Open Access Article
Open Access ArticleMolecular sonification for molecule to music information transfer†
Babak Mahjour a,
Jordan Benchab,
Rui Zhang
a,
Jordan Benchab,
Rui Zhang b,
Jared Frazier
b,
Jared Frazier ac and
Tim Cernak
ac and
Tim Cernak *ab
*ab
aDepartment of Medicinal Chemistry, University of Michigan, Ann Arbor, MI, USA. E-mail: tcernak@umich.edu
bDepartment of Chemistry, University of Michigan, Ann Arbor, MI, USA
cDepartment of Computer Science, Middle Tennessee State University, Murfreesboro, TN, USA
First published on 14th March 2023
Abstract
Organic chemical structures encode information about a molecule's atom and bond arrangement. The most established way to encode a molecular structure is through line drawing, although other representations based on graphs, strings, one-hot encoded labels, or fingerprint arrays are critical to the computational study of molecules. Here we show that music is a highly dimensional information storage medium that can be used to encode molecular structure. The resultant method allows a molecular structure to be heard as a musical composition, where the key of the music is based on the molecular properties and the melody is based on the atom and bond arrangement. This allows for a molecular generation approach that leverages modern artificial intelligence tactics for music generation.
Introduction
The representation of chemical structures is critical to the study and invention of functional molecules. Organic molecules are classically described as line drawings,1 where all atoms and their corresponding bonds are drawn on paper or on a computer. Other simple molecular representations or identifiers include molecular formulae, IUPAC names or CAS numbers, which require little memory and are machine readable, but carry minimal information. Molecules can also be represented as graphs, with atoms as nodes and bonds as edges. By encoding atomic coordinates and connectivities line by line, the topology of molecules can be embedded as a graph on a computer for rendering, editing and analysis. The transmission of molecular information into machine-readable formats has invited new molecular structure representations, such as SMILES,2 SMARTS,3 InChI keys,4 DeepSMILES,5 and SELFIES.6 These representations are cheap to store in memory and provide valuable structural information for rapid lookup and comparison. While these aforementioned representations have been useful for inputting molecules into computers, and encoding structural and stereochemical information, they are one-dimensional string representations that are more difficult for human users to interpret and interact with than the classic line drawing representation of molecules. To adapt them for machine learning and data science algorithms, SMILES and other strings are typically converted to vector representations via molecular fingerprints such as Extended Connectivity Fingerprints (ECFP),7 Morgan Fingerprints,8 atom-pair fingerprints,9 and others. This dimensionality expansion is a core tactic in the analysis of virtual chemical libraries or predictions of molecular properties. Other high-dimensionality fingerprint representations, such as physics-based descriptors10 or physicochemical descriptors,11,12 are also common. While computers can easily parse molecular information from these representations, interactivity with human users is difficult with the fingerprint-based information media. In addition, once converted to such a fingerprint, the molecule is typically no longer uniquely revertible to its atom-bond representation.13Music is a high-dimensional information storage medium that maximizes both human and computer interactivity, interpretability, and creativity. We considered that music could be used for storage of molecular information. While many aspects of a molecule are readily visible to the trained eye, a soundbite may be able transmit more information about the molecule into the mind. The encoding of molecules as music is particularly intriguing since the multiple dimensions of music can allow encoding of many molecular properties.14 Music is also highly interactive both for humans and for computers. Musicians can control many parameters that can embed information about a molecule, such as tempo, rhythm, notes, key, instrument, effects, etc. If molecules could be encoded as music, opportunities would emerge for visual-to-audio sensory substitution, for instance providing blind chemists new ways to interact with molecules.15 Contemporary chemistry and drug discovery leverage artificial intelligence16 (AI) and there has meanwhile been an explosion of AI methods in the study and creation of music,17 so we were excited by the prospects of merging modern chemistry machine learning (ML) techniques with recent ML techniques for music. Our initial impetus was to explore how music could be used as a creative medium to generate new molecules, but in the course of our studies we have learned that molecules likewise can provide an inspiration and creative outlet for the generation of new music.
