Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
ChemDataWriter: a transformer-based toolkit for auto-generating books that summarise research†
Shu
Huang
 a and
Jacqueline M.
Cole
a and
Jacqueline M.
Cole
 *ab
*ab
aCavendish Laboratory, Department of Physics, University of Cambridge, J. J. Thomson Avenue, Cambridge CB3 0HE, UK. E-mail: jmc61@cam.ac.uk
bISIS Neutron and Muon Source, Rutherford Appleton Laboratory, Harwell Science and Innovation Campus, Didcot, Oxfordshire OX11 0QX, UK
First published on 4th October 2023
Abstract
Since the number of scientific papers has grown substantially over recent years, scientists spend much time searching, screening, and reading papers to follow the latest research trends. With the development of advanced natural-language-processing (NLP) models, transformer-based text-generation algorithms have the potential to summarise scientific papers and automatically write a literature review from numerous scientific publications. In this paper, we introduce a Python-based toolkit, ChemDataWriter, which auto-generates books about research in a completely unsupervised fashion. ChemDataWriter adopts a conservative book-generation pipeline to automatically write the book by suggesting potential book content, retrieving and re-ranking the relevant papers, and then summarising and paraphrasing the text within the paper. To the best of our knowledge, ChemDataWriter is the first open-source toolkit in the area of chemistry to be able to compose a literature review entirely via artificial intelligence once one has suggested a broad topic. We also provide an example of a book that ChemDataWriter has auto-generated about battery-materials research. To aid the use of ChemDataWriter, its code is provided with associated documentation to serve as a user guide.
1 Introduction
The world has witnessed a significantly growing corpus of scientific papers over recent years, through which scientists publish their research progress as a means of communication within the scientific community.1 However, this large volume of scientific publications also makes it more difficult for researchers to follow research trends and gain insights into the latest scientific findings. In addition, writing a literature review based on numerous scientific papers is becoming very time-consuming. Thus, there is an urgent need to find an efficient way to read, review, and summarise scientific publications.With the development of deep-learning and natural-language-processing (NLP) technologies, research efforts have been invested in the text mining of scientific publications. For example, literature-mining techniques have been used in the biomedical area to identify chemical records,2,3 extract relational biochemical data,4,5 and summarise the biomedical literature.6 In chemistry and materials science, researchers have used NLP to perform data extraction,7–9 create databases,10–17 and make predictions out of the extracted data.18–22 To enhance the text-mining performance, many NLP toolkits and models have been created over the past few years, such as ChemDataExtractor,23,24 BatteryDataExtractor,25 MatBERT,26 MatSciBERT,27 and BatteryBERT.28
While most NLP-related research in chemistry and materials science focuses on natural-language understanding (NLU) and data extraction,29 another main branch of NLP, natural-language generation (NLG),30 is almost neglected in the text mining of chemical literature; even though such methods could significantly reduce the time for scientists who need to review the literature. Yet, NLG could be used to generate scientific text if it is tailored to sophisticated scientific concepts and content. By contrast, other fields have already seen many applications of such forms of text generation; see, for example, the automatic generation of fiction,31 sports news,32 and dialogue conversations.33 The slow progress in applying text generation to the scientific literature might be due to the difficulty of understanding sophisticated scientific concepts and content. The need to resolve chemical names from their associated labels that express the identity of a chemical in a scientific paper (chemical named entity recognition) is also a crucial consideration. Moreover, scientific writing requires high precision and a formal academic style compared to other types of writing. To automatically write a research book is still a challenging task, research on which is still in its infancy, especially for chemistry and material science.
Mishra et al. studied the first application of text-summarisation application in materials science using deep-learning methods.34 The scientific literature of a specific area, friction stir-welded magnesium alloys, was summarised using a text-generation NLP algorithm. However, its data sets were only based on abstracts of research papers, which inevitably causes information loss from the full text during the text summarisation process. The first machine-generated research book was published in 2019,35 which provides a brief overview of Li-ion batteries that had been summarised from research papers. With certain controls from human users, the book was written relatively conservatively to preserve the original meaning of the source text and ensure scientific accuracy. However, the source code and toolkit that achieved this book generation are not open source, thereby posing the difficulty in using their technology within the academic community. In addition, the book-generation algorithm was mainly based on traditional NLP algorithms, while there is potential to improve its performance by introducing deep-learning models. Recently, Taylor et al. released a huge deep-learning-based language model for science that is called Galactica.36 Galactica is the first tool to generate a literature review automatically; it is trained on a large scientific corpus of research papers, reference materials, knowledge bases and many other sources. However, the model demonstration of Galactica was removed soon after its release owing to controversial issues surrounding the potential to generate inaccurate and unreliable output as well as causing inadvertent plagiarism. ChatGPT37 has also been used to auto-generate literature reviews about “digital twins” in the healthcare sector. The review was generated by asking ChatGPT questions that it answered based on the inputted abstracts. The academic validity of the ChatGPT content is yet to be evaluated.38 Language models that are pre-trained unidirectionally, such as generative pre-trained transformer (GPT),39 face the disadvantage that its token representation only encodes the leftward context, while bidirectional models such as bidirectional encoder representations from transformers (BERT)40 and bidirectional and auto-regressive transformers (BART)41 have stronger language representations and are more suited to tasks that require a deeper comprehension of context.42,43 While it is true that unidirectional language models can show better performance when the model size is much larger than that of bidirectional language models, larger models have demonstrated only a marginal advantage over smaller models while requiring much greater training resources.44
This paper releases an open-source Python toolkit, ChemDataWriter, the first toolkit in the area of chemistry to automatically generate research books that summarises the literature according to an input corpus that has been selected by the user. The core of the tool adopts state-of-the-art transformer models, including text clustering, text retrieval and re-ranking, text summarisation, and paraphrasing. Our toolkit enables users to generate research books in a completely unsupervised fashion: users only need to provide candidate research papers for ChemDataWriter to review and then produce a research book for the user about the summary of the input corpus. In the following sections, we will provide implementation details of ChemDataWriter, as well as three case studies about the analysis of critical parts of our toolkit. We also provide an example of a book about battery research that ChemDataWriter has auto-generated. While ChemDataWriter offers several advantages, we also recognise the importance of reflecting upon the moral and philosophical implications of our toolkit. As AI continues to evolve, it is important to navigate its application responsibly. Meanwhile, we provide some recommendations for good practice when using ChemDataWriter.
