---|---|---|
| NTERA-2 | HCT-15 | BxPC3 | U-1285 | PSN-1 | A549 | MDA-MB-231 | |
| Cells (3–8 × 103 cells per well) were treated for 72 h with increasing concentrations of tested compounds. Cytotoxicity was assessed by the MTT test. The IC50 values were calculated by the four-parameter logistic model (p < 0.05). | |||||||
| HLCF3 | 21.1 ± 0.9 | 43.5 ± 8.2 | >50 | 44.8 ± 5.8 | >50 | >50 | >50 |
| NaLCF3 | ND | ND | ND | ND | ND | ND | ND |
| [Cu(LCF3)(PPh3)2] | 1.3 ± 0.4 | 2.0 ± 0.5 | 1.2 ± 0.4 | 1.5 ± 0.5 | 4.4 ± 0.3 | 5.4 ± 0.9 | 5.3 ± 1.2 |
| [Ag(LCF3)(PPh3)2] | 3.5 ± 0.6 | 2.1 ± 0.4 | 2.9 ± 0.1 | 2.6 ± 0.2 | 6.3 ± 1.2 | 11.1 ± 1.5 | 11.3 ± 2.2 |
| HLMes | 3.6 ± 0.1 | 5.3 ± 0.7 | 7.5 ± 1.2 | 8.1 ± 1.1 | 5.2 ± 0.8 | 12.5 ± 3.6 | ND |
| NaLMes | 5.7 ± 0.8 | 4.6 ± 1.4 | 4.3 ± 0.9 | 5.1 ± 0.5 | 4.1 ± 0.3 | 9.1 ± 2.3 | ND |
| [Cu(LMes)(PPh3)2] | 0.9 ± 0.3 | 1.2 ± 0.5 | 1.1 ± 0.4 | 1.2 ± 0.7 | 1.3 ± 0.5 | 5.6 ± 1.1 | 5.5 ± 0.8 |
| [Ag(LMes)(PPh3)2] | 1.7 ± 0.6 | 2.5 ± 1.0 | 1.7 ± 0.5 | 2.7 ± 0.9 | 2.1 ± 0.7 | 7.6 ± 1.4 | 9.5 ± 2.3 |
| [Ag(LMes)(PTA)] | 12.8± 2.5 | 19.5 ± 3.1 | 14.9 ± 2.2 | 17.3 ± 2.7 | 15.7 ± 1.9 | 22.8 ± 3.3 | 28.1 ± 3.4 |
| HLPh | >50 | >50 | >50 | 39.5 ± 2.8 | 38.5 ± 4.1 | >50 | >50 |
| NaLPh | 29.7 ± 5.4 | 25.2 ± 2.9 | 27.5 ± 0.1 | 29.2 ± 3.4 | 28.8 ± 4.1 | >50 | >50 |
| [Cu(LPh)(PPh3)2] | 5.1 ± 0.6 | 8.5 ± 1.5 | 6.2 ± 0.8 | 12.3 ± 2.2 | 9.8 ± 2.1 | 15.2 ± 2.8 | 25.3 ± 3.1 |
| [Ag(LPh)(PPh3)2] | 3.0 ± 0.4 | 1.9 ± 0.8 | 3.1 ± 0.8 | 4.2 ± 1.1 | 2.4 ± 0.1 | 8.3 ± 1.9 | 9.1 ± 2.1 |
| [Ag(LPh)(PTA)] | 35.8 ± 5.8 | 37.2 ± 2.5 | 40.7 ± 2.9 | 28.6 ± 4.1 | 25.3 ± 4.2 | >50 | >50 |
| Cisplatin | 14.6 ± 3.0 | 13.9 ± 1.6 | 11.9 ± 1.3 | 2.1 ± 0.8 | 12.1 ± 2.8 | 9.1 ± 1.4 | 21.5 ± 4.1 |
NaLPh/HLPh and NaLCF3/HLCF3 ligands proved to be hardly effective against all tested cancer cell lines. In contrast, NaLMes/HLMes ligands were quite effective in inhibiting cancer cell growth, with IC50 values in the low micromolar range. It is interesting to note that in the case of PPh3-containing Cu(I) and Ag(I) complexes, metal coordination significantly improved the cytotoxic potency compared to the free ligand, conversely, for [Ag(LMes)(PTA)] sensibly higher IC50 values were always obtained, confirming that the bioactivity of the metal complex depends on the full set of coordinating ligands. All tested complexes demonstrated a marked cytotoxic activity towards cancer cell lines belonging to the in-house cancer cell panel, showing IC50 values in the low/sub micromolar range. On average, the PPh3 derivatives were more effective than cisplatin, whereas the two Ag(I) complexes bearing the PTA moiety were less effective than the reference metallodrug cisplatin. Among the PPh3 derivatives, Cu and Ag complexes bearing the LMes ligand were the most effective derivatives, with average IC50 values of 2.4 and 4.0 μM, respectively. It is noteworthy that against testicular carcinoma NTERA-2 cells, [Cu(LMes)(PPh3)2] was up to 16-fold more efficacious than cisplatin in decreasing cell proliferation. Conversely, the weakest PPh3 derivatives were those bearing the LPh ligand. Interestingly, Cu(I) complexes containing the LCF3 and LMes ligands were much more effective (about 2-fold) than the corresponding Ag(I) derivatives whereas in the case of the LPh ligand, the Ag(I) complex was about 2.7 times more effective than the corresponding copper derivative.
The in vitro antitumor activity of the newly developed Cu(I) and Ag(I) derivatives was also assayed in 3D cell culture models of human colon cancer cells. Although the two-dimensional cell cultures are the most employed assays for in vitro screening (due to the low cost, simplicity, and reliability), 2D methods are unable to mimic the in vivo properties of solid tumor models. On the other hand, 3D cell cultures are much more effective in closely mimicking the heterogeneity and complexity of the tumor mass, and therefore are more predictive for in vivo results than conventional 2D cell cultures.115 On these bases, we tested the activity of the newly developed complexes on spheroids obtained from human HCT-15 tumor cells. HCT15 cells are known for their ability to form spheroids, which makes them a valuable tool for studying the interactions between cancer cells and the surrounding microenvironment. Human colon cancer cell spheroids were treated with the investigated compounds for 72 h, and cell viability was assessed by means of the acid phosphatase (APH) assay and the results are reported in Table 2. Results, reported in Table 2, were completely different from those obtained by 2D screening, and clearly showed that among the new Cu(I) and Ag(I) complexes, only derivative [Cu(LCF3)(PPh3)2] possessed an antiproliferative activity against 3D tumor spheroids comparable to that of the reference drug cisplatin.
| IC50 (μM) ± S.D. | |
|---|---|
| HCT-15 | |
| Cells (2.5 × 103 cells per well) were treated for 72 h with increasing concentrations of tested compounds. Cytotoxicity was assessed by the APH assay. The IC50 values were calculated using the four-parameter logistic model (p < 0.05). | |
| [Cu(LCF3)(PPh3)2] | 58.5 ± 5.8 |
| [Ag(LCF3)(PPh3)2] | >100 |
| [Cu(LMes)(PPh3)2] | 86.6 ± 6.7 |
| [Ag(LMes)(PPh3)2] | >100 |
| [Ag(LMes)(PTA)] | >100 |
| [Cu(LPh)(PPh3)2] | >100 |
| [Ag(LPh)(PPh3)2] | 82.5 ± 5.8 |
| [Ag(LPh)(PTA)] | >100 |
| Cisplatin | 59.5 ± 3.3 |
Conclusions
In this study, we report the synthesis, characterization and biological evaluation of Cu(I) and Ag(I) phosphane complexes supported by the anion of sterically hindered β-diketone ligands. In particular, 1,3-dimesitylpropane-1,3-dione (HLMes) and 1,3-bis(3,5-bis(trifluoromethyl)phenyl)-3-hydroxyprop-2-en-1-one (HLCF3) characterized by the presence of trifluoromethyl or methyl groups on the phenyl moieties were employed for the preparation of the complexes 4–11, using the lipophilic PPh3 and the hydrophilic PTA as co-ligands to stabilize the metal in the +1 oxidation state. The analogous complexes of the anion of the parent 1,3-diphenylpropane-1,3-dione (HLPh) ligand were also synthesized and evaluated for their antitumor activity to compare the biological effects of substituents on the phenyl moieties.The compounds were fully characterized both in the solid state and in solution. The X-ray crystal structures of [Cu(LCF3)(PPh3)2] and [Ag(LPh)(PPh3)2] show that both the copper and silver complexes adopt a distorted tetrahedral geometry. The diketonate based CuO2C3 metallacycle is essentially planar in [Cu(LCF3)(PPh3)2], while the AgO2C3 core of [Ag(LPh)(PPh3)2] adopts a half-boat conformation with the silver atom residing out of the O2C3 plane.
Biological studies highlighted that both Ag(I) and Cu(I) complexes containing PPh3 as the phosphane coligand were more potent than cisplatin in inhibiting cancer cell growth. Among them, Cu complexes bearing the LCF3 and LMes ligands were the most effective derivatives, in particular [Cu(LMes)(PPh3)2] was up to 16-fold more efficacious than cisplatin in decreasing the cell proliferation of 2D testicular carcinoma cell cultures. Cytotoxicity experiments performed exploiting the proclivity of HCT-15 cells to form spheroids showed that [Cu(LCF3)(PPh3)2] possessed a marked antiproliferative activity against 3D tumor spheroids, confirming the ability of this derivative to penetrate across the entire spheroid domain and reach the inner hypoxic core.
