Open Access Article
Open Access ArticleImportance of isothermal titration calorimetry for the detection of the direct binding of metal ions to mismatched base pairs in duplex DNA†
Hidetaka
Torigoe
 * and
Fumihiro
Arakawa
* and
Fumihiro
Arakawa
Department of Applied Chemistry, Faculty of Science, Tokyo University of Science, 1-3 Kagurazaka, Shinjuku-ku, Tokyo 162-8601, Japan. E-mail: htorigoe@rs.tus.ac.jp; Fax: +81-3-5261-4631; Tel: +81-3-5228-8259
First published on 25th August 2023
Abstract
Metal ion–nucleic acid interactions contribute substantially to the structure and biological activity of nucleic acids and have a wide range of potential applications in nanotechnology. In this study, we examined the interactions between metal ions and mismatched base pairs in duplex DNA to reveal the underlying molecular mechanism. UV melting analyses showed that the melting temperature (Tm) of a 21-base pair duplex DNA with each of the C–A, C–C and C–T mismatched base pairs increased upon the addition of Ag+. However, isothermal titration calorimetry (ITC) demonstrated that Ag+ only bound to the C–C mismatched base pair of the duplex DNA to form C–Ag–C bonds, without binding to the C–A and C–T mismatches. These results showed that Tm increased even when metal ions did not bind to the mismatched base pairs of the duplex DNA. Although the increase in Tm upon the addition of the metal ions is often used to detect metal ion binding to mismatched base pairs of duplex DNA, these results indicated that UV melting analyses are unable to detect the direct binding of metal ions to the mismatched base pairs. Because ITC analyses directly detect the heat derived from metal ion binding to mismatched base pairs of duplex DNA, we concluded that this may be an effective detection approach.
Introduction
Binding between metal ions and nucleic acids contributes to the structure and folding of nucleic acids, attenuation of electrostatic repulsion among nucleic acid phosphate backbones,1–3 and the biological functions of nucleic acids, such as enzymatic activity of ribozymes and DNAzymes.4,5 This binding also has a wide range of potential applications for nanotechnology, such as the development of biomolecular nanomaterials, nanomachines and nanodevices.6–8The structural and thermodynamic properties of binding to duplex DNA with only perfectly matched base pairs have been studied for many metal ions, such as Cr3+,9 Cr6+,9 Tl+,10 Fe2+,11 Fe3+,11 Al3+,12 Cd2+,13 and Mn2+.14 Recently, the binding between a metal ion, Hg2+, and duplex DNA with a mismatched base pair has also been analyzed.15–22 UV melting analyses have shown that the melting temperature (Tm) of duplex DNA with a T–T mismatched base pair increased significantly in response to Hg2+ addition,15,16,18 with no significant change for the corresponding duplex DNA with perfectly matched base pairs or other mismatched base pairs.18 No significant increase in the Tm of the duplex DNA with the T–T mismatched base pair was observed upon the addition of other metal ions (Mg2+, Ca2+, Mn2+, Fe2+, Fe3+, Co2+, Ni2+, Cu2+, Zn2+, Ru3+, Pd2+, Ag+, Cd2+, and Pb2+).15,16 Thus, the increase in the Tm of the duplex DNA with the T–T mismatch upon the addition of Hg2+ was quite specific. The Tm value of the duplex DNA with the T–T mismatched base pair upon the addition of Hg2+ was comparable to that with the corresponding T–A or A–T perfectly matched base pair. Isothermal titration calorimetry (ITC)23 has shown that Hg2+ binds directly to the T–T mismatched base pair in duplex DNA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a binding constant on the order of 105 M−1 to form a T–Hg–T base pair.18,20 The direct binding of Hg2+ increased the Tm value of the duplex DNA with the T–T mismatched base pair.
1 with a binding constant on the order of 105 M−1 to form a T–Hg–T base pair.18,20 The direct binding of Hg2+ increased the Tm value of the duplex DNA with the T–T mismatched base pair.
The interaction between another metal ion, Ag+, and duplex DNA with a mismatched base pair has been examined.19,22,24–30 The increase in the Tm of a mismatched base pair duplex DNA was observed upon Ag+ addition.24,26,30 The Tm values of duplex DNA with each of the C–A, C–C and C–T mismatched base pairs increased upon the addition of Ag+, although the magnitude of the increase was the highest in the case of the C–C mismatched base pair. The increase in the Tm is often used to detect the direct binding of metal ions to mismatched base pairs.31–38 However, it is not clear whether the increase in the Tm values of duplex DNA with the C–A, C–C or C–T mismatched base pairs upon the addition of Ag+ indicates the direct binding of the metal ions to the mismatched base pairs. In the present study, we performed ITC analyses to examine whether Ag+ directly binds to each of the C–A, C–C and C–T mismatched base pairs in duplex DNA to form C–Ag–A, C–Ag–C and C–Ag–T metal-mediated base pairs. Unexpectedly, ITC results revealed that Ag+ is unable to bind to C–A and C–T mismatched base pairs, but binds directly to the C–C mismatched base pair in the duplex DNA to form a C–Ag–C metal-mediated base pair. We discuss the reason why Ag+ was unable to bind to the C–A and C–T mismatched base pairs in duplex DNA, despite the increase in Tm upon the addition of Ag+.