2 Implementation details
2.1 System overview
Fig. 1 outlines the pipeline of ChemDataWriter, which includes seven main stages: paper downloading, paper screening, topic modelling, text retrieval & re-ranking, text summarisation, content organisation, and reference auto-generation. The final output is a research book about a specific topic. We employ a relatively conservative approach, by which we mean the generated summary is extracted and re-organised from multiple original sentences rather than written in a new and creative form; this ensures that ChemDataWriter generates an accurate and reliable book. Implementation details of each stage can be found below. | ||
| Fig. 1 Operational pipeline of ChemDataWriter showing each of the seven steps with the specific models used in each stage being described as footnotes in each box. | ||
2.2 Paper downloading
ChemDataWriter uses the same web scrapers that are embedded within BatteryDataExtractor25 to download papers from three publishers (the Royal Society of Chemistry, Elsevier, and Springer), as well as the same document processors to pre-process the HTML/XML files into plain text. Web scrapers allow users to download multiple papers on a specific topic, over a specific date range, or from a particular set of journals. ChemDataWriter also includes logic that differentiates and categorises sections of research papers, such as the abstract, introduction, conclusions, and references. Users can also use their own data sources for book generation by providing files in a certain format, i.e. a complete JSON file including the title, abstract, citation information, and full text (optional).2.3 Paper screening
Papers are retrieved according to input keywords, through a query in web scrapers, but the downloaded corpus can contain irrelevant papers where the keyword of the query is usually mentioned in the original paper but does not belong to that exact topic. Hence, a paper-screening step must be completed to filter out irrelevant papers before generating a research book.Since a high precision is preferred over a high recall for scientific book generation, ChemDataWriter adopts a prompt-based learning strategy to classify relevant and irrelevant papers. Prompt-based learning calculates the probability of a given text option, by directly modifying the original input with a prompt template, and can be used in a “few-shot” or “zero-shot” scenario.45 For example, Yin et al. used a prompt template, “the topic of this document is [Z]”, which was then inputted into masked pre-trained language models, to predict text that fills the slot [Z].46 In our study, we also used masked language models, such as BERT or domain-specific BERT, to screen papers according to their abstracts. A prompt template, “A paper in the area of [MASK] with an abstract…”, is fed into the language model to predict the [MASK] word. For instance, if we want to obtain a collection of research papers about batteries, all the papers with “battery” or “batteries” as the masked output word will be saved. This way, some papers about battery research may be unintentionally filtered out, but with a key benefit that the resulting corpus will be very clean with few noisy data.
2.4 Topic modelling
In this stage, ChemDataWriter provides suggestions on potential topics that can be written based on text that is contained within the screened set of scientific papers. According to the output of suggested research topics, users can define titles and sub-titles of each chapter, after which ChemDataWriter will produce a full table of contents for the entire book. ChemDataWriter can also generate text in a mode that automatically provides chapter titles based on the output of suggested research topics, albeit that the auto-generated titles are a list of words rather than a full sentence. Since topic modelling can be time-consuming when the number of papers is large, we set up this step as an optional stage. Users who want to control the content of the auto-generated book themselves can instead manually provide the full table of contents to ChemDataWriter.We use the BERTopic algorithm as the default topic model to cluster papers in ChemDataWriter.47 BERTopic generates topics through several independent but sequential steps: creating document embeddings using a transformer-based language model; reducing the dimensionality of these embeddings; creating semantically similar clusters, and using a class-based version of a term frequency-inverse document frequency (TF-IDF) model to extract the topic representation from each topic. The output of the topic model is a list of keywords that represent the topic, where each paper is also categorised into a certain topic. While users can choose various alternative embeddings or models for each process, default models are embedded in ChemDataWriter as follows: document embeddings (sentence transformer48), dimensionality reduction (UMAP49), clustering (HDBSCAN50), topic representations (c-TF-IDF51).
2.5 Text retrieval & re-ranking
The text-retrieval step retrieves relevant papers according to the inputted topic words, i.e. the names of chapters and sub-chapters, which are then re-ranked according to their relevance, from high to low. We embedded Haystack's retriever software module52 into ChemDataWriter in order to perform the semantic search. The collection of papers is first saved into a database (in the form of a document store within Haystack, default: InMemoryDocumentStore), from which the retriever can quickly identify the relevant documents that need to be summarised in the next step and dismiss the irrelevant ones. The retriever that employs the TF-IDF model is the default text retriever in ChemDataWriter in order to maintain a good search efficiency, while language models are also accessible and can be used for embedding retrieval. Users can specify the number of relevant documents that need to be found by the text retriever, and the final output will automatically re-rank the extracted papers in terms of their relevance scores.2.6 Text summarisation
Text summarisation is the core part of ChemDataWriter. The text summarisation algorithms in NLP include extractive summarisation and abstractive summarisation.53 Extractive summarisation is a relatively conservative way to summarise text by selecting and combining the most important part of text or sentences from the original corpus without adding or modifying any information. By contrast, abstractive summarisation is used to rephrase the original text and generate new sentences based on machine comprehension. Abstractive summarisation can create more creative text than extractive summarisation, but this greater creativity comes at the expense of a more error-prone summarisation process.In order to preserve the original meaning of the text that needs to be summarised, we tested several transformer-based extractive-summarisation language models. We selected the fine-tuned DistilBART that is available on Hugging Face54 as our chosen model. DistilBART is a smaller version of BART, a transformer-based encoder-decoder model that has been pretrained for natural-language generation.55 BART models combine the features of BERT40 and GPT39 models and are particularly effective when they are fine-tuned for text generation. In this study, the CNN Dailymail data set56,57 was used to fine-tune DistilBART, as this data set offers a diverse range of topics and writing styles, to aid improvements in the generalisation capabilities of our model.54 The fine-tuned text-summarisation model results in a distillation of the most relevant information from each paragraph within original papers into several sentences, thus preserving the original content and meaning, and reducing the risk of creating mistakes.
