 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fine-tuning the cytotoxicity of ruthenium(II) arene compounds to enhance selectivity against breast cancers†
Sarah A. P.
Pereira‡
ab,
Jan
Romano-deGea‡
 b,
Ana Isabel
Barbosa
b,
Ana Isabel
Barbosa
 a,
Sofia A.
Costa Lima
a,
Paul J.
Dyson
a,
Sofia A.
Costa Lima
a,
Paul J.
Dyson
 *b and
M. Lúcia M. F. S.
Saraiva
*b and
M. Lúcia M. F. S.
Saraiva
 *a
*a
aLAQV, REQUIMTE, Departamento de Ciências Químicas, Faculdade de Farmácia, Universidade do Porto, Rua Jorge Viterbo Ferreira, no 228, 4050-313 Porto, Portugal. E-mail: lsaraiva@ff.up.pt
bInstitut des Sciences et Ingénierie Chimiques, École Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland. E-mail: paul.dyson@epfl.ch
First published on 28th July 2023
Abstract
Ruthenium-based complexes have been suggested as promising anticancer drugs exhibiting reduced general toxicity compared to platinum-based drugs. In particular, Ru(η6-arene)(PTA)Cl2 (PTA = 1,3,5-triaza-7-phosphaadamantane), or RAPTA, complexes have demonstrated efficacy against breast cancer by suppressing metastasis, tumorigenicity, and inhibiting the replication of the human tumor suppressor gene BRCA1. However, RAPTA compounds have limited cytotoxicity, and therefore comparatively high doses are required. This study explores the activity of a series of RAPTA-like ruthenium(II) arene compounds against MCF-7 and MDA-MB-231 breast cancer cell lines and [Ru(η6-toluene)(PPh3)2Cl]+ was identified as a promising candidate. Notably, [Ru(η6-toluene)(PPh3)2Cl]Cl was found to be remarkably stable and highly cytotoxic, and selective to breast cancer cells. The minor groove of DNA was identified as a relevant target.
Introduction
Breast cancer is the most common malignant diagnosed tumor in women and the leading cause of cancer death in the world. Several factors contribute to its high mortality, e.g., acquired resistance during chemotherapy, active invasion, and advanced metastasis.1,2 Despite drug resistance, chemotherapy is the main treatment option for breast cancer once it is in an advanced state, and cisplatin is the most frequently used drug used to treat malignant breast cancers, both as a single agent or in combination with other chemotherapeutics.3–5 Despite the undoubted success of cisplatin and its derivatives in breast cancer treatment, platinum-based compounds exhibit severe side effects due to their lack of selectivity.6Ruthenium complexes have attracted interest as promising alternatives to platinum-based anticancer drugs, apparently presenting lower side effects, and some show promising activity against different breast cancer cell lines.7–9 Although ruthenium-based compounds are not currently used in the clinic, two ruthenium(III) compounds, NAMI A (ImH[trans-RuCl4(DMSO-S)(Im)], Im = imidazole)10 and KP1019 (IndH[trans-RuCl4(Ind)2], Ind = indazole),11 have completed phase I clinical trials. Recently, an analog of KP1019 with a different counterion (Na[trans-RuCl4(Ind)2], known as NKP-1339, IT-139, or BOLD-100) has recently been approved for clinical use as an orphan drug for the treatment of gastric cancers.12
Ruthenium(II)-arene complexes, Ru(η6-arene)(PTA)Cl2 (PTA = 1,3,5-triaza-7-phosphaadamantane), frequently called RAPTA compounds, and related compounds, have been extensively studied for their anticancer properties.13 Although RAPTA compounds tend to display limited cytotoxicity to many cancer cell lines, RAPTA compounds bearing p-cymene (RAPTA-C) and toluene (RAPTA-T) have an impact on metastatic tumors in vivo,14,15 also exerting an antiangiogenic effect16 and normalizing blood vessels in the tumor environment.17 Subsequently, it has been shown that RAPTA-C reduces the growth of primary tumors in preclinical models for ovarian and colorectal carcinomas.18 RAPTA-T also sensitizes cancer cells towards other drugs when combined, and several promising in vivo studies against primary tumors were reported.19–21
Recently, BRCA1 gene damage in cancer cells has received much attention as a potential molecular target for metal-based anticancer compounds,22,23 and RAPTA-T has been shown to inhibit BRCA1 replication in a dose-dependent manner, causing more BRCA1 damage in BRCA1-defective breast cancer (HCC1937 – BRCA1 mutant, triple-negative breast cancer) compared to BRCA1-competent adenocarcinoma (MCF-7) at equal doses.24 RAPTA-T exhibits greater effects on highly invasive metastatic MDA-MB-231 breast cancer cells compared to non-invasive MCF-7 cells or non-tumorigenic HBL-100 cells,25 selectively inhibiting several steps involved in metastasis progression, i.e., detachment, migration, invasion, and re-adhesion.15 Polymeric micelles containing RAPTA-C inhibit the growth of MCF-7 and MD-MB-231 cells grown in both 2D and 3D cultures.26 Note that many other ruthenium complexes also display activity against breast cancers in vivo.27–29
Since RAPTA compounds display promising activity in MCF-7 and MDA-MB-231 breast cancer cells but are not strongly cytotoxic, we hypothesized that more cytotoxic RAPTA-like complexes might offer enhanced activity against breast cancers while potentially retaining selectivity. Hence, we report a study assessing the in vitro effects of a series of ruthenium–arene (RAPTA-type or RAPTA-like) compounds on MCF-7 and MDA-MB-231 breast cancer cell lines and non-tumorigenic L929 fibroblasts. For a promising cationic complex, i.e. [Ru(η6-toluene)(PPh3)2Cl]+, the impact of different counter-anions on the cytotoxicity was explored – an approach rarely explored in inorganic medicinal chemistry beyond changing the counterion to facilitate synthesis, purification, or crystallization.30–32
Results and discussion
Synthesis and characterization of the compounds
A collection of ruthenium(II)–arene compounds (Fig. 1) was used in the study. Compounds 1,332,333,344,145,356,357,348,36 and 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 have been reported previously and were prepared according to the literature procedures. Compounds 10, 11NO3, 11BF4, 11OTf, 11PF6, 11SbF6, and 11BPh4 were prepared by the treatment of [Ru(η6-toluene)(PPh3)Cl2] with PTA (10) or triphenylphosphine (11+) in the presence of the appropriate silver or sodium counterion salt. Compound 11Cl was prepared by resin anion exchange. Compound 10 is similar to [Ru(η6-p-cymene)(PTA)(PPh3)Cl] reported previously.38 Compounds 11BF4
37 have been reported previously and were prepared according to the literature procedures. Compounds 10, 11NO3, 11BF4, 11OTf, 11PF6, 11SbF6, and 11BPh4 were prepared by the treatment of [Ru(η6-toluene)(PPh3)Cl2] with PTA (10) or triphenylphosphine (11+) in the presence of the appropriate silver or sodium counterion salt. Compound 11Cl was prepared by resin anion exchange. Compound 10 is similar to [Ru(η6-p-cymene)(PTA)(PPh3)Cl] reported previously.38 Compounds 11BF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 and 11PF6
39 and 11PF6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 have been reported previously, but in this work, they were prepared following a different synthetic route (see Materials and methods).
37 have been reported previously, but in this work, they were prepared following a different synthetic route (see Materials and methods).
31P NMR spectroscopy confirmed the successful synthesis of 10 and 11CA, which show an upfield shift of the triphenylphosphine singlet in [Ru(η6-toluene)(PPh3)Cl2]. The spectrum of the asymmetrical bis-phosphine complex 10 exhibits two doublets of equal intensity with large 2JPP couplings of ca. 54 Hz, confirming the presence of the two phosphines coupling to each other. Chemical shifts of the coordinated PPh3 ligands in 11CA range from 22.1 to 22.8 ppm. No major differences were observed in the chemical shifts of cation 11+ in the 1H and 13C NMR spectra of 11CA. Additionally, complexes 10 and 11CA were analyzed by ESI-MS in positive ion mode, which gave intense peaks with the expected isotopic patterns corresponding to molecular species at m/z 648 and 753, respectively. Fragmentation of the peak at m/z 753 corresponding to intact 11+ revealed that a phosphine ligand is lost in preference to the arene or the chloride, followed by the loss of the halide and then arene at higher dissociation energies.
Single crystal X-ray structures of 10, 11Cl, 11OTf, 11PF6, 11SbF6, and 11BPh4 were obtained and are shown in Fig. 2 (see ESI† for crystal growth, data collection, and refinement details). All the complexes have the expected piano-stool-type conformation with the methyl group of the coordinated arene locked in the same plane as the Ru–Cl bond. No significant structural differences were observed in the selected bond lengths and angles of the solid-state structures of the cation in 11CA (CA – counter-anion) (Table S1†), highlighting the absence of an effect in the crystal due to the different counterions.