Experimental section
Preparation of oligonucleotides
We synthesized 21-mer complementary DNA oligonucleotides, F21X (5′-d(GCCCATTGGAXTGACGCTCTG)-3′) (X = A, C, G and T) (Fig. 1) and R21Y (5′-d(CAGAGCGTCAYTCCAATGGGC)-3′) (Y = A, C, G and T) (Fig. 1), and other 25-mer complementary DNA oligonucleotides, F25Z (5′-d(GCCCTGCCTGTCZCCCAGATCACTG)-3′) (Z = A, C, G and T) (Fig. 1) and R25W (5′-d(CAGTGATCTGGGWGACAGGCAGGGC)-3′) (W = A, C, G and T) (Fig. 1), using a DNA synthesizer by a solid-phase cyanoethyl phosphoramidite method. The oligonucleotides were purified by reverse-phase high-performance liquid chromatography (HPLC) on a Wakosil DNA column. The concentration of all purified oligonucleotides was determined by UV absorbance. The complementary strands, F21X (X = A, C, G and T) and R21Y (Y = A, C, G and T), and other complementary strands, F25Z (Z = A, C, G and T) and R25W (W = A, C, G and T), were annealed by heating up to 90 °C, followed by gradual cooling to room temperature. The annealed sample was applied on a hydroxyapatite column (Bio-Rad Inc., Hercules, CA, USA) to remove the unpaired single strands. The concentration of the duplex DNAs, F21X:R21Y and F25Z:R25W, was determined by UV absorption, assuming that an absorbance of 1.0 at 260 nm corresponds to 50 μg ml−1.UV melting
UV melting experiments were carried out using a DU-640 spectrophotometer (Beckman Inc., Brea, CA, USA) equipped with a Peltier cell holder. The cell path length was 1 cm. The UV melting profiles were measured in 10 mM sodium cacodylate–cacodylic acid (pH 6.8) and 100 mM NaClO4 (buffer A) without or with 1 μM AgNO3 at a scan rate of 0.2 °C min−1 with detection at 260 nm. The peak temperatures for the first derivative calculated from the UV melting profile were designated as Tm. The concentration of the duplex DNAs, F21X:R21Y and F25Z:R25W, was 1 μM. Since the water solubility of AgCl is very low, buffers containing NaCl are not suitable for the present study involving Ag+. Instead, a buffer containing NaClO4 (buffer A) was chosen in the present study due to the high water solubility of AgClO4.CD spectroscopy
CD spectra were recorded at 25 °C in buffer A (see the section UV melting) without or with 1 μM AgNO3 using a JASCO J-725 spectropolarimeter interfaced with a microcomputer. The cell path length was 1 cm. The concentration of the duplex DNAs, F21X:R21Y and F25Z:R25W, was 1 μM.Isothermal titration calorimetry (ITC)
ITC experiments were carried out using the Microcal ITC200 system (Malvern Inc., Malvern, UK). The duplex DNA solutions were prepared by extensive dialysis against buffer A (see the section UV melting). AgNO3 was dissolved in the dialysis buffer. The AgNO3 solution in buffer A was injected 20 times in 2 μl increments at 3 min intervals at 25 °C into the duplex DNA solutions without changing the reaction conditions. The heat for each injection was subtracted by the heat of dilution of the injectant, which was measured by injecting the AgNO3 solution into the same buffer. Each corrected heat value was divided by the moles of AgNO3 injected and analyzed using Microcal Origin supplied by the manufacturer.Results
UV melting analyses of duplex DNA containing each single mismatched base pair and the corresponding perfectly matched base pair without or with Ag+
Previous studies have reported that the Tm values of duplex DNA with each of the C–A, C–C and C–T mismatched base pairs increased upon the addition of Ag+, although the magnitude of the increase was the highest in the case of the C–C mismatched base pair.24 To examine whether the similar increase of the Tm values in the case of C–A, C–C and C–T mismatched base pairs upon the addition of Ag+ is observed in the other base sequences, the thermal stability of a series of duplex DNAs, F21X:R21Y (X–Y = C–A, C–C, C–G and C–T) (Fig. 1), was examined in 10 mM sodium cacodylate–cacodylic acid (pH 6.8) and 100 mM NaClO4 (buffer A) either without or with AgNO3 by UV melting (Table 1). Without AgNO3, the Tm value of the F21C:R21G duplex DNA (68.9 °C) with the perfectly matched C–G was significantly higher than those of the F21C:R21A, F21C:R21C, and F21C:R21T duplex DNAs with a single mismatched base pair (Table 1). The addition of AgNO3 significantly increased the Tm values of all duplex DNA samples. It should be noted that the magnitude of the increase in Tm by the addition of AgNO3 was the highest in the case of F21C:R21C (Table 1). These results indicated that the thermal stability of duplex DNA increased substantially and the F21C:R21C duplex DNA was the most highly stabilized by the addition of Ag+.| X–Y | T m (−Ag+) (°C) | T m (+Ag+) (°C) | ΔTm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (°C) a (°C) |
T m (+2Ag+) (°C) | ΔTm2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b (°C) b (°C) |
|---|---|---|---|---|---|
| a ΔTm = Tm (+Ag+) − Tm (−Ag+). b ΔTm2 = Tm (+2Ag+) − Tm (−Ag+). | |||||
| C–A | 59.2 ± 0.1 | 61.3 ± 0.2 | 2.1 | 62.2 ± 0.3 | 3.0 |
| C–C | 57.5 ± 0.7 | 60.3 ± 0.7 | 2.8 | 63.0 ± 0.8 | 5.5 |
| C–G | 68.9 ± 0.8 | 70.0 ± 0.9 | 1.1 | 71.8 ± 0.8 | 2.9 |
| C–T | 58.7 ± 0.6 | 60.6 ± 0.8 | 1.9 | 62.5 ± 0.6 | 3.8 |
In addition, the thermal stability of a series of duplex DNAs, F21X:R21Y with the 12 other base pairs (X–Y = A–A, A–C, A–G, A–T, G–A, G–C, G–G, G–T, T–A, T–C, T–G, and T–T) (Fig. 1), was examined in buffer A either without or with AgNO3 by UV melting (Table S1†). The addition of Ag+ increased the Tm values of all duplex DNA samples. Combination of the results of Table 1 and Table S1† indicated that in all of the 16 base pairs, the case of the C–C base pair was the most highly stabilized by the addition of Ag+.