One problem that conservative text-summarisation models of the papers face is that the summarised output can be very similar or even the same as the original sentence, thus increasing the risk of inadvertent plagiarism. We mitigated this issue by paraphrasing the text of the summary before it is written into the book. To this end, we adopted a “back-translation” paraphrasing approach in order to ensure semantic and syntactic correctness. The “back-translation” model in ChemDataWriter employs two transformer-based language models that have been fine-tuned for translation: one to translate English into another languages and another to convert the translated text back into English. We evaluated the performance of ChemDataWriter in paraphrasing text using four different foreign languages in the back-translation model, and selected English-to-German and German-to-English as the default paraphrasing models. Users can choose any language models and parameters for back-translation and paraphrasing in order to fulfil their specific needs.
2.7 Content organisation
The content-organisation stage of ChemDataWriter automatically organises the auto-generated text summary into a complete book with a certain format. ChemDataWriter contains pre-defined content selection and organisational logic in order to auto-generate a research book, the nature of which is best explained by illustration. For example, the auto-generation of a research book about “Na-ion batteries” will require the inclusion of several chapters that belong to this specific topic, with each chapter representing a sub-area such as anodes, cathodes, or electrolytes. Within each chapter, we generate an introduction, a literature-review section that is sourced from several sub-sections of each inputted paper, and a Conclusion section. The Introduction section is summarised from every abstract of each article in order to provide an overview; while the sub-section of the main text (i.e. the literature review) is generated from the Introduction sections of each paper. Likewise, the Conclusion section of each book chapter consists of the summarised conclusion of each paper. Note that similar sentences that have been summarised from different papers will be merged into one, whereby they have been identified using a similarity measure.For each Literature-review section between the Introduction and Conclusion of a book chapter, we further simplify the title of each chapter sub-section using a transformer-based title-generation model to provide a clear view of the summarised paper. The default title-generation model in ChemDataWriter is the T5 model58 which has been fine-tuned on the TitleWave59 data set. By inputting the original long title, the fine-tuned T5 model will output a short title as the title of each chapter sub-section. Overall, this content-organisation approach of combining the summarised text of each paper separately is not ideal for combining the summary of research outputs, but its conservative nature significantly increases scientific correctness as it does not produce abstractive text.
2.8 Reference auto-generation
The last part of the machine-generated book is a list of references that comprises a bibliography. We include a reference auto-generator in ChemDataWriter for each publisher (the RSC, Elsevier, Springer) to produce an academic-style reference list. The format is “authors, title, journals, date, volume, issue, page, DOI”. The relevant reference information is extracted from the metadata of the HTML/XML file. If users provide their own paper files, we suggest that they also provide the reference data in the correct format.3 Results and discussion
We performed three case studies to evaluate the performance of ChemDataWriter. In case study 1, we assessed the performance of the topic modelling stage of ChemDataWriter, where topics were extracted from three corpora of papers about battery research. Case study 2 compared and contrasted different paraphrasing models to improve the quality of book writing and to avoid potential plagiarism issues. Our third case study focused on providing scientific insights into an entire book about recent battery research that ChemDataWriter auto-generated.3.1 Case study 1: suggesting chapter titles based on the topic modelling capabilities of ChemDataWriter
ChemDataWriter suggests the potential content to be written, using the BERTopic model to extract features and identify topics that are present in the original text. In this case study, we inputted three different corpora of scientific papers into ChemDataWriter to test the performance of the topic model, including (1) the entire corpus of battery-research papers with title, abstract, and conclusion, (2) the full-text corpus of papers about Na-ion batteries as defined such that “Na-ion” is part of the title of a paper, and (3) the full-text corpus of papers about “Li-metal” batteries. Each topic consists of five words, and the minimum number of documents per topic was set as 10.Table 1 lists the corpus size, the number of extracted topics, and two evaluation metrics (topic coherence and topic diversity) for each data set. Topic coherence is calculated based on the normalised pointwise mutual information (NPMI).60 The topic-coherence score ranges from −1 to 1, where a high coherence score close to 1 means that the words in a topic are semantically similar. In contrast, a coherence score of −1 indicates that words are semantically dissimilar, while a zero coherence score means that no clear semantic relationship has been found within a topic. Topic diversity is the percentage of unique words across all topics within a data set, in which a higher score indicates that topics are varied and words do not overlap between classes of topics. Table 1 shows that topic coherence and topic diversity performance metrics are similar across different data sets. In general, a high topic coherence score ensures that our tool conveys information in a clear and understandable manner, while a high diversity is often required when the objective is to generate more creative content. Compared to the reported topic coherence and topic diversity scores in the original BERTopic paper,47 the best topic coherence score in our study (0.178) is only slightly lower than the best value in the original paper (0.192), which indicates a reasonable coherence of the generated scientific-related topics. However, the best topic diversity score is much lower (0.669 < 0.886), which is as expected as ChemDataWriter's ability to convey information is more valued than its creative abilities in the generation of scientific documents. In contrast, the number of generated topics is more varied owing to the difference in the input corpus size and paper types. For example, the entire mixed battery-paper corpus can generate 141 topics, whereas only 26 topics can be found for the Li-metal battery-paper corpus. Even though it involves 7 million words, the Na-ion battery-paper corpus only generates 58 topics, much less than that of the full corpus which possesses 2.34 million more words.
| Battery research types | Corpus size (number of words) | Number of topics | Topic coherence score | Topic diversity score |
|---|---|---|---|---|
| Na-ion batteries (full text) | 7.00 million | 58 | 0.149 | 0.648 |
| Li-metal batteries (full text) | 3.22 million | 26 | 0.178 | 0.669 |
| All (title, abstract, conclusion) | 9.34 million | 141 | 0.144 | 0.655 |
The importance of the data set that is used for input can also be reflected in Fig. 2, whereby representative topics in each corpus are illustrated. From the entire corpus of battery papers, we can observe that the topic-modelling process found some topics that are varied and diverse, such as “Li–S batteries”, “supercapacitors”, “Li–O2 batteries”, and “SOC estimation”; taking these topics as an example, one choice would be to write a book about the different kinds of battery applications. For the Na-ion and Li-metal battery-paper corpora, specific topics were more likely to be found, such as a particular material (TiO2, MoS2, ScO2) or an application (electrolyte, cathode, impedance). ChemDataWriter offers suggestions of topics, based on the results of the topic-modelling process, and, by default, lets users define the titles of each chapter in the auto-generated research book. However, it can also produce a table of contents entirely automatically using the list of topic words as titles directly.
