Open Access Article
Open Access ArticleThe laboratory of Babel: highlighting community needs for integrated materials data management
Brenden G.
Pelkie
 and
Lilo D.
Pozzo
*
and
Lilo D.
Pozzo
*
Department of Chemical Engineering, University of Washington, Seattle, Washington 98195, USA. E-mail: dpozzo@uw.edu
First published on 27th March 2023
Abstract
Automated experimentation methods are unlocking a new data-rich research paradigm in materials science that promises to accelerate the pace of materials discovery. However, if our data management practices do not keep pace with progress in automation, this revolution threatens to drown us in unusable data. In this perspective, we highlight the need to update data management practices to track, organize, process, and share data collected from laboratories with deeply integrated automation equipment. We argue that a holistic approach to data management that integrates multiple scales (experiment, group and community scales) is needed. We propose a vision for what this integrated data future could look like and compare existing work against this vision to find gaps in currently available data management tools. To realize this vision, we believe that development of standard protocols for communicating with equipment and data sharing, the development of new open-source software tools for managing data in research groups, and leadership and direction from funding agencies and other organizations are needed.
Introduction
Automated experimentation methods are rapidly transitioning from being research subjects themselves to serving as indispensable tools in materials research. The availability of relatively affordable off the shelf hardware, the spread of data-hungry machine learning methods to materials science, and the ever-pressing need to accelerate the pace of materials innovation to meet a changing climate have all contributed to the adoption of automated experimental methods in our laboratories.1–3 A dizzying array of recent research has contributed tools that enable this paradigm shift, including new open hardware platforms,4 optimization and experiment planning methods,5 and methods for sharing procedures across different laboratories.6–8 This newfound ability to generate vast troves of experimental data comes as new machine learning and data science methods build off that data,9,10 turning it into a first-class research product in itself.11 However, comparably little effort has been expended on systems to collect, organize, store, and share this data effectively. As a research community, we've largely applied the existing data management methods and culture that developed around manual experimentation to automated workflows. This worked fine for initial demonstration projects and forays into the field, but as the field matures and continues producing valuable data with automated platforms, we need to adopt better data management practices. The data management path we are currently following reminds us of the ‘Library of Babel’ imagined by J. L. Borges in the namesake short story.12 This vast library contains an enumeration of all possible past and future human knowledge, with the catch that all of the valuable information is hidden amongst a sea of utter gibberish. In this library, generations of librarians are driven mad trying to find meaning in the expanse of text. If we continue advancing the state of automated experimentation without overhauling how we collect, organize, and share our data, we will find ourselves lost and isolated standing in an analogous ‘Laboratory of Babel’. We'll know that the data we need to support our next materials innovation is out there somewhere, held in an unknown dataset in a distant repository, mixed into an expanse of useless untracked and unexplainable data. We believe that now is the right time to make technical and cultural shifts in how we handle this problem, so that we help future generations of researchers to develop an ‘index’ to effectively extract value from abundant ‘gibberish’ in materials data held in this global ‘Laboratory’.We envision a data management future where collecting and organizing experimental data and metadata is automated and effortless, data provenance is fully tracked, access to up to the minute data is enabled, and new community data sharing platforms make all experimental data findable. Having such a data management practice in place would streamline the development of machine learning models for accelerating materials discovery, allow researchers to check the reproducibility of their results, and improve the quality and trustworthiness of the data we generate. An agreed upon system for data management would allow data related issues to fade into the background, allowing scientists to focus on the science that ultimately motivates us.
In this perspective we propose a holistic vision for how data might be collected, organized, and shared, focusing on laboratories that have adopted automated equipment. This vision is based on our research group's experience and challenges in setting up automated experimentation workflows from scratch, and from ideas proposed in the literature. We compare the current state of the field against this vision to elucidate opportunities for improvement, but also to highlight current successes. We intentionally discuss these topics at a high level and ground them in examples, rather than getting into details of technical implementations. While many recent works have explored various aspects of the data management problem,13,14 and several projects implement isolated items that are needed to make data management work,8,15 we believe that discussion about how disparate pieces of data management tooling fit together to form an integrated system is missing. Our hope is to start an accessible conversation around what our data management systems should do, not how they go about doing this. We believe this will provide useful guidance for future work on technical solutions to this problem and provide motivation for a cultural shift around integrated and holistic data management. While we hope our perspective helps guide future work on research data infrastructure, it should not replace formal customer development or user requirement scoping processes, such as those used in technology and entrepreneurship (e.g. NSF Innovation Corps).16,17 Developers of new data management tools should thoroughly evaluate the needs of the scientists who will be using them, so that these tools are a simple and valuable addition to research workflows.
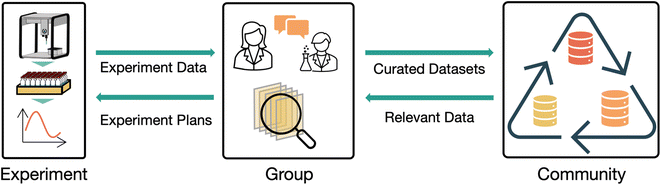
Throughout our discussion, we talk about data management ‘systems’ or ‘platforms’ in somewhat abstract terms. Because we aim to discuss our vision in terms of capabilities rather than specific implementations, we avoid discussing how these aspects of our vision could or should be implemented. If specific implementation strategies are of interest to the reader, we recommend the tutorial perspective on databases for chemistry by Duke et al.18 Any implementation of the ideas we discuss here would be intimately related to laboratory automation initiatives such as laboratory scheduling tools, remote equipment control capabilities, or full self-driving laboratories. The ‘ideal’ data management software implementation would likely include these capabilities, but these topics are out of scope for this perspective. To frame the field of experimental data management into a structured discussion, we break the task into three scales: experimental data collection, group data management, and community data sharing. Experimental data collection concerns the collection and management of data from individual experiments. Group data management concerns management of data within a laboratory, research group, collaboration, or organization. Community data sharing concerns the sharing of data among the broader community in a manner that makes it broadly accessible and reusable. Each scale has unique requirements and challenges but relies on integration with the other two scales to realize its full potential, as illustrated in Fig. 1.
Experiment-scale data management
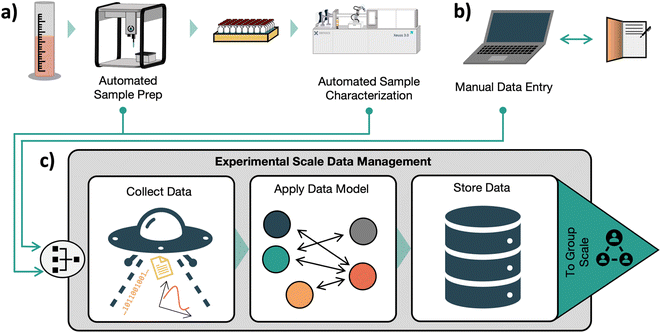
In our three-part organization framework, the task of accurately and completely gathering experiment data and metadata is handled at the experiment level. Here we consider an ‘experiment’ to be the collection of preparation, processing, and characterization steps performed on a sample or group of samples prepared in the same campaign, and ‘data’ to be any recorded information associated with an experiment, including characterization and preparation information. This is the minimum granularity of data that provides context to enable downstream use of the data. For example, in a synthesis experiment with characterization by nuclear magnetic resonance (NMR), the NMR results on their own are meaningless without the context of how the sample was prepared. In our framework, Experiment-scale data management tools are primarily concerned with correctly recording data, and may have limited support for enforcing quality of data, tracking the motivation behind an experiment, or otherwise providing context beyond the boundaries of an individual experiment. These tasks are mainly addressed at higher levels of our framework. An Experiment-scale data management tool should ensure that any point of data recorded in a laboratory is surrounded by the context needed to interpret it. To maintain this standard, such a system must be capable of maintaining a complete record of provenance, processing, and characterization steps applied to any sample. For example, in a battery electrolyte screening study, a sample record should contain the source of the stock solutions and components a sample is made from, a detailed log of the liquid or solid handling steps used in the preparation of the sample, and the results of any characterization processes. To the greatest extent possible, the collection and organization of this data should be automated. Manual data transfer slows research and introduces opportunities for error.19 However, many laboratories may never be fully automated, and some experimental steps may always require human interaction. The manual entry of data and notes by researchers needs to be well supported. A graphical user interface could provide this support. Recent advances in natural language processing technologies such as GPT-4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 may also enable new ways of recording data, such as a voice-assistant based lab notebook. Additionally, recording exploratory experiments without a preplanned structure should be straightforward. The intended steps in an experiment (e.g. target weights) should be captured for comparison to the actual executed experiment. This would allow for any deviations from the plan to be automatically flagged, aiding in the identification of systemic issues with an experiment or in the hardware that is used in its execution. To enable data-driven workflows like closed-loop optimization experiments, data should be made available in real time as it is collected. Once data from an experiment is collected, it should be stored in a flat structure to enable direct access to data attributes without parsing individual files. While data quality control is not a primary focus of an experimental data collection system in our vision, identifying ‘bad’ data as early as possible can avoid wasted time and effort. Thus, quality control should happen whenever possible. Users should be able to flag experiments and data points with known issues. Automatic real-time data validation could help catch mistakes as they happen to prevent wasted time and effort. However, these data collection systems should still log and store ‘bad’ data so that a complete record of experiments is obtained, and any data validation checks should pose a minimal interruption to the user. Implementation of a system that manages all these tasks needs to be simple to use so that adoption in the laboratory does not pose an undue burden on researchers. Such a system would streamline the collection and organization of data in the laboratory. We believe this would reduce errors associated with incorrect data recording and save researcher's time. Simpler data recording could facilitate the collection of data from experiments that ‘fail’ in the eyes of researchers, contributing to a more balanced record of experimental data that would better support machine learning use cases.21 This system would provide a single complete record of entire experiments, ensure data that might be relevant for future data science initiatives or repurposing of results is collected, and lay the foundation for the organization of data at the group and community scales. Fig. 2 illustrates how this system could interact with laboratory processes and equipment.