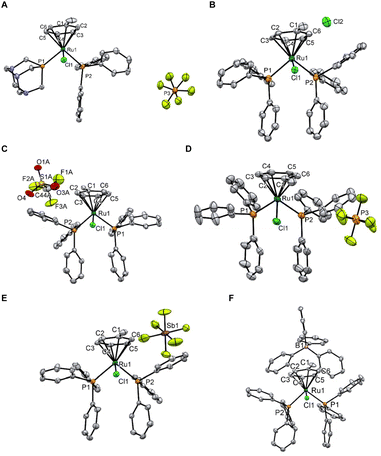 | ||
| Fig. 2 ORTEP diagrams of 10 (A), 11Cl (B), 11OTf (C), 11PF6 (D), 11SbF6 (E), and 11BPh4 (F). Thermal ellipsoids are drawn with a 50% probability. Solvates and disorders have been omitted for clarity. | ||
Compounds containing labile metal–halide bonds, such as cisplatin or RAPTA-C, undergo hydrolysis and/or ligand substitution as a key activation hallmark of their therapeutical activity.40 Among other methods, NMR spectroscopy and mass spectrometry are widely employed to study these reactions and elucidate the fate of a compound in a physiological environment. The stability of 11CA under pseudo-physiological conditions was evaluated by ESI-MS with speciation and the relative intensity of the different ruthenium-containing ions recorded over 72 h. All complexes remained stable under the conditions (Fig. S2–S8†), and no noticeable differences were observed as a consequence of the different counterions. As 11Cl displays higher solubility than the other salts in aqueous media (Table S3†), its stability was additionally studied by NMR spectroscopy in neutral conditions. No signs of hydrolysis or decomposition were observed (Fig. S9 and S10†). To further confirm the inertness of the Ru–Cl bond, 11Cl and 11BPh4 were incubated for 72 h in DMSO, a strongly coordinating solvent. No changes were observed, confirming the overall resistance of 11CA to solvolysis (Fig. S11–S14†). The inertness of 11CA is likely due to the difficulty of eliminating a negatively charged Cl− ligand from a cationic complex to afford a doubly charged species. For example, note that the second hydrolysis of RAPTA-C is only facilitated after the deprotonation of first-coordinated water.40
In vitro studies
The cytotoxicity of 1–11CA was assessed against human breast cancer MCF-7 and MDA-MB-231 cells, and non-tumorigenic adherent mouse fibroblast connective tissue L929 cells using the MTT assay after 24 and 72 h (Tables S5† and Table 1, respectively). The dose–response curves of compounds 10 and 11CA at 24 h and 72 h are presented in Fig. S15.†| Compound | IC50 (μM) (average ± standard deviation) 72 h | SIa | ||
|---|---|---|---|---|
| MCF-7 | MDA-MB-231 | L929 | ||
| a The selectivity index (SI) was calculated as IC50 non-cancerous L929 cells divided by the average of the IC50 of the breast cancer cell lines. | ||||
| 1 | 48 ± 10 | 41 ± 3 | 58 ± 4 | 1.3 |
| 2 | 51 ± 6 | 63 ± 3 | 64 ± 4 | 1.1 |
| 3 | 51 ± 4 | 60 ± 3 | 84 ± 5 | 1.5 |
| 4 | 68 ± 7 | 83 ± 4 | 79 ± 4 | 1.0 |
| 5 | 57 ± 5 | 71 ± 4 | 84 ± 4 | 1.3 |
| 6 | 74 ± 7 | 63 ± 5 | 82 ± 3 | 1.2 |
| 7 | 62 ± 4 | 83 ± 5 | 104 ± 4 | 1.4 |
| 8 | 41 ± 5 | 50 ± 2 | 50 ± 3 | 1.1 |
| 9 | 52 ± 7 | 44 ± 3 | 70 ± 4 | 1.5 |
| 10 | 3.1 ± 0.2 | 2.8 ± 0.2 | 31 ± 3 | 10.5 |
| 11Cl | 0.061 ± 0.003 | 0.086 ± 0.006 | 0.87 ± 0.05 | 11.8 |
| 11NO3 | 0.078 ± 0.005 | 0.109 ± 0.009 | 1.04 ± 0.05 | 11.1 |
| 11BF4 | 0.14 ± 0.01 | 0.19 ± 0.01 | 1.13 ± 0.05 | 6.8 |
| 11OTf | 0.60 ± 0.07 | 1.4 ± 0.1 | 1.8 ± 0.2 | 1.8 |
| 11PF6 | 0.15 ± 0.01 | 0.16 ± 0.01 | 0.17 ± 0.01 | 1.1 |
| 11SbF6 | 0.08 ± 0.01 | 0.131 ± 0.009 | 1.35 ± 0.09 | 12.8 |
| 11BPh4 | 0.52 ± 0.06 | 1.7 ± 0.3 | 3.9 ± 0.3 | 3.5 |
There is a direct correlation between cell growth inhibition and the lipophilicity of the complexes (Table S3†). Compounds 1–9 exhibit IC50 values >40 μM and low selectivity indexes. The presence of hydrophobic triphenylphosphine (PPh3) ligands increases the cytotoxicity of the complexes. Compound 10, bearing a hydrophilic PTA and a hydrophobic PPh3 ligand, is considerably more cytotoxic than 1–9 and shows good cancer cell selectivity. Compounds 11CA with two PPh3 ligands even reach cytotoxicity values in the nanomolar range with some having a selectivity index (between non-tumoral and breast cancer cell lines) above 10. Note that since the sodium salt of BPh4− presented the lowest IC50 values in all the cell lines at both incubation times (see Table S7 in the ESI†), it might be expected that 11BPh4 would be the most cytotoxic in the series, but that was not the case. Indeed, 11BPh4 exhibits the lowest cytotoxicity among all the 11CA compounds (cf. IC50 of 11BPh4 and 11Cl in MCF-7 cells 0.52 ± 0.06 μM and 0.061 ± 0.003 μM, respectively), highlighting that the activity of these salts is not correlated with the intrinsic toxicity of the counterions. Possibly, the toxicity is much more affected by the solvation and dissociation of the complex in aqueous media and the availability of the complex to interact within the cell environment as highlighted by the conductivity measurements and calculated hydration energies (Tables S3 and S4†).
Compounds 11CA are more active against breast cancer cell lines than platinum-based chemotherapeutics currently used in the clinic (cisplatin, 28 ± 6 μM (ref. 41) and 38.70 ± 0.03 μM;42 carboplatin, 27.3 μM (ref. 43) and 15.4 ± 3.3 μM (ref. 44) for MCF-7 and MDA-MB-231 cells, respectively). Compared to previously reported complexes tested against breast cancers, the most active representatives of the 11CA series (11Cl, 11NO3, and 11SbF6) are comparable to the most cytotoxic ruthenium-based45 or metal drugs46 reported for breast cancers. Furthermore, 11CA are more cytotoxic than RAPTA-EA, a selective glutathione transferase (GSTP-1) inhibitor, which shows good cytotoxicity profiles against breast cancer cell lines (20.0 ± 2.2 μM and 10.5 ± 0.5 μM for MCF-7 and MDA-MB-231 cells, respectively).47
The most active and selective compounds identified from the cytotoxicity studies are 10 and 11CA and were therefore tested for their ability to induce cell death via apoptosis, the preferred mechanism of cell death48 and sometimes associated with multidrug-resistance.49 The compounds were incubated with MCF-7 and MDA-MB-231 cells at their 24 h IC50 dose and Annexin V caspase assay was used to determine the extent of apoptosis (Fig. 3).