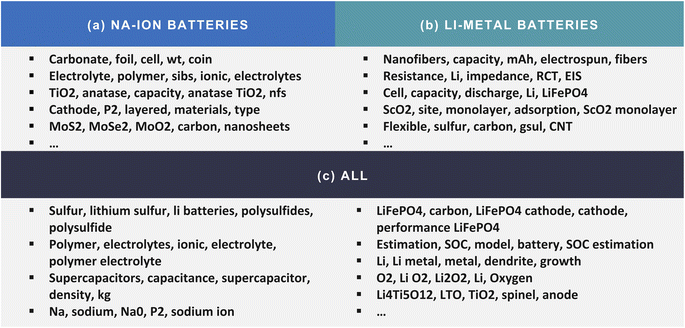 | ||
| Fig. 2 Keywords in representative topics that were extracted from the corpus of research papers about (a) Na-ion batteries, (b) Li-metal batteries, and (c) all batteries. | ||
This case study demonstrates that hidden topics can be found using the BERTopic model, based on which ChemDataWriter can suggest the potential book content that can be auto-generated. The topic model works especially well on a large, diverse text-based data set, such as the full corpus of battery papers. However, this approach also has several weaknesses, which is why this step is optional in ChemDataWriter by default. First, the long computational time required for the topic-modelling stage prevents ChemDataWriter from finding optimal results, as the model needs a few hours to realise the necessary inference from an input corpus that contains around 10 million words. The long-running execution time makes it impossible to fine-tune parameters in each stage of the BERTopic modelling operational process (document embedding, dimensionality reduction, clustering, and generating topic representations). In addition, since the BERTopic model only finds topics in terms of the importance of words, as judged by their frequency of appearance in the text, a list of topics may contain words that are very different from each other. In Fig. 2c, for example, the first topic list consists of both broad topics about Li–S batteries (sulphur, lithium–sulphur, Li batteries) and very specific topics about the material polysulfide (polysulfides, polysulfide). This issue can lead to a difficult topic interpretation; as such, the default option, to use human intervention in helping to choose section topics, will tend to afford an auto-generated document with a higher-quality output. Creating a more diverse table of contents is a subject in its own right that still under development.
3.2 Case study 2: introducing paraphrasing control to reduce text similarity
We mitigated the issue of conservative extractive summarisation, in that the summary can be very similar to the original text, by adopting a “back-translation” strategy to paraphrase text that has been summarised. This strategy follows the notion that one can keep the meaning of a sentence but alter its original wording by translating it into a foreign language and back again into english text. We tested five language models that are used as machine-translation tools for back-translation, all of which convert english text to and (back) from a foreign language; specifically, we employ French, Russian, Arabic, and two German machine translation models (trained on different data sets).61 These models have been developed by the Helsinki-NLP group on open world translation data sets,61 except for the German2 model, which was the same as the German model except that it has trained on different data set: Facebook's news translation data set.62The performance of our paraphrasing models was evaluated using two well-accepted automatic quantitative evaluation metrics: the Bilingual Evaluation Understudy (BLEU) and Recall-Oriented Understudy for Gisting Evaluation (ROUGE) score. Both metrics compare the generated text with a reference text from a gold-standard data set. BLEU compares two texts by counting the number of words in the generation that appear in the reference, where a high precision is preferred to a high recall. By contrast, ROUGE is a recall-oriented metric that checks how much text in the reference also occurs in the generated text. Common ROUGE metrics include ROUGE-1 (unigram/individual words co-occurrence statistics), ROUGE-2 (bigram overlap), and ROUGE-L (longest common subsequence, LCS). The LCS can be calculated for any pair of strings. For example, the LCS for “abcde” and “ace” would be “ace” with a length of 3. The BLEU and ROUGE scores range between 0 and 1, but are often represented as percentages with a range from 0 to 100, as is shown in Table 2.
| Languages | BLEU | ROUGE-1 | ROUGE-2 | ROUGE-l |
|---|---|---|---|---|
| German | 25.46 | 47.04 | 25.13 | 43.60 |
| French | 23.61 | 46.79 | 23.22 | 42.95 |
| Russian | 13.78 | 37.45 | 14.50 | 33.81 |
| Arabic | 6.90 | 32.38 | 11.34 | 29.12 |
| German2 | 20.15 | 43.21 | 20.61 | 39.56 |
Finding a suitable gold-standard data set of reference text determines whether or not the evaluation process can indeed reflect the model behaviour. In order to test the performance of paraphrasing in the scientific domain, we evaluated our models on the ParaSCI data set, the first large-scale scientific paraphrase data set that has been extracted mainly from arXiv papers.63 The ParaSCI data set contains common patterns and characteristics of scientific papers. A high BLEU and ROUGE score could indicate that the paraphrasing models can work well in the scientific area of interest.
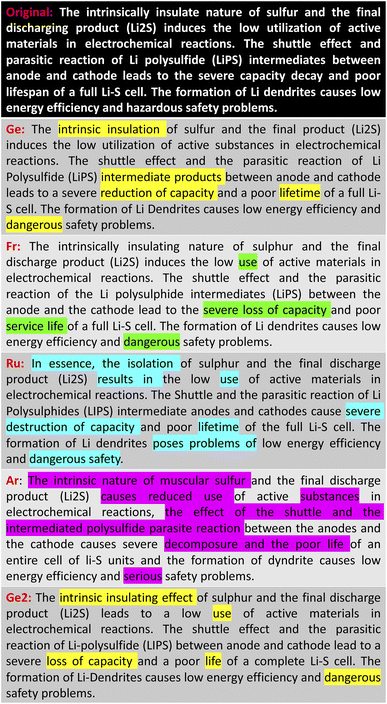
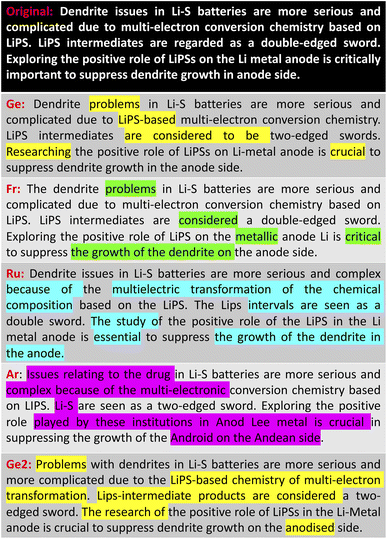
Table 2 lists the performance of our back-translation paraphrasing models on the ParaSCI data set. The English-to-German and German-to-English model called “German” scores the highest on all metrics, which are slightly better than those of the English-French back-translation model. Russian and Arabic paraphrasing models showed much worse performance due to the larger language difference between each of them and the English language. This result is also consistent with our evaluation of the battery-related text summarisation (Fig. 3 and 4). Both examples demonstrate that paraphrasing models using German and French only change several words or phrases compared to the original text, while there are major differences between the outputs of the Russian and Arabic language models and their original text. In addition, the Russian and Arabic back-translation models are more likely to produce paraphrased text with incorrect meanings, especially when the scientific term is mentioned (e.g. “insulate nature”, “the shuttle effect”, “LiPS”).