To investigate the effect of the addition of other metal ions on the thermal stability of duplex DNA, a series of duplex DNAs, F21X:R21Y with 16 base pairs (X–Y = A–A, A–C, A–G, A–T, C–A, C–C, C–G, C–T, G–A, G–C, G–G, G–T, T–A, T–C, T–G, and T–T) (Fig. 1), was examined in buffer A either without or with Mn(NO3)2, Co(NO3)2, Ni(NO3)2, Zn(NO3)2, Cd(NO3)2, TlNO3, and Pb(NO3)2 by UV melting (Tables S2–S8†). The Tm values of the duplex DNA samples with any kind of base pair were not significantly changed by the addition of Mn2+, Co2+, Ni2+, Zn2+, Cd2+, Tl+, and Pb2+.
To reveal whether the similar stabilization by the addition of Ag+ is observed in the duplex DNAs with other base sequences and lengths, the thermal stability of a series of duplex DNAs with other base sequences and lengths, F25Z:R25W (Z–W = A–A, A–C, A–G, A–T, C–A, C–C, C–G, C–T, G–A, G–C, G–G, G–T, T–A, T–C, T–G, and T–T) (Fig. 1), was examined in buffer A either without or with AgNO3 by UV melting (Table S9†). By the addition of Ag+, the Tm values of all of the 16 duplex DNA samples were increased, and the case of the C–C base pair was the most highly stabilized, which is quite similar to the results obtained from F21X:R21Y. We conclude that, although all of the duplex DNAs were stabilized by the addition of Ag+, the case of the C–C base pair was the most highly stabilized.
CD spectroscopy of duplex DNA containing each single mismatched base pair and the corresponding perfectly matched base pair without or with Ag+
To examine the effect of Ag+ on the higher-order structure of duplex DNA, the CD spectra of the two above-described series of 1 μM duplex DNA, F21X:R21Y (X–Y = C–A, C–C, C–G and C–T) (Fig. 1) and F25Z:R25W (Z–W = C–A, C–C, C–G, and C–T) (Fig. 1), were measured in buffer A (see the section UV melting) without or with 1 μM AgNO3 at 25 °C (Fig. 2 and Fig. S1†). The CD profiles of each duplex DNA with AgNO3 were quite similar to those observed without AgNO3. These results indicated that there was no significant change in the higher-order structure of all duplex DNA upon the addition of Ag+.ITC analyses of the interaction between Ag+ and each of the C–C mismatched base pair duplex DNAs and the corresponding C–G perfectly matched duplex DNAs
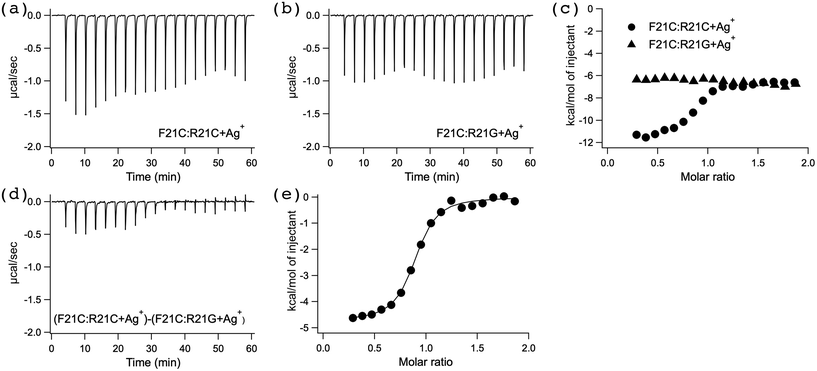
To explore the mechanism underlying the high stabilization of the F21C:R21C duplex DNA (Fig. 1) by the addition of Ag+ (Table 1), we examined the thermodynamic properties of the interaction between AgNO3 and F21C:R21C duplex DNA in buffer A (see the section UV melting) at 25 °C by ITC (Fig. 3). Fig. 3a shows a typical ITC profile of the interaction between AgNO3 and F21C:R21C. An exothermic heat pulse was observed each time AgNO3 was injected into F21C:R21C. The magnitude of each peak decreased gradually with each new injection, and a peak was still observed at the molar ratio of the last injection. The area under each peak was integrated, and the heat of the dilution of AgNO3 measured in a separate experiment by injecting AgNO3 into buffer A (see the section UV melting) was subtracted from the integrated values. The corrected heat was divided by the moles of the injected solution. The resulting values were plotted as a function of a molar ratio of [Ag+]/[F21C:R21C] (circles in Fig. 3c). The resultant titration plot was sigmoidal, indicating that Ag+ specifically bound to F21C:R21C.To explore the mechanism underlying the stabilization of the F21C:R21G duplex DNA (Fig. 1) by the addition of Ag+, we investigated the thermodynamic properties of the interaction between AgNO3 and F21C:R21G duplex DNA in buffer A (see the section UV melting) at 25 °C by ITC (Fig. 3). Fig. 3b shows a typical ITC profile for the interaction between AgNO3 and F21C:R21G. Although an exothermic heat pulse was observed after each injection of AgNO3 into F21C:R21G, the magnitude of each peak did not change significantly after each new injection, in sharp contrast with the ITC profile observed for the interaction between AgNO3 and F21C:R21C (Fig. 3a). The titration plot obtained from Fig. 3b (triangles in Fig. 3c) in the same way as that obtained from Fig. 3a (circles in Fig. 3c) was not sigmoidal, indicating that Ag+ bound nonspecifically to F21C:R21G.