Experimental section
Chemistry
NaLCF3 (1). The ligand HLCF3 (3.000 mmol, 1.489 g) and NaOH (3.000 mmol, 0.120 g) were dissolved in ethanol (100 mL), obtaining an opalescent yellow solution. The reaction mixture was stirred at room temperature for 4 hours. The solution was then dried at reduced pressure giving the orange solid product NaLCF3 in 85% yield. M.p.: 226–230 °C. FT-IR (cm−1, Fig. S4†): 3099wbr (C–H); 1628m, 1592m (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1580sh, 1511m, 1471m, 1421m, 1360s, 1275s, 1240m, 1189m; 1163s, 1121vs (CF3); 1044m, 955m, 905s, 887m, 845m, 787s, 702m, 681s. 1H-NMR (DMSO-d6, 293 K, Fig. S5†): δ 6.57 (s, 1H, COCHCO), 8.08 (s, 2H, p-CHar), 8.48 (s, 4H, o-CHar). 13C{1H}-NMR (DMSO-d6, 293 K, Fig. S6†): δ 90.91 (COCHCO); 123.96 (q, 1JCF = 273 Hz, CF3); 130.40 (q, 2JCF = 33 Hz, CCF3); 122.88, 127.68, 146.16 (CHar and Car); 179.21 (CO). 19F{1H}-NMR (DMSO-d6, 293 K, Fig. S7†): δ −61.21 (s). ESI-MS (major positive ions, CH3CN), m/z (%): 541 (40) [NaLCF3 + Na]+. Elemental analysis calculated for C19H7F12NaO2: C 44.04, H 1.36; found: C 43.89, H 1.34.
O); 1580sh, 1511m, 1471m, 1421m, 1360s, 1275s, 1240m, 1189m; 1163s, 1121vs (CF3); 1044m, 955m, 905s, 887m, 845m, 787s, 702m, 681s. 1H-NMR (DMSO-d6, 293 K, Fig. S5†): δ 6.57 (s, 1H, COCHCO), 8.08 (s, 2H, p-CHar), 8.48 (s, 4H, o-CHar). 13C{1H}-NMR (DMSO-d6, 293 K, Fig. S6†): δ 90.91 (COCHCO); 123.96 (q, 1JCF = 273 Hz, CF3); 130.40 (q, 2JCF = 33 Hz, CCF3); 122.88, 127.68, 146.16 (CHar and Car); 179.21 (CO). 19F{1H}-NMR (DMSO-d6, 293 K, Fig. S7†): δ −61.21 (s). ESI-MS (major positive ions, CH3CN), m/z (%): 541 (40) [NaLCF3 + Na]+. Elemental analysis calculated for C19H7F12NaO2: C 44.04, H 1.36; found: C 43.89, H 1.34.
NaLMes (2). To the ligand HLMes (2.00 mmol, 0.617 g) solubilized in methanol (15 mL), NaOH (2.000 mmol, 0.080 g) was added. The reaction mixture was stirred under reflux for 24 hours. The solution was dried at reduced pressure and the residue was recrystallized by diethyl ether and filtered. From the mother liquors, dried at reduced pressure, the light orange whitish compound NaLMes has been obtained in 74% yield. M.p.: 102–104 °C. FT-IR (cm−1, Fig. S8†): 3281wbr, 3147wbr, 2971wbr, 2951wbr, 2918w, 2858w (C–H); 1611m, 1565br (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1557m, 1499m, 1417sbr, 1373s, 1298w, 1271m, 1164m, 1110m, 1028mbr, 955w, 926w, 882w, 848m, 791m, 779m, 718m. 1H-NMR (CDCl3, 293 K, Fig. S9†): δ 2.23 (s, 6H, p-CH3), 2.24 (s, 12H, o-CH3), 5.29 (s, 1H, COCHCO), 6.72 (s, 4H, m-CH). 13C{1H}-NMR (CDCl3, 293 K, Fig. S10†): δ 19.58 (o-CCH3); 20.96 (p-CCH3); 103.85 (COCHCO); 127.85, 132.65, 133.20, 136.10 (CHar and Car); 191.24 (CO). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 331 (100) [NaLMes + H]+, 353 (40) [NaLMes + Na]+, 683 (10) [2NaLMes + Na]+. ESI-MS(−) (major negative ions, CH3OH), m/z (%): 307 (100) [LMes]−, 637 (10) [2LMes + Na]−. Elemental analysis (%) calculated for C21H23NaO2: C 76.32, H 7.02; found C 74.09, H 7.09.
O); 1557m, 1499m, 1417sbr, 1373s, 1298w, 1271m, 1164m, 1110m, 1028mbr, 955w, 926w, 882w, 848m, 791m, 779m, 718m. 1H-NMR (CDCl3, 293 K, Fig. S9†): δ 2.23 (s, 6H, p-CH3), 2.24 (s, 12H, o-CH3), 5.29 (s, 1H, COCHCO), 6.72 (s, 4H, m-CH). 13C{1H}-NMR (CDCl3, 293 K, Fig. S10†): δ 19.58 (o-CCH3); 20.96 (p-CCH3); 103.85 (COCHCO); 127.85, 132.65, 133.20, 136.10 (CHar and Car); 191.24 (CO). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 331 (100) [NaLMes + H]+, 353 (40) [NaLMes + Na]+, 683 (10) [2NaLMes + Na]+. ESI-MS(−) (major negative ions, CH3OH), m/z (%): 307 (100) [LMes]−, 637 (10) [2LMes + Na]−. Elemental analysis (%) calculated for C21H23NaO2: C 76.32, H 7.02; found C 74.09, H 7.09.
NaLPh (3). The ligand HLPh (10.000 mmol, 2.243 g) and NaOH (10.000 mmol, 0.400 g) were solubilized in methanol (60 mL) and the reaction was stirred at room temperature for 24 hours under constant magnetic stirring. The precipitate formed was filtered and the mother liquors were evaporated at reduced pressure. The white product NaLPh was obtained in 80% yield. M.p.: 300–304 °C. FT-IR (cm−1, Fig. S11†): 3302wbr, 3160wbr, 3058w, 2939wbr, 2827wbr; 1643w, 1596s (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1556vs, 1510s, 1452vs, 1426vs, 1383s, 1297s, 1276s, 1219s, 1176m, 1114m, 1072m, 1044s, 1012s, 999m, 946m, 937m, 851w, 814w, 785s, 767s, 730vs. 1H-NMR (CD3OD, 293 K, Fig. S12†): δ 7.39–7.40 (m, 6H, CHar), 7.84–7.85 (m, 4H, CHar). 1H-NMR (acetone-d6, 293 K, Fig. S13†): δ 6.51 (s, 1H, COCHCO), 7.31–7.37 (m, 6H, CHar), 7.92–7.94 (m, 4H, CHar). 13C{1H}-NMR (acetone-d6, 293 K, Fig. S14†): δ 91.47 (COCHCO); 126.92, 127.69, 129.04, 143.6 (CHar and Car); 183.64 (CO). ESI-MS(+) (major positive ions, EtOH), m/z (%): 247 (100) [NaLPh + H]+. Elemental analysis calculated for C15H11NaO2: C 73.17, H 4.50; found: C 72.51, H 4.54.
O); 1556vs, 1510s, 1452vs, 1426vs, 1383s, 1297s, 1276s, 1219s, 1176m, 1114m, 1072m, 1044s, 1012s, 999m, 946m, 937m, 851w, 814w, 785s, 767s, 730vs. 1H-NMR (CD3OD, 293 K, Fig. S12†): δ 7.39–7.40 (m, 6H, CHar), 7.84–7.85 (m, 4H, CHar). 1H-NMR (acetone-d6, 293 K, Fig. S13†): δ 6.51 (s, 1H, COCHCO), 7.31–7.37 (m, 6H, CHar), 7.92–7.94 (m, 4H, CHar). 13C{1H}-NMR (acetone-d6, 293 K, Fig. S14†): δ 91.47 (COCHCO); 126.92, 127.69, 129.04, 143.6 (CHar and Car); 183.64 (CO). ESI-MS(+) (major positive ions, EtOH), m/z (%): 247 (100) [NaLPh + H]+. Elemental analysis calculated for C15H11NaO2: C 73.17, H 4.50; found: C 72.51, H 4.54.
[Cu(LCF3)(PPh3)2] (4). Tetrakis(acetonitrile)copper(I) hexafluorophosphate (0.500 mmol, 0.186 g) and triphenylphosphine (1.000 mmol, 0.262 g,) were solubilized in CH3CN (40 mL). The reaction mixture was kept under constant magnetic stirring at room temperature overnight. Then, ligand NaLCF3 (0.500 mmol, 0.259 g,) was added to the solution. After 5 hours of stirring, the mixture was evaporated and the solid formed was washed in Et2O. From the mother liquors an orange solid was precipitated, and it was crystallized using Et2O and n-hexane to give complex 4 in 60% yield. A batch of good quality crystals of Cu(LCF3)(PPh3)2, suitable for X-ray analysis, was obtained by slow evaporation of an ethanol/acetone solution of 4. M.p.: 109–112 °C. FT-IR (cm−1, Fig. S15†): 3056wbr (C–H); 1626w, 1582m (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1539w, 1519w, 1501w, 1480s, 1435m, 1417m, 1360s, 1275vs, 1247m; 1171s, 1125vs (CF3); 1097s, 1027m, 998w, 952m, 903m, 844m, 790w, 778m, 744s, 693s, 680s, 663m. 1H-NMR (CDCl3, 293 K, Fig. S16†): δ 6.17 (s, 1H, COCHCO), 7.22–7.41 (m, 30H, CHar), 7.90 (s, 2H, p-CHar), 8.11 (s, 4H, o-CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S17†): δ 6.72 (s, 1H, COCHCO), 7.31–7.65 (m, 30H, CHar), 8.19 (s, 2H, p-CHar), 8.38 (s, 4H, o-CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S18†): δ 92.03 (COCHCO); 123.42 (q, 1JCF = 273 Hz, CF3); 131.23 (q, 2JCF = 33 Hz, CCF3); 123.09, 126.95, 128.43, 129.58, 133.41, 133.60, 133.79, 133.88, 144.36 (CHar and Car); 181.65 (CO). 31P{1H}-NMR (CDCl3, 223 K, Fig. S19†): δ −3.68 (s). ESI-MS (major positive ions, CH3CN), m/z (%): 366 (40) [Cu(PPh3) + CH3CN]+, 587 (100) [Cu(PPh3)2]+. 19F{1H}-NMR (CDCl3, 293 K, Fig. S20†): δ −62.64 (s). ESI-MS (major negative ions, CH3CN), m/z (%): 495 (100) [LCF3]−. Elemental analysis (%) calculated for C55H37CuF12O2P2: C 60.98, H 3.44; found: C 59.95, H 3.36.