We also observed that paraphrasing models can perform differently even when using the same foreign language during back-translation, if they are trained on different data sets. We selected the best performing model (“German”) to investigate its model behaviour when trained on a different data set: Facebook's news translation data set.62 As is shown in Table 2, the BLEU and ROUGE scores of the alternative model, which we call “German2”, are lower than both the “German” and “French” back-translation models. However, “German2” also showed more differences between the paraphrased and original text (Fig. 3 and 4), while the scientific meaning still remains correct. Therefore, if users want to find models that differ more substantially from the original text, they could test this text on paraphrasing models which feature the same back-translation language but have been trained on different data sets.
To summarise, our paraphrasing models enable ChemDataWriter to produce text that differs significantly from the extractive text summary to reduce the risk of inadvertent plagiarism that has been reported in the use of other text-generation tools. Thereby, English–German back-translation models, “German”, showed the best performance on the ParaSCI data set. Users can easily control the paraphrased output by changing the language models or the training sets that are associated with the back-translation process. However, problems also exist in this approach, such as a relatively high similarity between the original text and the paraphrased output. This text similarity issue is inevitable due to the nature of the extractive summarisation algorithm. A hybrid approach that employs a rule-based and transformer-based model could further improve model performance in this regard, but that would involve considerable human input, while our objective is to focus on achieving an automatic pipeline without any human effort. Abstractive summarisation can also mitigate the similarity problem, but the development of this methodology is not yet sufficiently mature in order to produce reliable scientific content.
3.3 Case study 3: analysis of an example auto-generated book
In this case study, we will analyse an example of a research book that ChemDataWriter has auto-generated. This book is entitled “Literature Summary of Recent Research About Na-ion, Li–S, and Li–O2 Battery Materials”. It was generated by summarising text from 152 scientific papers about battery materials. These papers had been downselected from 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 736 research papers about battery materials, each of which had been classified as relevant battery papers by the prompt-based binary classifier. Each file must also include the necessary information to auto-generate a book, including a valid title, abstract, introduction, conclusion, and bibliography. As a result, 152 scientific papers were extracted to compose the final book about the three battery applications. A copy of the full book can be found in ESI.† Note that we have also obtained explicit permission from the publishers of the 152 papers to allow us to reproduce textual content based on the work of the original authors, just to safeguard ChemDataWriter from any inadvertent plagiarism.
736 research papers about battery materials, each of which had been classified as relevant battery papers by the prompt-based binary classifier. Each file must also include the necessary information to auto-generate a book, including a valid title, abstract, introduction, conclusion, and bibliography. As a result, 152 scientific papers were extracted to compose the final book about the three battery applications. A copy of the full book can be found in ESI.† Note that we have also obtained explicit permission from the publishers of the 152 papers to allow us to reproduce textual content based on the work of the original authors, just to safeguard ChemDataWriter from any inadvertent plagiarism.
Fig. 5 shows the high-level table of contents of this book, including three main parts (Na-ion batteries, Li–S batteries, and Li–O2 batteries). Each part consists of three chapters on: cathode materials, anode materials, and electrolytes. The auto-selection of these chapter contents follows the popular writing style of review articles on battery materials, such as a literature review of sodium-ion batteries.64 The difference between machine-generated and human-written books lies in the title of the sub-section of each chapter. While ChemDataWriter can only name sub-section titles according to the original title of papers, humans can summarise them more abstractly. For example, Hwang et al. named the titles of sub-sections of “Anode materials” in terms of the reaction mechanisms: insertion materials, conversion materials, and alloying reaction materials.64 The ability to achieve this high-level title generation using machines requires the further development of NLP algorithms, by understanding and uncovering the hidden meaning from the scientific text.
 | ||
| Fig. 5 Table of contents for the example of the auto-generated book: “Literature summary of recent research about Na-ion, Li–S, and Li–O2 battery materials”. | ||
The introduction of each book chapter was summarised from the abstract of each paper that is cited in that chapter. Fig. 6 shows one of these abstracts together with a paragraph of summarised text that is afforded as part of the auto-generated book.65 The first three sentences in the original form of this abstract were extracted as sentences that contain important information, such as the background, current issue, and the main objective of the paper. The structure of sentences and several words were changed with help from the paraphrasing model, and the term “we” was automatically transformed into “the authors” by ChemDataWriter. Similarly, the “Conclusion” section in each chapter was also written in the same way as the Introduction.
 | ||
| Fig. 6 Extractive text summarisation from the original abstract of an example paper to afford a paragraph of the Introduction section of the auto-generated book. | ||
More detailed scientific content of the input papers is provided in the second section, “Literature reviews”, where the content for each chapter is summarised from introductions of each original paper. The “Literature reviews” section consists of multiple sub-sections, representing the summary of an individual paper. The title of each sub-section is a simplified version of the full title of each paper, achieved by the transformer-based title generation algorithm.58 An example of the title of a paper can be “Layered tin sulphide and selenide anode materials for Li- and Na-ion batteries”, which can be simplified to a sub-section title “Layered tin sulphide and selenide anode materials” by ChemDataWriter. Since the title-generation algorithm was not trained on a data set of scientific text,59 there is a risk that the simplification process could cause the loss of crucial information in a full title. Hence, users may need to refer to full-title information and other metadata in the last part of the book: the auto-generated references.