 | ||
Fig. 3 Induction of apoptosis by 10 and 11CA on breast cancer cell lines. Determination of live and apoptotic events treated with 10 and 11CA for 24 h. (a), MCF-7 and (b), MDA-MB-231;  viable and viable and  apoptotic events. The flow cytometry graphs for the tested compounds are provided in Fig. S16 and the IC50 values at 24 h are provided in Table S5 of the ESI.† apoptotic events. The flow cytometry graphs for the tested compounds are provided in Fig. S16 and the IC50 values at 24 h are provided in Table S5 of the ESI.† | ||
With the exception of 11PF6, the tested compounds induce a slight increase in the number of apoptotic MCF-7 cells to approximately 10% compared to the control. Induction of apoptosis was more pronounced in the MDA-MB-231 cells, especially for 10, 11OTf, 11SbF6, and 11BPh4, presenting average apoptosis percentages of 41, 40, 95, and 33%, respectively. Negligible necrosis (<2%) was observed for all the compounds (Fig. S3†). Ruthenium compounds inducing >60% of apoptotic cells are rare, but not unreported. For example, cationic ruthenium complexes containing heterocyclic thioamidates induce nearly 75% of cell apoptosis at 24 h and 82% at 48 h in a hepatocarcinoma cell line.50 Some cationic chloroquine-functionalized ruthenium complexes also induce apoptosis in ca. 90% after 16 h of incubation in acute T-cells leukemia Jurkat cells.51 Conversely, the high apoptotic cell values of 11SbF6 could be related to the antimony counterion as other antimony-containing compounds, including sodium stibogluconate (an antileishmanial drug), SbCl3, or Sb2O3, induce significant apoptosis in acute promyelocytic leukemia cell lines.52,53 Hence, the antiproliferative activity of 11SbF6 (Table 1) might be partially linked to apoptosis induction of the antimony counterion. However, the induction of apoptosis of the remaining complexes does not necessarily correlate with the higher cytotoxicity or the selectivity of the complexes (Table 1), indicating that other cell death mechanisms may be involved.54,55 Compounds that are able to induce programmed cell death via alternative mechanisms might be able to overcome apoptosis-related resistance, suggesting that 11Cl and 11BF4 could have potential applications in cancers that develop apoptosis resistance.56
Interaction of 11CA with biomolecules
Nucleophilic model biomolecules were initially employed to explore the reactivity between 11Cl and potential biological targets. Compound 11Cl was incubated with three nucleophilic amino acids, i.e., glutamic acid (Glu), histidine (His), and sulfur-bearing cysteine (Cys), and the nucleotide deoxyguanosine monophosphate (dGMP) and the reactions were monitored by NMR spectroscopy and mass spectrometry. No changes in the model biomolecules, the metal complex, or adduct formation were observed during the incubation of 11Cl with any of the model compounds (Fig. S17–S28†), suggesting that the 11+ cation does not exert its anticancer effect via covalent binding. To exclude the possibility of a catalytic mode of action, common to other ruthenium complexes,57–5911Cl was evaluated as a catalyst for the oxidation of GSH and in the transfer of hydrogenation of NAD+ to 1,4-NADH. No catalytic or stoichiometric conversion was observed, and the compound remained unchanged (Fig. S29–S32†).These results indicate that 11Cl might engage in non-covalent interactions with biomolecules, similar to other inert ruthenium complexes.60–62 Hence, the interaction between 11Cl and bovine serum albumin (BSA), employed as a model plasma protein, was evaluated by measuring the fluorescence quenching of tryptophan. The calculated value for the bimolecular quenching rate due to short-range interactions, kq, is 1.9 × 1012 M−1 s−1. This value is higher than the average diffusion-controlled quenching rates of representative biomolecules in aqueous media (2 × 1010 M−1 s−1),63 indicating that fluorescence quenching is due to specific interactions between 11Cl and the protein. The calculated value for the bimolecular quenching rate due to short-range interactions, kq, indicates that the decrease in fluorescence is a result of specific interactions between 11Cl and the protein. The binding constant of 11Cl to BSA (5.6 × 105 M−1) is moderate compared to the reported binding constants of other ruthenium(II) arene complexes.64,65 Furthermore, the estimated value of n (∼1) supports the likelihood of an unimolecular single binding site for 11Cl in BSA (Table S8 in ESI†).
To gain insights into potential interactions between 11Cl and DNA, a fluorescence-based competitive binding assay was conducted using three DNA probes (Table 2), i.e., DAPI (minor groove binder), MG (major groove binder), and PI (intercalator). From the obtained fluorescence data, apparent DNA binding constants, Kapp, were calculated according to the competitive binding model described by Tse and Boger.66 A decrease in the emission intensity of DAPI and MG was observed as the concentration of 11Cl increased, without their respective emission wavelengths changing. In contrast, no linear quenching was observed with PI. This data suggests that 11Cl does not intercalate DNA but interacts with the major or minor groove of DNA via non-covalent interactions. The difference between the obtained apparent binding constants, Kapp, (4.4 × 105 M−1 and 0.43 × 105 M−1, calculated from the competitive binding of MG and DAPI, respectively) suggests that 11Cl shows a 10-fold preference towards the minor groove (Table S9 in ESI†). The calculated binding constants are in line with those reported for other DNA-binding ruthenium(II)–arene complexes65,67 and for ruthenium(II) groove-binding compounds.68,69
| PDB structure | N minG (%) | ΔGminG (kcal mol−1) | N MajG (%) | ΔGMajG (kcal mol−1) | Other | N other (%) | ΔGother (kcal mol−1) |
|---|---|---|---|---|---|---|---|
| 1BNA (native) | 59 | −5.62 (−4.65) | 41 | −5.38 (−4.77) | 0 | — | — |
| 5T4W (minor groove) | 79 | −6.41 (−5.39) | 21 | −5.68 (−5.12) | 0 | — | — |
| 1G3X (intercalator) | 23 | −8.89 (−8.23) | 0 | — | Int. + minG | 76 | −8.95 (−8.63) |
| Int. + MajG | 1 | −6.42 |
The interaction of 11Cl with plasmid DNA was also assessed by visualizing the relaxation of the plasmid using agarose gel electrophoresis (Fig. 4). Different concentrations of the 11Cl were incubated with pBR322 DNA at 37 °C for 24 h and then analyzed by gel electrophoresis (Fig. 4). Compound 11Cl did not induce the stabilization of the linear form, further corroborating that 11Cl does not interact covalently with DNA.70 However, at ratios from 0.2, 11Cl produces alterations in the supercoiled structure of the plasmid in the form of tailing and a slight retardation on the bands, which might be an indication of non-covalent interactions with the DNA.71 Notably, no precipitation was observed in any of the studied concentrations, indicating that the 11Cl charge does not neutralize DNA and, therefore, implying that strong electrostatic binding and aggregation of DNA by 11CA as the mechanism of action is unlikely.72,73
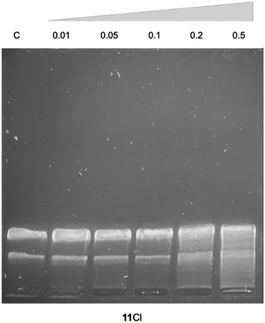 | ||
| Fig. 4 Gel electrophoresis of 11Cl with pBR322 DNA. C – control; 11Cl at ri 0.01; 0.05; 0.1; 0.2 and 0.5 (ri = [11Cl]/[DNA bp]). | ||
Circular dichroism (CD) was used to identify the non-covalent binding mode of 11Cl.74 The CD profile of right-handed B-DNA (its most common form) displays two positive (220 and 268 nm) and two negative (210 and 246 nm) elliptical signals.75 Compound 11Cl induced changes in the intensity of the bands of ctDNA (Fig. 5), characterized by a decrease in the ellipticity of the bands at 210, 246 and 270 nm, and an increase in the circularity of the band at 220 nm. A small bathochromic (red) shift was also observed for the band at 220 nm, whereas the maximum wavelength remained unchanged for the other the bands. This data indicates that 11Cl induces changes in the circularity of DNA and distorts its secondary structure.
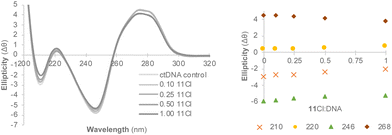 | ||
| Fig. 5 Circular dichroism spectra of 11Cl (left) and changes in the intensity of the main bands (right). | ||
The CD spectra of 11Cl with B-DNA were compared to the spectra of the probes DAPI (minG), MG (MajG) and PI (Int.) (Fig. S33 and Table S10 in the ESI†) indicate that 11Cl interacts with both the minor and major grooves of DNA. This finding is further supported by the induced circular dichroism (ICD) spectra of the DNA probes before and after the addition of 11Cl (Fig. S34†). The intensity of the ICD bands of DAPI and MG decreased by 35% and 20%, respectively, following treatment with 11Cl. Moreover, the difference in quenching between these probes confirms that 11Cl prefers to interact with the minor groove. Additionally, the induced changes in the CD spectra were not coherent with those observed for previously reported DNA condensation agents,76 further rejecting DNA aggregation as a mode of action.
Tandem mass spectrometry was used to investigate the binding mode interaction of 11Cl with single- (ss) and double-strand (ds) DNA oligomers.77,78 After incubation, 43 and 23% of the identified ssDNA and dsDNA ions contained ruthenium, respectively, with [Ru(η6-toluene)(PPh3)]2+ as the main ruthenium adduct in the gas phase (in agreement with the fragmentation behavior observed during the characterization of the complexes, see above). The interaction between the oligonucleotides and 11Cl is non-covalent in nature (in agreement with fluorescence and circular dichroism data) as loss of the ruthenium from the isolated adduct can be observed even when no collision-induced dissociation energy (to fragment the ions) is applied. Fragmentation of 14-mer ssDNA ions containing [Ru(η6-toluene)(PPh3)]2+ resulted in longer nucleotide fragments compared to the intact oligomer (12 ± 2 vs. 6 ± 4 nucleotides, Fig. S35 and S36†), arising from the stabilization of the ruthenium-bound DNA,79 likely through the interaction of the cationic 11+ with negatively-charged phosphate groups.80 Likewise, fragmentation of 6-mer dsDNA ruthenium-bound ions led to a similar outcome (5 ± 1 vs. 3 ± 1 nucleotides, Fig. S35 and S37†). In both studied oligonucleotides, w-type fragmentation, and base loss remained the main fragmentation mechanisms in intact DNA (native) and metal-free (fragments originating from the isolated ruthenium-DNA adduct but not containing the metal) ions.81,82 However, ruthenium-containing ions displayed a decrease in the number of fragments (more unfragmented ions were present) and in the number of internal fragments, as well as enhancing base-loss fragmentation pathways (Fig. S38 and S39†). This behavior indicates that 11Cl influences the fragmentation behavior of the oligonucleotides, supporting the stabilization of DNA by the ruthenium complex through an interaction with its secondary structure. Additionally, the lack of an apparent nucleotide selectivity or sequence binding preference,83,84 and the promotion of base-loss pathways,85 while retaining the backbone and sugar fragments, are coherent with the absence of covalent interactions between the nucleobases and 11Cl, further confirming its interaction with the phosphate backbone in the DNA grooves.