The nonspecific binding between Ag+ and F21C:R21G judged from the ITC titration plots (triangles in Fig. 3c) may be explained by the electrostatic attraction between the positive charge of Ag+ and the negative charge of the DNA phosphate backbones. In contrast, the specific binding between Ag+ and F21C:R21C judged from the ITC titration plot (circles in Fig. 3c) suggests that Ag+ binds specifically to the C–C mismatched base pair of F21C:R21C in addition to the nonspecific binding between Ag+ and the DNA phosphate backbones of F21C:R21C. Thus, the net heat derived from the specific binding between Ag+ and the C–C mismatched base pair of F21C:R21C should be estimated by subtracting the heat observed for F21C:R21G from that observed for F21C:R21C. Accordingly, to analyze the thermodynamic parameters of the specific binding between Ag+ and the C–C mismatched base pair of F21C:R21C, the ITC profile observed for F21C:R21G in Fig. 3b was subtracted from that observed for F21C:R21C in Fig. 3a to obtain the profile shown in Fig. 3d. The area under each peak in Fig. 3d was integrated, and the integrated values were divided by the moles of the injected solution. The resulting values were plotted as a function of the molar ratio of [Ag+]/[duplex DNA] (Fig. 3e). The resultant titration plot was fitted to a sigmoidal curve by a nonlinear least-squares method. The stoichiometry (n), binding constant (Ka) and the enthalpy change (ΔH) for the specific binding between Ag+ and the C–C mismatched base pair were obtained from the fitted curve.23 The Gibbs free energy change (ΔG) and the entropy change (ΔS) were calculated from the equation ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = ΔH − TΔS, where R is the gas constant and T is the temperature.23
Ka = ΔH − TΔS, where R is the gas constant and T is the temperature.23
Table 2 summarizes the thermodynamic parameters for the specific binding between Ag+ and the C–C mismatched base pair, obtained from Fig. 3e. The obtained value of n was nearly 1, indicating that Ag+ bound to the C–C mismatched base pair at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Although the sign of ΔH was negative, the sign of ΔS was positive. Both the observed negative ΔH and positive ΔS were favorable for the specific binding between Ag+ and the C–C mismatched base pair. The magnitudes of the observed Ka and ΔG were significantly larger than those previously reported for the nonspecific interaction between metal ions and duplex DNA,9–14 indicating that Ag+ specifically bound to the C–C mismatched base pair in duplex DNA.
1. Although the sign of ΔH was negative, the sign of ΔS was positive. Both the observed negative ΔH and positive ΔS were favorable for the specific binding between Ag+ and the C–C mismatched base pair. The magnitudes of the observed Ka and ΔG were significantly larger than those previously reported for the nonspecific interaction between metal ions and duplex DNA,9–14 indicating that Ag+ specifically bound to the C–C mismatched base pair in duplex DNA.