O); 1539w, 1519w, 1501w, 1480s, 1435m, 1417m, 1360s, 1275vs, 1247m; 1171s, 1125vs (CF3); 1097s, 1027m, 998w, 952m, 903m, 844m, 790w, 778m, 744s, 693s, 680s, 663m. 1H-NMR (CDCl3, 293 K, Fig. S16†): δ 6.17 (s, 1H, COCHCO), 7.22–7.41 (m, 30H, CHar), 7.90 (s, 2H, p-CHar), 8.11 (s, 4H, o-CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S17†): δ 6.72 (s, 1H, COCHCO), 7.31–7.65 (m, 30H, CHar), 8.19 (s, 2H, p-CHar), 8.38 (s, 4H, o-CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S18†): δ 92.03 (COCHCO); 123.42 (q, 1JCF = 273 Hz, CF3); 131.23 (q, 2JCF = 33 Hz, CCF3); 123.09, 126.95, 128.43, 129.58, 133.41, 133.60, 133.79, 133.88, 144.36 (CHar and Car); 181.65 (CO). 31P{1H}-NMR (CDCl3, 223 K, Fig. S19†): δ −3.68 (s). ESI-MS (major positive ions, CH3CN), m/z (%): 366 (40) [Cu(PPh3) + CH3CN]+, 587 (100) [Cu(PPh3)2]+. 19F{1H}-NMR (CDCl3, 293 K, Fig. S20†): δ −62.64 (s). ESI-MS (major negative ions, CH3CN), m/z (%): 495 (100) [LCF3]−. Elemental analysis (%) calculated for C55H37CuF12O2P2: C 60.98, H 3.44; found: C 59.95, H 3.36.
[Ag(LCF3)(PPh3)2] (5). Silver nitrate (0.500 mmol, 0.085 g,) and PPh3 (1.000 mmol, 0.262 g) were solubilized in a methanol/acetonitrile solution (20/20 mL). The reaction mixture was stirred for 3 hours at room temperature in the dark. Subsequently, the HLCF3 ligand (0.500 mmol, 0.248 g) was added and after 1 hour NaOH (0.500 mmol, 0.020 g) was added to the reaction solution. The reaction was run for 3 hours at room temperature in the dark. The yellow opalescent precipitate was filtered off; the solution was evaporated to dryness and the yellow solid product was solubilized in diethyl ether, and purified by filtration to give compound 5 in 40% yield. M.p.: 140–143 °C. FT-IR (cm−1, Fig. S21†): 3056wbr, 2985wbr, 2855wbr (CH); 1626m, 1591m (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1524w, 1504w, 1504w, 1471m, 1456sh, 1446w, 1434m, 1425m, 1361s, 1327w, 1276vs, 1232sh, 1220m; 1168s, 1123vs (CF3); 1095sh, 1027m, 997m, 972w, 949m, 940sh, 924m, 902m, 845m, 794m, 778m, 742s, 704sh, 692vs, 683vs. 1H-NMR (CDCl3, 293 K, Fig. S22†): δ 6.21 (s, 1H, COCHCO), 7.27–7.50 (m, 30H, CHar), 7.94 (s, 2H, p-CHar), 8.23 (s, 4H, o-CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S23†): δ 6.88 (s, 1H, COCHCO), 7.37–7.50 (m, 30H, CHar), 8.23 (s, 2H, p-CHar), 8.51 (s, 4H, o-CHar).13C{1H}-NMR (CDCl3, 293 K, Fig. S24†): δ 92.35 (COCHCO); 123.38 (q, 1JCF = 273 Hz, CF3); 123.42, 127.13, 128.66, 128.74, 130.08, 131.33, 132.56, 132.77, 133.91, 134.04, 144.84 (CCF3, CHar and Car); 182.48 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S25†): δ 8.88 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S26†): δ 8.53 (d, 1J(107Ag–31P) = 430 Hz and d, 1J(109Ag–31P) = 496 Hz). 19F{1H}-NMR (CDCl3, 223 K, Fig. S27†): δ −62.66 (s). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 633 (100) [Ag(PPh3)2]+. ESI-MS(−) (major negative ions, CH3CN), m/z (%): 495 (100) [LCF3]−. Elemental analysis (%) calculated for C55H37AgF12O2: C 58.58, H 3.31; found: C 57.61, H 3.49.
O); 1524w, 1504w, 1504w, 1471m, 1456sh, 1446w, 1434m, 1425m, 1361s, 1327w, 1276vs, 1232sh, 1220m; 1168s, 1123vs (CF3); 1095sh, 1027m, 997m, 972w, 949m, 940sh, 924m, 902m, 845m, 794m, 778m, 742s, 704sh, 692vs, 683vs. 1H-NMR (CDCl3, 293 K, Fig. S22†): δ 6.21 (s, 1H, COCHCO), 7.27–7.50 (m, 30H, CHar), 7.94 (s, 2H, p-CHar), 8.23 (s, 4H, o-CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S23†): δ 6.88 (s, 1H, COCHCO), 7.37–7.50 (m, 30H, CHar), 8.23 (s, 2H, p-CHar), 8.51 (s, 4H, o-CHar).13C{1H}-NMR (CDCl3, 293 K, Fig. S24†): δ 92.35 (COCHCO); 123.38 (q, 1JCF = 273 Hz, CF3); 123.42, 127.13, 128.66, 128.74, 130.08, 131.33, 132.56, 132.77, 133.91, 134.04, 144.84 (CCF3, CHar and Car); 182.48 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S25†): δ 8.88 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S26†): δ 8.53 (d, 1J(107Ag–31P) = 430 Hz and d, 1J(109Ag–31P) = 496 Hz). 19F{1H}-NMR (CDCl3, 223 K, Fig. S27†): δ −62.66 (s). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 633 (100) [Ag(PPh3)2]+. ESI-MS(−) (major negative ions, CH3CN), m/z (%): 495 (100) [LCF3]−. Elemental analysis (%) calculated for C55H37AgF12O2: C 58.58, H 3.31; found: C 57.61, H 3.49.
[Cu(LMes)(PPh3)2] (6). Triphenylphospine (1.000 mmol, 0.262 g) and tetrakis(acetonitrile)copper(I) hexafluorophosphate (0.500 mmol, 0.186 g) were solubilized in a mixture of CH2Cl2/CH3OH (15
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 mL). The colourless clear solution was stirred at room temperature for 1 hour. Successively, the ligand NaLMes (0.500 mmol, 0.165 g) was added and the green-yellow solution was stirred at room temperature for 3 hours. The mixture was then dried at reduced pressure giving a light green solid product. The product was washed firstly in Et2O and in a second step in CH3OH, giving a light green precipitate that was filtered and dried under reduced pressure. The light green complex 6 was obtained in 86% yield. A batch of poor quality crystals of [Cu(LMes)(PPh3)2], suitable for X-ray analysis, was obtained by slow evaporation of a CH2Cl2 solution of 6. M.p.: 210–215 °C. FT-IR (cm−1, Fig. S28†): 3050wbr, 2953wbr, 2917wbr, 2855wbr (C–H); 1613w, 1555s (C
5 mL). The colourless clear solution was stirred at room temperature for 1 hour. Successively, the ligand NaLMes (0.500 mmol, 0.165 g) was added and the green-yellow solution was stirred at room temperature for 3 hours. The mixture was then dried at reduced pressure giving a light green solid product. The product was washed firstly in Et2O and in a second step in CH3OH, giving a light green precipitate that was filtered and dried under reduced pressure. The light green complex 6 was obtained in 86% yield. A batch of poor quality crystals of [Cu(LMes)(PPh3)2], suitable for X-ray analysis, was obtained by slow evaporation of a CH2Cl2 solution of 6. M.p.: 210–215 °C. FT-IR (cm−1, Fig. S28†): 3050wbr, 2953wbr, 2917wbr, 2855wbr (C–H); 1613w, 1555s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1504m, 1479m, 1434s, 1395vs, 1372s, 1311m, 1281m, 1185w, 1167m, 1113m, 1093s, 1071m, 1027m, 997m, 956w, 924w, 849s, 880m, 777m, 742s, 724m, 693vs, 608m, 541m, 525s, 503vs. 1H-NMR (CDCl3, 293 K, Fig. S29†): δ 2.13–2.34 (s, 18H, o- and p-CH3), 5.34 (s, 1H, COCHCO), 6.74 (s, 4H, m-CHar), 7.21–7.39 (m, 30H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S30†): δ 2.05–2.16 (s, 18H, o- and p-CH3), 5.05 (s, 1H, COCHCO), 6.71 (s, 4H, m-CHar), 7.28–7.63 (m, 30H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S31†): δ 19.72 (o-CCH3); 20.96 (p-CCH3); 103.75 (COCHCO); 127.98, 128.49, 129.36, 131.93, 132.17, 133.92, 134.04, 136.11, 141.64 (CHar and Car); 189.37 (CO). 31P{1H}-NMR (CD2Cl2, 293 K, Fig. S32†): δ −5.41 (sbr). ESI-MS (+) (major positive ions, CH2Cl2/CH3CN) m/z (%): 587 (100) [Cu(PPh3)2]+. ESI-MS (−) (major negative ions, CH2Cl2/CH3CN) m/z (%): 307 (100) [LMes]−, 341 (100) [LMes + Cl]−, 396 (65) [Cu(PPh3) + 2Cl]−. Elemental analysis calculated for C57H53CuO2P2: C 76.45, H 5.97; found: C 75.97, H 5.88.