Molecular docking studies were performed to probe the nature of the interactions of 11+ with the major and minor grooves of DNA. Since 11+ is chemically inert, standard biomolecule-ligand docking methodologies were used as the metal mainly plays a structural role and does not require a precise electronic description.86 The molecular docking studies support the experimental results, confirming that both the major and the minor grooves in B-DNA are likely binding sites for 11+ (Fig. 6. The binding of the minor groove is slightly favored, supported by the larger number of poses and lower binding energies (Table 2). This preference is also observed in other B-DNA structures (Fig. S40†). The nature of the interaction of 11+ with DNA depends principally on the hydrophobic interactions and electrostatic attraction between the ruthenium cation and the negatively charged DNA phosphate backbone (Fig. S41†). The electrostatic component only represents an average of 15% or 8% of the total binding energy, for the minor groove and the major groove poses respectively. To a lesser extent, these conformations are also favored by π-interactions between the aromatic rings on 11+ and the nucleobases.
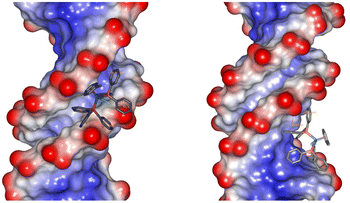 | ||
| Fig. 6 Docked conformations of 11+ with the minor (right) and major (left) grooves of 1BNA. | ||
The performed studies have helped elucidate the possible biological targets of 11CA. The hydrolysis-resistant complexes interact with biomolecules in non-covalent fashion, deviating from the more typical mechanisms associated with metallic drugs. 11Cl binds both proteins and nucleotides and, as expected from the highly lipophilic character of the compound, it displays a preference for binding the minor groove of DNA. It can be expected that these interactions are fundamental for the high antiproliferative activity displayed by the developed cationic complexes.
Conclusions
From a collection of ruthenium(II)–arene compounds a hydrophobic, cationic complex, 11Cl, was identified as a promising candidate for the treatment of breast cancers, presenting inhibitory growth rates in the nanomolar range, and displaying around a 10-fold selectivity for the breast cancer cell lines compared to the healthy ones. The interaction studies confirmed that 11Cl interacts with biomolecules in a non-covalent fashion with a preference for binding the minor groove of DNA.Experimental
[Ru(η6-toluene)Cl2]2,87 [Ru(η6-toluene)(PPh3)Cl2]37 and 1,332,333,344,145,356,357,348![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 88 and 9
88 and 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 were prepared according to reported procedures and their spectroscopic data are in agreement with those reported. Compounds 10 and 11CA (CA = counteranion, 7 examples) were prepared by the treatment of [Ru(η6-toluene)(PPh3)Cl2] with PTA or triphenylphosphine in the presence of the appropriate silver or sodium counterion salt, except in the case of 11Cl which was prepared by resin anion exchange. Compounds 11BF4
37 were prepared according to reported procedures and their spectroscopic data are in agreement with those reported. Compounds 10 and 11CA (CA = counteranion, 7 examples) were prepared by the treatment of [Ru(η6-toluene)(PPh3)Cl2] with PTA or triphenylphosphine in the presence of the appropriate silver or sodium counterion salt, except in the case of 11Cl which was prepared by resin anion exchange. Compounds 11BF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 and 11PF6
39 and 11PF6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 37 were previously reported, but in this work, they were prepared following a different synthetic route. Full experimental details and characterization data are provided in the ESI (Fig. S1 and Tables S1 and S2).†
37 were previously reported, but in this work, they were prepared following a different synthetic route. Full experimental details and characterization data are provided in the ESI (Fig. S1 and Tables S1 and S2).†
Adherent mouse fibroblast connective tissue cells (L929) and human breast cancer cells (MCF-7 and MDA-MB-231) were obtained from the European Collection of Authenticated Cell Cultures (ECACC), Public Health England, Salisbury, UK and ATCC, Middlesex, UK, respectively. All cell culture reagents i.e., Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin–streptomycin mixture, and trypsin-EDTA 0.25% (v/v) were purchased from Gibco® (Invitrogen Corporation, UK). Deoxyribonucleic acid (calf thymus) and Dimethyl sulfoxide (DMSO) were purchased from Merck® (EMD Millipore Corp., USA). FITC Annexin V Apoptosis Detection Kit with 7-AAD was purchased from BioLegend®. Agarose basic was purchased from PanReac AppliChem®. pBR322 DNA, GeneRuler 1 kb Plus DNA ladder, and SYBR™ Safe DNA Gel Stain were purchased from Thermo Fisher Scientific®. Methyl green, DAPI, and propidium iodide 95% were purchased from Sigma-Aldrich®, Ivalua®, and Acros Organics®, respectively.
Stability in solution and reactivity with model biomolecules
The stability of 11CA under pseudo-physiological conditions was assessed using ESI-MS and 1H and 31P NMR spectroscopy. Furthermore, the interaction between 11Cl and different model biomolecules (amino acids: glutamic acid, histidine and cysteine, and nucleotide, dGMP), was also studied. The catalytic activity of 11Cl in transfer hydrogenation and glutathione (GSH) oxidation reactions was monitored by 1H and 31P NMR spectroscopy under previously reported biologically relevant conditions.57Preparation of stock solutions of the compounds
Stock solutions were prepared in supplemented DMEM with 10% DMSO to ensure solubility. Then, necessary dilutions were made using the supplement DMEM, to prepare the range of tested concentrations. Since DMSO was used to solubilize the compounds, a supplemented DMEM + DMSO (at the same percentage as in the sequential dilutions) was considered as a control in the 3-[4,5-dimetylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay.Cell lines and culture conditions
L929, MCF-7, and MDA-MB-231 cell lines were cultured in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) of penicillin–streptomycin. The cells were incubated in a humidified chamber at 37 °C under saturated air with a 5% CO2 atmosphere. Cells were passed at 80%–90% of confluency with 0.25% (w/v) trypsin and the cell culture media was changed every 2–3 days.Cell viability assessment
The compounds were tested on two breast cancer cell lines (MCF-7 and MDA-MB-231) and L929 fibroblasts, used as a reference for the safety evaluation according to ISO 10993-1:2009 “Biological evaluation of medical devices”. Cells were seeded on sterile 96-well plates at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (L929) and 20
000 cells per well (L929) and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (MCF-7 and MDA-MB-231) in supplemented DMEM medium (100 μL) for 24 h. The culture medium was removed and new media containing different concentrations of the compounds (0–1000 μM) was added to the wells (100 μL). Control cells were incubated under the same conditions in the absence of a compound. Cells were incubated for 24 and 72 h under the same conditions described above. Then, the medium was removed, and the cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 0.5 mg mL−1) for 3 h. After this time, the medium was removed, and formazan crystals were solubilized with DMSO (100 μL per well). The absorbance in each well was read on a SynergyTM HT Multimode microplate reader (BioTek Instruments; Winooski, VT, USA) at 570 and 630 nm (for background correction), and the cell viability was calculated according to the following equation (eqn (1)):
000 cells per well (MCF-7 and MDA-MB-231) in supplemented DMEM medium (100 μL) for 24 h. The culture medium was removed and new media containing different concentrations of the compounds (0–1000 μM) was added to the wells (100 μL). Control cells were incubated under the same conditions in the absence of a compound. Cells were incubated for 24 and 72 h under the same conditions described above. Then, the medium was removed, and the cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 0.5 mg mL−1) for 3 h. After this time, the medium was removed, and formazan crystals were solubilized with DMSO (100 μL per well). The absorbance in each well was read on a SynergyTM HT Multimode microplate reader (BioTek Instruments; Winooski, VT, USA) at 570 and 630 nm (for background correction), and the cell viability was calculated according to the following equation (eqn (1)): | (1) |
Analysis of cell apoptosis
Apoptosis in MCF-7 and MDA-MB-231 cells was determined using FITC Annexin V with a 7-aminoactinomycin D (7-AAD) staining kit (BioLegend®, California, US) according to the manufacturer's instructions. Briefly, cells (120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were incubated in sterile 24-well plates with the IC50 concentration of the compounds for 24 h. Following incubation, the cells were washed with PBS twice and detached using 0.25% trypsin. After obtaining cell pellets by centrifugation (300g for 5 min), the cells were resuspended in 100 μL of PBS, followed by the addition of 10 μL of FITC Annexin V and 7-AAD, and then incubated for 15 min at room temperature while protected from light. After the incubation period, 200 μL of binding buffer was added to the cell suspension and the samples were analyzed immediately using a BD Accuri C6 flow cytometer with a minimum of 10
000 cells per well) were incubated in sterile 24-well plates with the IC50 concentration of the compounds for 24 h. Following incubation, the cells were washed with PBS twice and detached using 0.25% trypsin. After obtaining cell pellets by centrifugation (300g for 5 min), the cells were resuspended in 100 μL of PBS, followed by the addition of 10 μL of FITC Annexin V and 7-AAD, and then incubated for 15 min at room temperature while protected from light. After the incubation period, 200 μL of binding buffer was added to the cell suspension and the samples were analyzed immediately using a BD Accuri C6 flow cytometer with a minimum of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events. The assays were performed in quadruplicate and the results were analyzed with Accuri C6 analysis software.