| Profile | N | K a (M−1) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) |
|---|---|---|---|---|---|
| Fig. 3e | 0.96 ± 0.01 | (9.18 ± 1.17) × 105 | −8.13 ± 0.08 | −4.74 ± 0.07 | 11.4 ± 0.5 |
| Fig. S2e† | 1.06 ± 0.03 | (5.86 ± 1.29) × 105 | −7.87 ± 0.15 | −2.37 ± 0.07 | 18.4 ± 0.7 |
Furthermore, to investigate whether the similar specific binding of Ag+ to the C–C mismatched base pair is observed in the duplex DNAs with other base sequences and lengths, we examined the thermodynamic properties of the interaction between AgNO3 and each of the F25C:R25C and F25C:R25G duplex DNAs in buffer A (see the section UV melting) at 25 °C by ITC (Fig. S2†). Fig. S2a and S2b† show typical ITC profiles of the interaction between AgNO3 and each of the F25C:R25C and F25C:R25G duplex DNAs, respectively. Although the titration plot obtained from Fig. S2b† (triangles in Fig. S2c†) was not sigmoidal, that obtained from Fig. S2a† (circles in Fig. S2c†) was sigmoidal. The ITC profile observed for F25C:R25G in Fig. S2b† was subtracted from that observed for F25C:R25C in Fig. S2a† to obtain the profile shown in Fig. S2d,† which corresponds to only the specific binding between Ag+ and the C–C mismatched base pair. The titration plot obtained from Fig. S2d† (Fig. S2e†) was fitted to a sigmoidal curve by a nonlinear least-squares method to obtain the thermodynamic parameters of the specific binding between Ag+ and the C–C mismatched base pair of F25C:R25C (Table 2). The thermodynamic parameters obtained from Fig. S2e† were similar in magnitude to those obtained from Fig. 3e (Table 2). The difference of the base sequences and lengths of the C–C mismatched duplex DNA did not significantly influence the thermodynamic parameters of the specific binding between Ag+ and the C–C mismatched base pair.
ITC analyses of the interaction between Ag+ and each of the C–A and C–T mismatched base pair duplex DNAs
To examine the mechanism underlying the stabilization of the F21C:R21A and F21C:R21T duplex DNAs (Fig. 1) by the addition of Ag+ (Table 1), we investigated the thermodynamic properties of the interaction between AgNO3 and each of the F21C:R21A and F21C:R21T duplex DNAs in buffer A (see the section UV melting) at 25 °C by ITC (Fig. 4). Fig. 4a and c show typical ITC profiles for the interaction between AgNO3 and each of the F21C:R21A and F21C:R21T DNAs, respectively. Although an exothermic heat pulse was observed after each injection of AgNO3 into F21C:R21A and F21C:R21T, the magnitude of each peak did not change significantly after each new injection, in sharp contrast to the ITC profile observed for the interaction between AgNO3 and F21C:R21C (Fig. 3a). The titration plots obtained from Fig. 4a for F21C:R21A (circles in Fig. 4b) and Fig. 4c for F21C:R21T (circles in Fig. 4d), as in Fig. 3c, were not sigmoidal, and they were quite similar in magnitude to the titration plot obtained from Fig. 3b for F21C:R21G (triangles in Fig. 3c, 4b and d), indicating that Ag+ showed nonspecific binding to F21C:R21A and F21C:R21T. The nonspecific binding between Ag+ and each of the F21C:R21A and F21C:R21T DNAs judged from the ITC titration plots (circles in Fig. 4b and d) may be attributed to the electrostatic attraction between the positive charge of Ag+ and the negative charge of the DNA phosphate backbones, similar to the case of perfectly matched F21C:R21G (triangles in Fig. 3c, 4b and d). In conclusion, the ITC titration plots for the C–A (circles in Fig. 4b) and C–T (circles in Fig. 4d) mismatched duplex DNAs indicated that Ag+ may not bind to the C–A and C–T mismatched base pairs of the duplex DNA, although it may bind to the phosphate backbones of the duplex DNA. | ||
| Fig. 4 Thermodynamic analyses of the interaction between Ag+ and the C–A or C–T mismatches (F21C:R21A or F21C:R21T) or the perfectly matched (F21C:R21G) duplex DNA. (a) Typical ITC profile for the interaction between AgNO3 and F21C:R21A at 25 °C and pH 6.8 in buffer A (see the section UV melting). AgNO3 solution (800 μM in buffer A) was injected 20 times in 2 μl increments into F21C:R21A solution (80 μM in buffer A). Injections were administered over 4 s at 3 min intervals. (b) Titration plot against the molar ratio of [Ag+]/[duplex DNA], obtained from the ITC profiles for F21C:R21A in (a) and for F21C:R21G (triangles in Fig. 3c). (c) Typical ITC profile for the interaction between AgNO3 and F21C:R21T at 25 °C and pH 6.8 in buffer A (see the section UV melting). AgNO3 solution (800 μM in buffer A) was injected 20 times in 2 μl increments into F21C:R21T solution (80 μM in buffer A). Injections were administered over 4 s at 3 min intervals. (d) Titration plot against the molar ratio of [Ag+]/[duplex DNA], obtained from the ITC profiles for F21C:R21T in (c) and for F21C:R21G (triangles in Fig. 3c). | ||
Discussion