O); 1504m, 1479m, 1434s, 1395vs, 1372s, 1311m, 1281m, 1185w, 1167m, 1113m, 1093s, 1071m, 1027m, 997m, 956w, 924w, 849s, 880m, 777m, 742s, 724m, 693vs, 608m, 541m, 525s, 503vs. 1H-NMR (CDCl3, 293 K, Fig. S29†): δ 2.13–2.34 (s, 18H, o- and p-CH3), 5.34 (s, 1H, COCHCO), 6.74 (s, 4H, m-CHar), 7.21–7.39 (m, 30H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S30†): δ 2.05–2.16 (s, 18H, o- and p-CH3), 5.05 (s, 1H, COCHCO), 6.71 (s, 4H, m-CHar), 7.28–7.63 (m, 30H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S31†): δ 19.72 (o-CCH3); 20.96 (p-CCH3); 103.75 (COCHCO); 127.98, 128.49, 129.36, 131.93, 132.17, 133.92, 134.04, 136.11, 141.64 (CHar and Car); 189.37 (CO). 31P{1H}-NMR (CD2Cl2, 293 K, Fig. S32†): δ −5.41 (sbr). ESI-MS (+) (major positive ions, CH2Cl2/CH3CN) m/z (%): 587 (100) [Cu(PPh3)2]+. ESI-MS (−) (major negative ions, CH2Cl2/CH3CN) m/z (%): 307 (100) [LMes]−, 341 (100) [LMes + Cl]−, 396 (65) [Cu(PPh3) + 2Cl]−. Elemental analysis calculated for C57H53CuO2P2: C 76.45, H 5.97; found: C 75.97, H 5.88.
[Ag(LMes)(PPh3)2] (7). Silver nitrate (0.500 mmol, 0.085 g) was added to a methanol solution (20 mL) of triphenylphosphine (1.000 mmol, 0.262 g). The reaction mixture was stirred at room temperature and in the dark for one hour. Successively, the ligand NaLMes (0.500 mmol, 0.165 g) was added, and the reaction was left under constant magnetic stirring at room temperature for 4 hours. The precipitate formed was filtered and dried under reduced pressure to give whitish complex 7 in a 40% yield. M.p.: 194–198 °C. FT-IR (cm−1, Fig. S33†): 3054wbr, 2989wbr, 2953wbr, 2915wbr, 2851wbr (C–H); 1612w, 1582m, 1557m (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1492m, 1479m, 1435m, 1397s, 1372m, 1266m, 1182w, 1166w, 1111w, 1096s, 1071w, 1027m, 996w, 846m, 787w, 743s, 722m. 1H-NMR (CDCl3, 293 K, Fig. S34†): δ 2.22 (s, 18H, o- and p-CH3), 5.26 (s, 1H, COCHCO), 6.73 (s, 4H, m-CHar), 7.29–7.45 (m, 30H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S35†): δ 2.19 (s, 18H, o- and p-CH3), 5.45 (s, 1H, COCHCO), 6.73 (s, 4H, m-CHar), 7.41–7.46 (m, 30H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S36†): δ 19.57 (o-CCH3); 20.98, 21.11 (p-CCH3); 102.76 (COCHCO); 127.88, 128.46, 128.55, 128.71, 128.78, 129.87, 132.08, 132.16, 132.99, 133.18, 133.27, 133.97, 134.10, 135.67, 142.72 (CHar and Car); 190.51 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S37†): δ 6.73 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S38†): δ 6.22 (d, 1J(107Ag–31P) = 408 Hz, and d, 1J(109Ag–31P) = 472 Hz). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 633 (100) [Ag(PPh3)2]+. Elemental analysis calculated for C57H53AgO2P2: C 74.84, H 5.68; found: C 73.91, H 5.62.
O); 1492m, 1479m, 1435m, 1397s, 1372m, 1266m, 1182w, 1166w, 1111w, 1096s, 1071w, 1027m, 996w, 846m, 787w, 743s, 722m. 1H-NMR (CDCl3, 293 K, Fig. S34†): δ 2.22 (s, 18H, o- and p-CH3), 5.26 (s, 1H, COCHCO), 6.73 (s, 4H, m-CHar), 7.29–7.45 (m, 30H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S35†): δ 2.19 (s, 18H, o- and p-CH3), 5.45 (s, 1H, COCHCO), 6.73 (s, 4H, m-CHar), 7.41–7.46 (m, 30H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S36†): δ 19.57 (o-CCH3); 20.98, 21.11 (p-CCH3); 102.76 (COCHCO); 127.88, 128.46, 128.55, 128.71, 128.78, 129.87, 132.08, 132.16, 132.99, 133.18, 133.27, 133.97, 134.10, 135.67, 142.72 (CHar and Car); 190.51 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S37†): δ 6.73 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S38†): δ 6.22 (d, 1J(107Ag–31P) = 408 Hz, and d, 1J(109Ag–31P) = 472 Hz). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 633 (100) [Ag(PPh3)2]+. Elemental analysis calculated for C57H53AgO2P2: C 74.84, H 5.68; found: C 73.91, H 5.62.
[Ag(LMes)(PTA)] (8). To a methanol (30 mL) solution of 1,3,5-triazaphosphaadamantane (2.000 mmol, 0.314 g), AgNO3 (1.000 mmol, 0.170 g) was added, and the solution was stirred at room temperature in the dark for 2 hours. Successively, the ligand NaLMes was added (1000 mmol, 0.330 g) and the reaction mixture was left under magnetic stirring in the dark for 4 hours. The mixture was then filtered, and the precipitate was dried under reduced pressure, obtaining the orange complex 8 in 51% yield. M.p.: 193–197 °C. FT-IR (cm−1, Fig. S39†): 3176wbr, 2948wbr, 2917wbr, 2861wbr (C–H); 1610m, 1574mbr, 1558mbr (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1497m, 1418sbr, 1373s, 1351s, 1286mbr, 1241m, 1164m, 1104m, 1039m, 1013m, 972s, 949m, 902wbr, 849m, 827mbr, 792m, 746m, 718s. 1H-NMR (CD3OD, 293 K, Fig. S40†): δ 2.24 (s, 6H, p-CCH3), 2.32 (s, 12H, o-CCH3), 4.28 (d, 6H, PCH2N), 4.01–4.93 (m, 6H, NCH2N), 6.79 (s, 4H, m-CHar).1H-NMR (DMSO-d6, 293 K, Fig. S41†): δ 2.18–2.28 (d, 18H, p- and o-CCH3), 4.15 (s, 6H, PCH2N), 4.41–4.57 (m, 6H, NCH2N), 4.81 (s, 1H, COCHCO), 6.72 (s, 4H, m-CHar). 13C{1H}-NMR (DMSO-d6, 293 K, Fig. S42†): δ 19.69, 21.06 (CH3); 51.06 (PCH2N); 72.68, 72.73 (NCH2N); 101.66 (COCHCO), 127.88, 132.87 (CHar and Car), 188.37 (CO). 31P{1H}-NMR (CD3OD, 293 K, Fig. S43†): δ −83.73 (s). 31P{1H}-NMR (D2O, 293 K, Fig. S44†): δ −81.50 (s). 31P{1H}-NMR (CD3OD, 223 K, Fig. S45†): δ −81.2 (br). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 158 (100) [PTA + H]+, 420 (36) [Ag(PTA)2]+. Elemental analysis (%) calculated for C27H35AgN3O2P: N 7.34, C 56.65, H 6.16; found: N 6.95, C 55.66, H 6.03.
O); 1497m, 1418sbr, 1373s, 1351s, 1286mbr, 1241m, 1164m, 1104m, 1039m, 1013m, 972s, 949m, 902wbr, 849m, 827mbr, 792m, 746m, 718s. 1H-NMR (CD3OD, 293 K, Fig. S40†): δ 2.24 (s, 6H, p-CCH3), 2.32 (s, 12H, o-CCH3), 4.28 (d, 6H, PCH2N), 4.01–4.93 (m, 6H, NCH2N), 6.79 (s, 4H, m-CHar).1H-NMR (DMSO-d6, 293 K, Fig. S41†): δ 2.18–2.28 (d, 18H, p- and o-CCH3), 4.15 (s, 6H, PCH2N), 4.41–4.57 (m, 6H, NCH2N), 4.81 (s, 1H, COCHCO), 6.72 (s, 4H, m-CHar). 13C{1H}-NMR (DMSO-d6, 293 K, Fig. S42†): δ 19.69, 21.06 (CH3); 51.06 (PCH2N); 72.68, 72.73 (NCH2N); 101.66 (COCHCO), 127.88, 132.87 (CHar and Car), 188.37 (CO). 31P{1H}-NMR (CD3OD, 293 K, Fig. S43†): δ −83.73 (s). 31P{1H}-NMR (D2O, 293 K, Fig. S44†): δ −81.50 (s). 31P{1H}-NMR (CD3OD, 223 K, Fig. S45†): δ −81.2 (br). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 158 (100) [PTA + H]+, 420 (36) [Ag(PTA)2]+. Elemental analysis (%) calculated for C27H35AgN3O2P: N 7.34, C 56.65, H 6.16; found: N 6.95, C 55.66, H 6.03.