The number of sub-sections is determined by human input. In the example of the book presented herein, the maximum number of sub-sections is the default value, 30. However, the exact number of sub-sections in “Literature reviews” is much lower than 30, owing to the conservative method of filtering out papers whereby only their title, including the query word, will be summarised. In this way, we ensure that each whole paper discusses the specific topic, e.g. anodes of Na-ion batteries. Once candidate papers are found, they are re-ranked according to their relevance scores before being written into the book.
Overall, ChemDataWriter provides an approach to automatically generate books that review a scientific area by merging text summaries from multiple papers on a given topic. Users only need to input the candidate paper corpus and, if they wish, a list of topics to form a table of contents, based on which ChemDataWriter can write a book in a completely unsupervised way. The current logic of automatically writing a book is to merge the single-document summary from individual papers. However, we also see opportunities to further improve the text summarisation model by introducing multi-document summarisation. Large-scale data sets such as Multi-XScience have been created recently so that machines can auto-generate a single summary paragraph from multiple paper sources.66 Multi-document summarisation models are still under development, and we expect more domain-specific data sets will be created, in due course, that can improve the text summarisation performance on the scientific area. However, the creation of a custom annotated dataset is a very time-consuming process that requires careful curation and substantial human resources to ensure quality and consistency – it is essentially a major research study in its own right. We expect further advances in this area, once more data have been curated and the technology matures.
4 Conclusions and outlook
ChemDataWriter is the first open-source transformer-based toolkit for auto-generating books that summarise research in the area of chemistry. The book-writing pipeline involves conservative text summarisation approaches to ensure the correctness and trustworthiness of the auto-generated book. The book-generation process is implemented in a completely unsupervised way, where users only need to provide the corpus of research papers in order to generate a literature summary. ChemDataWriter can identify hidden topics from a large corpus of data and suggest the book content in terms of extracted topics. ChemDataWriter embeds a “back-translation” model to paraphrase the summarised text in order to alleviate the text similarity issue. We believe that ChemDataWriter has the potential to help scientists accelerate their literature-searching and screening processes. Researchers can also find the most recent and relevant information about research progress in a specific field using our toolkit.While ChemDataWriter offers promising potential in scientific research, it is incumbent upon us to use such tools responsibly. There are two key issues to address: mitigating plagiarism and the role of review writing as a form of education.
As is mentioned in the paper, our tool has the potential to be misappropriated as a plagiarism mechanism. The paraphrasing style that ChemDataWriter embues, as demonstrated in Fig. 3 and 4, could be reasonably viewed as patchwork plagiarism (sometimes called mosaic plagiarism).67,68 Such plagiarism is considered to be just as wrong as any other form of plagiarism.
It is therefore crucial that any users of ChemDataWriter solicit a priori explicit permission to reproduce text from the papers that they provide as input to our tool, as we have done through the Copyright Clearance Centre69 for all 152 papers that fed our case study that produced a book. Otherwise, the result can be furnished as plagiarism, as we exemplify by showing the patchwork plagiarism that would appear by applying a plagiarism checker70 to Chapter 4 of our book, had we not sought and obtained reproduction rights (see ESI†). While we appreciate that it might feel laborious to seek such reproduction permissions for so many papers, it is imperative since it is the only way to ensure that the result is lawful; besides, this laborious process is still far quicker than writing the review manually.
Considering the role of review writing as a form of education, ChemDataWriter does not stop this process, although it may of course tempt some to avoid learning via this type of education. It is perhaps better to view this as responsible AI in the sense that AI can actively assist humans in writing a review. Indeed, the intention behind ChemDataWriter is not to supplant human authors, but to support them in navigating and consolidating vast amounts of data. While we recognise that paper reviewing can be an important process for a researcher who is new to the field, such as a PhD student, our tool equally enables many senior scientists, especially those in industry, to speed up the paper review process by implementing this latest technology to assist them. It normally requires several months for a human researcher to write a scientific review article in a given field, while ChemDataWriter can achieve this in only a few hours. The automation capabilities of our tool could relieve researchers from the burdensome and lengthy process of literature reviews, allowing them to dedicate more time to devising hypotheses, conducting experiments, and pursuing innovative ideas. Moreover, the advent of advanced AI tools like this can speed up the pace of scientific discovery and contribute to the democratisation of knowledge by making complex scientific literature more accessible.
More generally, the development and deployment of AI tools like ChemDataWriter are inevitable as we move forward in the era of AI. Thus, we consider it our responsibility to take the lead in presenting new tools, such as ChemDataWriter, together with clear recommendations in their utility, as we have provided here, before others set inappropriate trends that could carry forward irrecoverably. The world has seen recent examples of this already. Indeed, the line between leveraging technology and maintaining human oversight remains a delicate balance that the scientific community must continue to navigate.
Regarding ChemDataWriter specifically, it is important to remember that its input is known and provided by the user. The user therefore has full control over their input decisions, and their copyright choices, which determine their output. This is a key ethical difference to other tools that have been released in the public media recently. Moreover, we have released ChemDataWriter as an open-source tool, and have likewise provided its code, via this publication. This will allow others to expand upon our work as well as simply use it. In turn, this will democratise the development of these methods; a greater diversity of developers will encourage ethical behaviour and best practice by notion of a collective effort in this research field.
Challenges still exist in terms of needing more data and more advanced models in order to further improve the book auto-generation pipeline. Most of the transformer-based models in our toolkit were trained on general English-language data sets, while the use of domain-specific data sets will likely enhance the performance of book-generation algorithms in generating scientific text. We also encourage the creation of more gold-standard chemistry data sets for the purpose of evaluation. In terms of models, multi-document summarisation models could be introduced in order to update the content organisation of an auto-generated book. In addition, a hybrid approach of transformer- and rule-based methods could syntactically and semantically improve the quality of the generated text, although such an approach would currently necessitate a considerable human effort; thereby limiting its level of automation. Another future work might be to incorporate creative writing in the book-generation process while preserving the text trustworthiness.
Data availability
The source code of ChemDataWriter can be found at https://github.com/ShuHuang/chemdatawriter. The documentation of the software is available at https://chemdatawriter.readthedocs.io/. The website https://www.chemdatawriter.org collates this information and provides a full overview of the tool.Author contributions
JMC and SH conceived the overarching project and designed the study. SH performed ChemDataWriter software design and development and case studies under the PhD supervision of JMC. SH drafted the manuscript with assistance from JMC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
JMC is grateful for the BASF/Royal Academy of Engineering Research Chair in Data-Driven Molecular Engineering of Functional Materials, which is partly supported by the STFC via the ISIS Neutron and Muon Source. SH is indebted to Christ's College, Cambridge, for a graduate bursary. The authors thank the Argonne Leadership Computing Facility, which is a DOE Office of Science Facility, for use of its research resources, under contract no. DE-AC02-06CH11357.References
- L. Bornmann and R. Mutz, J. Assoc. Inf. Sci. Technol., 2015, 66, 2215–2222 CrossRef CAS.
- I. Korvigo, M. Holmatov, A. Zaikovskii and M. Skoblov, J. Cheminf., 2018, 10, 1–10 Search PubMed.
- L. Luo, Z. Yang, P. Yang, Y. Zhang, L. Wang, J. Wang and H. Lin, J. Cheminf., 2018, 10, 1–10 Search PubMed.
- D. Sousa, A. Lamurias and F. M. Couto, Artificial Neural Networks, Springer, 2021, pp. 289–305 Search PubMed.
- J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim, C. H. So and J. Kang, Bioinformatics, 2020, 36, 1234–1240 CrossRef CAS PubMed.
- M. Wang, M. Wang, F. Yu, Y. Yang, J. Walker and J. Mostafa, J. Am. Med. Inf. Assoc., 2021, 28, 2287–2297 CrossRef PubMed.
- Z. Jensen, E. Kim, S. Kwon, T. Z. Gani, Y. Roman-Leshkov, M. Moliner, A. Corma and E. Olivetti, ACS Cent. Sci., 2019, 5, 892–899 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Tomala, S. Matthews, E. Strubell, A. Saunders, A. McCallum and E. Olivetti, Sci. Data, 2017, 4, 1–9 Search PubMed.
- R. Mahbub, K. Huang, Z. Jensen, Z. D. Hood, J. L. Rupp and E. A. Olivetti, Electrochem. Commun., 2020, 121, 106860 CrossRef CAS.
- C. J. Court and J. M. Cole, Sci. Data, 2018, 5, 180111 CrossRef CAS PubMed.
- S. Huang and J. M. Cole, Sci. Data, 2020, 7, 1–13 CrossRef PubMed.
- J. Zhao and J. M. Cole, Sci. Data, 2022, 9, 192 CrossRef PubMed.
- Q. Dong and J. M. Cole, Sci. Data, 2022, 9, 193 CrossRef PubMed.
- O. Sierepeklis and J. M. Cole, Sci. Data, 2022, 9, 648 CrossRef PubMed.
- P. Kumar, S. Kabra and J. M. Cole, Sci. Data, 2022, 9, 292 CrossRef CAS.
- E. J. Beard, G. Sivaraman, Á. Vázquez-Mayagoitia, V. Vishwanath and J. M. Cole, Sci. Data, 2019, 6, 1–11 CrossRef PubMed.
- E. J. Beard and J. M. Cole, Sci. Data, 2022, 9, 329 CrossRef CAS PubMed.
- J. Zhao and J. M. Cole, J. Chem. Inf. Model., 2022, 62, 2670–2684 CrossRef CAS PubMed.
- V. Tshitoyan, J. Dagdelen, L. Weston, A. Dunn, Z. Rong, O. Kononova, K. A. Persson, G. Ceder and A. Jain, Nature, 2019, 571, 95–98 CrossRef CAS PubMed.
- C. J. Court, A. Jain and J. M. Cole, Chem. Mater., 2021, 33, 7217–7231 CrossRef CAS.
- C. B. Cooper, E. J. Beard, Á. Vázquez-Mayagoitia, L. Stan, G. B. Stenning, D. W. Nye, J. A. Vigil, T. Tomar, J. Jia, G. B. Bodedla, S. Chen, L. Gallego, S. Franco, A. Carella, K. R. J. Thomas, S. Xue, X. Zhu and J. M. Cole, Adv. Energy Mater., 2019, 9, 1802820 CrossRef.
- L. R. Devereux, Á. Vázquez-Mayagoitia, M. G. Sternberg and J. M. Cole, Adv. Energy Mater., 2023, 13, 2203536 CrossRef CAS.
- M. C. Swain and J. M. Cole, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- J. Mavračić, C. J. Court, T. Isazawa, S. R. Elliott and J. M. Cole, J. Chem. Inf. Model., 2021, 61, 4280–4289 CrossRef PubMed.
- S. Huang and J. M. Cole, Chem. Sci., 2022, 13, 11487–11495 RSC.
- A. Trewartha, N. Walker, H. Huo, S. Lee, K. Cruse, J. Dagdelen, A. Dunn, K. A. Persson, G. Ceder and A. Jain, Patterns, 2022, 3, 100488 CrossRef PubMed.
- T. Gupta, M. Zaki and N. Krishnan, et al. , npj Comput. Mater., 2022, 8, 1–11 CrossRef.
- S. Huang and J. M. Cole, J. Chem. Inf. Model., 2022, 62, 6365–6377 CrossRef CAS PubMed.
- E. A. Olivetti, J. M. Cole, E. Kim, O. Kononova, G. Ceder, T. Y.-J. Han and A. M. Hiszpanski, Appl. Phys. Rev., 2020, 7, 041317 CAS.
- C. Dong, Y. Li, H. Gong, M. Chen, J. Li, Y. Shen and M. Yang, ACM Comput. Surv., 2023, 55, 173 CrossRef.
- D. Yang, Y. Zhou, Z. Zhang, T. J. Li and R. LC, Joint Proceedings of the IUI 2022 Workshops: APEx-UI, HAI-GEN, HEALTHI, HUMANIZE, TExSS, SOCIALIZE co-located with the ACM International Conference on Intelligent User Interfaces (IUI 2022), Virtual Event, Helsinki, Finland, 2022, pp. 56–65 Search PubMed.
- J. Kanerva, S. Rönnqvist, R. Kekki, T. Salakoski and F. Ginter, Proceedings of the 22nd Nordic Conference on Computational Linguistics, Turku, Finland, 2019, pp. 242–252 Search PubMed.
- J. Juraska, K. Bowden and M. Walker, Proceedings of the 12th International Conference on Natural Language Generation, Tokyo, Japan, 2019, pp. 164–172 Search PubMed.
- A. Mishra, Int. J. Interact. Des. Manuf., 2022, 1–7 Search PubMed.
- B. Writer, Lithium-ion Batteries: A Machine-generated Summary of Current Research, Springer, 2019 Search PubMed.
- R. Taylor, M. Kardas, G. Cucurull, T. Scialom, A. Hartshorn, E. Saravia, A. Poulton, V. Kerkez and R. Stojnic, arXiv, 2022, preprint, arXiv:2211.09085, DOI:10.48550/arXiv.2211.09085.
- C. Leiter, R. Zhang, Y. Chen, J. Belouadi, D. Larionov, V. Fresen and S. Eger, arXiv, 2023, preprint, arXiv:2302.13795, DOI:10.48550/arXiv.2302.13795.
- Ö. Aydın and E. Karaarslan, available at SSRN 4308687, 2022.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, Advances in Neural Information Processing Systems, 2020, pp. 1877–1901 Search PubMed.
- J. Devlin, M. Chang, K. Lee and K. Toutanova, Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019, Minneapolis, MN, USA, 2019, vol. 1, pp. 4171–4186 Search PubMed.
- M. Lewis, Y. Liu, N. Goyal, M. Ghazvininejad, A. Mohamed, O. Levy, V. Stoyanov and L. Zettlemoyer, Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, ACL 2020, 2020, pp. 7871–7880 Search PubMed.
- X. Qiu, T. Sun, Y. Xu, Y. Shao, N. Dai and X. Huang, Sci. China: Technol. Sci., 2020, 63, 1872–1897 CrossRef.
- A. Patel, B. Li, M. S. Rasooli, N. Constant, C. Raffel and C. Callison-Burch, arXiv, 2022, preprint, arXiv:2209.14500, DOI:10.48550/arXiv.2209.14500.
- Z. Hong, A. Ajith, J. G. Pauloski, E. Duede, C. Malamud, R. Magoulas, K. Chard and I. T. Foster, arXiv, 2022, preprint, arXiv:2205.11342, DOI:10.48550/arXiv.2205.11342.
- P. Liu, W. Yuan, J. Fu, Z. Jiang, H. Hayashi and G. Neubig, ACM Comput. Surv., 2023, 55, 1–35 Search PubMed.
- W. Yin, J. Hay and D. Roth, Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing, EMNLP-IJCNLP 2019, Hong Kong, China, 2019, pp. 3912–3921 Search PubMed.
- M. Grootendorst, arXiv, 2022, preprint, arXiv:2203.05794, DOI:10.48550/arXiv.2203.05794.
- N. Reimers and I. Gurevych, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing, 2020 Search PubMed.
- L. McInnes and J. Healy, arXiv, 2018, preprint, arXiv:1802.03426, DOI:10.48550/arXiv.1802.03426.
- L. McInnes, J. Healy and S. Astels, J. Open Source Softw., 2017, 2, 205 CrossRef.
- T. Joachims, Proceedings of the Fourteenth International Conference on Machine Learning, San Francisco, CA, USA, 1997, pp. 143–151 Search PubMed.
- Haystack: end-to-end python framework for building natural language search interfaces to data, 2021, https://haystack.deepset.ai.
- V. Dalal and L. G. Malik, 6th International Conference on Emerging Trends in Engineering and Technology, ICETET 2013, Nagpur, India, 2013, pp. 109–110 Search PubMed.
- The fine-tuned DistilBART model checkpooint in Hugging Face, https://huggingface.co/sshleifer/distilbart-cnn-12-6, last accessed 25 September 2023.
- M. Lewis, Y. Liu, N. Goyal, M. Ghazvininejad, A. Mohamed, O. Levy, V. Stoyanov and L. Zettlemoyer, Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, 2020, pp. 7871–7880 Search PubMed.
- A. See, P. J. Liu and C. D. Manning, Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics, Vancouver, Canada, 2017, vol. 1, pp. 1073–1083 Search PubMed.
- K. M. Hermann, T. Kociský, E. Grefenstette, L. Espeholt, W. Kay, M. Suleyman and P. Blunsom, Neural Information Processing Systems, 2015, pp. 1693–1701 Search PubMed.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, J. Mach. Learn. Res., 2020, 21, 1–67 Search PubMed.
- TitleWave, 2021, https://github.com/tennessejoyce/TitleWave.
- G. Bouma, Proc. GSCL, 2009, 30, 31–40 Search PubMed.
- J. Tiedemann and S. Thottingal, Proceedings of the 22nd Annual Conferenec of the European Association for Machine Translation (EAMT), Lisbon, Portugal, 2020 Search PubMed.
- N. Ng, K. Yee, A. Baevski, M. Ott, M. Auli and S. Edunov, Proc. WMT, 2020, 314–319 Search PubMed.
- Q. Dong, X. Wan and Y. Cao, Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, 2021, pp. 424–434 Search PubMed.
- J.-Y. Hwang, S.-T. Myung and Y.-K. Sun, Chem. Soc. Rev., 2017, 46, 3529–3614 RSC.
- H. Cheng and K. Scott, J. Power Sources, 2013, 235, 226–233 CrossRef CAS.
- Y. Lu, Y. Dong and L. Charlin, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2020, pp. 8068–8074 Search PubMed.
- N. Das and M. Panjabi, Perspect. Clin. Res., 2011, 2, 67 CrossRef PubMed.
- N. Das, Perspect. Clin. Res., 2018, 9, 56–57 CrossRef PubMed.
- Copyright Clearance Center - Copyright & Licensing Experts, https://www.copyright.com/, last accessed 31 July 2023.
- Turnitin UK, http://uk.turnitincn.com, last accessed 31 July 2023.
Footnote |
| † Electronic supplementary information (ESI) available: An example book that ChemDataWriter auto-generates: literature summary of recent research about Na-ion, Li–S, and Li–O2 battery materials. Results from applying a plagiarism checker to chapter 4 of this example book. See DOI: https://doi.org/10.1039/d3dd00159h |
| This journal is © The Royal Society of Chemistry 2023 |