UV melting analyses showed that among mismatches, the C–C mismatched base pair (F21C:R21C and F25C:R25C) was most highly stabilized by the addition of Ag+ (Tables 1, S1 and S9†). We previously reported that a C–C mismatch in duplex DNA with another base sequence was also highly stabilized by the Ag+ addition,24,26,30 consistent with the present results. ITC analyses (Fig. 3, Fig. S2,† and Table 2) showed that Ag+ may specifically bind to the C–C mismatched base pair of the duplex DNA in addition to nonspecific binding between Ag+ and the DNA phosphate backbones of the duplex DNA, although only the nonspecific binding between Ag+ and the DNA phosphate backbones was observed for the other duplex DNA. The two types of the binding may explain the high stability of the C–C mismatch in duplex DNAs (F21C:R21C and F25C:R25C) upon the addition of Ag+. These results demonstrated that Ag+ may stabilize the C–C mismatched base pair in duplex DNA by direct binding to the mismatched base pair.ITC analyses of the interaction between Ag+ and the C–C mismatched base pairs (F21C:R21C and F25C:R25C) revealed that Ag+ bound to the C–C mismatched base pair in the duplex DNA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 3, Fig. S2,† and Table 2). The Ka and ΔG values for the specific binding between Ag+ and the C–C mismatched base pair were nearly 106 M−1 and −8.13 or −7.87 kcal mol−1, respectively (Table 2). These values were significantly greater than previously reported estimates for the nonspecific interaction between metal ions and DNA,9–14 supporting the specific binding between Ag+ and the C–C mismatched base pair. The observed ΔG resulted from both the negative ΔH and positive ΔS (Table 2). The positive ΔS for the specific binding between Ag+ and the C–C mismatched base pair measured by ITC (Table 2) can be explained, in large part, by a positive dehydration entropy change from the release of structured water molecules surrounding Ag+ and the duplex DNA and a conformational entropy change derived from the conformational change of the duplex DNA upon binding to Ag+. The CD spectra showed that the higher-order structure of the duplex DNA was not significantly distorted by the specific binding of Ag+ (Fig. 2), suggesting that the conformational entropy change did not contribute substantially to the observed positive ΔS (Table 2). Thus, the positive ΔS (Table 2) may result mainly from the positive dehydration entropy change from the release of structured water molecules surrounding Ag+ and the duplex DNA. In fact, a previously reported positive dehydration entropy change of Ag+ (18 cal mol−1 K−1)39 was similar in magnitude to the observed positive ΔS (Table 2). The negative ΔH for the specific binding between Ag+ and the C–C mismatched base pair measured by ITC (Table 2) reflects the positive dehydration enthalpy change of Ag+,40 and a negative binding enthalpy change derived from the bond formation between Ag+ and the Ag+ binding positions in the two cytosine bases to form the C–Ag–C metal-mediated base pair. Since the sign of the binding enthalpy change upon bond formation was negative and the sign of the dehydration enthalpy change was positive, the observed negative ΔH (Table 2) might have been driven mainly by the negative binding enthalpy change upon the bond formation of the C–Ag–C metal-mediated base pair. Based on these findings, we propose a possible scheme for the specific binding between Ag+ and the C–C mismatched base pair (Fig. 5). Ag+ surrounded by structured water molecules may be dehydrated, and this may be closely related to the positive dehydration entropy change. The dehydrated Ag+ may bind to the two cytosine bases to form the C–Ag–C metal-mediated base pair, and this may be closely related to the negative binding enthalpy change.
1 (Fig. 3, Fig. S2,† and Table 2). The Ka and ΔG values for the specific binding between Ag+ and the C–C mismatched base pair were nearly 106 M−1 and −8.13 or −7.87 kcal mol−1, respectively (Table 2). These values were significantly greater than previously reported estimates for the nonspecific interaction between metal ions and DNA,9–14 supporting the specific binding between Ag+ and the C–C mismatched base pair. The observed ΔG resulted from both the negative ΔH and positive ΔS (Table 2). The positive ΔS for the specific binding between Ag+ and the C–C mismatched base pair measured by ITC (Table 2) can be explained, in large part, by a positive dehydration entropy change from the release of structured water molecules surrounding Ag+ and the duplex DNA and a conformational entropy change derived from the conformational change of the duplex DNA upon binding to Ag+. The CD spectra showed that the higher-order structure of the duplex DNA was not significantly distorted by the specific binding of Ag+ (Fig. 2), suggesting that the conformational entropy change did not contribute substantially to the observed positive ΔS (Table 2). Thus, the positive ΔS (Table 2) may result mainly from the positive dehydration entropy change from the release of structured water molecules surrounding Ag+ and the duplex DNA. In fact, a previously reported positive dehydration entropy change of Ag+ (18 cal mol−1 K−1)39 was similar in magnitude to the observed positive ΔS (Table 2). The negative ΔH for the specific binding between Ag+ and the C–C mismatched base pair measured by ITC (Table 2) reflects the positive dehydration enthalpy change of Ag+,40 and a negative binding enthalpy change derived from the bond formation between Ag+ and the Ag+ binding positions in the two cytosine bases to form the C–Ag–C metal-mediated base pair. Since the sign of the binding enthalpy change upon bond formation was negative and the sign of the dehydration enthalpy change was positive, the observed negative ΔH (Table 2) might have been driven mainly by the negative binding enthalpy change upon the bond formation of the C–Ag–C metal-mediated base pair. Based on these findings, we propose a possible scheme for the specific binding between Ag+ and the C–C mismatched base pair (Fig. 5). Ag+ surrounded by structured water molecules may be dehydrated, and this may be closely related to the positive dehydration entropy change. The dehydrated Ag+ may bind to the two cytosine bases to form the C–Ag–C metal-mediated base pair, and this may be closely related to the negative binding enthalpy change.