20 may also enable new ways of recording data, such as a voice-assistant based lab notebook. Additionally, recording exploratory experiments without a preplanned structure should be straightforward. The intended steps in an experiment (e.g. target weights) should be captured for comparison to the actual executed experiment. This would allow for any deviations from the plan to be automatically flagged, aiding in the identification of systemic issues with an experiment or in the hardware that is used in its execution. To enable data-driven workflows like closed-loop optimization experiments, data should be made available in real time as it is collected. Once data from an experiment is collected, it should be stored in a flat structure to enable direct access to data attributes without parsing individual files. While data quality control is not a primary focus of an experimental data collection system in our vision, identifying ‘bad’ data as early as possible can avoid wasted time and effort. Thus, quality control should happen whenever possible. Users should be able to flag experiments and data points with known issues. Automatic real-time data validation could help catch mistakes as they happen to prevent wasted time and effort. However, these data collection systems should still log and store ‘bad’ data so that a complete record of experiments is obtained, and any data validation checks should pose a minimal interruption to the user. Implementation of a system that manages all these tasks needs to be simple to use so that adoption in the laboratory does not pose an undue burden on researchers. Such a system would streamline the collection and organization of data in the laboratory. We believe this would reduce errors associated with incorrect data recording and save researcher's time. Simpler data recording could facilitate the collection of data from experiments that ‘fail’ in the eyes of researchers, contributing to a more balanced record of experimental data that would better support machine learning use cases.21 This system would provide a single complete record of entire experiments, ensure data that might be relevant for future data science initiatives or repurposing of results is collected, and lay the foundation for the organization of data at the group and community scales. Fig. 2 illustrates how this system could interact with laboratory processes and equipment.
Current data management workflows typically rely on manual record organization and usually fail to fully integrate automation and digitalization. 15 years ago, Shankar found that record keeping in research tends to be left up to individual researchers.22 In our experience, not much has changed since. Individuals are left to find a system they feel comfortable with. Experimental procedures and some results are usually stored in a researcher's paper laboratory notebook, in varying levels of detail. Data files from characterization instruments are generally organized in a directory and file naming structure set up by the researcher, and are manually copied from instruments to a central location like the researcher's PC or a cloud storage provider. Log files recording processing steps on automated equipment are stored in a similar fashion, if they are retained. These files are linked together manually using a laboratory notebook as an index.23 Records of provenance for results reside across an array of files, notes, and the researcher's memory. Extracting data from these results for future work tends to require bespoke file and data processing and can be an onerous task. There are many examples of projects and tools that address these limitations and implement aspects of our vision. A core component of an automated data management system is the ability to retrieve data from experimental equipment. A common approach for this task is to write a series of custom scripts that collect and aggregate data.24,25 This approach is effective but can require extensive effort to implement and may be impacted by small changes in the laboratory environment, such as updated equipment configurations. Laboratory orchestration software packages and standards that enable automated experimentation already interact directly with equipment, which provides an opportunity to leverage existing capabilities to automate data capture. The Bluesky family of python packages allows users to specify and execute experiments and collect data by directly interfacing with hardware that uses the EPICS protocol as well as a few other hardware interface protocols.26 This project was developed to standardize experiment specification and data collection from synchrotron light sources. It is widely used at the National Synchrotron Light Source II (NSLS-II) as well as other US and international light sources.27 The Standardization in Lab Automation (SiLA) standard and the Laboratory and Analytical Device Standard (LADS) are two competing standards that seek to provide a unified application programming interface (API) for interacting with lab equipment. Both are primarily targeted at life science laboratories, and both build from existing network communication protocols to provide lab-specific features. SiLA has seen adoption among equipment manufacturers,28 and LADS is scheduled to be released in late 2023.29,30 Collaborative development of competing lab equipment standards could lead to a set of widely adopted interfaces to equipment that each have specialized support for a particular use case. This would allow experimenters to pick the best tools for particular experimental tasks. For example, an automated flow-through nanoparticle synthesis experiment could communicate with a bank of syringe pumps over SiLA to control experimental conditions and a beamline with BlueSky to manage sample characterization. Each of these standards fulfills the needs of the application it is used for and alleviates the need for a single monolithic standard to handle every research task imaginable. However, development of many overlapping standards also has the potential to fracture the ecosystem for managing hardware and software, and preclude straightforward digital data management and communication. Care should be taken in standards development and adoption to avoid this.
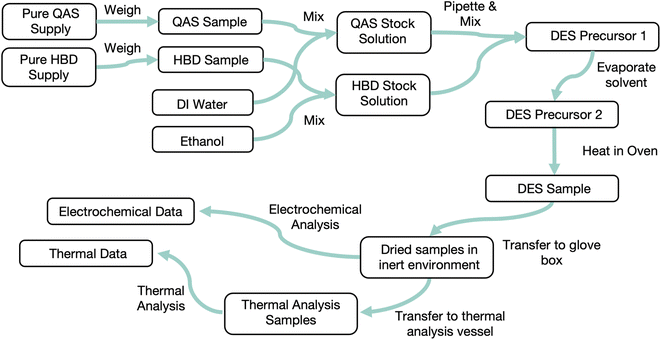
Data collected from an experiment needs to be validated, organized, and stored. Data validation checks that collected data is in an expected format and an expected range. For example, a simple validation on the recorded mass of a sample could check first that the entry is numeric and not a text string, then that the value is within the measurable range of the balance used. This approach does not verify that the recorded number is correct but can catch major issues with data. As discussed above, invalid data should still be recorded but also flagged for review. Several data models for organizing experimental data have been proposed. A common theme among many of them is to represent each sample in an experiment as a graph of sample states connected by procedures. This is an intuitive way to represent an experiment: in the lab, a sample starts from some feedstock materials (an initial state) before a number of processing steps are applied, each of which generates a new sample state. At states of interest, characterizations on the sample are performed. Fig. 3 illustrates the application of a graph data model to an experimental procedure for one sample.31 Projects or works that use this form of data model include the Event-Sourced Architecture for Materials Provenance (ESAMP),32 Cript,15 and Citrine Informatics' GEMD data model.33 Explicitly organizing experimental data in a format that follows this structure would make tracking the provenance of any piece of data straightforward: given a sample or measurement, the chain of processes and samples can be traced backwards to determine where the sample came from, or forwards to see how a future step turned out. This data model can be implemented in any database or storage format, and each of the mentioned projects has built its own version. A prerequisite to using such a data model is a schema to describe what data is stored and how it is related. Tools to parse data from its source and transform it into the data model are also needed to implement the data models we describe. Developing these items can be a significant challenge. A potential opportunity exists to establish a standardized representation for experiments that can be shared and reused between different software systems.
Once infrastructure is in place to collect data from equipment and a conceptual model for organizing that data is agreed on, these must be implemented into a piece of useable software. The Experiment Specification, Capture, and Laboratory Automation Technology (ESCALATE) ontology and software package implements tools to specify experiment plans, record experimental execution, and link resulting files to samples in a database.34 ESCALATE provides both a data model for organizing experimental data as well as a software implementation. It provides capabilities to specify experiment plans, record experimental executions, and manage files. Users interact with the software either through a graphical web interface or an API. Bespoke solutions in this space are also common. Several organizations have discussed the design and implementation of custom in-house data management systems. When implementing their internal data system, The Joint Center for Artificial Photosynthesis developed a lightweight system of file types and scripts to track sample data and automate the recording of data when possible.35 Data from this system eventually made it into the Materials Experiment and Analysis Database (discussed below). The National Renewable Energy Laboratory (NREL) has implemented a similar file and script based workflow that enables tracking sample preparation and characterization data by having users load data files into a centralized warehouse.25
Electronic lab notebooks (ELNs) also fit into this section of our framework. Traditional ELNs sought to entirely replace paper lab notebooks with a direct translation to a digital document. While this provides major improvements for data searchability, shareability, and security, it does not enable the data collection infrastructure we envision. More modern ELNs incorporate more extensive data management features, like recording data directly from instruments or supporting inline data analysis.36,37 Modern ELNs can also interact with a vendor's laboratory information management system (LIMS) product to enable organization of data across experiments and laboratories. LIMS are discussed below. Given the wide selection of ELN systems tailored for diverse types of lab work, the lack of adoption in academic laboratories38 raises questions. While a full exploration of issues associated with the limited adoption of ELNs is beyond the scope of this work, high cost, significant effort, low community expectations, and traditionalist attitudes are contributing factors.23
Group scale data management
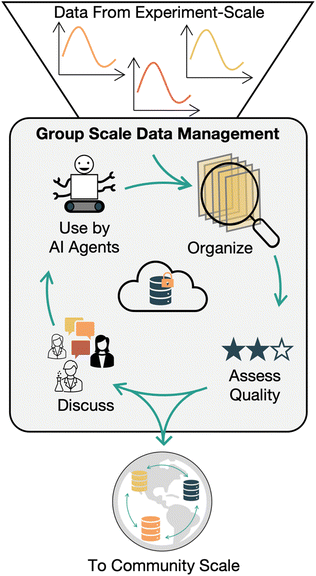
In our vision, the goal of a Group scale data management platform is to organize individual experiments across research projects and other group objectives. In this discussion a group is a collection of researchers actively collaborating on a project, such as an academic research group, a department/unit, or multiple collaborators spanning different institutions. Elements of editorial discretion are also introduced at this level. Group members will know and trust each other, understand the context around the collection of data, and agree on how it will be used. They will also be involved in discussions to assess the quality of collected data prior to broadly disseminating it or reporting major outcomes in the scientific literature. Effective Group scale data management builds from and complements strong experimental-scale data collection and management. In practice, the distinction between experimental and group scales is ‘thin’ and may not be apparent in real world data management systems. However, the disparate goals of these two data management scales merit separate treatments.To support the goals of group data management, tools should enable linking data from related experiments and samples. In turn, groups of experiments should be linkable into project campaigns. For example, in an automated sample synthesis process, multiple experiment campaigns might be run, with multiple replicates of a material in each campaign. Samples that are replicates of one material should be linked to that material, as well as to the campaign where they were generated. In our examples, multiple high-throughput synthesis campaigns that are all part of the same project should be linked and accessible as one combined project. Data from different but loosely related experiments should also be grouped together. A user should be able to view all the work done with a particular sample precursor, grouped across different sample preparations and experiments. Computational results should be included in this grouping to broaden the scope of available information. Having all this data in one place will enable anyone involved in a project, be they researchers, supervisors, or artificial intelligence agents, to have access to up-to date versions of data which will enable faster and smarter decision making around future experimental plans. It is important that editorial tasks and human data interpretation be supported. Capturing the motivation and intent behind running an experiment can give context to a group of experiments in a project. Low quality or compromised data could still be relevant to a project at this scale, but quality issues should be flagged. Tracking the quality of data should be supported. This could involve automated or human data review. Backups of data should be automated so data isn't lost by accident, and modifications to data should be tracked and version controlled so data isn't manipulated by malice or by mistake.39 Preparation of data for downstream uses, like machine learning initiatives or for ‘export’ to community level databases, should be straightforward and automatable to prevent errors and to remove data processing bottlenecks (Fig. 4).
As with experimental data collection, a common approach to managing data at the group scale is to design a custom system. This can either be a software system, or a manual workflow. Building a custom software system requires the expertise and effort to set up physical and digital data management infrastructure but can yield a system that better complements a laboratory's experimental workflows. The internal systems developed at NREL and JCAP, both discussed above, also provide group data management capabilities. They allow for linking individual experiments into campaigns, sorting experiments by criteria like experimental method, and sharing up to date data among a group.25,35 These one-off systems can work well for groups with the resources to fully implement them but are out of reach for most researchers. Many laboratories have pieced together a manually updated data management system centered around a commercial cloud storage provider such as Google Drive or Dropbox. These platforms are attractive to use as they facilitate simple data sharing amongst laboratory members, provide a degree of data versioning and backup, and are often provided via an institutional license making them free to use. Before relying on third party cloud storage solutions, researchers need to consider the appropriateness of a particular offering for the sensitivity of the data they work with. As an example, storing protected health information on a consumer Google Drive account would violate HIPAA.40 The vendors of these products may also choose to make changes to either the product itself or the terms of use of the product that can be disruptive to how they are used in a laboratory or group, forcing researchers to make disruptive changes to their workflows.
Commercially available systems for managing data at the group scale are commonly referred to as Laboratory Information Management Systems (LIMS). Traditional LIMS systems provide sample tracking and provenance management capabilities, a centralized store of experimental data and other information, some level of integration with instruments for data collection, capabilities to manage experimental workflows, and a user interface. Some systems integrate with automated equipment, providing capabilities we classify as experiment management. These systems usually work in concert with a vendor's ELN solution. Like ELNs, LIMS are available from a robust array of vendors,37,41–43 and at least one open source option is available.44 Many available LIMS systems are designed for life science or commercial laboratories, but there are options targeted at materials science research. Dotmatics offers a platform with LIMS capabilities that is designed for materials science and chemistry laboratories.45 Citrine informatics' data management system is based on their GEMD data model (discussed above) and targeted at materials laboratories. This system has the benefit that data is extracted from files and stored directly in their data model, which makes the data more searchable and useable.46 As with ELNs, adoption in academic laboratories is limited, likely for the same reasons (e.g. cost and/or complexity).
We believe that there is a notable lack of open-source software options that provide the capabilities we envision for laboratory data management, especially in materials science fields. An open-source software tool, built off a robust experiment capture infrastructure as described above, would make the group data management we envision accessible and customizable for a wide array of groups. An important criterion for this software will be its useability and ease of adoption, in addition to how it handles technical data management tasks. Adoption of a group data infrastructure with the attributes we envision that is integrated with experimental data collection tools would revolutionize data management in most academic laboratories, and in our opinion would be a worthwhile investment on its own. However, even greater benefits can be realized by using this infrastructure to share research data with the broader community.
Community scale data management
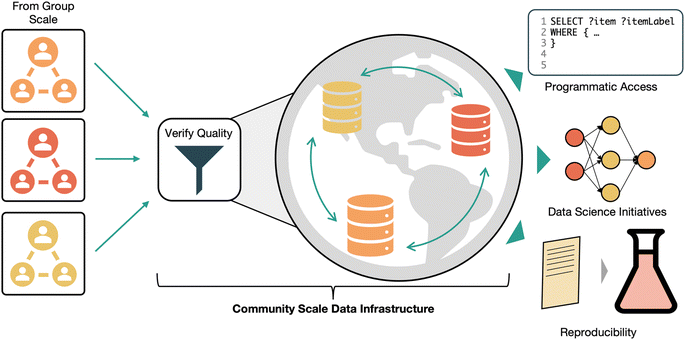
Community data sharing has always been at the core of scientific communication. Traditionally, data has been shared through plots and tables in manuscripts, with important context embedded in manuscript text. While recent efforts make it possible to extract information from these documents using natural language processing methods,47–50 traditional publications are not an efficient way of transmitting data. Fortunately, a slow shift towards more open data sharing is underway. The importance of accessible data sharing has gained broad acceptance. References to making data FAIR (originally defined as findable, accessible, interoperable, and repurpose-able)51 are common in the literature.50,52–54 Well-designed community data sharing practices enable the aggregation and dissemination of data from multiple research groups and heterogeneous projects in a unified fashion, allowing existing data to drive unforeseen future works. Ultimately the goal of openly and accessibly sharing research data is to make it reusable in future research. Effective data sharing can enable new machine learning initiatives, make comparing new results to existing values simple, or prevent the unnecessary reproduction of existing work. What exactly this future reuse looks like is difficult to define, which is part of what makes establishing robust and useful data sharing infrastructure difficult. Thus, community data sharing initiatives should be built to be as generally useful as possible, rather than optimizing for a particular downstream use case. To support data findability, data should be stored in curated, focused databases. The domain scope for these databases should be tuned so that relevant materials or experiments are stored together, without fractioning the ecosystem into hyper-specific datasets. Choosing a level of specificity for a database is an important consideration that impacts how data is likely to be re-used in the future. Specialized databases may make re-use simple for new applications that are similar to the original use of the data. However, being too specific can limit community contributions and engagement that is needed to sustain a database after initial support runs out, and can make it difficult to find databases that contain the desired information. Conversely, databases that are too broad in scope might not support the level of detail needed for some downstream use cases. In an example from computational materials science, the Catalysis Hub surface reactions database provides a home for relaxation and chemisorption energies of reactants on catalyst surfaces obtained from electronic structure calculations.55 This focus strikes a balance that makes it specific enough to be useful, but broad enough to have over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 entries. A critical aspect of making data accessible is ensuring the long-term existence of databases and their accessibility over the internet. Shared data is an important part of the scientific record, so community databases should be administered in a way that guarantees long term availability. To make data interoperable, data needs to be accessible as database records rather than as groups of files. This makes searching and filtering data using standard query tools like SQL or SPARQL possible. However, existing community and domain specific standards should be respected where applicable. For example, the small angle scattering (SAXS, SANS) community has standardized beamline data around the CANSAS standard.56 Any database dealing in a particular type of data needs to support data retrieval in the agreed standard to facilitate use with domain-specific tools. Data format exchange tools like Tiled57 can facilitate data access in preferred formats. Human readability should be enabled through well designed user interfaces. This would make exploratory data analysis easier and enable access by users without a coding background. To make data reusable, it is necessary to maintain a minimum standard of quality and completeness for data. Low quality data can't be tolerated at this scale because users lack the context to critically evaluate data quality and experimental nuances that is present at smaller scales. Peer-reviewing data submissions to these databases as part of journal manuscript submissions could help curate this data quality. To enable reproducibility of data (another interpretation of the ‘R’ in FAIR), information about how a sample was prepared and characterized should be shared in a standardized format. Intermediate data points should be shared alongside final results when possible, to support use cases that rely on them. Fig. 5 illustrates the process for sharing data in this envisioned system. Once one establishes a new community data repository that satisfies all the points of our vision, attention needs to be turned to the social aspects of managing a community data resource. Use of these databases needs to be incentivized for researchers. Mechanisms for encouraging use might involve mandates from publishers or funding agencies, the generation of a citable digital object identifier (DOI) and other mechanisms for claiming credit for data submissions, and a seamless interface with lab data management systems to take the pain out of publishing data. When broadly sharing data as we advocate for, export controls may need to be considered. Current US export controls don't generally restrict the public sharing of basic research outputs.58 However the US government has recently announced efforts to limit foreign access to US technology59 and influence over research,60 so future controls on research sharing may become more restrictive. Researchers outside of the US will need to consider how their government's export control policies might impact how they share research data.
000 entries. A critical aspect of making data accessible is ensuring the long-term existence of databases and their accessibility over the internet. Shared data is an important part of the scientific record, so community databases should be administered in a way that guarantees long term availability. To make data interoperable, data needs to be accessible as database records rather than as groups of files. This makes searching and filtering data using standard query tools like SQL or SPARQL possible. However, existing community and domain specific standards should be respected where applicable. For example, the small angle scattering (SAXS, SANS) community has standardized beamline data around the CANSAS standard.56 Any database dealing in a particular type of data needs to support data retrieval in the agreed standard to facilitate use with domain-specific tools. Data format exchange tools like Tiled57 can facilitate data access in preferred formats. Human readability should be enabled through well designed user interfaces. This would make exploratory data analysis easier and enable access by users without a coding background. To make data reusable, it is necessary to maintain a minimum standard of quality and completeness for data. Low quality data can't be tolerated at this scale because users lack the context to critically evaluate data quality and experimental nuances that is present at smaller scales. Peer-reviewing data submissions to these databases as part of journal manuscript submissions could help curate this data quality. To enable reproducibility of data (another interpretation of the ‘R’ in FAIR), information about how a sample was prepared and characterized should be shared in a standardized format. Intermediate data points should be shared alongside final results when possible, to support use cases that rely on them. Fig. 5 illustrates the process for sharing data in this envisioned system. Once one establishes a new community data repository that satisfies all the points of our vision, attention needs to be turned to the social aspects of managing a community data resource. Use of these databases needs to be incentivized for researchers. Mechanisms for encouraging use might involve mandates from publishers or funding agencies, the generation of a citable digital object identifier (DOI) and other mechanisms for claiming credit for data submissions, and a seamless interface with lab data management systems to take the pain out of publishing data. When broadly sharing data as we advocate for, export controls may need to be considered. Current US export controls don't generally restrict the public sharing of basic research outputs.58 However the US government has recently announced efforts to limit foreign access to US technology59 and influence over research,60 so future controls on research sharing may become more restrictive. Researchers outside of the US will need to consider how their government's export control policies might impact how they share research data.
The current state of experimental data sharing in materials science has been described as ‘critical’.61 A report on the materials genome initiative by the (US) National Science Foundation points to data sharing as a bottleneck in the materials innovation process, and suggests that national agencies should establish data sharing infrastructure like what we envision above.62 We think this is an appropriately dire assessment of the current situation, but there is a lot of work in this space that gives cause for hope. Perhaps the most widespread form of data sharing currently is through journal article electronic supplementary information (ESI) and general data repositories like Zenodo63 and Dryad.64 Repositories allow users to upload their data files along with a description of what the data contains, and then generate a unique identifier for that data. Materials focused repositories include the Materials Data Facility65 and Citrination.66 Sharing data in this form at least makes it available but doesn't address the spirit of the FAIR philosophy. Assembling datasets from files shared as ESI, or via a repository, requires searching the literature for relevant publications and datasets, checking that they contain the desired data, then manually collecting and parsing files that likely use unique (i.e. non-standardized) formatting. Poor metadata annotation often means one needs to read the original journal publication to understand the data. Curated databases provide solutions to these issues. These databases aggregate data from multiple experiments and projects into one database with a specific domain focus. Examples in the materials science field are limited. The NREL High Throughput Experimental Materials (HTEM) database contains records for over 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique samples of inorganic thin film materials. This database is populated using the NREL internal research data infrastructure described in the experimental data section and includes characterization data as well as some sample synthesis metadata.67 The Materials Experiment and Analysis Database (MEAD) was hosted by the Joint Center for Artificial Photosynthesis and populated by their internal experimental data collection system.35 Unfortunately, this database was not accessible at the time of writing, which highlights the need to plan for the long-term stability and availability of community data resources. The inorganic crystal structure database (ICSD) provides a database of inorganic crystal structures compiled from literature.68 This database is neither open to community contributions, nor open access. Outside of experimental materials science, many more examples show the promise of shared community data sharing. The Materials Project is a widely known community database for computational materials data. It contains properties for over 154
000 unique samples of inorganic thin film materials. This database is populated using the NREL internal research data infrastructure described in the experimental data section and includes characterization data as well as some sample synthesis metadata.67 The Materials Experiment and Analysis Database (MEAD) was hosted by the Joint Center for Artificial Photosynthesis and populated by their internal experimental data collection system.35 Unfortunately, this database was not accessible at the time of writing, which highlights the need to plan for the long-term stability and availability of community data resources. The inorganic crystal structure database (ICSD) provides a database of inorganic crystal structures compiled from literature.68 This database is neither open to community contributions, nor open access. Outside of experimental materials science, many more examples show the promise of shared community data sharing. The Materials Project is a widely known community database for computational materials data. It contains properties for over 154![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 materials69 and has over 200
000 materials69 and has over 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 registered users.70 This database makes computed properties of materials available via both an easy to navigate web page and an API. Arguably the best success story of community data sharing is the Protein Data Bank (PDB), a database of protein structures. This database has lasted for over 50 years and has grown to over 30
000 registered users.70 This database makes computed properties of materials available via both an easy to navigate web page and an API. Arguably the best success story of community data sharing is the Protein Data Bank (PDB), a database of protein structures. This database has lasted for over 50 years and has grown to over 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 data contributors and over a million site views per year. This database provides curated, validated data in a consistent format. It has become a core part of research in its field, as submitting a new protein structure for publication practically requires submission of that structure to the PDB.71 Access to the PDB has also enabled groundbreaking advancements such as the accurate prediction of protein folding and de novo structures based on sequence, with machine learning models.9,72
000 data contributors and over a million site views per year. This database provides curated, validated data in a consistent format. It has become a core part of research in its field, as submitting a new protein structure for publication practically requires submission of that structure to the PDB.71 Access to the PDB has also enabled groundbreaking advancements such as the accurate prediction of protein folding and de novo structures based on sequence, with machine learning models.9,72
Several projects and organizations are working toward new data sharing platforms that provide many of the capabilities we described above. The FAIRmat consortium is a German initiative to realize many of the goals for community data sharing that we describe here. This project aims to build a series of domain specific data repositories, following similar criteria as we propose to establish “as few as possible but as many are needed” to support diverse needs of different fields. Their proposal, which is to create a federated network of databases with centrally searchable metadata, has the potential to enable domain specific databases that are still findable and reusable for applications in other contexts. This project has also considered the needs of experimental and laboratory data management infrastructure to feed these community repositories.73 The development of knowledge graphs to store and structure experimental data is a promising approach to implanting data sharing infrastructure. Knowledge graphs structure information as a connected graph, with data points and entities represented as graph nodes and properties or relationships represented as edges.74 This is similar to the graph data models discussed in the experimental data collection section. When coupled with a standardized definition of properties and relationships, known as an ontology, knowledge graphs promise enhanced interoperability between databases and flexibility in defining data schemas. Knowledge graphs can be queried using standard query languages such as SPARQL. Bai et al. envision a knowledge-graph based approach to managing data, and more broadly, laboratory automation. In their vision, computational agents maintain the state of the knowledge graph and use it to drive closed-loop experimentation.14 The Mat-o-lab initiative seeks to develop domain-specific ontologies for materials research, and corresponding knowledge graph representations of data.54 This initiative envisions a network of knowledge graphs using compatible ontologies to represent data and enable re-use across projects. The OPTIMADE consortium aims to solve the data interoperability challenge by providing an API specification for materials databases.75 This specification has been adopted by several databases including the Materials Project.76 The wide range of solutions under development to address the community data sharing problem shows that this issue is well understood by the community and makes us hopeful that truly FAIR data sharing is near.
Common obstacles and recommendations
As shown above, data management is an active area of research, and the need for the capabilities we describe herein are recognized by the community. So, what motivated this perspective? Most of these initiatives are carried out as individual efforts to solve a small subset of the problems facing the field. While this bottom-up approach is leading to innovative and exciting tools, these individual efforts generally don't integrate with other tools and do little to reduce the fractured nature of the data management field. The NREL internal research data infrastructure team recognized these limitations, and called for a top-down approach to design data management infrastructure with a holistic vision in mind.25 While this is a noble goal, building an entire research data infrastructure from scratch as one project is a major undertaking. Further, one organization is likely unable to anticipate and build for the diverse use cases and requirements of such a tool. Rather than leave the development of future tools to one entity, we believe that future development should continue to be undertaken by diverse community projects, but with a stronger eye towards how projects will inter-operate to enable the seamless data management system we envision. To enable this interoperability between different tools and software, we should define and adopt standards for how data is represented as a community. As we discussed in the experimental data collection section, interfacing lab equipment with any data collection software is a challenge due to vendor-specific protocols and data formats. Developing a standardized API for communicating with laboratory equipment would resolve this challenge. A standardized data model for representing collected experimental data would enable greater interoperability between competing solutions. Agreeing on a common implementation and specification of a graph data model for sharing between data management tools would enable easier data sharing and make data more reusable. Alongside data, information needed to reproduce experiments needs to be shared. The χ-DL project seeks to develop a ‘compilable’ language for specifying synthesis steps.6,7 A standard means to specifying experimental procedures promises to make experiments reproducible on heterogeneous automated experimentation platforms.We noted that there is an acute lack of software tools to organize and store data at the laboratory or group scale. This gap in the data management infrastructure compounds shortfalls in experimental data collection and community data sharing. Without an effective means to organize and use large amounts of experimental data and metadata, little motivation exists to expend effort to collect data beyond what is immediately needed. And if collected data is scattered across a wide range of files and locations, preparing data for submission to a community database can be a herculean task. A robust, user-friendly, and generalizable implementation of a laboratory data management system would bridge the gap between the two other levels of data management, encouraging wider experimental data collection and facilitating rapid dissemination of data to community databases. An open-source software tool for managing laboratory data that implements the data management standards we describe should be created, either by establishing a new project or extending an existing one. This tool should be generalizable to different laboratory environments, but customizable to provide the specificity and workflow efficiencies needed for any given laboratory. This tool should provide both a graphical user interface for easy data management, as well as programmatic access via (at least) a python library so that use of the tool can be integrated into existing data-generating processes controlled by python scripts. In practice, such a tool could also fulfill our vision for experimental data collection and management as the two are closely related. We believe the availability of an open-source tool (as opposed to a proprietary one) in this space is critical. While proprietary software can solve many data management problems, it raises issues with vendor lock-in, laboratory equipment support, and custom use cases. Automation in research laboratories involves prototyping new hardware and workflows, so it is virtually impossible for a commercial vendor to envision and support all the possible use cases. A community-driven open-source tool could be more responsive to new applications, and individual laboratories would have a fair shot at adapting an open-source tool to creative new workflows or use cases. An open-source tool would also remove financial hurdles to using a data management infrastructure.
At present, implementing data management infrastructure requires a high level of proficiency in software engineering and systems administration. Most academic laboratories do not have access to personnel with these skills. Tasking grad students with learning them is problematic as it adds to their already full workloads. Expecting laboratories to establish their own research data infrastructure13,18 will lead to low adoption rates, poor quality systems, and frazzled graduate students. We do not think it is fair to ask this of junior researchers. Community organizations should establish a leadership stance on this front and guide the development of both software tools for use at the experimental and group scale, and community databases for data sharing. These organizations could be federal agencies like the NSF or NIST, existing community organizations like the Materials Research Society, or newly formed consortia with data management as their express goal. For their part, the NSF has recognized the need for a leader in this space, and the role they could play.62 The FAIRmat initiative has been referenced as a model for what that role could look like.62
A common theme across all areas of data management is the need for broad community buy-in. Building the perfect data management tool won't make a difference if nobody chooses to use it. Getting researchers to move beyond paying lip service to data as a first-class research product is a major barrier to our envisioned data management future. Funding agencies should make effective data sharing a core project requirement by seriously considering data management plans in grant application evaluations and following up to ensure they are followed. The extra overhead this creates for researchers should also be recognized and accommodated. Publishers could require data sharing to community databases as a condition of publication.77 Mechanisms for giving credit to researchers for their data contributions would provide further incentives. Success stories shared by early adopters of time saved, mistakes avoided, and discoveries enabled by new data management tools and practices would also help show that taking data management seriously is a worthwhile endeavor.
Conclusion
Materials science and chemistry are entering a new phase where automated and autonomous experimentation methods will multiply the capabilities of researchers and make new data driven research paradigms commonplace. To make the most out of these new research paradigms, the community needs to overhaul how data is handled at all scales of the research process. However, we are concerned that the moment to make these changes is being missed. Rather than enabling new research approaches based on easy access to data, new automated methods may lead to an incomprehensible data landscape and siloed research projects. We believe that effective data management practices for this new era must consider the entire data lifecycle across scales from individual experiments to broad community dissemination. We envision one possible set of capabilities and norms that could contribute to such a multiscale data management system. However, this is certainly not the only vision that should be considered. Our hope is that this perspective encourages more researchers to participate in the discussion around data management by making it accessible and by presenting a tantalizing potential future where data management ‘just works’ and can fade into the background. We are excited to hear alternative visions from other researchers and to collaborate towards a future that embraces digital data management methods and prevents us from getting lost in the ‘Laboratory of Babel’.Data availability
As this is a perspective article, no primary research results, data, software or code have been included.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is primarily supported by the National Science Foundation through NSF-CBET Grant No. 1917340, which directly supported the background research and writing work of B. Pelkie. We also acknowledge support from the US Department of Energy (DOE) Office of Energy Sciences (BES) under Award Number DE-SC0019911 and the UW Molecular Engineering Materials Center, a Materials Research Science and Engineering Center (Grant No. DMR-1719797), which supported L. D. Pozzo and enabled broad understanding of common data management needs across AI-driven projects and platforms.References
- M. L. Green, C. L. Choi, J. R. Hattrick-Simpers, A. M. Joshi, I. Takeuchi, S. C. Barron, E. Campo, T. Chiang, S. Empedocles, J. M. Gregoire, A. G. Kusne, J. Martin, A. Mehta, K. Persson, Z. Trautt, J. Van Duren and A. Zakutayev, Fulfilling the Promise of the Materials Genome Initiative with High-Throughput Experimental Methodologies, Appl. Phys. Rev., 2017, 4(1), 011105, DOI:10.1063/1.4977487.
- C. Ashraf, N. Joshi, D. A. C. Beck and J. Pfaendtner, Data Science in Chemical Engineering: Applications to Molecular Science, Annu. Rev. Chem. Biomol. Eng., 2021, 12(1), 15–37, DOI:10.1146/annurev-chembioeng-101220-102232.
- M. Seifrid, J. Hattrick-Simpers, A. Aspuru-Guzik, T. Kalil and S. Cranford, Reaching Critical MASS: Crowdsourcing Designs for the next Generation of Materials Acceleration Platforms, Matter, 2022, 5(7), 1972–1976, DOI:10.1016/j.matt.2022.05.035.
- PhasIR: An Instrumentation and Analysis Software for High-throughput Phase Transition Temperature Measurements, https://openhardware.metajnl.com/articles/10.5334/joh.39/, accessed 2022-11-17 Search PubMed.
- F. Häse, M. Aldeghi, R. J. Hickman, L. M. Roch and A. Aspuru-Guzik, Gryffin: An Algorithm for Bayesian Optimization of Categorical Variables Informed by Expert Knowledge, Appl. Phys. Rev., 2021, 8(3), 031406, DOI:10.1063/5.0048164.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson, D. Angelone and L. Cronin, Organic Synthesis in a Modular Robotic System Driven by a Chemical Programming Language, Science, 2019, 363(6423), eaav2211, DOI:10.1126/science.aav2211.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, A Universal System for Digitization and Automatic Execution of the Chemical Synthesis Literature, Science, 2020, 370(6512), 101–108, DOI:10.1126/science.abc2986.
- C. J. Leong, K. Y. A. Low, J. Recatala-Gomez, P. Quijano Velasco, E. Vissol-Gaudin, J. D. Tan, B. Ramalingam, R. I Made, S. D. Pethe, S. Sebastian, Y.-F. Lim, Z. H. J. Khoo, Y. Bai, J. J. W. Cheng and K. Hippalgaonkar, An Object-Oriented Framework to Enable Workflow Evolution across Materials Acceleration Platforms, Matter, 2022, 5(10), 3124–3134, DOI:10.1016/j.matt.2022.08.017.
- M. Baek, F. DiMaio, I. Anishchenko, J. Dauparas, S. Ovchinnikov, G. R. Lee, J. Wang, Q. Cong, L. N. Kinch, R. D. Schaeffer, C. Millán, H. Park, C. Adams, C. R. Glassman, A. DeGiovanni, J. H. Pereira, A. V. Rodrigues, A. A. van Dijk, A. C. Ebrecht, D. J. Opperman, T. Sagmeister, C. Buhlheller, T. Pavkov-Keller, M. K. Rathinaswamy, U. Dalwadi, C. K. Yip, J. E. Burke, K. C. Garcia, N. V. Grishin, P. D. Adams, R. J. Read and D. Baker, Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network, Science, 2021, 373(6557), 871–876, DOI:10.1126/science.abj8754.
- L. Chanussot, A. Das, S. Goyal, T. Lavril, M. Shuaibi, M. Riviere, K. Tran, J. Heras-Domingo, C. Ho, W. Hu, A. Palizhati, A. Sriram, B. Wood, J. Yoon, D. Parikh, C. L. Zitnick and Z. Ulissi, Open Catalyst 2020 (OC20) Dataset and Community Challenges, ACS Catal., 2021, 2020, 6059–6072, DOI:10.1021/acscatal.0c04525.
- D. A. Beck, J. M. Carothers, V. R. Subramanian, and J. Pfaendtner, Data Science: Accelerating Innovation and Discovery in Chemical Engineering, 2016, Vol. 62, pp 1402–1416.
- J. L. Borges, The Library of Babel, The Garden of Forking Paths, Editorial Sur, 1941 Search PubMed.
- J. Medina, A. W. Ziaullah, H. Park, I. E. Castelli, A. Shaon, H. Bensmail and F. El-Mellouhi, Accelerating the Adoption of Research Data Management Strategies, Matter, 2022, 5(11), 3614–3642, DOI:10.1016/j.matt.2022.10.007.
- J. Bai, L. Cao, S. Mosbach, J. Akroyd, A. A. Lapkin and M. Kraft, From Platform to Knowledge Graph: Evolution of Laboratory Automation, JACS Au, 2022, 2(2), 292–309, DOI:10.1021/jacsau.1c00438.
- D. Walsh, W. Zou, L. Schneider, R. Mello, M. Deagen, J. Mysona, T.-S. Lin, J. Pablo, K. Jensen, D. Audus, and B. Olsen, CRIPT: A Scalable Polymer Material Data Structure, 2022, DOI:10.26434/chemrxiv-2022-xpz37.
- L. Bosman and J. Garcia-Bravo, Lessons Learned: Research Benefits and Beyond Associated with Participating in the NSF I-Corps™ Customer Discovery Program, Technol. Innovation, 2021, 22(1), 41–54, DOI:10.21300/21.4.2021.5.
- C. C. Nnakwe, N. Cooch and A. Huang-Saad, Investing in Academic Technology Innovation and Entrepreneurship: Moving Beyond Research Funding through the NSF I-CORPS™ Program, Technol. Innovation, 2018, 19(4), 773–786, DOI:10.21300/19.4.2018.773.
- R. Duke, V. Bhat and C. Risko, Data Storage Architectures to Accelerate Chemical Discovery: Data Accessibility for Individual Laboratories and the Community, Chem. Sci., 2022, 13, 13646–13656, 10.1039/D2SC05142G.
- S. Seidel, M. N. Cruz-Bournazou, S. Groß, J. K. Schollmeyer, A. Kurreck, S. Krauss, and P. Neubauer, A Comprehensive IT Infrastructure for an Enzymatic Product Development in a Digitalized Biotechnological Laboratory, Advances in Biochemical Engineering/Biotechnology, in Smart Biolabs of the Future, ed. S. Beutel and F. Lenk, Springer International Publishing, Cham, 2022, pp. 61–82, DOI:10.1007/10_2022_207.
- OpenAI, GPT-4 Technical Report, arXiv March 16, 2023, DOI:10.48550/arXiv.2303.08774.
- J. M. Cole, The Chemistry of Errors, Nat. Chem., 2022, 14(9), 973–975, DOI:10.1038/s41557-022-01028-6.
- K. Shankar, Order from Chaos: The Poetics and Pragmatics of Scientific Recordkeeping, J. Am. Soc. Inf. Sci. Technol., 2007, 58(10), 1457–1466, DOI:10.1002/asi.20625.
- S. Kanza, C. Willoughby, N. Gibbins, R. Whitby, J. G. Frey, J. Erjavec, K. Zupančič, M. Hren and K. Kovač, Electronic Lab Notebooks: Can They Replace Paper?, J. Cheminf., 2017, 9(1), 31, DOI:10.1186/s13321-017-0221-3.
- N. Schwarz, S. Veseli and D. Jarosz, Data Management at the Advanced Photon Source, Synchrotron Radiat. News, 2019, 32(3), 13–18, DOI:10.1080/08940886.2019.1608120.
- K. R. Talley, R. White, N. Wunder, M. Eash, M. Schwarting, D. Evenson, J. D. Perkins, W. Tumas, K. Munch, C. Phillips and A. Zakutayev, Research Data Infrastructure for High-Throughput Experimental Materials Science, Patterns, 2021, 2(12), 100373, DOI:10.1016/j.patter.2021.100373.
- Hardware Interface Packages — bluesky 1.10.0.post14+gfc4204d4 documentation, https://blueskyproject.io/bluesky/hardware-interfaces.html, accessed 2022-11-17 Search PubMed.
- D. Allan, T. Caswell, S. Campbell and M. Rakitin, Bluesky's Ahead: A Multi-Facility Collaboration for an a La Carte Software Project for Data Acquisition and Management, Synchrotron Radiat. News, 2019, 32(3), 19–22, DOI:10.1080/08940886.2019.1608121.
- D. Juchli, SiLA2: The Next Generation Lab Automation Standard, Advances in Biochemical Engineering/Biotechnology, in Smart Biolabs of the Future, ed. S. Beutel, and F. Lenk, Springer International Publishing, Cham, 2022, pp. 147–174, DOI:10.1007/10_2022_204.
- A. Brendel, F. Dorfmüller, A. Liebscher, P. Kraus, K. Kress, H. Oehme, M. Arnold, and R. Koschitzki, Laboratory and Analytical Device Standard (LADS): A Communication Standard Based on OPC UA for Networked Laboratories, Advances in Biochemical Engineering/Biotechnology, in Smart Biolabs of the Future, ed. S. Beutel, and F. Lenk, Springer International Publishing, Cham, 2022, pp. 175–194, DOI:10.1007/10_2022_209.
- Networked laboratory equipment. SPECTARIS – Deutscher Industrieverband für Optik, Photonik, Analysen-und Medizintechnik. https://www.spectaris.de/en/association/thespectarisindustries/networked-laboratory-equipment/, accessed 2023-02-01 Search PubMed.
- High-throughput and data driven strategies for the design of deep-eutectic solvent electrolytes – Molecular Systems Design & Engineering (RSC Publishing), https://pubs.rsc.org/en/content/articlehtml/2022/me/d2me00050d, accessed 2023-02-1, 10.1039/D2ME00050D.
- ESAMP: Event-Sourced Architecture for Materials Provenance Management and Application to Accelerated Materials Discovery|Materials Chemistry|ChemRxiv|Cambridge Open Engage, https://chemrxiv.org/engage/chemrxiv/article-details/60c73cbf842e650956db1678, accessed 2022-10-04 Search PubMed.
- GEMD Documentation, https://citrineinformatics.github.io/gemd-docs/, accessed 2022-11-18 Search PubMed.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, Experiment Specification, Capture and Laboratory Automation Technology (ESCALATE): A Software Pipeline for Automated Chemical Experimentation and Data Management, MRS Commun., 2019, 9(3), 846–859, DOI:10.1557/mrc.2019.72.
- E. Soedarmadji, H. S. Stein, S. K. Suram, D. Guevarra and J. M. Gregoire, Tracking Materials Science Data Lineage to Manage Millions of Materials Experiments and Analyses, npj Comput. Mater., 2019, 5(1), 1–9, DOI:10.1038/s41524-019-0216-x.
- Electronic Lab Notebook (ELN), Labfolder, https://labfolder.com/, accessed 2022-12-23 Search PubMed.
- Inc, B. Laboratory Information Management System | LIMS|Labguru, https://www.labguru.com/lims, accessed 2022-11-21 Search PubMed.
- N. Argento, Institutional ELN/LIMS Deployment, EMBO Rep., 2020, 21(3), e49862, DOI:10.15252/embr.201949862.
- E. M. Bik, A. Casadevall and F. C. Fang, The Prevalence of Inappropriate Image Duplication in Biomedical Research Publications, mBio, 2016, 7(3), e00809–e00816, DOI:10.1128/mBio.00809-16.
- HIPAA Compliance with Google Workspace and Cloud Identity – Google Workspace Admin Help, https://support.google.com/a/answer/3407054?hl=en accessed 2023-02-15 Search PubMed.
- Inc, L. Automate Your Laboratory with the Global Leader for LIMS and ELN, https://www.labware.com, accessed 2022-11-21 Search PubMed.
- Cloud-based platform for biotech R&D|Benchling, https://www.benchling.com/, accessed 2022-11-21 Search PubMed.
- LIMS-Laboratory Information Management Systems – US, https://www.thermofisher.com/us/en/home/digital-solutions/lab-informatics/lab-information-management-systems-lims.html, accessed 2022-11-21 Search PubMed.
- SENAITE⋯Enterprise Open Source Laboratory System, https://github.com/senaite/senaite.github.io/, accessed 2022-11-21 Search PubMed.
- Entity Registration|Dotmatics, https://www.dotmatics.com/capabilities/entity-registration, accessed 2022-11-21 Search PubMed.
- Data Management. Citrine Informatics, https://citrine.io/product/what-is-the-citrine-platform/data-management/, accessed 2022-11-21 Search PubMed.
- R. Yan, X. Jiang, W. Wang, D. Dang and Y. Su, Materials Information Extraction via Automatically Generated Corpus, Sci. Data, 2022, 9(1), 401, DOI:10.1038/s41597-022-01492-2.
- M. C. Swain and J. M. Cole, ChemDataExtractor: A Toolkit for Automated Extraction of Chemical Information from the Scientific Literature, J. Chem. Inf. Model., 2016, 56(10), 1894–1904, DOI:10.1021/acs.jcim.6b00207.
- V. Venugopal, S. Sahoo, M. Zaki, M. Agarwal, N. N. Gosvami and N. M. A. Krishnan, Looking through Glass: Knowledge Discovery from Materials Science Literature Using Natural Language Processing, Patterns, 2021, 2(7), 100290, DOI:10.1016/j.patter.2021.100290.
- T. J. Jacobsson, A. Hultqvist, A. García-Fernández, A. Anand, A. Al-Ashouri, A. Hagfeldt, A. Crovetto, A. Abate, A. G. Ricciardulli, A. Vijayan, A. Kulkarni, A. Y. Anderson, B. P. Darwich, B. Yang, B. L. Coles, C. A. R. Perini, C. Rehermann, D. Ramirez, D. Fairen-Jimenez, D. Di Girolamo, D. Jia, E. Avila, E. J. Juarez-Perez, F. Baumann, F. Mathies, G. S. A. González, G. Boschloo, G. Nasti, G. Paramasivam, G. Martínez-Denegri, H. Näsström, H. Michaels, H. Köbler, H. Wu, I. Benesperi, M. I. Dar, I. Bayrak Pehlivan, I. E. Gould, J. N. Vagott, J. Dagar, J. Kettle, J. Yang, J. Li, J. A. Smith, J. Pascual, J. J. Jerónimo-Rendón, J. F. Montoya, J.-P. Correa-Baena, J. Qiu, J. Wang, K. Sveinbjörnsson, K. Hirselandt, K. Dey, K. Frohna, L. Mathies, L. A. Castriotta, M. H. Aldamasy, M. Vasquez-Montoya, M. A. Ruiz-Preciado, M. A. Flatken, M. V. Khenkin, M. Grischek, M. Kedia, M. Saliba, M. Anaya, M. Veldhoen, N. Arora, O. Shargaieva, O. Maus, O. S. Game, O. Yudilevich, P. Fassl, Q. Zhou, R. Betancur, R. Munir, R. Patidar, S. D. Stranks, S. Alam, S. Kar, T. Unold, T. Abzieher, T. Edvinsson, T. W. David, U. W. Paetzold, W. Zia, W. Fu, W. Zuo, V. R. F. Schröder, W. Tress, X. Zhang, Y.-H. Chiang, Z. Iqbal, Z. Xie and E. Unger, An Open-Access Database and Analysis Tool for Perovskite Solar Cells Based on the FAIR Data Principles, Nat. Energy, 2022, 7(1), 107–115, DOI:10.1038/s41560-021-00941-3.
- M. D. Wilkinson, M. Dumontier, Ij. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, The FAIR Guiding Principles for Scientific Data Management and Stewardship, Sci. Data, 2016, 3(1), 160018, DOI:10.1038/sdata.2016.18.
- C. Draxl and M. Scheffler, The NOMAD Laboratory: From Data Sharing to Artificial Intelligence, J. Phys.: Mater., 2019, 2(3), 036001, DOI:10.1088/2515-7639/ab13bb.
- L. C. Brinson, L. M. Bartolo, B. Blaiszik, D. Elbert, I. Foster, A. Strachan and P. W. Voorhees, FAIR Data Will Fuel a Revolution in Materials Research, arXiv April 6, 2022, DOI:10.48550/arXiv.2204.02881.
- B. Bayerlein, T. Hanke, T. Muth, J. Riedel, M. Schilling, C. Schweizer, B. Skrotzki, A. Todor, B. Moreno Torres, J. F. Unger, C. Völker and J. Olbricht, A Perspective on Digital Knowledge Representation in Materials Science and Engineering, Adv. Eng. Mater., 2022, 24(6), 2101176, DOI:10.1002/adem.202101176.
- K. T. Winther, M. J. Hoffmann, J. R. Boes, O. Mamun, M. Bajdich and T. Bligaard, Catalysis-Hub.Org, an Open Electronic Structure Database for Surface Reactions, Sci. Data, 2019, 6(1), 1–10, DOI:10.1038/s41597-019-0081-y.
- canSAS.org., https://www.cansas.org/, accessed 2022-11-22.
- "Tiled" — tiled 0.1.0a87 documentation, https://blueskyproject.io/tiled/, accessed 2023-03-20.
- Export Administration Regulations, vol. 15, p. 734, 8.
- Implementation of Additional Export Controls: Certain Advanced Computing and Semiconductor Manufacturing Items; Supercomputer and Semiconductor End Use; Entity List Modification. Federal Register, https://www.federalregister.gov/documents/2022/10/13/2022-21658/implementation-of-additional-export-controls-certain-advanced-computing-and-semiconductor, accessed 2023-02-09 Search PubMed.
- An Update on Research Security: Streamlining Disclosure Standards to Enhance Clarity, Transparency, and Equity|OSTP. The White House, https://www.whitehouse.gov/ostp/news-updates/2022/08/31/an-update-on-research-securitystreamlining-disclosure-standards-to-enhance-clarity-transparency-and-equity/, accessed 2023-02-09 Search PubMed.
- M. K. Horton and R. Woods-Robinson, Addressing the Critical Need for Open Experimental Databases in Materials Science, Patterns, 2021, 2(12), 100411, DOI:10.1016/j.patter.2021.100411.
- National Academies of Sciences, NSF Efforts to Achieve the Nation's Vision for the Materials Genome Initiative: Designing Materials to Revolutionize and Engineer Our Future (DMREF), 2022, DOI:10.17226/26723.
- Zenodo, Research Shared, https://zenodo.org/, accessed 2023-02-02.
- Dryad, Our mission, https://datadryad.org/stash/our_mission, accessed 2023-02-02 Search PubMed.
- B. Blaiszik, K. Chard, J. Pruyne, R. Ananthakrishnan, S. Tuecke and I. Foster, The Materials Data Facility: Data Services to Advance Materials Science Research, JOM, 2016, 68(8), 2045–2052, DOI:10.1007/s11837-016-2001-3.
- Search Citrination, https://citrination.com/search/simple?searchMatchOption=fuzzyMatch, accessed 2022-11-22 Search PubMed.
- A. Zakutayev, J. Perkins, M. Schwarting, R. White, K. Munch, W. Tumas, N. Wunder and C. Phillips, High Throughput Experimental Materials Database, 2017, 2 files, DOI:10.7799/1407128.
- Home|ICSD, https://icsd.products.fiz-karlsruhe.de/, accessed 2022-11-22 Search PubMed.
- Materials Project – Materials Explorer. Materials Project, https://materialsproject.org/materials, accessed 2023-02-14 Search PubMed.
- Materials Project – Community. Materials Project, https://materialsproject.org/community, accessed 2023-02-14 Search PubMed.
- S. K. Burley, H. M. Berman, C. Christie, J. M. Duarte, Z. Feng, J. Westbrook, J. Young and C. Zardecki, RCSB Protein Data Bank: Sustaining a Living Digital Data Resource That Enables Breakthroughs in Scientific Research and Biomedical Education, Protein Sci., 2018, 27(1), 316–330, DOI:10.1002/pro.3331.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly Accurate Protein Structure Prediction with AlphaFold, Nature, 2021, 596(7873), 583–589, DOI:10.1038/s41586-021-03819-2.
- M. Scheffler, M. Aeschlimann, M. Albrecht, T. Bereau, H.-J. Bungartz, C. Felser, M. Greiner, A. Groß, C. T. Koch, K. Kremer, W. E. Nagel, M. Scheidgen, C. Wöll and C. Draxl, FAIR Data Enabling New Horizons for Materials Research, Nature, 2022, 604(7907), 635–642, DOI:10.1038/s41586-022-04501-x.
- P. A. Hitzler, Review of the Semantic Web Field, Commun. ACM, 2021, 64(2), 76–83, DOI:10.1145/3397512.
- C. W. Andersen, R. Armiento, E. Blokhin, G. J. Conduit, S. Dwaraknath, M. L. Evans, Á. Fekete, A. Gopakumar, S. Gražulis, A. Merkys, F. Mohamed, C. Oses, G. Pizzi, G.-M. Rignanese, M. Scheidgen, L. Talirz, C. Toher, D. Winston, R. Aversa, K. Choudhary, P. Colinet, S. Curtarolo, D. Di Stefano, C. Draxl, S. Er, M. Esters, M. Fornari, M. Giantomassi, M. Govoni, G. Hautier, V. Hegde, M. K. Horton, P. Huck, G. Huhs, J. Hummelshøj, A. Kariryaa, B. Kozinsky, S. Kumbhar, M. Liu, N. Marzari, A. J. Morris, A. A. Mostofi, K. A. Persson, G. Petretto, T. Purcell, F. Ricci, F. Rose, M. Scheffler, D. Speckhard, M. Uhrin, A. Vaitkus, P. Villars, D. Waroquiers, C. Wolverton, M. Wu and X. Yang, OPTIMADE, an API for Exchanging Materials Data, Sci. Data, 2021, 8(1), 217, DOI:10.1038/s41597-021-00974-z.
- OPTIMADE, materials-consortia.github.io, https://optimade.org/, accessed 2023-02-22 Search PubMed.
- Empty Rhetoric over Data Sharing Slows Science, Editorial, Nature, 2017, 546(7658), 327, DOI:10.1038/546327a.
| This journal is © The Royal Society of Chemistry 2023 |