[Cu(LPh)(PPh3)2] (9). Triphenylphosphine (2.000 mmol, 0.524 g) was solubilized in CH2Cl2 (25 mL) and copper(I) chloride (1.000 mmol, 0.099 g) was added. The solution was stirred for 3 hours at room temperature. Successively, ligand NaLPh (1.000 mmol, 0.246 g) was added, and the reaction was left under constant magnetic stirring for 2 hours. The mixture was filtered, and the mother liquors were evaporated at reduced pressure. The residue was washed in EtOH, filtered and dried at reduced pressure. The yellow complex 9 was obtained in 34% yield. M.p.: 209–213 °C. FT-IR (cm−1, Fig. S46†): 3072w, 3046wbr, 3003w (C–H); 1596m, 1548s (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1511s, 1477m, 1455s, 1434s, 1403sbr, 1304m, 1273m, 1222m, 1179m, 1160m, 1092m, 1068m, 1022m, 997m, 936m, 922mbr, 840w, 806w, 784m, 741s, 719s. 1H-NMR (CDCl3, 293 K, Fig. S47†): δ 6.40 (s, 1H, COCHCO), 7.24–7.44 (m, 36H, CHar), 7.79 (d, 4H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S48†): δ 6.47 (s, 1H, COCHCO), 7.29–7.42 (m, 36H, CHar), 7.83 (d, 4H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S49†): δ 93.06 (COCHCO); 126.91, 127.80, 128.35, 128.42, 128.46, 128.55, 129.32, 132.08, 132.16, 133.93, 134.05, 134.14 (CHar and Car); 184.59 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S50†): δ −3.96 (s). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 587 (100) [(LPh)Cu(PPh3) + K]+. ESI-MS(−) (major negative ions, CH3CN), m/z (%): 223 (100) [LPh]−. Elemental analysis calculated for C51H41CuO2P2: C 75.50, H 5.09; found: C 74.11, H 4.62.
O); 1511s, 1477m, 1455s, 1434s, 1403sbr, 1304m, 1273m, 1222m, 1179m, 1160m, 1092m, 1068m, 1022m, 997m, 936m, 922mbr, 840w, 806w, 784m, 741s, 719s. 1H-NMR (CDCl3, 293 K, Fig. S47†): δ 6.40 (s, 1H, COCHCO), 7.24–7.44 (m, 36H, CHar), 7.79 (d, 4H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S48†): δ 6.47 (s, 1H, COCHCO), 7.29–7.42 (m, 36H, CHar), 7.83 (d, 4H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S49†): δ 93.06 (COCHCO); 126.91, 127.80, 128.35, 128.42, 128.46, 128.55, 129.32, 132.08, 132.16, 133.93, 134.05, 134.14 (CHar and Car); 184.59 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S50†): δ −3.96 (s). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 587 (100) [(LPh)Cu(PPh3) + K]+. ESI-MS(−) (major negative ions, CH3CN), m/z (%): 223 (100) [LPh]−. Elemental analysis calculated for C51H41CuO2P2: C 75.50, H 5.09; found: C 74.11, H 4.62.
[Ag(LPh)(PPh3)2] (10). Triphenylphosphine (2.000 mmol, 0.524 g) was solubilized in methanol (50 mL) and AgNO3 (1.000 mmol, 0.170 g) was added. The reaction mixture was stirred in the dark for 2 hours. Successively, the ligand NaLPh (1.000 mmol, 0.246 g) and the mixture was left under constant magnetic stirring in the dark for 4 hours. The solution was then filtered, and the precipitate was dried under reduced pressure to give the yellow-orange complex 10 in 54% yield. A batch of good quality crystals of [Ag(LPh)(PPh3)2], suitable for X-ray analysis, was obtained by slow evaporation of a chloroform/hexane solution of 10. M.p.: 193–197 °C. FT-IR (cm−1, Fig. S51†): 3071mbr, 3045mbr, 3004br (CH); 1598m, 1557m (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1505s, 1477m, 1453s, 1434s, 1407sbr, 1333m, 1300m, 1258m, 1216m, 1178m, 1160m, 1093m, 1066m, 1026m, 1019m, 966m, 933m, 933m, 921m, 912m, 841m, 780m, 737s, 720s, 705s. 1H-NMR (CDCl3, 293 K, Fig. S52†): δ 6.41 (s, 1H, COCHCO), 7.29–7.40 (m, 23H, CHar), 7.46–7.50 (m, 13H, CHar), 7.85 (t, 4H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S53†): δ 6.41 (s, 1H, COCHCO), 7.34–7.38 (m, 23H, CHar), 7.39–7.47 (m, 13H, CHar), 7.84 (t, 4H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S54†): δ 93.38 (COCHCO); 127.02, 127.18, 127.78, 128.45, 128.55, 128.69, 128.71, 132.08, 132.16, 133.93, 134.05, 134.14 (CHar and Car); 186.02 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S55†): δ 7.24 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S56†): δ 7.21 (dbr, 1J(Ag–31P) = 439 Hz). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 633 (100) [Ag(PPh3)2]+. Elemental analysis calculated for C51H41AgO2P2: C 71.59, H 4.89; found: C 71.14, H 4.58.
O); 1505s, 1477m, 1453s, 1434s, 1407sbr, 1333m, 1300m, 1258m, 1216m, 1178m, 1160m, 1093m, 1066m, 1026m, 1019m, 966m, 933m, 933m, 921m, 912m, 841m, 780m, 737s, 720s, 705s. 1H-NMR (CDCl3, 293 K, Fig. S52†): δ 6.41 (s, 1H, COCHCO), 7.29–7.40 (m, 23H, CHar), 7.46–7.50 (m, 13H, CHar), 7.85 (t, 4H, CHar). 1H-NMR (DMSO-d6, 293 K, Fig. S53†): δ 6.41 (s, 1H, COCHCO), 7.34–7.38 (m, 23H, CHar), 7.39–7.47 (m, 13H, CHar), 7.84 (t, 4H, CHar). 13C{1H}-NMR (CDCl3, 293 K, Fig. S54†): δ 93.38 (COCHCO); 127.02, 127.18, 127.78, 128.45, 128.55, 128.69, 128.71, 132.08, 132.16, 133.93, 134.05, 134.14 (CHar and Car); 186.02 (CO). 31P{1H}-NMR (CDCl3, 293 K, Fig. S55†): δ 7.24 (s). 31P{1H}-NMR (CDCl3, 223 K, Fig. S56†): δ 7.21 (dbr, 1J(Ag–31P) = 439 Hz). ESI-MS(+) (major positive ions, CH3CN), m/z (%): 633 (100) [Ag(PPh3)2]+. Elemental analysis calculated for C51H41AgO2P2: C 71.59, H 4.89; found: C 71.14, H 4.58.
[Ag(LPh)(PTA)]·H2O (11). 1,3,4-Triazaphosphaadamantane (2.000 mmol, 0.314 g) was solubilized in methanol (30 mL) and AgNO3 (1.000 mmol, 0.170 g) was added. The reaction mixture was stirred in the dark at room temperature for 2 hours. Successively, the ligand NaLPh (1.000 mmol, 0.246 g) was added and the solution was stirred in the dark at room temperature for 4 hours. The mixture was then filtered and the precipitate was dried under reduced pressure. The orange product was recovered to give complex 11 in 81% yield. M.p.: 198–203 °C. FT-IR (cm−1, Fig. S57†): 3225mbr (O–H); 3060mbr, 2942mbr (C–H); 1598s, 1555s (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1513s, 1466s, 1419vs, 1345vsbr, 1289vs, 1242s, 1225s, 1180mbr, 1102s, 1069m, 1039m, 1013s, 971vs, 947vs, 898s, 843m, 828m, 804m, 790m, 745vs, 712vs. 1H-NMR (CD3OD, 293 K, Fig. S58†): δ 4.25 (d, 6H, PCH2N), 4.56–4.67 (m, 6H, NCH2N), 4.86 (H2O), 6.78 (br, 1H, COCHCO), 7.48 (s, 5H, CHar), 7.95 (s, 5H, CHar). 1H-NMR (D2O, 293 K, Fig. S59†): δ 4.08 (d, 6H, PCH2N), 4.45–4.51 (m, 6H, NCH2N), 6.32 (s, 1H, COCHCO), 7.42–7.43 (s, 5H, CHar), 7.73 (s, 5H, CHar). 1H-NMR (DMSO, 293 K, Fig. S60†): δ 4.13 (d, 6H, PCH2N), 4.39–4.55 (m, 6H, NCH2N), 6.45 (s, 1H, COCHCO), 7.42 (s, 5H, CHar), 7.90 (s, 5H, CHar). 13C{1H}-NMR (CD3OD, 293 K, Fig. S61†): 50.10 (PCH2N); 72.00, 72.05 (NCH2N); 90.35 (COCHCO); 126.76, 127.28, 128.07, 128.29, 128.36, 128.84 (CHar and Car), 179.93 (CO). 31P{1H}-NMR (CD3OD, 293 K, Fig. S62†): δ −83.82 (s). 31P{1H}-NMR (CD3OD, 223 K, Fig. S63†): δ −81.5 (br). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 158 (100) [PTA + H]+, 421 (8) [Ag(PTA)2]+. Elemental analysis (%) calculated for C21H25AgN3O3P: N 8.30, C 49.82, H 4.98; found: N 8.95, C 48.93, H 4.88.