UV melting analyses showed that the addition of Ag+ increased the Tm of the C–A (F21C:R21A) and C–T (F21C:R21T) mismatched base pairs in duplex DNA (Table 1). These findings were consistent with our previous analyses, showing that the C–A and C–T mismatches in duplex DNA with other base sequences were also highly stabilized by the Ag+ addition.30 The increase in the Tm of duplex DNA with mismatched base pairs upon the addition of metal ions frequently results from direct binding of the metal ions to the mismatched base pair. In fact, Tm for the T–T mismatch in duplex DNA increased upon the addition of Hg2+ due to direct binding.15,16,18 However, in ITC analyses of the interaction between Ag+ and the C–A and C–T mismatches, the titration plots for F21C:R21A (circles in Fig. 4b) and for F21C:R21T (circles in Fig. 4d) were quite similar in magnitude to that for the perfectly matched duplex DNA (F21C:R21G) (triangles in Fig. 3c, 4b and d). These results indicate that the positive charge of Ag+ may bind to the negative charge of the phosphate backbones of F21C:R21A and F21C:R21T in a nonspecific manner without the specific binding to the C–A and C–T mismatches of F21C:R21A and F21C:R21T. The nonspecific interaction between the positive charge of Ag+ and the negative charge of the phosphate backbones of F21C:R21A and F21C:R21T may increase the Tm value and the thermal stability of F21C:R21A and F21C:R21T.
UV melting analyses are often used to examine the binding of metal ions to the mismatched base pair in duplex DNA.31–38 The increase in Tm observed by UV melting analyses may result from stabilization by the two types of the binding: (1) the specific binding between metal ions and mismatched base pairs and (2) the nonspecific binding between the positive charge of metal ions and the negative charge of DNA phosphate backbones of the duplex DNA. Since UV melting analyses of the change in the thermal stability of the duplex DNA are unable to capture the direct binding between metal ions and mismatches in duplex DNA, they cannot be used to discriminate between the two types of the binding. On the other hand, because ITC analyses directly detect the heat derived from binding,23 they can be used to discriminate between the two types of binding. Accordingly, we conclude that ITC analyses may be more effective than UV melting analyses for the detection of the specific binding of metal ions to the mismatched base pairs in duplex DNA.
Conclusions
Our results demonstrated that Ag+ binds specifically to the C–C mismatched base pair of the duplex DNA without binding to the C–A and C–T mismatched base pairs, although Ag+ may bind to the phosphate backbones of the duplex DNA nonspecifically. Furthermore, the Tm value of the mismatched duplex DNA increased upon the addition of metal ions, even when the metal ions were unable to bind to the mismatched base pair, as in the case of the C–A and C–T mismatches. Thus, the increase in the Tm of the mismatched duplex DNA upon the addition of metal ions cannot be used to detect the specific binding between the metal ion and the mismatched base pair. Alternatively, by the direct detection of the heat derived from the binding between the metal ions and the mismatched base pair,23 ITC analyses can be used to evaluate specific binding. UV melting analyses are often used to propose novel binding schemes between metal ions and an artificially designed mismatched base pair of the duplex DNA.31–38 Owing to the low reliability of UV melting analyses, previously proposed binding schemes between metal ions and artificially designed mismatched base pairs should be reexamined by ITC analyses. The formation of metal-mediated base pairs by the specific binding between metal ions and mismatches has various types of applications,22 including applications in metal (Hg2+ and Ag+) sensors, Hg2+ trapping, single nucleotide polymorphism detection, and DNA nanomachines (DNA tweezers, DNA walkers and logic gates). Taken together, we conclude that ITC analyses are important for the detection of the specific binding of metal ions to mismatched base pairs in duplex DNA and may expand the applications of metal-mediated base pairs in various fields.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported in part by the Grant-in-Aid for Challenging Exploratory Research (18K19308) and the Grant-in-Aid for Scientific Research (B) (17H03033) from the Japan Society for the Promotion of Science.References
- Y. Hu, A. Cecconello, A. Idili, F. Ricci and I. Willner, Angew. Chem., Int. Ed., 2017, 56, 15210–15233 CrossRef CAS PubMed
.
- S. Naskar, R. Guha and J. Muller, Angew. Chem., Int. Ed., 2020, 59, 1397–1406 CrossRef CAS PubMed
.
- M. Nishio, K. Tsukakoshi and K. Ikebukuro, Biosens. Bioelectron., 2021, 178, 113030 CrossRef CAS PubMed
.
- P. J. Huang and J. Liu, ChemistryOpen, 2020, 9, 1046–1059 CrossRef CAS PubMed
.
- L. Ma and J. Liu, iScience, 2020, 23, 100815 CrossRef CAS PubMed
.
- R. Saran, Y. Wang and I. T. S. Li, Sensors, 2020, 20, 7019 CrossRef CAS PubMed
.
- A. Heuer-Jungemann and V. Linko, ACS Cent. Sci., 2021, 7, 1969–1979 CrossRef CAS PubMed
.
- S. Lu, J. Shen, C. Fan, Q. Li and X. Yang, Adv. Sci., 2021, 8, 2100328 CrossRef CAS PubMed
.
- H. Arakawa, R. Ahmad, M. Naoui and H. A. Tajmir-Riahi, J. Biol. Chem., 2000, 275, 10150–10153 CrossRef CAS PubMed
.
- A. A. Ouameur, S. Nafisi, N. Mohajerani and H. A. Tajmir-Riahi, J. Biomol. Struct. Dyn., 2003, 20, 561–565 CrossRef CAS PubMed
.