000 events. The assays were performed in quadruplicate and the results were analyzed with Accuri C6 analysis software.
BSA binding assay
BSA (16 μM) in 10 mM Tris-HCl pH 7.6 + 1 mM EDTA solution and varying concentrations of 11Cl (0–32 μM) were incubated at 37 °C for 24 h. Afterward, the fluorescence spectra were measured for each solution between 300 and 500 nm (λex = 280 nm). The intensity at λmax (340 nm) was recorded. Quenching parameters were obtained by applying the Stern–Volmer equation (eqn (2)): | (2) |
 | (3) |
Competitive DNA binding assay
Emission intensity measurements were performed using a SpectraMax M5e reader at room temperature. A 50 μM (bp) of ct-DNA was pre-incubated with the groove binding agents (MG and DAPI) (2 μM) and the intercalating agent propidium iodide (2 μM) for 30 min at 37 °C, to allow full interaction of the dyes into the ctDNA. Increasing amounts of 11Cl (0–100 μM) were added to this mixture and the final samples were incubated for 24 h at 37 °C. After incubation, the emission spectra were recorded at excitations of 633, 358, and 535 nm for the measurements of MG, DAPI, and PI, respectively. Quenching parameters were obtained by applying the Stern–Volmer equation (eqn (2)). The apparent binding constants, Kapp, were calculated according to the competitive binding model described by Tse and Boger66 using eqn (4):90| Kprobe[probe] = Kappc50 | (4) |
Interaction of 11Cl with plasmid DNA by gel electrophoresis
Assays were performed using 1.2% (w/v) agarose gels. A plasmid pBR322 DNA was purchased from Thermofisher Scientific® at 0.5 μg μL−1 concentration in phosphate buffer 50 mM (pH 7.4). A 20 μL portion containing 0.125 μg μL−1 of DNA in 10 mM Tris-HCl (pH 7.6) and 1 mM EDTA was incubated with increasing volumes of the stock solution of 11Cl (1 mM in DMSO) at ri values ranging from 0.05 to 0.5 (ri = [complex]/[no. nucleotide]). Samples were incubated at 37 °C for 24 h, after which 2 μL of the TriTrack DNA loading dye buffer was added. 20 μL of the sample was loaded in the agarose gel (1.2% w/v), and electrophoresis was carried out for a period of 100 min at 40 V in TAE 1× (Tris-acetate/EDTA) buffer. After electrophoresis, the gel was immersed in 200 mL of TAE 1× buffer containing 20 μL of a stock solution of SYBR safe for 30 min to stain the DNA. Then, the gel was washed with Mili Q water for 10 min and the stained gel was analyzed with a blue light in a Safe Imager™ 2.0 (Invitrogen).Interaction of 11Cl with calf thymus DNA by circular dichroism
Calf thymus (ct) DNA (Merck®) was used to perform the circular dichroism assay to detect conformational changes in the ctDNA induced by the compounds. CD spectral data was collected on a Chirascan™ V100 (Applied Photophysics) spectropolarimeter equipped with a large area Avalanche photodiodes detector and computer-controlled thermoelectrical cell holder using a 1 cm path length quartz cuvette. CD spectra of ctDNA in the absence and presence of different concentrations of 11Cl were recorded at wavelengths ranging from 320 to 190 nm under constant nitrogen flush at 25 °C. For each CD spectrum, the number of scans was set to 5, and spectral data were collected at 0.1 ms intervals. Additionally, methyl green (MG) and 4′,6-diamidino-2-phenylindole (DAPI), and propidium iodide (PI) were analyzed as minor and major groove binders and as an intercalator, respectively, to help identify the binding mode of 11Cl to DNA. Samples were prepared in 10 mM Tris-HCl buffer (pH 7.6) with 1 mM EDTA. The concentration of ctDNA was fixed at 100 μM (concerning base pairs) and the ctDNA to 11Cl ratio was varied between 0 and 1. The samples were incubated for 24 h at 37 °C and then analyzed in the CD spectropolarimeter.Tandem mass spectrometry of 11Cl and DNA oligomers
11Cl was incubated with 14-mer ssDNA 5′–3′ (ATACATGGTACATA) and 6-mer dsDNA 5′–3′ (AGGCAG) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at 37 °C in MilliQ water. After 24 h the solution was centrifuged with an Amicon® Ultra 0.5 mL centrifugal filter 3 kDa MWCO to remove the unreacted metal complex and excess salts. Mass spectra were recorded in an LTQ Orbitrap operating in negative mode and analyzed using Analysis of Oligonucleotide Modifications from Mass Spectra (Aom2s).93
1 ratio at 37 °C in MilliQ water. After 24 h the solution was centrifuged with an Amicon® Ultra 0.5 mL centrifugal filter 3 kDa MWCO to remove the unreacted metal complex and excess salts. Mass spectra were recorded in an LTQ Orbitrap operating in negative mode and analyzed using Analysis of Oligonucleotide Modifications from Mass Spectra (Aom2s).93
Molecular docking studies
Molecular docking studies were performed using AutoDock 4.2.94 The crystal structure of a B-DNA dodecamer d(CGCGAATTCGCG)2, 1BNA,95 the 12-mer with DAPI bound in the minor groove, 5T4W,96 and the 12-mer with an acridine-based intercalator, 1G3X,97 were obtained from the Protein Data Bank (PDB). The structure for 11Cl was obtained from the X-ray crystal structure with the counterion and solvent molecules removed to model 11+.Statistical analysis
Each cell viability result represents the mean ± standard deviation (SD) for n = 4 replicates in each of 3 independent assays, resulting in 12 independent values. Each apoptosis value determination represents the mean ± standard deviation of 2 independent assays. The IC50 values were obtained using GraphPad Prism software and ANOVA (analysis of variance) was performed to determine the statistical difference between the IC50 values of different cell lines.Author contributions
Sarah A. P. Pereira and Jan Romano-DeGea: Conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing – original draft, visualization. Ana Isabel Barbosa: Methodology, formal analysis, investigation, writing – original draft. Sofia A. Costa Lima: Methodology, writing – review & editing. Paul J. Dyson: Conceptualization, resources, writing – review & editing, supervision, funding acquisition. M. Lúcia M. F. S. Saraiva: Conceptualization, resources, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors that developed this work acknowledge the financial support from the Swiss National Science Foundation (SNSF) and PT national funds (FCT/MCTES, Fundação para a Ciência e Tecnologia and Ministério da Ciência, Tecnologia e Ensino Superior) through grant UIDB/QUI/50006/2020 and UIDP/50006/2020. S. A. P. P. acknowledges FCT for her Ph.D. Grant (SFRH/BD/138835/2018). J. R-dG. thanks to the Swiss National Science Foundation (SNSF) for financial support. A. I. B acknowledges her funding from FCT/MEC (SFRH/BD/147038/2019), and S. A. C. L thanks her assistant research grant (CEECIND/01620/2017) funded by National Funds, respectively.References
- X. Wu, X. Liu, J. Sengupta, Y. Bu, F. Yi, C. Wang, Y. Shi, Y. Zhu, Q. Jiao and F. Song, Silencing of Bmi-1 gene by RNA interference enhances sensitivity to doxorubicin in breast cancer cells, Indian J. Exp. Biol., 2011, 49, 105–112 CAS.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- S. Dasari and P. B. Tchounwou, Cisplatin in cancer therapy: molecular mechanisms of action, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- N. Koshy, D. Quispe, R. Shi, R. Mansour and G. V. Burton, Cisplatin-gemcitabine therapy in metastatic breast cancer: Improved outcome in triple negative breast cancer patients compared to non-triple negative patients, Breast, 2010, 19, 246–248 CrossRef PubMed.
- T. Byrski, R. Dent, P. Blecharz, M. Foszczynska-Kloda, J. Gronwald, T. Huzarski, C. Cybulski, E. Marczyk, R. Chrzan, A. Eisen, J. Lubinski and S. A. Narod, Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer, Breast Cancer Res., 2012, 14, R110 CrossRef CAS PubMed.
- R. Oun, Y. E. Moussa and N. J. Wheate, The side effects of platinum-based chemotherapy drugs: a review for chemists, Dalton Trans., 2018, 47, 6645–6653 RSC.
- C. A. Vock, W. H. Ang, C. Scolaro, A. D. Phillips, L. Lagopoulos, L. Juillerat-Jeanneret, G. Sava, R. Scopelliti and P. J. Dyson, Development of ruthenium antitumor drugs that overcome multidrug resistance mechanisms, J. Med. Chem., 2007, 50, 2166–2175 CrossRef CAS PubMed.
- D.-L. Ma, C. Wu, S.-S. Cheng, F.-W. Lee, Q.-B. Han and C.-H. Leung, Development of Natural Product-Conjugated Metal Complexes as Cancer Therapies, Int. J. Mol. Sci., 2019, 20, 341 CrossRef PubMed.