O); 1513s, 1466s, 1419vs, 1345vsbr, 1289vs, 1242s, 1225s, 1180mbr, 1102s, 1069m, 1039m, 1013s, 971vs, 947vs, 898s, 843m, 828m, 804m, 790m, 745vs, 712vs. 1H-NMR (CD3OD, 293 K, Fig. S58†): δ 4.25 (d, 6H, PCH2N), 4.56–4.67 (m, 6H, NCH2N), 4.86 (H2O), 6.78 (br, 1H, COCHCO), 7.48 (s, 5H, CHar), 7.95 (s, 5H, CHar). 1H-NMR (D2O, 293 K, Fig. S59†): δ 4.08 (d, 6H, PCH2N), 4.45–4.51 (m, 6H, NCH2N), 6.32 (s, 1H, COCHCO), 7.42–7.43 (s, 5H, CHar), 7.73 (s, 5H, CHar). 1H-NMR (DMSO, 293 K, Fig. S60†): δ 4.13 (d, 6H, PCH2N), 4.39–4.55 (m, 6H, NCH2N), 6.45 (s, 1H, COCHCO), 7.42 (s, 5H, CHar), 7.90 (s, 5H, CHar). 13C{1H}-NMR (CD3OD, 293 K, Fig. S61†): 50.10 (PCH2N); 72.00, 72.05 (NCH2N); 90.35 (COCHCO); 126.76, 127.28, 128.07, 128.29, 128.36, 128.84 (CHar and Car), 179.93 (CO). 31P{1H}-NMR (CD3OD, 293 K, Fig. S62†): δ −83.82 (s). 31P{1H}-NMR (CD3OD, 223 K, Fig. S63†): δ −81.5 (br). ESI-MS(+) (major positive ions, CH3OH), m/z (%): 158 (100) [PTA + H]+, 421 (8) [Ag(PTA)2]+. Elemental analysis (%) calculated for C21H25AgN3O3P: N 8.30, C 49.82, H 4.98; found: N 8.95, C 48.93, H 4.88.
X-ray structure determination
A suitable crystal covered with a layer of hydrocarbon/Paratone-N oil was selected and mounted on a Cryo-loop, and immediately placed in a low temperature nitrogen stream. The X-ray intensity data of HLCF3 and [Cu(LCF3)(PPh3)2] were measured at 100 K on a SMART APEX II CCD area detector system while X-ray intensity data of [Ag(LPh)(PPh3)2] were measured at 100(2) K on a Bruker D8 Quest equipped with a PHOTON II 7 CPAD detector. These systems were equipped with an Oxford Cryosystems 700 series cooler, a graphite monochromator, and a Mo Kα fine-focus sealed tube (λ = 0.71073 Å). Intensity data were processed using the Bruker Apex program suite. Absorption corrections were applied by using SADABS.116 Initial atomic positions were located using SHELXT,117 and the structures of the compounds were refined by the least-squares method using SHELXL118 within Olex2 GUI.119 All the non-hydrogen atoms were refined anisotropically. Hydrogen atoms were included at calculated positions and refined using appropriate riding models. The molecule HLCF3 sites on a two-fold rotation axis and the O–H position are therefore disordered over two oxygen sites. [Cu(LCF3)(PPh3)2] crystallizes in the P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with two chemically similar molecules in the asymmetric unit. X-ray structural figures were generated using Olex2. CCDC 2279297–2279299 files contain the supplementary crystallography data. Additional details are provided in the ESI.† We have also recorded single crystal X-ray data of [Cu(LMes)(PPh3)2] and obtained its molecular structure. Unfortunately, the crystal quality is poor and also suffers due to twinning. As a result, the structure is not of sufficient quality for a detailed analysis of metrical parameters. The atom connectivity and basic features of the molecule are however clear from the data (see the ESI, Tables S1–S9†).
space group with two chemically similar molecules in the asymmetric unit. X-ray structural figures were generated using Olex2. CCDC 2279297–2279299 files contain the supplementary crystallography data. Additional details are provided in the ESI.† We have also recorded single crystal X-ray data of [Cu(LMes)(PPh3)2] and obtained its molecular structure. Unfortunately, the crystal quality is poor and also suffers due to twinning. As a result, the structure is not of sufficient quality for a detailed analysis of metrical parameters. The atom connectivity and basic features of the molecule are however clear from the data (see the ESI, Tables S1–S9†).
Experiments with cultured human cancer cells
Ag(I) and Cu(I) complexes and the corresponding uncoordinated ligands and related salts were dissolved in DMSO just before the experiment, and a calculated volume of the drug solution was added to the cell growth medium to a final solvent concentration of 0.5%, which had no detectable effects on cell viability. Cisplatin was dissolved in 0.9% sodium chloride solution. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and cisplatin were obtained from Sigma Chemical Co, St Louis, MO, USA.Cell cultures
Human SCLC (U1285), NSCLC (A549), testicular (NTERA-2), colon (HCT-15), and pancreatic (PSN-1 and BxPC3) carcinoma cell lines were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). Cell lines were maintained in the logarithmic phase at 37 °C under a 5% carbon dioxide atmosphere using the RPMI-1640 medium (EuroClone, Milan, Italy) containing 10% fetal calf serum (EuroClone, Milan, Italy), antibiotics (50 units per mL penicillin and 50 μg mL−1 streptomycin) and 2 mM L-glutamine.MTT assay
The growth inhibitory effect toward tumor cells was evaluated by means of the MTT assay as previously described.85 IC50 values, the drug concentrations that reduce the mean absorbance at 570 nm to 50% of those in the untreated control wells, were calculated using the four-parameter logistic (4-PL) model. The evaluation was based on means from at least three independent experiments.Spheroid cultures and acid phosphatase (APH) assay
Spheroid cultures were obtained by seeding 2.5 × 103 HCT-15 human cancer cells per well in a round-bottom non-treated tissue culture 96-well plate (Greiner Bio-one, Kremsmünster, Austria) in a phenol red free RPMI-1640 medium (Sigma Chemical Co., St Louis, MO, USA) containing 10% fetal calf serum supplemented with 20% methyl cellulose stock solution. An APH modified assay was employed for evaluating cell viability in 3D spheroids, as previously described.85 IC50 values (drug concentrations that reduce the mean absorbance at 405 nm to 50% of those in the untreated control wells) were calculated using the 4-PL model. The evaluation was based on means from at least three independent experiments.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by Unione Europea - NextGenerationEU (MUR-Fondo Promozione e Sviluppo - D.M. 737/2021, INVIRCuM, University of Camerino, FAR 2022 PNR, and NGEU PNRR, D.M. n. 351/2022 M4C1 I4.1) and by the University of Padova (PRID BIRD225980). H.V.R.D. acknowledges the support by the National Science Foundation through grant CHE-1954456.References
- K. D. Mjos and C. Orvig, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS PubMed.
- N. P. E. Barry and P. J. Sadler, Chem. Commun., 2013, 49, 5106–5131 RSC.
- Metal-based Anticancer Agents, ed. A. Casini, A. Vessières and S. M. Meier-Menches, The Royal Society of Chemistry, Croydon, UK, 2019 Search PubMed.
- Metallo-Drugs: Development and Action of Anticancer Agents, ed. A. Sigel, E. Freisinger and R. K. O. Sigel, De Gruyter, 2018 Search PubMed.
- S. Medici, M. Peana, V. M. Nurchi and M. A. Zoroddu, J. Med. Chem., 2019, 62, 5923–5943 CrossRef CAS PubMed.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed.
- S. Nobili, E. Mini, I. Landini, C. Gabbiani, A. Casini and L. Messori, Med. Res. Rev., 2010, 30, 550–580 CrossRef CAS PubMed.
- J. A. Joule and K. Mills, Heterocyclic Chemistry, Wiley, Chichester (UK), Fifth edn, 2010 Search PubMed.
- C. Y. Chen, J. C. Lien, C. Y. Chen, C. C. Hung and H. C. Lin, Int. J. Mol. Sci., 2021, 22, 12171 CrossRef CAS PubMed.
- C. Pettinari, F. Marchetti and A. Drozdov, in Compr. Coord. Chem. II, ed. J. A. McCleverty and T. J. Meyer, Pergamon, Oxford, 2003, pp. 97–115 Search PubMed.
- A. R. Siedle, in Compr. Coord. Chem, ed. G. Wilkinson, R. D. Gillard and J. A. McCleverty, Pergamon, Oxford, UK, 1987, pp. 365–412 Search PubMed.
- A. S. Crossman and M. P. Marshak, in Compr. Coord. Chem. III, ed. E. C. Constable, G. Parkin and L. Que Jr, Elsevier, Oxford, 2021, pp. 331–365 Search PubMed.
- M. Stalpaert and D. De Vos, ACS Sustainable Chem. Eng., 2018, 6, 12197–12204 CrossRef CAS.
- A. S. Crossman, A. T. Larson, J. X. Shi, S. M. Krajewski, E. S. Akturk and M. P. Marshak, J. Org. Chem., 2019, 84, 7434–7442 CrossRef CAS PubMed.
- S. M. Krajewski, A. S. Crossman, E. S. Akturk, T. Suhrbier, S. J. Scappaticci, M. W. Staab and M. P. Marshak, Dalton Trans., 2019, 48, 10714–10722 RSC.
- E. S. Akturk, S. J. Scappaticci, R. N. Seals and M. P. Marshak, Inorg. Chem., 2017, 56, 11466–11469 CrossRef CAS PubMed.
- T. Schilling, K. B. Keppler, M. E. Heim, G. Niebch, H. Dietzfelbinger, J. Rastetter and A. R. Hanauske, Invest. New Drugs, 1995, 13, 327–332 CrossRef CAS PubMed.
- H. Bischoff, M. R. Berger, B. K. Keppler and D. Schmähl, J. Cancer Res. Clin. Oncol., 1987, 113, 446–450 CrossRef CAS PubMed.
- A. Wu, D. C. Kennedy, B. O. Patrick and B. R. James, Inorg. Chem. Commun., 2003, 6, 996–1000 CrossRef CAS.
- R. Fernández, M. Melchart, A. Habtemariam, S. Parsons and P. J. Sadler, Chem. – Eur. J., 2004, 10, 5173–5179 CrossRef PubMed.
- M. Melchart, A. Habtemariam, S. Parsons and P. J. Sadler, J. Inorg. Biochem., 2007, 101, 1903–1912 CrossRef CAS PubMed.