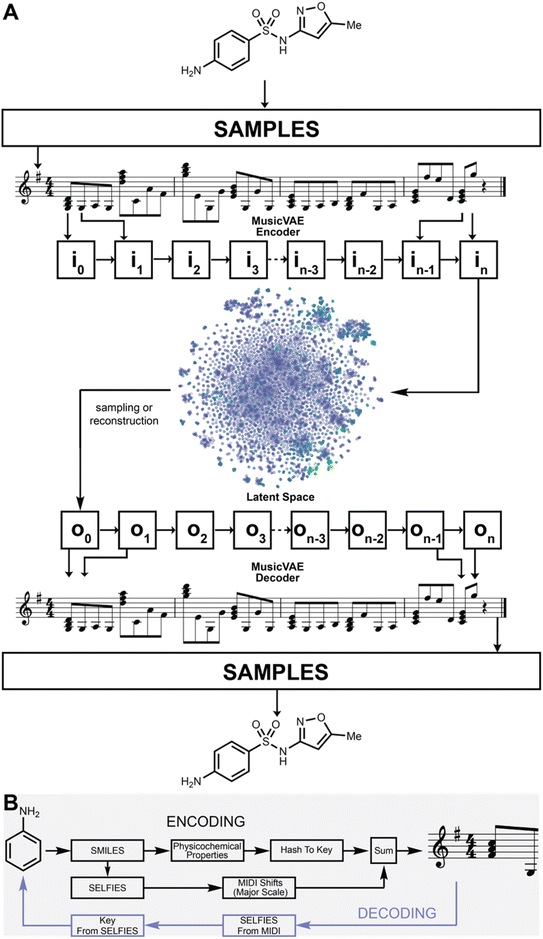
Sonification is the encoding of non-musical information as music and provides a means to encode information in many musical dimensions, while simultaneously providing a new means of interactivity.18 A variety of information sources have been sonified, such as visual art,19 the architecture of spider webs,20 infrared spectra of chemicals,21 amino acid sequences,22 air pollution,23 fire,24 and many more.25–32 The SELFIES representation provided a viable input for molecular sonification, both for the encoding of molecules into a melody and the construction of new molecules via performance on a musical instrument such as the piano. We developed a workflow for transferring molecules into music, and vice versa, which we call Sonic Architecture for Molecule Production and Live-Input Encoding Software, or SAMPLES (Fig. 1).
Methods and workflow
Encoding
To create a melody based on a molecular structure, the key and the sequence of notes are derived from its physicochemical properties and its SELFIES sequence, respectively. To determine the key, the physicochemical properties of a molecule—such as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, molecular weight, and number of hydrogen bond donors and acceptors—are summed, and the final number is linearly projected into the integer space between 1 and 12 from the minimum and maximum values found in the entire dataset (in this case, DrugBank), with each bin corresponding to a particular key. In our encoding scheme, the projection is largely dictated by the molecular weight due to its magnitude with respect to the other features. Due to low deviation, low mean, and outliers of high molecular weight, most molecules from the DrugBank dataset fall into the first bin. In our scheme, the keys represented by the bins are ordered by the popularity of keys found on Spotify, with the most popular bin being the key of G (see Fig. S1†). As such, most compounds in DrugBank are encoded into G and larger molecules are encoded into the less popular keys. The sequence of notes is determined from a one-to-one mapping between the SELFIES token of the molecule and multi-octave steps in the major scale (see ESI† for the specific mappings and key distribution used in this study). By adding the MIDI value of the melody's key to the MIDI shifts that correspond to notes of the major scale (derived from the SELFIES tokens of the molecule), the final melody is produced. In our case, the MIDI shifts correspond to SELFIES tokens as ranked by their popularity in DrugBank. As such, lower MIDI shifts away from the encoded key of the molecule indicate the more popular the SELFIES token. Every fourth note, starting from the first note, was converted to a major chord to increase the texture of the music. We envision polyphony as a potential avenue to encode atomic features. This algorithm could be extended by including more descriptors in the encoding and assigning distinct keys to clusters. As well, mapping more elements or structural subgroups to MIDI shifts or other musical parameters can enable the encoding of proteins, inorganic molecules, and other chemical phenomena.
