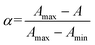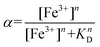Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of temporin L hydroxamate-based peptides and evaluation of their coordination properties with iron(III )†
Rosa
Bellavita
 a,
Linda
Leone
a,
Linda
Leone
 b,
Angela
Maione
c,
Lucia
Falcigno
a,
Gabriella
D'Auria
a,
Francesco
Merlino
b,
Angela
Maione
c,
Lucia
Falcigno
a,
Gabriella
D'Auria
a,
Francesco
Merlino
 a,
Paolo
Grieco
a,
Flavia
Nastri
a,
Paolo
Grieco
a,
Flavia
Nastri
 b,
Emilia
Galdiero
c,
Angela
Lombardi
b,
Emilia
Galdiero
c,
Angela
Lombardi
 b,
Stefania
Galdiero
b,
Stefania
Galdiero
 *a and
Annarita
Falanga
*a and
Annarita
Falanga
 *d
*d
aDepartment of Pharmacy, University of Naples “Federico II”, Via Domenico Montesano 49, 80138 Naples, Italy. E-mail: stefania.galdiero@unina.it
bDepartment of Chemical Sciences, University of Napoli “Federico II”, Napoli, Italy
cDepartment of Biology, University of Naples Federico II, Via Cinthia, 80126 Naples, Italy
dDepartment of Agricultural Sciences, University of Naples “Federico II”, via Università 100, 80055, Portici, Italy. E-mail: annarita.falanga@unina.it
First published on 25th January 2023
Abstract
Ferric iron is an essential nutrient for bacterial growth. Pathogenic bacteria synthesize iron-chelating entities known as siderophores to sequestrate ferric iron from host organisms in order to colonize and replicate. The development of antimicrobial peptides (AMPs) conjugated to iron chelators represents a promising strategy for reducing the iron availability, inducing bacterial death, and enhancing simultaneously the efficacy of AMPs. Here we designed, synthesized, and characterized three hydroxamate-based peptides Pep-cyc1, Pep-cyc2, and Pep-cyc3, derived from a cyclic temporin L peptide (Pep-cyc) developed previously by some of us. The Fe3+ complex formation of each ligand was characterized by UV-visible spectroscopy, mass spectrometry, and IR and NMR spectroscopies. In addition, the effect of Fe3+ on the stabilization of the α-helix conformation of hydroxamate-based peptides and the cotton effect were examined by CD spectroscopy. Moreover, the antimicrobial results obtained in vitro on some Gram-negative strains (K. pneumoniae and E. coli) showed the ability of each peptide to chelate efficaciously Fe3+ obtaining a reduction of MIC values in comparison to their parent peptide Pep-cyc. Our results demonstrated that siderophore conjugation could increase the efficacy and selectivity of AMPs used for the treatment of infectious diseases caused by Gram-negative pathogens.
Introduction
Iron is a vital metabolic cofactor for both humans and bacteria that compete for metal acquisition.1,2 Iron has a complex and multifaceted biological role in humans.3 When present at high concentrations,4 it might be toxic causing oxidative stress; thus, its bioavailability is strictly controlled by iron-binding proteins.5 Interestingly, iron scavenging is also key to limiting iron access to invading microorganisms, which also require iron to function properly. Generally, prokaryotes require ca. 10−6 M of iron to colonize and replicate, but the poor solubility of the metal at neutral pH limits its concentration to 10−18 M.6 As a matter of fact, bacteria use different strategies to acquire iron from the host environments. Among these, one strategy is the synthesis and secretion of small molecules known as siderophores,7,8 which acquire iron from tissues, fluids, and cells and translocate iron–siderophore complexes inside cells by exploiting porins, which are located in the bacterial outer membranes.9,10The large structural and functional diversity of siderophores secreted by bacteria inspired the design of new antimicrobial molecules able to capture iron with higher affinity, influencing iron acquisition, transport, and storage that are essential for bacterial survival.11,12 In particular, siderophores are commonly classified into catecholate, hydroxamate, carboxylate, and phenolate according to the moieties involved in iron chelation.13–15 Ferric iron (Fe3+) is coordinated by six donor atoms in an octahedral structure; further these ligands can be classified according to whether they have two (bidentate), three (tridentate), or six (hexadentate) donor atoms available for coordination to Fe3+.16 Hexadentate complexes present the greatest stability; in fact, most siderophores produced by microorganisms use hexadentate ligands.17,18
Given the different roles of siderophores in both the physiology and pathogenicity of bacteria, exploiting or targeting them represents a challenging strategy for the development of new antimicrobial molecules.19 Generally, siderophores are conjugated to currently used antibiotics to enhance their activity since siderophore–antibiotic conjugates can exploit the iron uptake pathways to transport the antibiotic into the bacterial cell, which then acts on its intracellular target.20 This strategy is known as the Trojan horse approach.21 Similarly, antimicrobial peptides (AMPs), which are widely studied and considered a promising alternative to traditional antibiotics for their mode of action,22–28 may be linked to siderophore–iron chelators to reduce the iron availability for bacterial growth and enhance their efficacy (Fig. 1).29–32
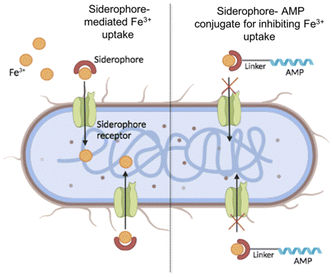 | ||
| Fig. 1 Schematic representation of Fe3+ uptake by natural siderophores (on the left) and its hypothetical inhibition by the siderophore–AMP conjugate. | ||
In this work, we introduced the hydroxamate moiety as a coordinating functional group into a cyclic AMP that was recently developed by some of us.33 This synthetic cyclic AMP, hereafter named Pep-cyc, belongs to temporins, which is one of the most abundant AMP families in nature.34,35 Among the different temporin isoforms, our research has been focused on the temporin L (TL) isoform, with the aim to introduce modifications that may lead to an improved therapeutic index as it is highly hemolytic in its native form.36–39Pep-cyc (FVPWF[KKFlE]RIL) is derived from a detailed structure–activity relationship (SAR) campaign conducted on the analogue [Pro3,DLeu9]TL, in which the α-helix conformation was stabilized through the application of different i,i + 4 intramolecular macrocyclization strategies.33 In particular, Pep-cyc was obtained by the introduction of a lactam bridge between positions 6 and 10 and was identified as the most promising antimicrobial candidate due to its potent activity against Gram-positive strains and some Candida species, with the minimal inhibitory concentration (MIC) ranging from 3.12 μM to 12.5 μM.40 Furthermore, it showed activity against A. baumannii (MIC of 3.12 μM), a Gram-negative strain.33 Herein, to improve the antimicrobial efficacy of Pep-cyc against Gram-negative superbugs, such as E. coli and K. pneumoniae, a hydroxamate chelating moiety was covalently bound through linkers of different lengths at its C-terminal helical region, resulting in being able to chelate and subtract iron from the bacteria and consequently reduce its growth. We reported the synthesis of three cyclic peptides Pep-cyc1, Pep-cyc2, and Pep-cyc3 containing hydroxamate chelating units anchored to their C-terminus as pendants. The Fe3+ complex formation was studied by UV-visible spectroscopy, mass spectrometry, and infrared (IR) and NMR spectroscopies. The effect of Fe3+ on the α-helix conformation of hydroxamate-based peptides was investigated by circular dichroism (CD); the Cotton effect in the near visible region induced by the interaction of peptides with Fe3+ was evaluated. In addition, the ability of hydroxamate-based peptides to chelate iron was assessed in vitro on some Gram-negative strains using iron-supplemented Mueller–Hinton broth. We found significant agreement between the antibacterial and iron coordination studies, which point to these peptides as effective Fe3+ chelators and new-generation antimicrobial molecules.