- C. A. Vock, A. K. Renfrew, R. Scopelliti, L. Juillerat-Jeanneret and P. J. Dyson, Eur. J. Inorg. Chem., 2008, 1661–1671 CrossRef CAS.
- C. Aliende, M. Pérez-Manrique, F. A. Jalón, B. R. Manzano, A. M. Rodríguez, J. V. Cuevas, G. Espino, M. Á. Martínez, A. Massaguer, M. González-Bártulos, R. De Llorens and V. Moreno, J. Inorg. Biochem., 2012, 117, 171–188 CrossRef CAS PubMed.
- S. Seršen, J. Kljun, K. Kryeziu, R. Panchuk, B. Alte, W. Körner, P. Heffeter, W. Berger and I. Turel, J. Med. Chem., 2015, 58, 3984–3996 CrossRef PubMed.
- J. Kljun and I. Turel, Eur. J. Inorg. Chem., 2017, 2017, 1655–1666 CrossRef CAS.
- J. Conradie and J. C. Swarts, Dalton Trans., 2011, 40, 5844–5851 RSC.
- J. Conradie and J. C. Swarts, Eur. J. Inorg. Chem., 2011, 2011, 2439–2449 CrossRef.
- J. Do Couto Almeida, I. M. Marzano, F. C. S. De Paula, M. Pivatto, N. P. Lopes, P. C. De Souza, F. R. Pavan, A. L. B. Formiga, E. C. Pereira-Maia and W. Guerra, J. Mol. Struct., 2014, 1075, 370–376 CrossRef CAS.
- D. I. Ugwu and J. Conradie, J. Inorg. Biochem., 2023, 112268 CrossRef CAS PubMed.
- A. Muscella, N. Calabriso, C. Vetrugno, F. P. Fanizzi, S. A. De Pascali, C. Storelli and S. Marsigliante, Biochem. Pharmacol., 2011, 81, 91–103 CrossRef CAS PubMed.
- A. Muscella, N. Calabriso, C. Vetrugno, F. P. Fanizzi, S. A. De Pascali and S. Marsigliante, Biochem. Pharmacol., 2011, 81, 1271–1285 CrossRef CAS PubMed.
- A. Muscella, N. Calabriso, C. Vetrugno, L. Urso, F. P. Fanizzi, S. A. De Pascali and S. Marsigliante, Br. J. Pharmacol., 2010, 160, 1362–1377 CrossRef CAS PubMed.
- A. Muscella, N. Calabriso, F. P. Fanizzi, S. A. De Pascali, L. Urso, A. Ciccarese, D. Migoni and S. Marsigliante, Br. J. Pharmacol., 2008, 153, 34–49 CrossRef CAS PubMed.
- K. Nakano, T. Nakayachi, E. Yasumoto, S. R. Morshed, K. Hashimoto, H. Kikuchi, H. Nishikawa, K. Sugiyama, O. Amano, M. Kawase and H. Sakagami, Anticancer Res., 2004, 24, 711–717 CAS.
- J. J. Wilson and S. J. Lippard, J. Med. Chem., 2012, 55, 5326–5336 CrossRef CAS PubMed.
- S. A. Gromilov and I. A. Baidina, J. Struct. Chem., 2004, 45, 1031–1081 CrossRef CAS.
- L. G. Bulusheva, A. V. Okotrub, T. I. Liskovskaya, S. A. Krupoder, A. V. Guselnikov, A. V. Manaev and V. F. Traven, J. Phys. Chem. A, 2001, 105, 8200–8205 CrossRef CAS.
- K. M. Chi, H. K. Shin, M. J. Hampden-Smith, E. N. Duesler and T. T. Kodas, Polyhedron, 1991, 10, 2293–2299 CrossRef CAS.
- A. T. Larson, A. S. Crossman, S. M. Krajewski and M. P. Marshak, Inorg. Chem., 2020, 59, 423–432 CrossRef CAS PubMed.
- H. Lang, M. Leschke, M. Melter, B. Walfort, K. Kohler, S. E. Schulz and T. Gessner, Z. Anorg. Allg. Chem., 2003, 629, 2371–2380 CrossRef CAS.
- H. K. Shin, K. M. Chi, J. Farkas, M. J. Hampden-Smith, T. T. Kodas and E. N. Duesler, Inorg. Chem., 1992, 31, 424–431 CrossRef CAS.
- K.-M. Chi, J. Farkas, M. J. Hampden-Smith, T. T. Kodas and E. N. Duesler, Dalton Trans., 1992, 3111–3117 RSC.
- H. K. Shin, M. J. Hampden-Smith, E. N. Duesler and T. T. Kodas, Can. J. Chem., 1992, 70, 2954–2966 CrossRef CAS.
- T. H. Baum and C. E. Larson, Chem. Mater., 1992, 4, 365–369 CrossRef CAS.
- S. K. Reynolds, C. J. Smart, E. F. Baran, T. H. Baum, C. E. Larson and P. Brock, Appl. Phys. Lett., 1991, 59, 2332 CrossRef CAS.
- D. B. Beach, F. K. LeGoues and C. K. Hu, Chem. Mater., 1990, 2, 216–219 CrossRef CAS.
- Y. L. Chow and G. E. Buono-Core, Can. J. Chem., 1983, 61, 795–800 CrossRef CAS.
- R. J. Restivo, A. Costin, G. Ferguson and A. J. Carty, Can. J. Chem., 1975, 53, 1949–1957 CrossRef CAS.
- W. A. Anderson, A. J. Carty, G. J. Palenik and G. Schreiber, Can. J. Chem., 1971, 49, 761–766 CrossRef.
- A. Jakob, C. C. Joubert, T. Rüffer, J. C. Swarts and H. Lang, Inorg. Chim. Acta, 2014, 411, 48–55 CrossRef CAS.
- P. Tasker, A. Parkin, T. C. Higgs, S. Parsons and D. Messanger, CSD communication (private communication), 2005.
- R.-N. Yang, D.-M. Wang, Y.-F. Liu and D.-M. Jin, Polyhedron, 2001, 20, 585–590 CrossRef CAS.
- S. A. Gulyaev, E. S. Vikulova, T. S. Sukhikh, I. Y. Ilyin and N. B. Morozova, J. Struct. Chem., 2021, 62, 1836–1845 CrossRef CAS.
- I. S. Fedoseev, E. S. Vikulova, I. Y. Il'in, A. I. Smolentsev, M. R. Gallyamov and N. B. Morozova, J. Struct. Chem., 2016, 57, 1667–1670 CrossRef CAS.
- K. Akhbari and A. Morsali, Cryst. Growth Des., 2007, 7, 2024–2030 CrossRef CAS.
- W. J. Evans, D. G. Giarikos, D. Josell and J. W. Ziller, Inorg. Chem., 2003, 42, 8255–8261 CrossRef CAS PubMed.
- C. Y. Xu, T. S. Corbitt, M. J. Hampdensmith, T. T. Kodas and E. N. Duesler, Dalton Trans., 1994, 2841–2849 RSC.
- E. S. Vikulova, T. S. Sukhikh, S. A. Gulyaev, I. Y. Ilyin and N. B. Morozova, Molecules, 2022, 27, 677 CrossRef CAS PubMed.
- F. Marandi, M. Ghadermazi, A. Marandi, I. Pantenburg and G. Meyer, J. Mol. Struct., 2011, 1006, 136–141 CrossRef CAS.
- A. J. Blake, N. R. Champness, S. M. Howdle, K. S. Morley, P. B. Webb and C. Wilson, CrystEngComm, 2002, 4, 88–92 RSC.
- Y. L. Shen, J. L. Jin, G. X. Duan, P. Y. Yu, Y. P. Xie and X. Lu, Chem. – Eur. J., 2021, 27, 1122–1126 CrossRef CAS PubMed.
- J. L. Jin, Y. P. Xie, H. Cui, G. X. Duan, X. Lu and T. C. W. Mak, Inorg. Chem., 2017, 56, 10412–10417 CrossRef CAS PubMed.
- H. Liu, S. Battiato, A. L. Pellegrino, P. Paoli, P. Rossi, C. Jimenez, G. Malandrino and D. Munoz-Rojas, Dalton Trans., 2017, 46, 10986–10995 RSC.
- A. O. Rybaltovskii, V. G. Arakcheev, A. N. Bekin, A. F. Danilyuk, V. I. Gerasimova, N. V. Minaev, E. N. Golubeva, O. O. Parenago and V. N. Bagratashvili, Russ. J. Phys. Chem. B, 2015, 9, 1137–1142 CrossRef CAS.
- G. Calabrese, S. Petralia, D. Franco, G. Nocito, C. Fabbi, L. Forte, S. Guglielmino, S. Squarzoni, F. Traina and S. Conoci, Mater. Sci. Eng., C, 2021, 118, 111394 CrossRef CAS PubMed.
- H. Schmidt, A. Jakob, T. Haase, K. Kohse-Hoinghaus, S. E. Schulz, T. Wachtler, T. Gessner and H. Lang, Z. Anorg. Allg. Chem., 2005, 631, 2786–2791 CrossRef CAS.
- D. A. Edwards, R. M. Harker, M. F. Mahon and K. C. Molloy, J. Mater. Chem., 1999, 9, 1771–1780 RSC.
- Z. Yuan, N. H. Dryden, J. J. Vittal and R. J. Puddephatt, Chem. Mater., 1995, 7, 1696–1702 CrossRef CAS.
- Z. Yuan, N. H. Dryden, J. J. Vittal and R. J. Puddephatt, Can. J. Chem., 1994, 72, 1605–1609 CrossRef CAS.
- N. H. Dryden, J. J. Vittal and R. J. Puddephatt, Chem. Mater., 1993, 5, 765–766 CrossRef CAS.
- W. Lin, T. H. Warren, R. G. Nuzzo and G. S. Girolami, J. Am. Chem. Soc., 1993, 115, 11644–11645 CrossRef CAS.