P, molecular weight, and number of hydrogen bond donors and acceptors—are summed, and the final number is linearly projected into the integer space between 1 and 12 from the minimum and maximum values found in the entire dataset (in this case, DrugBank), with each bin corresponding to a particular key. In our encoding scheme, the projection is largely dictated by the molecular weight due to its magnitude with respect to the other features. Due to low deviation, low mean, and outliers of high molecular weight, most molecules from the DrugBank dataset fall into the first bin. In our scheme, the keys represented by the bins are ordered by the popularity of keys found on Spotify, with the most popular bin being the key of G (see Fig. S1†). As such, most compounds in DrugBank are encoded into G and larger molecules are encoded into the less popular keys. The sequence of notes is determined from a one-to-one mapping between the SELFIES token of the molecule and multi-octave steps in the major scale (see ESI† for the specific mappings and key distribution used in this study). By adding the MIDI value of the melody's key to the MIDI shifts that correspond to notes of the major scale (derived from the SELFIES tokens of the molecule), the final melody is produced. In our case, the MIDI shifts correspond to SELFIES tokens as ranked by their popularity in DrugBank. As such, lower MIDI shifts away from the encoded key of the molecule indicate the more popular the SELFIES token. Every fourth note, starting from the first note, was converted to a major chord to increase the texture of the music. We envision polyphony as a potential avenue to encode atomic features. This algorithm could be extended by including more descriptors in the encoding and assigning distinct keys to clusters. As well, mapping more elements or structural subgroups to MIDI shifts or other musical parameters can enable the encoding of proteins, inorganic molecules, and other chemical phenomena.
Decoding
The MIDI shifts are reverse calculated for each key and converted into a molecular structure. As such, multiple structures are generated (one for each key) for the same MIDI sequence. Each structure is then hashed into a key using the original key encoding algorithm. If the hashed key matches the key used in the reverse calculation, the molecular structure is decoded. It is guaranteed that at least one decoding key will match a hashed key for any MIDI generated from SAMPLES.A demonstration of the SAMPLES encoding function is shown in Fig. 2. Ammonia (1) appears as a single note while benzene (2) generates a slightly more complex musical composition. The unity of these two molecules produces aniline (3) whose musical sequence highly resembles the concatenation of the two musical sequences of 1 and 2. Expansion of 3 into indole (4) creates a slightly more complex melody owing to both the increased molecular size and the additional information content required to describe a ring fusion between the 5- and 6-membered rings. In the reverse direction, songs are readily translated to molecules, such as 5, which is produced from the song “Twinkle, twinkle little star” when played using D flat as the reference note (the decoding key).