- A. A. Ouameur, H. Arakawa, R. Ahmad, M. Naoui and H. A. Tajmir-Riahi, DNA Cell Biol., 2005, 24, 394–401 CrossRef CAS PubMed
.
- J. Wu, F. Du, P. Zhang, I. A. Khan, J. Chen and Y. Liang, J. Inorg. Biochem., 2005, 99, 1145–1154 CrossRef CAS PubMed
.
- Y. Li, Y. L. Xia, Y. Jiang and X. P. Yan, Electrophoresis, 2008, 29, 1173–1179 CrossRef CAS PubMed
.
- K. Utsuno, Chem. Pharm. Bull., 2008, 56, 247–249 CrossRef CAS PubMed
.
- A. Ono and H. Togashi, Angew. Chem., Int. Ed., 2004, 43, 4300–4302 CrossRef CAS PubMed
.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa, T. Fujimoto, T. Machinami and A. Ono, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed
.
- Y. Tanaka, S. Oda, H. Yamaguchi, Y. Kondo, C. Kojima and A. Ono, J. Am. Chem. Soc., 2007, 129, 244–245 CrossRef CAS PubMed
.
- H. Torigoe, A. Ono and T. Kozasa, Chemistry, 2010, 16, 13218–13225 CrossRef CAS PubMed
.
- A. Ono, H. Torigoe, Y. Tanaka and I. Okamoto, Chem. Soc. Rev., 2011, 40, 5855–5866 RSC
.
- H. Torigoe, Y. Miyakawa, A. Ono and T. Kozasa, Thermochim. Acta, 2012, 532, 28–35 CrossRef CAS
.
- J. Kondo, T. Yamada, C. Hirose, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., Int. Ed., 2014, 53, 2385–2388 CrossRef CAS PubMed
.
- Y. Tanaka, J. Kondo, V. Sychrovsky, J. Sebera, T. Dairaku, H. Saneyoshi, H. Urata, H. Torigoe and A. Ono, Chem. Commun., 2015, 51, 17343–17360 RSC
.
- T. Wiseman, S. Williston, J. F. Brandts and L. N. Lin, Anal. Biochem., 1989, 179, 131–137 CrossRef CAS PubMed
.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 4825–4827, 10.1039/b808686a
.
- H. Torigoe, Y. Miyakawa, A. Ono and T. Kozasa, Nucleosides, Nucleotides Nucleic Acids, 2011, 30, 149–167 CrossRef CAS PubMed
.
- H. Torigoe, I. Okamoto, T. Dairaku, Y. Tanaka, A. Ono and T. Kozasa, Biochimie, 2012, 94, 2431–2440 CrossRef CAS PubMed
.
- J. Kondo, Y. Tada, T. Dairaku, H. Saneyoshi, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., Int. Ed., 2015, 54, 13323–13326 CrossRef CAS PubMed
.
- T. Dairaku, K. Furuita, H. Sato, J. Sebera, K. Nakashima, J. Kondo, D. Yamanaka, Y. Kondo, I. Okamoto, A. Ono, V. Sychrovsky, C. Kojima and Y. Tanaka, Chemistry, 2016, 22, 13028–13031 CrossRef CAS PubMed
.
- J. Kondo, Y. Tada, T. Dairaku, Y. Hattori, H. Saneyoshi, A. Ono and Y. Tanaka, Nat. Chem., 2017, 9, 956–960 CrossRef CAS PubMed
.
- T. Funai, M. Aotani, R. Kiriu, J. Nakamura, Y. Miyazaki, O. Nakagawa, S. I. Wada, H. Torigoe, A. Ono and H. Urata, ChemBioChem, 2020, 21, 517–522 CrossRef CAS PubMed
.
- H. Urata, E. Yamaguchi, Y. Nakamura and S. Wada, Chem. Commun., 2011, 47, 941–943 RSC
.
- T. Richters, O. Krug, J. Kosters, A. Hepp and J. Muller, Chem. – Eur. J., 2014, 20, 7811–7818 CrossRef CAS PubMed
.
- H. Z. Yang and F. Seela, Chem. – Eur. J., 2016, 22, 13336–13346 CrossRef CAS PubMed
.
- X. R. Guo and F. Seela, Chem. – Eur. J., 2017, 23, 11776–11779 CrossRef CAS PubMed
.
- B. Jash and J. Muller, Chem. – Eur. J., 2018, 24, 10636–10640 CrossRef CAS PubMed
.
- N. Sandmann, D. Defayay, A. Hepp and J. Muller, J. Inorg. Biochem., 2019, 191, 85–93 CrossRef CAS PubMed
.
- X. L. Zhou, D. Kondhare, P. Leonard and F. Seela, Chem. – Eur. J., 2019, 25, 10408–10419 CrossRef CAS PubMed
.
- I. Schonrath, H. Aukam, B. Jasper-Peter and J. Muller, Nucleosides, Nucleotides Nucleic Acids, 2022, 41, 23–35 CrossRef PubMed
.
- D. R. Rosseinsky, Chem. Rev., 1965, 65, 467–490 CrossRef CAS
.
- A. A. Rashin and B. Honig, J. Phys. Chem., 1985, 89, 5588–5593 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2dt02097a |
| This journal is © The Royal Society of Chemistry 2023 |