Experimental
Materials
Nα-Fmoc-protected conventional amino acids were purchased from GL Biochem Ltd (Shanghai, China). Unnatural amino acids such as Fmoc-L-Lys(Alloc)-OH and Fmoc-L-Glu(OAll)-OH, [2-[2-(Fmoc-amino)ethoxy]ethoxy]acetic acid (Fmoc-PEG2-OH), N,N-diisopropylethylamine (DIEA), piperidine, trifluoroacetic acid (TFA), (7-azabenzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyAOP), and the resin 2-chlorotrityl-N-Fmoc-hydroxylamine were acquired from Iris-Biotech GMBH. Coupling reagents such as N,N,N′,N′-tetramethyl-O-(1H-benzotriazol-1-yl) uranium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole (HOBt), (1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b] pyridinium 3-oxide hexafluorophosphate) (HATU), and 1-hydroxy-7-azabenzotriazole (HOAt) were commercially obtained from GL Biochem Ltd (Shanghai, China).6-(Fmoc-amino)hexanoic acid (Fmoc-6-Ahx)-OH, L-2-(Fmoc-amino)butyric acid (Fmoc-GABA-OH), 1,3-dimethylbarbituric acid, tetrakis(triphenylphosphine)palladium(0) [Pd(PPh3)4], and iron(III) chloride (FeCl3) were purchased from Sigma-Aldrich/Merck.
Methods
After the complete synthesis of the linear sequence, the resin was washed with dichloromethane (DCM), dried and the side-chain-to-side-chain lactamization was performed. Firstly, the allyl deprotection was performed on a resin using a solution of Pd(PPh3)4 (0.15 equiv.) and 1,3-dimethylbarbituric acid (3 equiv.) as an allyl scavenger in dry DCM/DMF (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v), and gently shaken for 1 h under argon.42 Then, the resin was washed with DMF and DCM, and the allyl deprotection was repeated for a second time. After these two allyl-deprotection steps, the resin was washed with 0.5% sodium N,N-diethyldithiocarbamate solution in DMF (30 min × 2) and the complete removal of the allyl groups was checked by LC-MS analysis. Then, the lactamization between the free amine and carboxylic groups was coupled using PyAOP (3 equiv.), HOAt (3 equiv.), and DIEA (6 equiv.) dissolved in DMF, for 12 h at rt. After washing the resin with DMF and DCM, this conversion in the cyclic product was ascertained by LC-MS.
2 v/v), and gently shaken for 1 h under argon.42 Then, the resin was washed with DMF and DCM, and the allyl deprotection was repeated for a second time. After these two allyl-deprotection steps, the resin was washed with 0.5% sodium N,N-diethyldithiocarbamate solution in DMF (30 min × 2) and the complete removal of the allyl groups was checked by LC-MS analysis. Then, the lactamization between the free amine and carboxylic groups was coupled using PyAOP (3 equiv.), HOAt (3 equiv.), and DIEA (6 equiv.) dissolved in DMF, for 12 h at rt. After washing the resin with DMF and DCM, this conversion in the cyclic product was ascertained by LC-MS.
Finally, the peptides were released from the resin and the protecting groups were removed through treatment with the cleavage cocktail of TFA/TIS/H2O (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5, v/v/v) for 3 h at rt. Then, the resin was removed by filtration and the crude peptides were precipitated using cold diethyl ether by centrifuging at 6000 rpm for 15 min × 2. Finally, the crude peptides were dissolved in 10% acetonitrile (MeCN 0.1% TFA) in H2O (0.1% TFA) and purified by reversed-phase high-pressure liquid chromatography (RP-HPLC) using a gradient of MeCN (0.1% TFA) in H2O (0.1% TFA) from 10 to 90% over 20 min. All peptides were characterized for purity by analytical RP-HPLC and the correct molecular mass was confirmed through high-resolution mass spectrometry (HRMS) (see ESI, Fig. S1–S3†).
2.5, v/v/v) for 3 h at rt. Then, the resin was removed by filtration and the crude peptides were precipitated using cold diethyl ether by centrifuging at 6000 rpm for 15 min × 2. Finally, the crude peptides were dissolved in 10% acetonitrile (MeCN 0.1% TFA) in H2O (0.1% TFA) and purified by reversed-phase high-pressure liquid chromatography (RP-HPLC) using a gradient of MeCN (0.1% TFA) in H2O (0.1% TFA) from 10 to 90% over 20 min. All peptides were characterized for purity by analytical RP-HPLC and the correct molecular mass was confirmed through high-resolution mass spectrometry (HRMS) (see ESI, Fig. S1–S3†).