- P. Starha and Z. Trávníček, Non-platinum complexes containing releasable biologically active ligands, Coord. Chem. Rev., 2019, 395, 130–145 CrossRef CAS.
- J. M. Rademaker-Lakhai, D. van den Bongard, D. Pluim, J. H. Beijnen and J. H. Schellens, A Phase I and pharmacological study with imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a novel ruthenium anticancer agent, Clin. Cancer Res., 2004, 10, 3717–3727 CrossRef CAS PubMed.
- C. G. Hartinger, M. A. Jakupec, S. Zorbas-Seifried, M. Groessl, A. Egger, W. Berger, H. Zorbas, P. J. Dyson and B. K. Keppler, KP1019, a new redox-active anticancer agent–preclinical development and results of a clinical phase I study in tumor patients, Chem. Biodivers., 2008, 5, 2140–2155 CrossRef CAS PubMed.
- NCT04421820, BOLD-100 in Combination With FOLFOX for the Treatment of Advanced Solid Tumours. 2021.
- B. S. Murray, M. V. Babak, C. G. Hartinger and P. J. Dyson, The development of RAPTA compounds for the treatment of tumors, Coord. Chem. Rev., 2016, 306, 86–114 CrossRef CAS.
- C. Scolaro, A. Bergamo, L. Brescacin, R. Delfino, M. Cocchietto, G. Laurenczy, T. J. Geldbach, G. Sava and P. J. Dyson, In Vitro and in Vivo Evaluation of Ruthenium(II)−Arene PTA Complexes, J. Med. Chem., 2005, 48, 4161–4171 CrossRef CAS PubMed.
- A. Bergamo, A. Masi, P. J. Dyson and G. Sava, Modulation of the metastatic progression of breast cancer with an organometallic ruthenium compound, Int. J. Oncol., 2008, 33, 1281–1289 CAS.
- P. Nowak-Sliwinska, J. R. van Beijnum, A. Casini, A. A. Nazarov, G. Wagnieres, H. van den Bergh, P. J. Dyson and A. W. Griffioen, Organometallic ruthenium(II) arene compounds with antiangiogenic activity, J. Med. Chem., 2011, 54, 3895–3902 CrossRef CAS PubMed.
- T. Riedel, S. Cavin, H. van den Bergh, T. Krueger, L. Liaudet, H. B. Ris, P. J. Dyson and J. Y. Perentes, Chemo-manipulation of tumor blood vessels by a metal-based anticancer complex enhances antitumor therapy, Sci. Rep., 2018, 8, 10263 CrossRef PubMed.
- A. Weiss, R. H. Berndsen, M. Dubois, C. Müller, R. Schibli, A. W. Griffioen, P. J. Dyson and P. Nowak-Sliwinska, In vivo anti-tumor activity of the organometallic ruthenium(II)-arene complex [Ru(η6-p-cymene)Cl2(pta)] (RAPTA-C) in human ovarian and colorectal carcinomas, Chem. Sci., 2014, 5, 4742–4748 RSC.
- R. H. Berndsen, A. Weiss, U. K. Abdul, T. J. Wong, P. Meraldi, A. W. Griffioen, P. J. Dyson and P. Nowak-Sliwinska, Combination of ruthenium(II)-arene complex [Ru(η(6)-p-cymene)Cl(2)(pta)] (RAPTA-C) and the epidermal growth factor receptor inhibitor erlotinib results in efficient angiostatic and antitumor activity, Sci. Rep., 2017, 7, 43005 CrossRef CAS PubMed.
- M. Rausch, A. Rutz, P. M. Allard, C. Delucinge-Vivier, M. Docquier, O. Dormond, P. J. Dyson, J. L. Wolfender and P. Nowak-Sliwinska, Drug Repurposing to Identify a Synergistic High-Order Drug Combination to Treat Sunitinib-Resistant Renal Cell Carcinoma, Cancers, 2021, 13, 3978 CrossRef CAS PubMed.
- T. Riedel, O. Demaria, O. Zava, A. Joncic, M. Gilliet and P. J. Dyson, Drug Repurposing Approach Identifies a Synergistic Drug Combination of an Antifungal Agent and an Experimental Organometallic Drug for Melanoma Treatment, Mol. Pharm., 2018, 15, 116–126 CrossRef CAS PubMed.
- K. Chakree, C. Ovatlarnporn, P. J. Dyson and A. Ratanaphan, Altered DNA binding and amplification of human breast cancer suppressor gene BRCA1 induced by a novel antitumor compound, [Ru(η(6)-p-phenylethacrynate)Cl(2)(pta)], Int. J. Mol. Sci., 2012, 13, 13183–13202 CrossRef CAS PubMed.
- A. Ratanaphan, P. Temboot and P. J. Dyson, In vitro Ruthenation of Human Breast Cancer Suppressor Gene 1 (BRCA1) by the Antimetastasis Compound RAPTA-C and Its Analogue CarboRAPTA-C, Chem. Biodiversity, 2010, 7, 1290–1302 CrossRef CAS PubMed.
- T. Nhukeaw, K. Hongthong, P. J. Dyson and A. Ratanaphan, Cellular responses of BRCA1-defective HCC1937 breast cancer cells induced by the antimetastasis ruthenium(II) arene compound RAPTA-T, Apoptosis, 2019, 24, 612–622 CrossRef CAS PubMed.
- A. Bergamo, A. Masi, P. J. Dyson and G. Sava, Modulation of the metastatic progression of breast cancer with an organometallic ruthenium compound, Int. J. Oncol., 2008, 33, 1281–1289 CAS.
- H. Lu, B. M. Blunden, W. Scarano, M. Lu and M. H. Stenzel, Anti-metastatic effects of RAPTA-C conjugated polymeric micelles on two-dimensional (2D) breast tumor cells and three-dimensional (3D) multicellular tumor spheroids, Acta Biomater., 2016, 32, 68–76 CrossRef CAS PubMed.
- O. A. Lenis-Rojas, C. Roma-Rodrigues, A. R. Fernandes, A. Carvalho, S. Cordeiro, J. Guerra-Varela, L. Sánchez, D. Vázquez-García, M. López-Torres, A. Fernández and J. J. Fernández, Evaluation of the In Vitro and In Vivo Efficacy of Ruthenium Polypyridyl Compounds against Breast Cancer, Int. J. Mol. Sci., 2021, 22, 8916 CrossRef CAS PubMed.
- M. Frik, A. Martínez, B. T. Elie, O. Gonzalo, D. Ramírez de Mingo, M. Sanaú, R. Sánchez-Delgado, T. Sadhukha, S. Prabha, J. W. Ramos, I. Marzo and M. Contel, In Vitro and in Vivo Evaluation of Water-Soluble Iminophosphorane Ruthenium(II) Compounds. A Potential Chemotherapeutic Agent for Triple Negative Breast Cancer, J. Med. Chem., 2014, 57, 9995–10012 CrossRef CAS PubMed.
- L. Côrte-Real, A. R. Brás, A. Pilon, N. Mendes, A. S. Ribeiro, T. D. Martins, J. P. S. Farinha, M. C. Oliveira, F. Gärtner, M. H. Garcia, A. Preto and A. Valente, Biotinylated Polymer-Ruthenium Conjugates: In Vitro and In Vivo Studies in a Triple-Negative Breast Cancer Model, Pharmaceutics, 2022, 14, 1388 CrossRef PubMed.
- M. M. Haghdoost, G. Golbaghi, J. Guard, S. Sielanczyk, S. A. Patten and A. Castonguay, Synthesis, characterization and biological evaluation of cationic organoruthenium(II) fluorene complexes: influence of the nature of the counteranion, Dalton Trans., 2019, 48, 13396–13405 RSC.
- D. L. Ma, C. Wu, K. J. Wu and C. H. Leung, Iridium(III) Complexes Targeting Apoptotic Cell Death in Cancer Cells, Molecules, 2019, 24, 2739 CrossRef CAS PubMed.
- P. C. Healy, B. T. Loughrey, M. L. Williams and P. G. Parsons, Synthesis, structure and cytotoxicity studies of four-coordinate bis (cis-bis(diphenylphosphino)ethene) gold(I) complexes, [Au(dppey)(2)]X, J. Inorg. Biochem., 2010, 104, 625–631 CrossRef CAS PubMed.
- D. N. Akbayeva, L. Gonsalvi, W. Oberhauser, M. Peruzzini, F. Vizza, P. Brüggeller, A. Romerosa, G. Sava and A. Bergamo, Synthesis, catalytic properties and biological activity of new water soluble ruthenium cyclopentadienyl PTA complexes [(C5R5)RuCl(PTA)2] (R=H, Me; PTA=1,3,5-triaza-7-phosphaadamantane), Chem. Commun., 2003, 2, 264–265 RSC.
- W. H. Ang, E. Daldini, C. Scolaro, R. Scopelliti, L. Juillerat-Jeannerat and P. J. Dyson, Development of Organometallic Ruthenium−Arene Anticancer Drugs That Resist Hydrolysis, Inorg. Chem., 2006, 45, 9006–9013 CrossRef CAS PubMed.