- S. Wanninger, V. Lorenz, A. Subhan and F. T. Edelmann, Chem. Soc. Rev., 2015, 44, 4986–5002 RSC.
- M. H. M. Leung, T. Harada and T. W. Kee, Curr. Pharm. Des., 2013, 19, 2070–2083 CAS.
- Y. Figueroa-DePaz, K. Resendiz-Acevedo, S. G. Dávila-Manzanilla, J. C. García-Ramos, L. Ortiz-Frade, J. Serment-Guerrero and L. Ruiz-Azuara, J. Inorg. Biochem., 2022, 231, 111772 CrossRef CAS PubMed.
- J. Serment-Guerrero, M. E. Bravo-Gomez, E. Lara-Rivera and L. Ruiz-Azuara, J. Inorg. Biochem., 2017, 166, 68–75 CrossRef CAS PubMed.
- I. Correia, S. Borovic, I. Cavaco, C. P. Matos, S. Roy, H. M. Santos, L. Fernandes, J. L. Capelo, L. Ruiz-Azuara and J. C. Pessoa, J. Inorg. Biochem., 2017, 175, 284–297 CrossRef CAS PubMed.
- L. Ruiz-Azuara and M. E. Bravo-Gomez, Curr. Med. Chem., 2010, 17, 3606–3615 CrossRef CAS PubMed.
- L. Polloni, A. C. D. Silva, S. C. Teixeira, F. V. D. Azevedo, M. A. P. Zoia, M. S. da Silva, P. Lima, L. I. V. Correia, J. D. Almeida, C. V. da Silva, V. D. R. Avila, L. R. F. Goulart, S. Morelli, W. Guerra and R. J. de Oliveira, Biomed. Pharmacother., 2019, 112, 108586 CrossRef PubMed.
- D. A. Paixao, B. C. A. de Oliveira, J. D. Almeida, L. M. Sousa, C. D. Lopes, Z. A. Carneiro, D. Y. Tezuka, J. C. T. Clavijo, J. Ellena, L. Polloni, P. H. A. Machado, S. de Albuquerque, R. J. de Oliveira, S. Guilardi and W. Guerra, Inorg. Chim. Acta, 2020, 499, 119164 CrossRef CAS.
- R. E. Malekshah, M. Salehi, M. Kubicki and A. Khaleghian, J. Coord. Chem., 2019, 72, 1697–1714 CrossRef.
- R. E. Malekshah, M. Salehi, M. Kubicki and A. Khaleghian, J. Coord. Chem., 2018, 71, 952–968 CrossRef.
- A. Pramanik, D. Laha, P. Pramanik and P. Karmakar, J. Drug Targeting, 2014, 22, 23–33 CrossRef CAS PubMed.
- D. F. Xu, Z. H. Shen, Y. Shi, Q. He and Q. C. Xia, Russ. J. Coord. Chem., 2010, 36, 458–462 CrossRef CAS.
- L. A. Khamidullina, I. S. Puzyrev, G. L. Burygin, P. V. Dorovatovskii, Y. V. Zubavichus, A. V. Mitrofanova, V. N. Khrustalev, T. V. Timofeeva, P. A. Slepukhin, P. D. Tobysheva, A. V. Pestov, E. Solari, A. G. Tskhovrebov and V. G. Nenajdenko, Molecules, 2021, 26, 6466 CrossRef CAS PubMed.
- M. Pellei, C. Santini, L. Bagnarelli, M. Caviglia, P. Sgarbossa, M. De Franco, M. Zancato, C. Marzano and V. Gandin, Int. J. Mol. Sci., 2023, 24, 4091 CrossRef CAS PubMed.
- F. Del Bello, M. Pellei, L. Bagnarelli, C. Santini, G. Giorgioni, A. Piergentili, W. Quaglia, C. Battocchio, G. Iucci, I. Schiesaro, C. Meneghini, I. Venditti, N. Ramanan, M. De Franco, P. Sgarbossa, C. Marzano and V. Gandin, Inorg. Chem., 2022, 61, 4919–4937 CrossRef CAS PubMed.
- M. Pellei, L. Bagnarelli, L. Luciani, F. Del Bello, G. Giorgioni, A. Piergentili, W. Quaglia, M. De Franco, V. Gandin, C. Marzano and C. Santini, Int. J. Mol. Sci., 2020, 21, 2616 CrossRef CAS PubMed.
- M. B. Morelli, C. Amantini, G. Santoni, M. Pellei, C. Santini, C. Cimarelli, E. Marcantoni, M. Petrini, F. Del Bello, G. Giorgioni, A. Piergentili and W. Quaglia, New J. Chem., 2018, 42, 11878–11887 RSC.
- V. Gandin, C. Ceresa, G. Esposito, S. Indraccolo, M. Porchia, F. Tisato, C. Santini, M. Pellei and C. Marzano, Sci. Rep., 2017, 7, 13936 CrossRef PubMed.
- F. Tisato, C. Marzano, V. Peruzzo, M. Tegoni, M. Giorgetti, M. Damjanovic, A. Trapananti, A. Bagno, C. Santini, M. Pellei, M. Porchia and V. Gandin, J. Inorg. Biochem., 2016, 165, 80–91 CrossRef CAS PubMed.
- G. Papini, G. Bandoli, A. Dolmella, G. Gioia Lobbia, M. Pellei and C. Santini, Inorg. Chem. Commun., 2008, 11, 1103–1106 CrossRef CAS.
- M. Inoue, Y. Sumii and N. Shibata, ACS Omega, 2020, 5, 10633–10640 CrossRef CAS PubMed.
- E. P. Gillis, K. J. Eastman, M. D. Hill, D. J. Donnelly and N. A. Meanwell, J. Med. Chem., 2015, 58, 8315–8359 CrossRef CAS PubMed.
- S. Purser, P. R. Moore, S. Swallow and V. Gouverneur, Chem. Soc. Rev., 2008, 37, 320–330 RSC.
- P. Shah and A. D. Westwell, J. Enzyme Inhib. Med. Chem., 2007, 22, 527–540 CrossRef CAS PubMed.
- L. H. Doerrer and H. V. R. Dias, Dalton Trans., 2023, 52, 7770–7771 RSC.
- S. Swallow, in Prog. Med. Chem, ed. G. Lawton and D. R. Witty, Elsevier, 2015, vol. 54, pp. 65–133 Search PubMed.
- I. M. Abdou, A. M. Saleh and H. F. Zohdi, Molecules, 2004, 9, 109–116 CrossRef CAS PubMed.
- P. N. Edwards, in Organofluorine Chemistry: Principles and Commercial Applications, ed. R. E. Banks, B. E. Smart and J. C. Tatlow, Springer US, Boston, MA, 1994, pp. 501–541 Search PubMed.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS PubMed.
- J. S. Lakhi, M. R. Patterson and H. V. R. Dias, New J. Chem., 2020, 44, 14814–14822 RSC.
- C. Zhang, P. Yang, Y. Yang, X. Huang, X.-J. Yang and B. Wu, Synth. Commun., 2008, 38, 2349–2356 CrossRef CAS.
- J. Emsley, in Complex Chemistry. Structure and Bonding, ed. J. Emsley, R. D. Ernst, B. J. Hathaway and K. D. Warren, Springer, Berlin, Heidelberg, 1984, vol. 57, pp. 147–191 Search PubMed.
- M. A. Halcrow, J.-S. Sun, J. C. Huffman and G. Christou, Inorg. Chem., 1995, 34, 4167–4177 CrossRef CAS.
- R. P. Dryden and A. Winston, J. Phys. Chem., 1958, 62, 635–637 CrossRef CAS.
- B. Cheng, H. Yi, C. He, C. Liu and A. Lei, Organometallics, 2014, 34, 206–211 CrossRef.
- C. He, G. Zhang, J. Ke, H. Zhang, J. T. Miller, A. J. Kropf and A. Lei, J. Am. Chem. Soc., 2013, 135, 488–493 CrossRef CAS PubMed.
- S. N. Slabzhennikov, O. B. Ryabchenko and L. A. Kuarton, Russ. J. Coord. Chem., 2006, 32, 545–551 CrossRef CAS.
- A. Guerriero, M. Peruzzini and L. Gonsalvi, Coord. Chem. Rev., 2018, 355, 328–361 CrossRef CAS.
- A. M. Kirillov, S. W. Wieczorek, M. F. C. Guedes da Silva, J. Sokolnicki, P. Smoleński and A. J. L. Pombeiro, CrystEngComm, 2011, 13, 6329–6333 RSC.
- H. V. R. Dias, J. A. Flores, M. Pellei, B. Morresi, G. Gioia Lobbia, S. Singh, Y. Kobayashi, M. Yousufuddin and C. Santini, Dalton Trans., 2011, 40, 8569–8580 RSC.
- M. Pellei, S. Alidori, G. Papini, G. Gioia Lobbia, J. D. Gorden, H. V. R. Dias and C. Santini, Dalton Trans., 2007, 4845–4853 RSC.
- H. V. R. Dias, S. Alidori, G. Gioia Lobbia, G. Papini, M. Pellei and C. Santini, Inorg. Chem., 2007, 46, 9708–9714 CrossRef CAS PubMed.
- V. Gandin, F. Tisato, A. Dolmella, M. Pellei, C. Santini, M. Giorgetti, C. Marzano and M. Porchia, J. Med. Chem., 2014, 57, 4745–4760 CrossRef CAS PubMed.
- S. El Harane, B. Zidi, N. El Harane, K.-H. Krause, T. Matthes and O. Preynat-Seauve, Journal, 2023, 12, 1001 CAS.
- L. Krause, R. Herbst-Irmer, G. M. Sheldrick and D. Stalke, J. Appl. Crystallogr., 2015, 48, 3–10 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2279297–2279299. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3dt02179c |
| This journal is © The Royal Society of Chemistry 2023 |