The absorbance at 580 nm, attributed to the Fe3+–hydroxamate complex, was used to calculate the fraction of the bound peptide at each point of the titration:
Data points were fit using the Hill equation
Cotton effects in the near UV-visible region from 350 nm to 700 nm were evaluated by dissolving the peptide in MeOH at a concentration of 100 μM and Fe3+ was added at the metal/peptide ratios of 0.1, 0.3 and 0.6.
Results and discussion
Grafting of iron chelators on the peptide Pep-cyc
During a bacterial infection, many Gram-negative pathogens synthesize small iron-sequestering molecules called siderophores as ferric iron is a key element for their colonization and replication.50 Siderophores are classified according to the functional group chelating Fe3+ and the most abundant families are catecholate, hydroxamate, and carboxylate. Among these, the carboxylate siderophores have a lower affinity towards Fe3+ at physiological pH compared to the catecholate and hydroxamate groups.51–53 Herein, we covalently bind hydroxamic acid as an iron-chelating siderophore at the C-terminal region of the cyclic peptide Pep-cyc to scavenge Fe3+ and induce the inhibition of the growth and replication of some Gram-negative pathogens. The peptide Pep-cyc, derived from a previous study performed on a temporin L analogue, is featured by a lactam bridge at positions 6 and 10 and assumes an amphipathic α-helix structure along residues Phe5–Leu13.33 The free N-terminus, which is random in aqueous solution is important for the interaction with the membrane bilayer and thus for the activity. Therefore, the conjugation of the hydroxamate unit was performed in its helical C-terminal region and linkers of different lengths were used to warrant the exposure of the chelator. In particular, we used linkers of 7, 12 and 16 atoms yielding Pep-cyc1, Pep-cyc2, and Pep-cyc3, respectively (Fig. 2).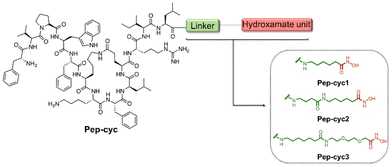 | ||
| Fig. 2 Sequences of the designed peptides Pep-cyc1, Pep-cyc2, and Pep-cyc3 featuring three different linkers bearing the hydroxamate unit. | ||
Hydroxamate-based cyclic peptides were assembled on the solid phase by using 2-chlorotrityl-N-Fmoc-hydroxylamine polymer-bound resin as the solid support to yield the C-terminal hydroxamic acid. Fmoc-Ahx-OH, Fmoc-GABA-OH, and Fmoc-PEG2-OH were chosen and introduced at C-terminus to have different linkers bearing the hydroxamate chelator after the peptide release from the resin. Before the peptide release, the lactam bridge was obtained on the resin by performing the palladium-catalyzed reaction between the residue Lys(Alloc) and Glu(OAll) at positions 6 and 10, respectively.
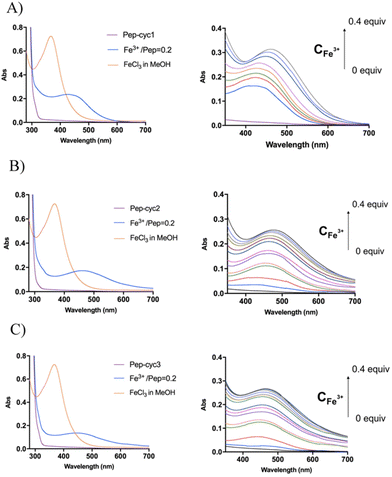
Binding of Fe3+ to hydroxamate-based peptides
The binding of Fe3+ to hydroxamate-based peptides designed in this study was investigated by UV-vis absorption spectroscopy. First, the UV-vis spectra of the peptides alone and FeCl3 were acquired and compared to those of the complexes, prepared using a [Fe3+]/[ligand] ratio of 0.22 (Fig. 3). The spectrum of FeCl3 in the absence of peptides shows an absorption maximum at 360 nm, while all the Fe3+–peptide complexes show a weak and broad absorption band between 450 and 550 nm. The latter was assigned to a ligand-to-metal charge transfer (LMCT) transition in the Fe3+–hydroxamate complexes.54 Slight differences in the spectra were observed among the three peptides, which are likely to be related to the length of the linker, which may cause a different metal environment, thus modulating the complex interaction between the hydroxamate group and the metal ion. Complex formation was then monitored at 580 nm, where the contribution of absorption by unbound ferric ions is negligible. The spectrum of peptide Pep-cyc, lacking the hydroxamate group, was also acquired under the same experimental conditions. The latter did not show any absorption in this spectral range, providing preliminary evidence that no other functional group in the peptides is able to bind iron (see ESI, Fig. S4†).Metal binding experiments were performed by titrating a solution containing a fixed ligand concentration (300 μM) with increasing amounts of Fe3+ in the range of 0–200 μM for Pep-cyc2 and Pep-cyc3 and of 0–500 μM for Pep-cyc1. As expected, the spectra showed a gradual increase in the absorbance at 480 nm for all the hydroxamate-based peptides, indicating the formation of iron complexes. For Pep-cyc2 and Pep-cyc3, no significant change in the absorbance was observed at [Fe3+] > 120 μM, indicating binding saturation. Instead, in the case of Pep-cyc1, [Fe3+] > 300 μM was necessary to achieve saturation. The stoichiometry of the complexes was first evaluated by plotting the fraction of iron-bound peptide (α) as a function of Fe3+ concentration and applying separate linear regression to the unsaturated and saturated regions of the data points (Fig. 4, panel A). The point of intersection between the linear fit of the two datasets occurred at [Fe3+] of 104 and 93 μM for Pep-cyc2 and Pep-cyc3, respectively, indicating the formation of FeL3 complexes, as expected for the bidentate hydroxamate ligands (Fig. 5, panel A). In the case of Pep-cyc1, the intersection was found at 275 μM [Fe3+], suggesting the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Fe3+–ligand complex (Fig. 5, panel B).