- C. S. Allardyce, P. J. Dyson, D. J. Ellis, P. A. Salter and R. Scopelliti, Synthesis and characterisation of some water soluble ruthenium(II)–arene complexes and an investigation of their antibiotic and antiviral properties, J. Organomet. Chem., 2003, 668, 35–42 CrossRef CAS.
- B. S. Murray, L. Menin, R. Scopelliti and P. J. Dyson, Conformational control of anticancer activity: the application of arene-linked dinuclear ruthenium(II) organometallics, Chem. Sci., 2014, 5, 2536–2545 RSC.
- A. B. Chaplin and P. J. Dyson, Catalytic Activity of Bis-phosphine Ruthenium(II)−Arene Compounds:Structure−Activity Correlations, Organometallics, 2007, 26, 2447–2455 CrossRef CAS.
- C. Scolaro, A. B. Chaplin, C. G. Hartinger, A. Bergamo, M. Cocchietto, B. K. Keppler, G. Sava and P. J. Dyson, Tuning the hydrophobicity of ruthenium(II)–arene (RAPTA) drugs to modify uptake, biomolecular interactions and efficacy, Dalton Trans., 2007, 43, 5065–5072 RSC.
- J. R. Polam and L. C. Porter, Arene complexes of Ru(II) Part I. Synthesis, characterization and X-ray crystal structure of [(η6-C6H5CH3)RuCl(PPh3)2][BF4], Inorg. Chim. Acta, 1993, 205, 119–121 CrossRef CAS.
- C. Scolaro, C. G. Hartinger, C. S. Allardyce, B. K. Keppler and P. J. Dyson, Hydrolysis study of the bifunctional antitumour compound RAPTA-C, [Ru(η6-p-cymene)Cl2(pta)], J. Inorg. Biochem., 2008, 102, 1743–1748 CrossRef CAS PubMed.
- S. Gama, F. Mendes, F. Marques, I. C. Santos, M. F. Carvalho, I. Correia, J. C. Pessoa, I. Santos and A. Paulo, Copper(II) complexes with tridentate pyrazole-based ligands: synthesis, characterization, DNA cleavage activity and cytotoxicity, J. Inorg. Biochem., 2011, 105, 637–644 CrossRef CAS PubMed.
- M. Montani, G. V. B. Pazmay, A. Hysi, G. Lupidi, R. Pettinari, V. Gambini, M. Tilio, F. Marchetti, C. Pettinari, S. Ferraro, M. Iezzi, C. Marchini and A. Amici, The water soluble ruthenium(II) organometallic compound [Ru(p-cymene)(bis(3,5 dimethylpyrazol-1-yl)methane)Cl]Cl suppresses triple negative breast cancer growth by inhibiting tumor infiltration of regulatory T cells, Pharmacol. Res., 2016, 107, 282–290 CrossRef CAS PubMed.
- N. M. Aborehab, M. R. Elnagar and N. E. Waly, Gallic acid potentiates the apoptotic effect of paclitaxel and carboplatin via overexpression of Bax and P53 on the MCF-7 human breast cancer cell line, J. Biochem. Mol. Toxicol., 2021, 35, e22638 CrossRef CAS PubMed.
- B. Corkery, J. Crown, M. Clynes and N. O'Donovan, Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer, Ann. Oncol., 2009, 20, 862–867 CrossRef CAS PubMed.
- G. Golbaghi and A. Castonguay, Rationally Designed Ruthenium Complexes for Breast Cancer Therapy, Molecules, 2020, 25, 265 CrossRef CAS PubMed.
- N. Nayeem and M. Contel, Exploring the Potential of Metallodrugs as Chemotherapeutics for Triple Negative Breast Cancer, Chem. – Eur. J., 2021, 27, 8891–8917 CrossRef CAS PubMed.
- K. Hongthong, T. Nhukeaw, P. Temboot, P. J. Dyson and A. Ratanaphan, Anticancer activity of RAPTA-EA1 in triple-negative BRCA1 proficient breast cancer cells: single and combined treatment with the PARP inhibitor olaparib, Heliyon, 2021, 7, e07749 CrossRef CAS PubMed.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Metallodrugs are unique: opportunities and challenges of discovery and development, Chem. Sci., 2020, 11, 12888–12917 RSC.
- F. H. Igney and P. H. Krammer, Death and anti-death: tumour resistance to apoptosis, Nat. Rev. Cancer, 2002, 2, 277–288 CrossRef CAS PubMed.
- S. P. Neves, N. C. de Carvalho, M. M. da Silva, A. C. B. C. Rodrigues, L. M. Bomfim, R. B. Dias, C. B. S. Sales, C. A. G. Rocha, M. B. P. Soares, A. A. Batista and D. P. Bezerra, Ruthenium Complexes Containing Heterocyclic Thioamidates Trigger Caspase-Mediated Apoptosis Through MAPK Signaling in Human Hepatocellular Carcinoma Cells, Front. Oncol., 2019, 9, 1–18 CrossRef PubMed.
- A. Martínez, C. S. K. Rajapakse, R. A. Sánchez-Delgado, A. Varela-Ramirez, C. Lema and R. J. Aguilera, Arene–Ru(II)–chloroquine complexes interact with DNA, induce apoptosis on human lymphoid cell lines and display low toxicity to normal mammalian cells, J. Inorg. Biochem., 2010, 104, 967–977 CrossRef PubMed.
- H. Huang, S. C. Shu, J. H. Shih, C. J. Kuo and I. D. Chiu, Antimony trichloride induces DNA damage and apoptosis in mammalian cells, Toxicology, 1998, 129, 113–123 CrossRef CAS PubMed.
- K. K. Mann, K. Davison, M. Colombo, A. L. Colosimo, Z. Diaz, A. M. S. Padovani, Q. Guo, P. J. Scrivens, W. Gao, S. Mader and W. H. Miller, Antimony trioxide-induced apoptosis is dependent on SEK1/JNK signaling, Toxicol. Lett., 2006, 160, 158–170 CrossRef CAS PubMed.
- D. Wernitznig, K. Kiakos, G. Del Favero, N. Harrer, H. Machat, A. Osswald, M. A. Jakupec, A. Wernitznig, W. Sommergruber and B. K. Keppler, First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro†, Metallomics, 2019, 11, 1044–1048 CrossRef CAS PubMed.
- C. Irace, G. Misso, A. Capuozzo, M. Piccolo, C. Riccardi, A. Luchini, M. Caraglia, L. Paduano, D. Montesarchio and R. Santamaria, Antiproliferative effects of ruthenium-based nucleolipidic nanoaggregates in human models of breast cancer in vitro: insights into their mode of action, Sci. Rep., 2017, 7, 45236 CrossRef CAS PubMed.
- M. Todaro, Y. Lombardo, M. G. Francipane, M. P. Alea, P. Cammareri, F. Iovino, A. B. Di Stefano, C. Di Bernardo, A. Agrusa, G. Condorelli, H. Walczak and G. Stassi, Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4, Cell Death Differ., 2008, 15, 762–772 CrossRef CAS PubMed.
- J. J. Soldevila-Barreda, I. Romero-Canelón, A. Habtemariam and P. J. Sadler, Transfer hydrogenation catalysis in cells as a new approach to anticancer drug design, Nat. Commun., 2015, 6, 6582 CrossRef CAS PubMed.
- J. J. Soldevila-Barreda and N. Metzler-Nolte, Intracellular Catalysis with Selected Metal Complexes and Metallic Nanoparticles: Advances toward the Development of Catalytic Metallodrugs, Chem. Rev., 2019, 119, 829–869 CrossRef CAS PubMed.
- F. Giannini, G. Süss-Fink and J. Furrer, Efficient Oxidation of Cysteine and Glutathione Catalyzed by a Dinuclear Areneruthenium Trithiolato Anticancer Complex, Inorg. Chem., 2011, 50, 10552–10554 CrossRef CAS PubMed.
- L. Cardo and M. J. Hannon, in Metallo-Drugs: Development and Action of Anticancer Agents, ed. S. Astrid, S. Helmut, F. Eva and K. O. S. Roland, De Gruyter, Berlin, Boston, 2018, pp. 303–324 Search PubMed.
- A. Merlino, Interactions between proteins and Ru compounds of medicinal interest: A structural perspective, Coord. Chem. Rev., 2016, 326, 111–134 CrossRef CAS.
- Y. Sheng, Z. Hou, S. Cui, K. Cao, S. Yuan, M. Sun, J. Kljun, G. Huang, I. Turel and Y. Liu, Covalent versus Noncovalent Binding of Ruthenium η6-p-Cymene Complexes to Zinc-Finger Protein NCp7, Chem. – Eur. J., 2019, 25, 12789–12794 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- C. P. Popolin, J. P. B. Reis, A. B. Becceneri, A. E. Graminha, M. A. P. Almeida, R. S. Corrêa, L. A. Colina-Vegas, J. Ellena, A. A. Batista and M. R. Cominetti, Cytotoxicity and anti-tumor effects of new ruthenium complexes on triple negative breast cancer cells, PLoS One, 2017, 12, e0183275 CrossRef PubMed.