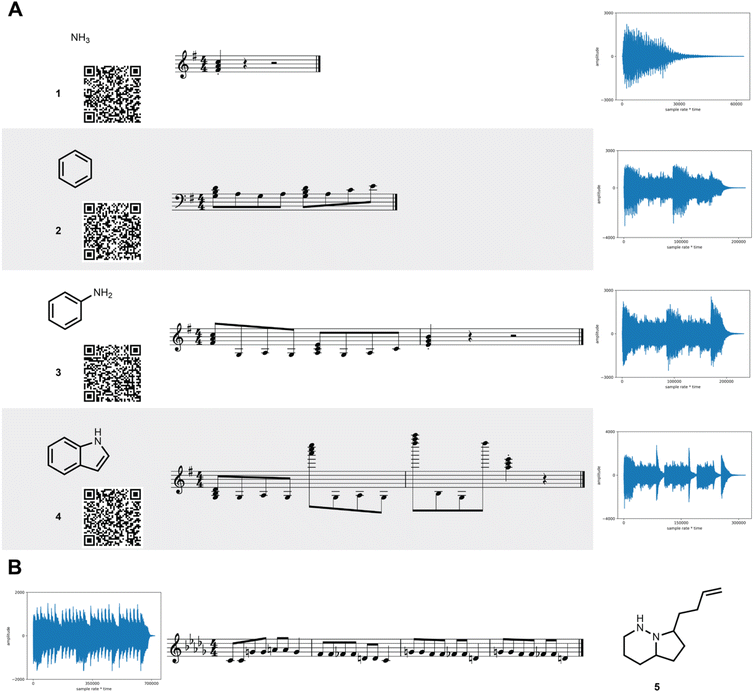 | ||
| Fig. 2 SAMPLES translates molecules into music. (A) The generation of increasingly complex molecules from 1 through 4 corresponds to increasing musical complexity. Each line shows the molecular structure, the corresponding musical score, and a waveform of the MIDI output. Audio recordings are available in the ESI† and can be quickly retrieved by scanning the QR code with a mobile device. (B) In the reverse direction, the song “Twinkle, Twinkle Little Star” produces molecule 5. | ||
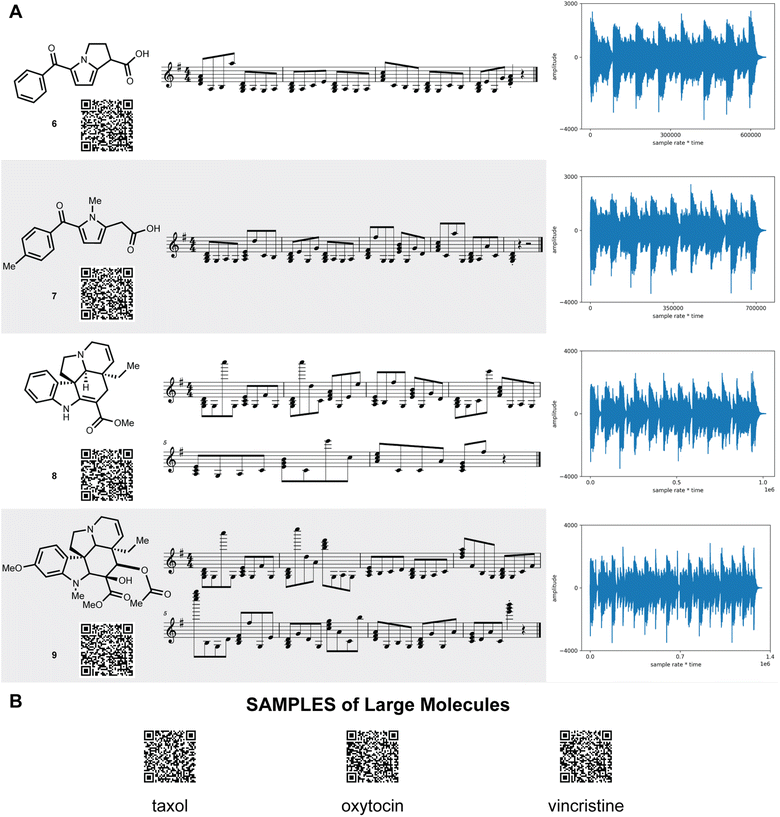
SAMPLES is readily scaled to more complex and drug-like molecules (Fig. 3). Tolmetin (6) and ketolorac (7) create a rich and textured musical composition. Meanwhile, tabersonine (8) and vindoline (9) provide complex melodies. Scaling to large complex molecules, such as taxol, oxytocin, or vincristine (see ESI†) required no modifications and generated nuanced euphonic melodies.
Case studies
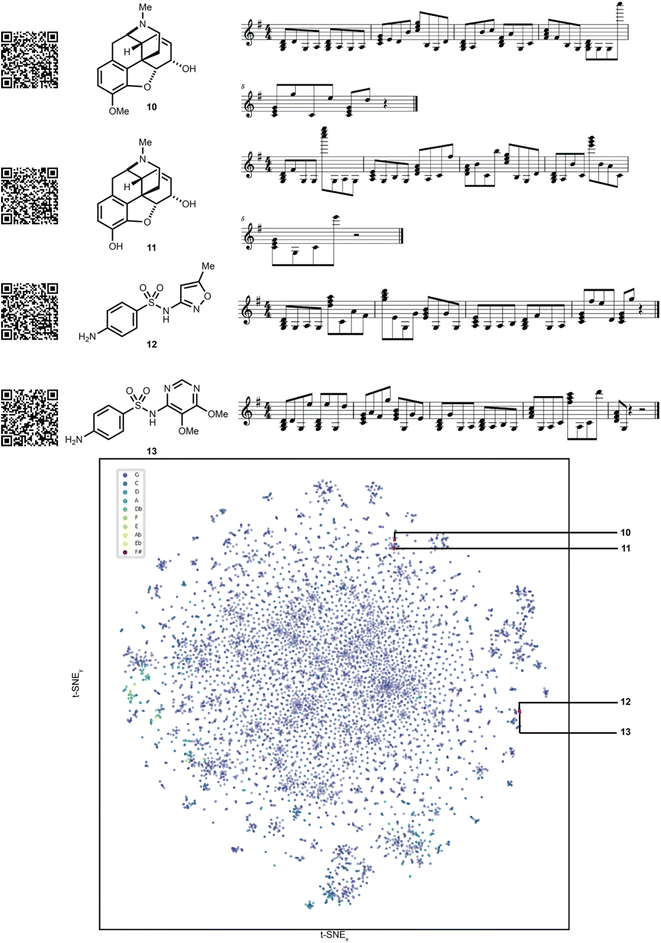
To showcase the utility of this novel algorithm, four experimental case studies are presented. Using our approach, molecular properties can be heard. For instance, the songs generated from molecules that pass the Lipinski rules33 can be auditorily distinguished from those that fail the Lipinski rules based on the musical key. This is largely because the molecule's aggregate physicochemical properties were hashed to the musical key, with the most popular physicochemical property fingerprints from the pharmaceutical database DrugBank hashing to the most popular song keys from the music database Spotify.34 The concept of molecular similarity is of high importance to molecular invention, such as in selecting molecules with comparable functional properties for drug discovery. We were curious to explore if SAMPLES generated from molecules with high Tanimoto similarity35 (fingerprint based) would sound similar, appreciating that both molecular similarity and musical similarity are difficult to define.36 Molecules represented in the t-SNE in Fig. 4 are more similar if closer to each other on the plot. Indeed, we deemed the SAMPLES of codeine (10) and morphine (11) to sound similar to each other while the SAMPLES of sulfamethoxazole (12) and sulfadoxin (13) likewise sound similar, while the pair of 10 and 11 sounded distinct from the pair of 12 and 13 (Fig. 4).Our second experiment investigates the generation of molecules via modification of the music domain. A key motivator for our research was the ability to generate new molecules through the interactivity of a piano keyboard, or other musical hardware or software. This was made possible in SAMPLES through the application of SELFIES, which enable editing of string bits while consistently producing valid molecular structures. Thus, starting from morphine (11), the musical score could be modified one note at a time (Fig. 5) to generate new chemical structures 14–16 bearing a clear relationship to 11 but with noticeably modified bond and atom architecture. Due to our encoding scheme, shifts further away from the melody's key result in SELFIES being modified into to atoms less commonly found in the DrugBank database. Random modification of SELFIES can result in drastic changes to molecular structure as seen in Fig. 5, due to the non-atom encoding SELFIES tokens that dictate the size of features such as branches and rings. Note that SAMPLES may generate undefined stereocenters.