1 Fe3+–ligand complex (Fig. 5, panel B).
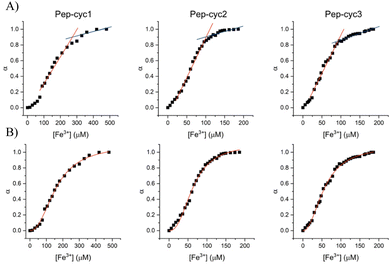 | ||
| Fig. 4 Linear regression (panel A) and Hill (panel B) analyses of the fraction of bound peptide (α) as a function of [Fe3+]. | ||
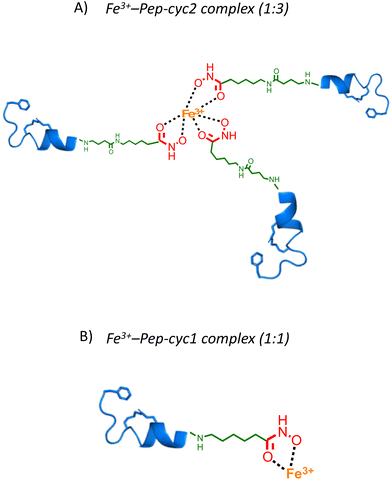 | ||
Fig. 5 Cartoon representing the coordination model structures of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Fe3+–Pep-cyc1 and 1 1 Fe3+–Pep-cyc1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 Fe3+–Pep-cyc2 complexes. 3 Fe3+–Pep-cyc2 complexes. | ||
This finding could be explained by considering that Pep-cyc1 contains the shortest linker to the hydroxamate group. Hence, the formation of the tris-chelate complex could be disfavored by steric interactions between the peptide ligands. The plot of α vs. [Fe3+] showed a slow increase in low Fe3+ concentration, followed by a fast increase before reaching saturation. Such a behavior suggests that complex formation occurs in a stepwise process, rather than in a two-state equilibrium. Hill analysis of the experimental data allowed us to obtain the apparent dissociation constants (KD) for the ferric complexes of all the hydroxamate-based peptides, which are 160 μM, 63 μM and 57 μM for Pep-cyc1, Pep-cyc2 and Pep-cyc3, respectively (Fig. 4, panel B). A Hill coefficient of n ≈ 2 was found in all cases, thus suggesting the presence of two different coordination sites in the iron complexes. Our hypothesis is that a cooperative effect would occur between the two donor oxygen atoms of the hydroxamate group, where the coordination of the carbonyl oxygen facilitates the deprotonation and, consequently, the binding of the carboxylate oxygen atom.
To further investigate the process of complex formation, reverse titration experiments were performed by treating a solution at a fixed Fe3+ concentration (60 μM) with increasing amounts of each peptide, ranging from 0.5 μM to 100 μM. The spectra obtained for Pep-cyc1 (Fig. 6, panel A) revealed the appearance of an absorption maximum at 316 nm, concurrent to the decrease in the band at 380 nm, corresponding to unbound ferric ions. An additional broad absorption band centered at 490 nm appeared upon increasing the peptide concentration, resembling that observed by adding iron to an excess of ligand (Fig. 3).
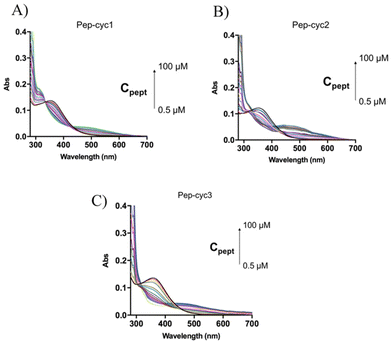 | ||
| Fig. 6 UV-vis spectra acquired during the titration of Fe3+ (black line) with increasing peptide concentrations ranging from 0.5 μM to 100 μM: (A) Pep-cyc1; (B) Pep-cyc2; (C) Pep-cyc3. | ||
The lack of isosbestic points between 360 and 490 nm indicates that multiple species are present in the solution at comparable concentrations of Fe3+ and peptide.
In the cases of Pep-cyc2 and Pep-cyc3, featuring the linker of intermediate and maximum lengths, respectively, the absorption band at 316 nm was not observed, while the broad absorption feature between 450 and 550 nm was still formed by increasing the peptide concentration (Fig. 6, panels B and C). The absence of isosbestic points further confirms the presence of multiple equilibria in the solution. These experiments were also performed on Pep-cyc and its linear analogue [Pro3, DLeu9]TL, displaying no complex formation (ESI Fig. S5†).
ESI-MS analysis of the Fe3+–peptide complexes
The formation of the Fe3+ complexes of hydroxamate-based peptides was also investigated by ESI-MS analysis. Table 1 shows the most abundant ions found in the mass spectrum of each sample. Complexes with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal
2 metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand stoichiometry were observed for all peptides, while the FeL3 complexes could not be detected in the experimental conditions of MS analysis.
ligand stoichiometry were observed for all peptides, while the FeL3 complexes could not be detected in the experimental conditions of MS analysis.