- R. Pettinari, F. Marchetti, A. Petrini, C. Pettinari, G. Lupidi, P. Smoleński, R. Scopelliti, T. Riedel and P. J. Dyson, From Sunscreen to Anticancer Agent: Ruthenium(II) Arene Avobenzone Complexes Display Potent Anticancer Activity, Organometallics, 2016, 35, 3734–3742 CrossRef CAS.
- D. L. Boger, B. E. Fink, S. R. Brunette, W. C. Tse and M. P. Hedrick, A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity, J. Am. Chem. Soc., 2001, 123, 5878–5891 CrossRef CAS PubMed.
- K. Chakree, C. Ovatlarnporn, P. J. Dyson and A. Ratanaphan, Altered DNA Binding and Amplification of Human Breast Cancer Suppressor Gene BRCA1 Induced by a Novel Antitumor Compound, [Ru(η6-p-phenylethacrynate)Cl2(pta)], Int. J. Mol. Sci., 2012, 13, 13183–13202 CrossRef CAS PubMed.
- L. Chen, H. Chao, Q. Zhao and H. Li, Unique Optical Oxygen-Sensing Performance of [Ru(IP)2(HNAIP)]2+during the Groove-Binding-Induced B-to-Z DNA Conformational Transition, Inorg. Chem., 2015, 54, 8281–8287 CrossRef CAS PubMed.
- D.-L. Ma, C.-M. Che, F.-M. Siu, M. Yang and K.-Y. Wong, DNA Binding and Cytotoxicity of Ruthenium(II) and Rhenium(I) Complexes of 2-Amino-4-phenylamino-6-(2-pyridyl)-1,3,5-triazine, Inorg. Chem., 2007, 46, 740–749 CrossRef CAS PubMed.
- R. Sáez, J. Lorenzo, M. J. Prieto, M. Font-Bardia, T. Calvet, N. Omeñaca, M. Vilaseca and V. Moreno, Influence of PPh3 moiety in the anticancer activity of new organometallic ruthenium complexes, J. Inorg. Biochem., 2014, 136, 1–12 CrossRef PubMed.
- B. R. Liu, Y.-W. Huang, R. S. Aronstam and H.-J. Lee, Identification of a Short Cell-Penetrating Peptide from Bovine Lactoferricin for Intracellular Delivery of DNA in Human A549 Cells, PLoS One, 2016, 11, e0150439 CrossRef PubMed.
- B. Sun, J.-X. Guan, L. Xu, B.-L. Yu, L. Jiang, J.-F. Kou, L. Wang, X.-D. Ding, H. Chao and L.-N. Ji, DNA Condensation Induced by Ruthenium(II) Polypyridyl Complexes [Ru(bpy)2(PIPSH)]2+and [Ru(bpy)2(PIPNH)]2+, Inorg. Chem., 2009, 48, 4637–4639 CrossRef CAS PubMed.
- J. Malina, N. P. Farrell and V. Brabec, Substitution-Inert Trinuclear Platinum Complexes Efficiently Condense/Aggregate Nucleic Acids and Inhibit Enzymatic Activity, Angew. Chem., Int. Ed., 2014, 53, 12812–12816 CrossRef CAS PubMed.
- A. Erxleben, Investigation of Non-covalent Interactions of Metal Complexes with DNA in Cell-free Systems, CHIMIA, 2017, 71, 102 CrossRef CAS PubMed.
- A. Kellett, Z. Molphy, C. Slator, V. McKee and N. P. Farrell, Molecular methods for assessment of non-covalent metallodrug–DNA interactions, Chem. Soc. Rev., 2019, 48, 971–988 RSC.
- B. I. Kankia, V. Buckin and V. A. Bloomfield, Hexamminecobalt(III)-induced condensation of calf thymus DNA: circular dichroism and hydration measurements, Nucleic Acids Res., 2001, 29, 2795–2801 CrossRef CAS PubMed.
- F. Rosu, V. Gabelica, C. Houssier and E. De Pauw, Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double–stranded oligodeoxynucleotides by electrospray ionization mass spectrometry, Nucleic Acids Res., 2002, 30, e82–e82 CrossRef PubMed.
- R. P. Eberle, Y. Hari and S. Schürch, Transition Metal-based Anticancer Drugs Targeting Nucleic Acids: A Tandem Mass Spectrometric Investigation, CHIMIA, 2017, 71, 120 CrossRef CAS PubMed.
- E. Stellwagen, J. M. Muse and N. C. Stellwagen, Monovalent Cation Size and DNA Conformational Stability, Biochemistry, 2011, 50, 3084–3094 CrossRef CAS PubMed.
- N. V. Hud and A. E. Engelhart, in Nucleic Acid–Metal Ion Interactions, ed. N. V. Hud, The Royal Society of Chemistry, 2008 Search PubMed.
- Z. Xu, J. B. Shaw and J. S. Brodbelt, Comparison of MS/MS Methods for Characterization of DNA/Cisplatin Adducts, J. Am. Soc. Mass Spectrom., 2013, 24, 265–273 CrossRef CAS PubMed.
- S. A. McLuckey, G. J. Van Berkel and G. L. Glish, Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides, J. Am. Soc. Mass Spectrom., 1992, 3, 60–70 CrossRef CAS PubMed.
- A. E. Egger, C. G. Hartinger, H. B. Hamidane, Y. O. Tsybin, B. K. Keppler and P. J. Dyson, High Resolution Mass Spectrometry for Studying the Interactions of Cisplatin with Oligonucleotides, Inorg. Chem., 2008, 47, 10626–10633 CrossRef CAS PubMed.
- L. K. Batchelor, D. Ortiz and P. J. Dyson, Histidine Targeting Heterobimetallic Ruthenium(II)–Gold(I) Complexes, Inorg. Chem., 2019, 58, 2501–2513 CrossRef CAS PubMed.
- R. P. Eberle, Y. Hari and S. Schürch, Specific Interactions of Antitumor Metallocenes with Deoxydinucleoside Monophosphates, J. Am. Soc. Mass Spectrom., 2017, 28, 1901–1909 CrossRef CAS PubMed.
- E. Ortega-Carrasco, A. Lledós and J.-D. Maréchal, Assessing protein–ligand docking for the binding of organometallic compounds to proteins, J. Comput. Chem., 2014, 35, 192–198 CrossRef CAS PubMed.
- M. A. Bennett and A. K. Smith, Arene ruthenium(II) complexes formed by dehydrogenation of cyclohexadienes with ruthenium(III) trichloride, J. Chem. Soc., Dalton Trans., 1974, 233–241 RSC.
- B. S. Murray, S. Crot, S. Siankevich and P. J. Dyson, Potential of Cycloaddition Reactions To Generate Cytotoxic Metal Drugs In Vitro, Inorg. Chem., 2014, 53, 9315–9321 CrossRef CAS PubMed.
- U. Anand and S. Mukherjee, Reversibility in protein folding: effect of β-cyclodextrin on bovine serum albumin unfolded by sodium dodecyl sulphate, Phys. Chem. Chem. Phys., 2013, 15, 9375–9383 RSC.
- M. Lee, A. L. Rhodes, M. D. Wyatt, S. Forrow and J. A. Hartley, GC base sequence recognition by oligoimidazolecarboxamide and C-terminus-modified analogs of distamycin deduced from circular dichroism, proton nuclear magnetic resonance, and methidiumpropylethylenediaminetetraacetate-iron(II) footprinting studies, Biochemistry, 1993, 32, 4237–4245 CrossRef CAS PubMed.
- B. Nordén and F. Tjerneld, Binding of methyl green to deoxyribonucleic acid analyzed by linear dichroism, Chem. Phys. Lett., 1977, 50, 508–512 CrossRef.
- G. Manzini, M. L. Barcellona, M. Avitabile and F. Quadrifoglio, Interaction of diamidino-2-phenylindole (DAPI) with natural and synthetic nucleic acids, Nucleic Acids Res., 1983, 11, 8861–8876 CrossRef CAS PubMed.
- D. Ortiz, N. Gasilova, F. Sepulveda, L. Patiny, P. J. Dyson and L. Menin, Aom2S: A new web-based application for DNA/RNA tandem mass spectrometry data interpretation, Rapid Commun. Mass Spectrom., 2020, 34, e8927 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- H. R. Drew, R. M. Wing, T. Takano, C. Broka, S. Tanaka, K. Itakura and R. E. Dickerson, Structure of a B-DNA dodecamer: conformation and dynamics, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 2179–2183 CrossRef CAS PubMed.
- H. I. Sbirkova-Dimitrova; and B. Shivachev, Crystal structure of the DNA sequence d(CGTGAATTCACG)2 with DAPI, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2017, 73, 500–504 CrossRef PubMed.
- L. Malinina, M. Soler-López, J. Aymamí and J. A. Subirana, Intercalation of an Acridine−Peptide Drug in an AA/TT Base Step in the Crystal Structure of [d(CGCGAATTCGCG)]2 with Six Duplexes and Seven Mg2+Ions in the Asymmetric Unit, Biochemistry, 2002, 41, 9341–9348 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2193598, 2201998, 2202259, 2210241, 2210245 and 2210246. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3dt02037a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |