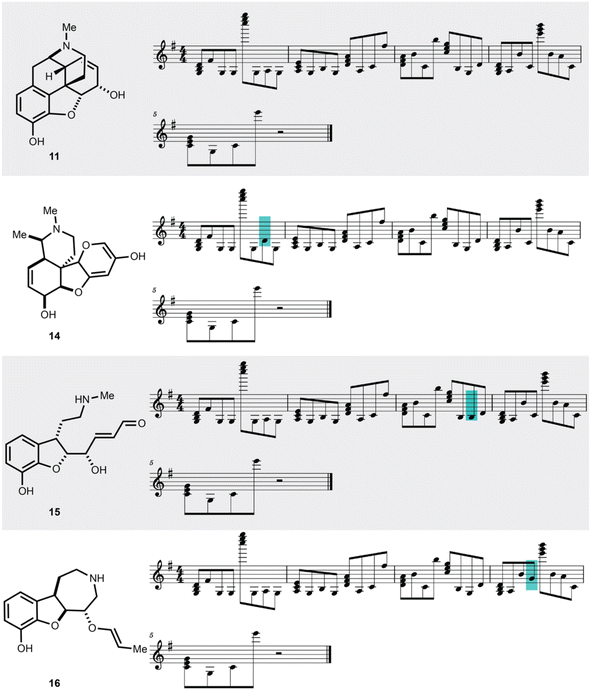 | ||
| Fig. 5 Molecular editing in SAMPLES generates distinct but related molecules. The manual editing of single notes in the SAMPLES of 11 leads to 14, 15, or 16. | ||
Having demonstrated the feasibility of molecular generation using SAMPLES, we explored the ability of modern machine learning methods developed for music generation as tools for molecule generation. In this third case study, we applied the melody mixing function of MusicVAE37 using MIDI melodies derived from SAMPLES as inputs. Using MusicVAE, two melodies could be blended to generate an interpolated melody, and that new melody could be translated back to a molecular structure using SAMPLES, thus creating a new molecule that was a “blend” of the two input molecules (Fig. 6). We call this function CROSSFADE. The blending of musical compositions is an established practice, with considerable hardware and software to support the musical blending process. While algorithms that generate new molecules by blending the structures or properties of input molecules are known,38 we are intrigued by the interactivity offered by CROSSFADE. As an example, glutamic acid (17) and acetylcholine (18) were CROSSFADEd to produce 19, 20 and 21 CROSSFADE to 22 and similar results are obtained for 23–28. A four-step interpolation is shown in the ESI.†
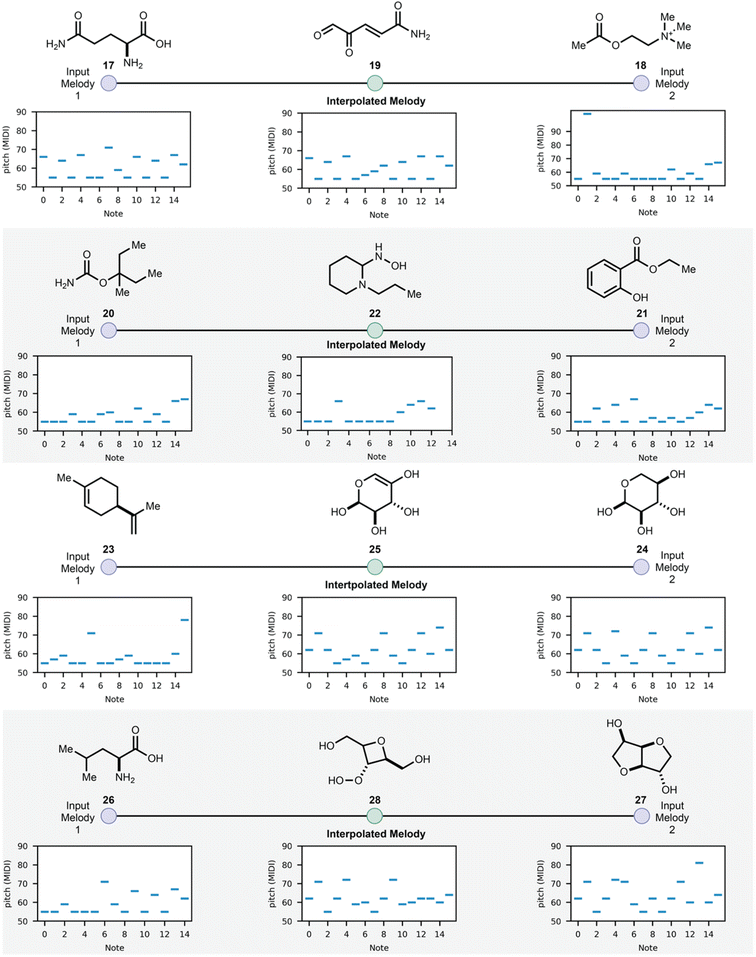 | ||
| Fig. 6 CROSSFADE merges SAMPLES with the melody mixing function of MusicVAE to create interpolated molecules based on two input molecules. | ||
As a final experiment, to take the editing of the molecules on the keyboard a step further, and to demonstrate the human-interactivity enabled by the SAMPLES algorithm, a human created a monophonic composition inspired by SAMPLES-generated music (Fig. 7), which was decoded to molecule 29. It was necessary to exert some human bias into the musical composition, based on the composer's knowledge of chemistry and SAMPLES, since generating a molecule that is as carboniferous as most drugs and natural products requires bias towards the key's tonic note, in this case C, since that is mapped to the carbon atom. In other words, the song was written to ensure that the most played note corresponded to the carbon atom to reflect the nature of organic molecules. This required a basic understanding of organic chemistry and musical theory. While this implementation of SAMPLES focuses on major scale notes, the embedding algorithm can be easily modified to include mappings for minor scale notes or accidentals.