| Ion | Calculated mass | Experimental mass |
|---|---|---|
| [Pep-cyc1 + 2H+]2+ | 917.64 | 918.00 |
| [Pep-cyc1 − H+ + Fe3+]2+ | 944.05 | 944.75 |
| [2Pep-cyc1 − H+ + Fe3+]2+ | 1861.1 | 1860.35 |
| [Pep-cyc2 + 2H+]2+ | 960.69 | 960.20 |
| [Pep-cyc2 − H+ + Fe3+]2+ | 986.54 | 986.85 |
| [2Pep-cyc2 + Fe3+ + H+ + H2O]4+ | 978.99 | 978.40 |
| [Pep-cyc3 + 2H+]2+ | 989.72 | 990.35 |
| [Pep-cyc3 − H+ + Fe3+]2+ | 1016.72 | 1017.05 |
| [2Pep-cyc3 + Fe3+]3+ | 1336.33 | 1337.85 |
FT-IR spectra analysis
The FTIR measurements were performed to identify the main functional groups of peptides involved in Fe3+ coordination. The transmittance band ranging from 4000 cm−1 to 400 cm−1 was recorded for the FTIR spectra as depicted in Fig. 7. In particular, the spectra collected for the peptide Pep-cyc showed no difference compared to the spectra recorded in the presence and in the absence of Fe3+, indicating that there are no interactions between Pep-cyc and Fe3+, in accordance with the other experimental data. The FTIR spectra collected for the peptide Pep-cyc3 in the presence and in the absence of Fe3+ showed significant differences. The –OH (alcohol) that arises at 2933 in Pep-cyc3 is shifted to 2927 cm−1 following the Fe3+ coordination. Furthermore, the frequency of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1178 cm−1 is shifted to 1192 and almost disappears after the Fe3+ coordination. All these data support iron coordination with hydroxamic acid.55
O at 1178 cm−1 is shifted to 1192 and almost disappears after the Fe3+ coordination. All these data support iron coordination with hydroxamic acid.55
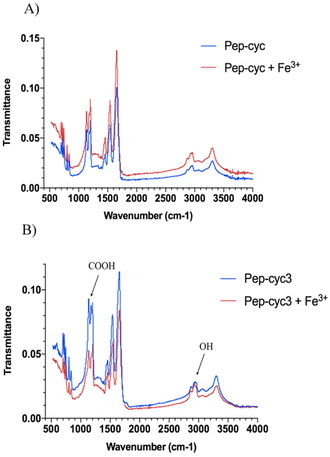 | ||
| Fig. 7 IR spectra of peptides Pep-cyc (panel A) and Pep-cyc3 (panel B) alone and their complexes with Fe3+. | ||
Circular dichroism (CD) spectroscopy
The peptide Pep-cyc adopts an α-helix structure along the residues 5–13 in the presence of micelles mimicking the bacterial membrane. We evaluated the effect of the metal binding on the secondary structure by using CD spectroscopy (Fig. 8).56 Each peptide alone in MeOH adopts an α-helix conformation as the peptide Pep-cyc without iron chelator. The helical content for each peptide increases upon increasing Fe3+ concentrations, as evidenced by Fig. 8. Regardless of the linker length, a significant increase in the helical content was observed at the Fe3+/peptide ratio of 0.3 for each peptide as detected by CD spectra.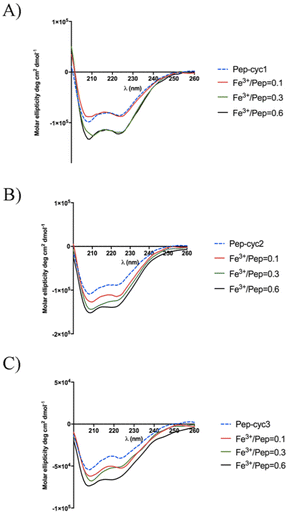 | ||
| Fig. 8 CD spectra of the titration of peptides with Fe3+ in the Fe3+/peptide ratios of 0.1, 0.3 and 0.6. | ||
CD spectroscopy was also used to explore the Cotton effect of the Fe3+–peptide complexes in the near UV and visible regions. Inspection of the CD spectra in the range 300–400 nm showed the absence of any peak (data not shown) suggesting the presence of a racemic mixture of the two enantiomeric forms lambda (Λ) and delta (Δ) that are typical of metal tris chelates.
NMR spectroscopy
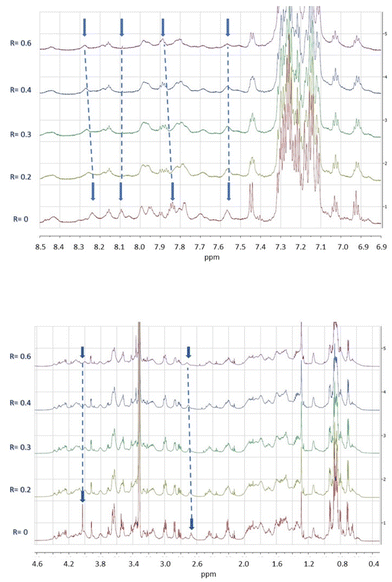
Among the peptides, Pep-cyc3 with the longest linker-bearing hydroxamate unit was analysed also by NMR. The measurements were performed in MeOH, the best solvent system among those tested. The analysis allowed us to verify the structure of the construct. The observation of the NOE effect between the δNH proton of Q10 and the εε′ protons of K6 confirms the condensation between the residue side chains and thus the cyclization of the [K6KFlQ10] segment. The linking to the C-terminal residue L13 of the tail formed by Ahx and PEG2 was verified by the presence of signals which, in terms of pattern and chemical shift values, correspond to those expected for them. The assignment of the terminal hydroxamate moiety, complicated by the unobservability of the NH-OH protons due to the chemical exchange phenomena (and tautomerism), was based on the presence of a singlet at 4.03 ppm consistent with the chemical shift expected for the hydroxamate adjacent CH2 protons. The line width and the scarcity of the NOE effects hindered the definition of the conformational organization of the peptide while suggesting aggregation phenomena at the concentrations used. Once confirmed that the structure of Pep-cyc3 corresponds to the design one, the aim of the investigation was also to test the binding properties of the system towards metal ions. In the first phase of the study, it was decided to evaluate the response of the peptide to the diamagnetic Zn2+ ion. The 1D and 2D spectra of the peptide were acquired in the presence of increasing amounts of Zn2+, up to a Zn2+/Pep-cyc3 ratio equal to 0.6 (Fig. 9). The spectral changes produced by the addition of metal ion are observable in Fig. 8. The mainly affected signals are those of the amide proton αNH of F8 (8.23 ppm), εNH of K6 (7.84 ppm), NH of PEG2 (8.09 ppm), 6-NH (7.56 ppm) of the amino-hexanoic acid segment, CH2 adjacent to the hydroxamate moiety (4.03 ppm), and δδ′ CH2 of K6 (2.71 ppm).The sensitivity of proton signals belonging to the C-terminal tail and the cycle [K6KFlQ10] to metal addition suggests that both the tail and the cycle may be sites of metal coordination. The identification of the tail site agrees with the results obtained through other techniques. The possibility that the cyclic structure could also function as a ligand is only observed by NMR. This is in line with the higher peptide concentration needed for this technique compared to that useful for the others, and possibly with the use of Zn2+ as an ersatz ion for the target paramagnetic Fe3+. However, it is interesting to note that since the addition of zinc ions to the peptide solution does not perturb all signals of the proton spectra to the same degree, thus indicating the occurrence of a region-specific interaction. The signals most affected by metal ions are those belonging to the most interacting peptide surfaces. We found that the diamagnetic analyses reported here confirm the binding properties of Pep-cyc3 and successfully localize the major metal ion binding site. The NMR data of Pep-cyc3 with Zn(II) diamagnetic ions have surprisingly shown that the adduct may room two coordination sites. This is a relevant piece of information for efficiently planning the NMR work with the paramagnetic Pep-cyc3/Fe(III) system.
Hydroxamate-based peptides inhibit bacterial growth
The metal binding of hydroxamate-based peptides was evaluated in vitro by determining the minimum inhibitory concentration (MIC) in the presence of iron-supplemented MHII agar (MHII + Fe3+) and in the absence of Fe3+ (MHII − Fe3+). In this study, the inhibition of the bacterial growth of two Gram-negative strains, E. coli ATCC 25922 and K. pneumoniae ATCC 13883, was evaluated (Table 2).| Bacteria | Pep-cyc | Pep-cyc1 | Pep-cyc2 | Pep-cyc3 | ||||
|---|---|---|---|---|---|---|---|---|
| MHII − Fe3+ | MHII + Fe3+ | MHII − Fe3+ | MHII + Fe3+ | MHII − Fe3+ | MHII + Fe3+ | MHII − Fe3+ | MHII + Fe3+ | |
| E. coli ATCC 25922 | 25 | >25 | >25 | 12.5 | >25 | 12.5 | >25 | 25 |
| K. pneumoniae ATCC 13883 | 12.5 | 12.5 | 12.5 | 3.12 | 12.5 | 3.12 | 25 | 6.25 |
The MIC values of peptide Pep-cyc were the same in the presence and in the absence of ferric iron confirming that the peptide did not chelate the metal. The conjugation of the hydroxamate chelating unit did not induce a significant modification in the activity of the native peptides as shown by the results obtained in the presence of MHII − Fe3+, characterized by the absence of iron. The MIC values against K. pneumoniae became remarkably lower in MHII + Fe3+, especially for the peptide Pep-cyc3 bearing the longest linker. In fact, Pep-cyc3 had a high MIC value of 25 μM in the absence of the metal, while a significant decrease in its MIC value (6.25 μM) was calculated in MHII + Fe3+ attributed to its strong affinity and ability to chelate Fe3+. Moreover, a reduction in the MIC value (3.12 μM) on K. pneumoniae was also observed for the peptide Pep-cyc2 featured by a linker of intermediate length and ability to chelate Fe3+ with the formation of a FeL3 complex as showed in UV measurements. Regarding the peptide Pep-cyc1 bearing the shortest linker and endowed with a lower capacity to chelate the metal (as shown by the KD), there was also a significant activity with a MIC of 3.12 μM in the presence of metal, indicating that the amount of iron subtracted from the bacteria is sufficient to induce an increase in activity.
However, we also observed a reduction in the MIC values of E. coli for all peptides in the presence of MHII + Fe3+, but they were less significant than those obtained for K. pneumoniae.
Overall, the reduction in the MIC values of hydroxamate-based peptides both on K. pneumoniae and E. coli is clearly due to their coordination of Fe3+ that negatively influences bacterial growth through the sequestration of available ferric iron away from bacteria.
Conclusions
Siderophore conjugation to antibacterial molecules represents a challenging research area to generate new molecules to treat Gram-negative infections. The presence of LPS in their outer membranes acts as a complex barrier against the penetration and diffusion of antibiotics into Gram-negative bacterial cells, making the development of new antibiotics more challenging. This work describes the effectiveness to conjugate small iron-chelator units to enhance the antimicrobial activity of AMPs against some Gram-negative pathogens. Here, we designed and synthesized three cyclic peptides Pep-cyc1, Pep-cyc2, and Pep-cyc3 by introducing in their C-terminus the hydroxamate siderophore bound to three linkers of lengths 7, 12, and 16 atoms, respectively.The UV-vis titration of each peptide with a solution of Fe3+ evidenced the presence of weak and broad absorption band complexes. By plotting the fraction of iron-bound peptide as a function of Fe3+ concentration, we observed that the complex formation occurs in a stepwise process. Moreover, we determined the formation of FeL3 complexes for the peptides Pep-cyc2 and Pep-cyc3 endowed with longer linkers, while we observed the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Fe3+–peptide for Pep-cyc1 with the shortest linker. Through the Hill equation, we also calculated KD values for the Fe3+–Pep-cyc2 and Fe3+–Pep-cyc3 complexes of 63 μM and 57 μM, respectively, while a higher KD of 160 μM was obtained for the Fe3+–Pep-cyc1 complex. Furthermore, we obtained a Hill coefficient of 2 for all the peptides suggesting a cooperative effect that could occur between two donor oxygen atoms of the hydroxamate group during the complex formation, where we supposed that the coordination of the carbonyl oxygen facilitates the deprotonation and, consequently, the binding of the carboxylate oxygen atom. The coordination of the hydroxamate unit was further confirmed by FTIR, where the frequency of the carbonyl group at 1178 cm−1 is shifted at 1192, while the signal of the hydroxyl group is shifted from 2933 to 2927 cm−1. In addition, in the CD spectra recorded in the visible region, no Cotton effect was observed during the complex formation and we hypothesized the formation of a racemic mixture of the enantiomeric forms Λ and Δ.
1 Fe3+–peptide for Pep-cyc1 with the shortest linker. Through the Hill equation, we also calculated KD values for the Fe3+–Pep-cyc2 and Fe3+–Pep-cyc3 complexes of 63 μM and 57 μM, respectively, while a higher KD of 160 μM was obtained for the Fe3+–Pep-cyc1 complex. Furthermore, we obtained a Hill coefficient of 2 for all the peptides suggesting a cooperative effect that could occur between two donor oxygen atoms of the hydroxamate group during the complex formation, where we supposed that the coordination of the carbonyl oxygen facilitates the deprotonation and, consequently, the binding of the carboxylate oxygen atom. The coordination of the hydroxamate unit was further confirmed by FTIR, where the frequency of the carbonyl group at 1178 cm−1 is shifted at 1192, while the signal of the hydroxyl group is shifted from 2933 to 2927 cm−1. In addition, in the CD spectra recorded in the visible region, no Cotton effect was observed during the complex formation and we hypothesized the formation of a racemic mixture of the enantiomeric forms Λ and Δ.
Our UV-vis and NMR data strongly support the antimicrobial results obtained in vitro on some Gram-negative strains, demonstrating the key role played by the efficacious metal coordination of all our hydroxamate-based peptides. In particular, in the presence of MHII + Fe3+, peptides Pep-cyc2 and Pep-cyc3 having better affinity towards Fe3+ caused the inhibition of the growth of K. pneumoniae and E. coli with MIC values ranging from 6.25 to 3.12 μM in comparison with those obtained in the absence of Fe3+.
In conclusion, siderophore conjugation may represent a valid opportunity for improving the activity and selectivity of some AMPs used for the treatment of infectious diseases caused especially by Gram-negative pathogens, acting as potential molecules to add to our current antibiotic arsenal.
Author contributions
The manuscript was written through the contributions of all authors. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Rosa Bellavita (R. B.) was supported by Fondazione Umberto Veronesi. R. B. thanks the Department of Pharmacy for supporting this work in the frame of the Department of Excellence. This work was partly supported by MISE-Contratto di Sviluppo (CdS) “Altergon Italia” project 000463. This research was funded by FRA (Finanziamento della Ricerca di Ateneo (FRA) linea B_2021), University of Naples, “Federico II”.References
- E. R. Frawley and F. C. Fang, Mol. Microbiol., 2014, 93, 609–616 CrossRef CAS PubMed.
- V. Braun and K. Hantke, Curr. Opin. Chem. Biol., 2011, 15, 328–334 CrossRef CAS PubMed.
- T. Goswami, A. Rolfs and M. A. Hediger, Biochem. Cell Biol., 2002, 80, 679–689 CrossRef CAS PubMed.
- P. L. Carver, Curr. Med. Chem., 2018, 25, 85–96 CrossRef CAS PubMed.
- K. N. Raymond, B. E. Allred and A. K. Sia, Acc. Chem. Res., 2015, 48, 2496–2505 CrossRef CAS PubMed.
- T. Zheng and E. M. Nolan, Metallomics, 2012, 4, 866–880 CrossRef CAS PubMed.
- B. C. Chu, A. Garcia-Herrero, T. H. Johanson, K. D. Krewulak, C. K. Lau, R. S. Peacock, Z. Slavinskaya and H. J. Vogel, Biometals, 2010, 23, 601–611 CrossRef CAS PubMed.
- J. Kramer, Ö. Özkaya and R. Kümmerli, Nat. Rev. Microbiol., 2020, 18, 152–163 CrossRef CAS PubMed.
- R. C. Hider and X. Kong, Nat. Prod. Rep., 2010, 27, 637–657 RSC.
- M. Miethke and M. A. Marahiel, Microbiol. Mol. Biol. Rev., 2007, 71, 413–451 CrossRef CAS PubMed.
- I. J. Schalk, M. Hannauer and A. Braud, Environ. Microbiol., 2011, 13, 2844–2854 CrossRef CAS PubMed.
- M. X. Zhang, C. F. Zhu, Y. J. Zhou, X. L. Kong, R. C. Hider and T. Zhou, Chem. Biol. Drug Des., 2014, 84, 659–668 CrossRef CAS PubMed.
- M. G. P. Page, Clin. Infect. Dis., 2019, 69, S529–S537 CrossRef CAS PubMed.
- R. C. Hider and X. Kong, Nat. Prod. Rep., 2010, 27, 637–657 RSC.
- G. Centola, F. Xue and A. Wilks, Metallomics, 2020, 12, 1863–1877 CrossRef CAS PubMed.
- S. A. Cotton, J. Coord. Chem., 2018, 71, 3415–3443 CrossRef CAS.
- H. Boukhalfa, S. D. Reilly, R. Michalczyk, S. Iyer and M. P. Neu, Inorg. Chem., 2006, 45, 5607–5616 CrossRef CAS PubMed.
- T. Palanché, S. Blanc, C. Hennard, M. A. Abdallah and A. M. Albrecht-Gary, Inorg. Chem., 2004, 43, 1137–1152 CrossRef PubMed.
- K. H. Negash, J. K. S. Norris and J. T. Hodgkinson, Molecules, 2019, 24, 3314 CrossRef CAS PubMed.
- A. C. Gomes, A. C. Moreira, G. Mesquita and M. S. Gomes, Pharmaceuticals, 2018, 11, 84 CrossRef CAS PubMed.
- S. Zhao, Z. P. Wang, X. Wen, S. Li, G. Wei, J. Guo and Y. He, Org. Lett., 2020, 22, 6632–6636 CrossRef CAS PubMed.
- R. Zhang, L. Xu and C. Dong, Protein Pept. Lett., 2022, 29, 641–650 CrossRef PubMed.
- V. Del Genio, A. Falanga, E. Allard-Vannier, K. Hervé-Aubert, M. Leone, R. Bellavita, R. Uzbekov, I. Chourpa and S. Galdiero, Pharmaceutics, 2022, 14, 1235 CrossRef CAS PubMed.
- C. Zhang and M. Yang, Antibiotics, 2022, 11, 349 CrossRef CAS PubMed.
- J. Talapko, T. Meštrović, M. Juzbašić, M. Tomas, S. Erić, L. Horvat Aleksijević, S. Bekić, D. Schwarz, S. Matić, M. Neuberg and I. Škrlec, Antibiotics, 2022, 11, 1417 CrossRef CAS PubMed.
- A. Maione, R. Bellavita, E. de Alteriis, S. Galdiero, L. Albarano, A. La Pietra, M. Guida, E. Parrilli, C. D'Angelo, E. Galdiero and A. Falanga, Int. J. Mol. Sci., 2022, 23, 2151 CrossRef CAS PubMed.
- A. Falanga, A. Maione, A. La Pietra, E. de Alteriis, S. Vitale, R. Bellavita, R. Carotenuto, D. Turrà, S. Galdiero, E. Galdiero and M. Guida, Pharmaceutics, 2022, 14, 1167 CrossRef CAS PubMed.
- R. Oliva, M. Chino, K. Pane, V. Pistorio, A. De Santis, E. Pizzo, G. D'Errico, V. Pavone, A. Lombardi, P. Del Vecchio, E. Notomista, F. Nastri and L. Petraccone, Sci. Rep., 2018, 8, 8888 CrossRef PubMed.
- K. A. Parsels, K. A. Mastro, J. M. Steele, S. J. Thomas and W. D. Kufel, J. Antimicrob. Chemother., 2021, 76, 1379–1391 CrossRef CAS PubMed.
- J. H. Boyce, B. Dang, B. Ary, Q. Edmondson, C. S. Craik, W. F. DeGrado and I. B. Seiple, J. Am. Chem. Soc., 2020, 142, 21310–21321 CrossRef CAS PubMed.
- D. Y. Kim and H. J. Kim, Org. Lett., 2021, 23, 5256–5260 CrossRef CAS.
- J. Oh, D. Kang, S. Hong, S. H. Kim, J. H. Choi and J. Seo, Dalton Trans., 2021, 50, 3459 RSC.
- R. Bellavita, B. Casciaro, S. Di Maro, D. Brancaccio, A. Carotenuto, A. Falanga, F. Cappiello, E. Buommino, S. Galdiero, E. Novellino, T. N. Grossmann, M. L. Mangoni, F. Merlino and P. Grieco, J. Med. Chem., 2021, 64, 11675–11694 CrossRef CAS PubMed.
- M. L. Mangoni, Cell. Mol. Life Sci., 2006, 63, 1060–1069 CrossRef CAS PubMed.
- S. M. Romero, A. B. Cardillo, M. C. Martínez Ceron, S. A. Camperi and S. L. Giudicessi, Surg. Infect., 2020, 21, 309–322 CrossRef PubMed.
- R. Bellavita, A. Vollaro, M. R. Catania, F. Merlino, L. De Martino, F. P. Nocera, M. DellaGreca, F. Lembo, P. Grieco and E. Buommino, Antibiotics, 2020, 9, 530 CrossRef CAS PubMed.
- E. Roscetto, R. Bellavita, R. Paolillo, F. Merlino, N. Molfetta, P. Grieco, E. Buommino and M. R. Catania, Antibiotics, 2021, 10, 1312 CrossRef CAS PubMed.
- C. Zannella, A. Chianese, L. Palomba, M. E. Marcocci, R. Bellavita, F. Merlino, P. Grieco, V. Folliero, A. De Filippis, M. L. Mangoni, L. Nencioni, G. Franci and M. Galdiero, Int. J. Mol. Sci., 2022, 23, 2060 CrossRef CAS PubMed.
- R. Bellavita, A. Falanga, F. Merlino, G. D'Auria, N. Molfetta, A. Saviano, F. Maione, U. Galdiero, M. R. Catania, S. Galdiero, P. Grieco, E. Roscetto, L. Falcigno and E. Buommino, J. Enzyme Inhib. Med. Chem., 2023, 38, 36–50 CrossRef CAS PubMed.
- R. Bellavita, A. Maione, F. Merlino, A. Siciliano, P. Dardano, L. De Stefano, S. Galdiero, E. Galdiero, P. Grieco and A. Falanga, Pharmaceutics, 2022, 14, 454 CrossRef CAS PubMed.
- Y. Sixto-López, J. A. Gómez-Vidal, N. de Pedro, M. Bello, M. C. Rosales-Hernández and J. Correa-Basurto, Sci. Rep., 2020, 10, 10462 CrossRef PubMed.
- A. M. Yousif, V. Ingangi, F. Merlino, D. Brancaccio, M. Minopoli, R. Bellavita, E. Novellino, M. V. Carriero, A. Carotenuto and P. Grieco, Eur. J. Med. Chem., 2018, 143, 348–360 CrossRef CAS PubMed.
- D. M. Al Shaer, F. Albericio and B. G. de la Torre, ChemistrySelect, 2021, 6, 7674–7681 CrossRef CAS.
- Y. Yang, J. Liu, L. Yang, K. Li, H. Zhang, S. Luo and L. Rao, Dalton Trans., 2015, 44, 8959–8970 RSC.
- G. De Tommaso, M. M. Salvatore, R. Nicoletti, M. DellaGreca, F. Vinale, A. Staropoli, F. Salvatore, M. Lorito, M. Iuliano and A. Andolfi, Toxics, 2021, 9, 19 CrossRef CAS PubMed.
- E. Orlowska, A. Roller, H. Wiesinger, M. Pignitter, F. Jirsa, R. Krachler, W. Kandioller and B. K. Kepplera, RSC Adv., 2016, 6, 40238–40249 RSC.
- T. A. Wencewicz, T. E. Long, U. Möllmann and M. J. Miller, Bioconjugate Chem., 2013, 24, 473–486 CrossRef CAS PubMed.
- CLSI, Methods for diluition antimicrobial susceptibility tests for bacteria that grow aerobically; approved standards-6, in Document m7-a6 Performance Standards for Antimicrobial Susceptibility Testing, CLSI, Wayne PA, USA, 2006 Search PubMed.
- E. de Alteriis, A. Maione, A. Falanga, R. Bellavita, S. Galdiero, L. Albarano, M. M. Salvatore, E. Galdiero and M. Guida, Antibiotics, 2021, 11, 26 CrossRef PubMed.
- A. Garénaux, M. Caza and C. M. Dozois, Vet. Microbiol., 2011, 153, 89–98 CrossRef PubMed.
- C. J. Carrano, H. Drechsel, D. Kaiser, G. Jung, B. Matzanke, G. Winkelmann, N. Rochel and A. M. Albrecht-Gary, Inorg. Chem., 1996, 35, 6429–6436 CrossRef CAS PubMed.
- F. Garzón-Posse, Y. Quevedo-Acosta, C. Mahecha-Mahecha and P. Acosta-Guzmán, Eur. J. Org. Chem., 2019, 48, 7747–7769 CrossRef.
- J. Keth, T. Johann and H. Frey, Biomacromolecules, 2020, 21, 2546–2556 CrossRef CAS PubMed.
- T. Inomata, H. Eguchi, Y. Funahashi, T. Ozawa and H. Masuda, Langmuir, 2012, 28, 1611–1617 CrossRef CAS PubMed.
- L. R. Hassan, E. H. Anouar, H. Bahron, F. Abdullah and A. Mohd Tajuddin, J. Biol. Inorg. Chem., 2020, 25, 239–252 CrossRef CAS PubMed.
- S. Futaki, T. Kiwada and Y. Sugiura, J. Am. Chem. Soc., 2004, 126, 15762–15769 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2dt04099a |
| This journal is © The Royal Society of Chemistry 2023 |