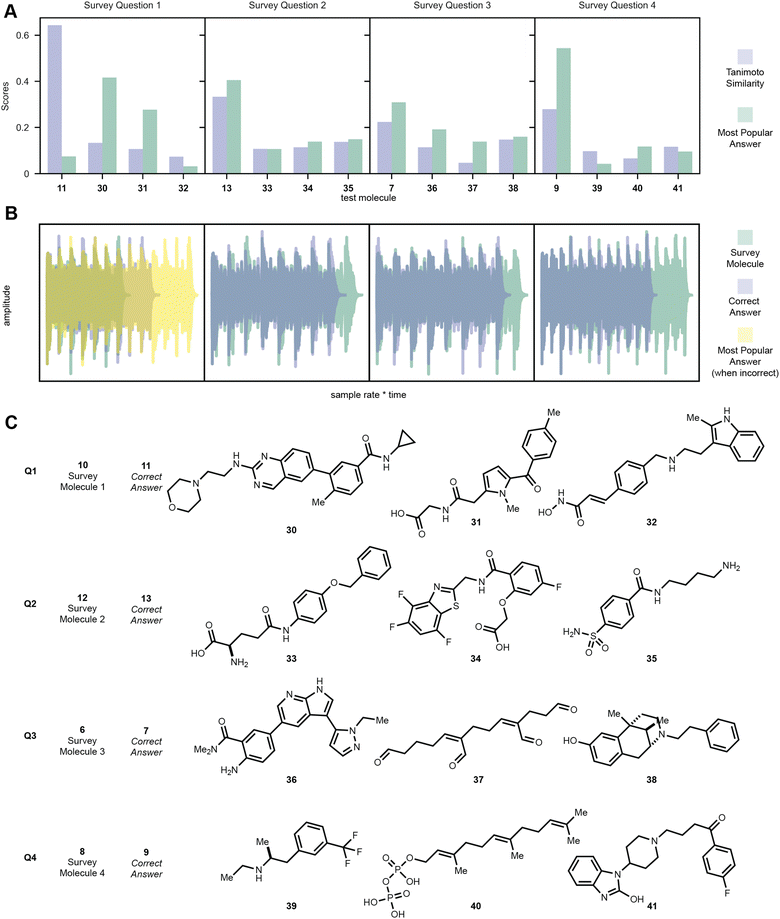
One goal of this embedding algorithm was to show that music can be used as a medium to encode molecular information and that similar molecules can produce similar sounding melodies. While this is largely dependent on the featurization and encoding of the molecules into music, our algorithm seemed to perform well for some druglike molecules as tested by the human ear. We surveyed 75 participants from a senior level undergraduate medicinal chemistry course with a blind multiple-choice poll. Students were given four questions, each of which asking the student to compare the melody of four encoded molecules to the melody of an encoded molecule given in the question. The student is prompted to select the melody that they believe to sound most similar to the given sample for each question without being shown any structures. In the first question, 10 was given to the student and the student was given a choice between the melodies of 11, 30, 31, and 32. In this case, despite the high Tanimoto similarity between codeine 10 and morphine 11, most students did not recognize similarity between the two encodings, instead hearing 30 and 31 to sound more like 10. However, in question two where 12 was compared to 13, 33, 34, and 35, most students correctly identified the melodies of sulfamethoxazole (12) and sulfadoxin (13) to be most similar. Students were also able to identify the most similar melodies and molecules as described by Tanimoto similarity in questions three and four, where melodies of 7 and 9 were correctly chosen to be similar to 6 and 8 over 36–38 and 39–41, respectively. Puzzlingly, question one had the highest similarity between the test molecule and correct answer molecule compared to the other question pairs (Fig. 8).
Conclusions
We report an alternative means of encoding organic molecules through music. The resultant melodies allow a human to interact with molecular structures through musical hardware and software via note editing, insertion and deletion, as well as produce molecular structures through original compositions. We note that far more molecular features can be encoded into a melody or even a short soundbite than can be visually represented by the 2D or 3D structure. Indeed, many richer possibilities can be enabled by encoding more features into musical elements such as other scales, chord compositions, rhythms, timings, timbres, etc. One transfer learning application for which the current study may be used is music generation. The motivation for machine learning for content generation is its generality, that is no formal grammar or rules must be specified for such a model to generate content.17 Transforming molecules into music provides a rich collection of musical data that can be used to train music generation models, as seen with MusicVAE. Particularly, sequence to sequence (seq2seq) models, such as recurrent neural networks,39 allow for the interconnection of domains containing data signals with variable lengths such as text, music, and machine-readable molecular representations based on structure. Seq2seq models can learn a fixed length embedding of variable length signals that can be used for classification tasks and direct mathematical comparison. For instance, word2vec40 and GloVe41 provide pretrained word embeddings that have been learned from massive text corpuses such as Wikipedia or Twitter. In a molecular context, variational autoencoders have been used to learn the distribution of molecular features, such as SELFIES tokens, to provide a continuous embedding of molecular space.42 SAMPLES provides an avenue to directly connect molecules to content-generating machine learning models in the music domain. Computational exploration and interpolation within the melodies described herein is possible, generating new molecules that sound and look similar to existing molecules. This highlights the possibility of leveraging music-based artificial intelligence for molecular design. An online implementation of the encoding portion of the SAMPLES algorithm can be accessed at http://samples.cernaklab.com.Materials
Sonification and visualization code was written in Python (version 3.7.12). All Python dependencies were installed using pip, version 21.1.3. SELFIES (version 1.0.0) was utilized to encode molecules into string format. RDKIT (version 2021.9.2.1) was utilized to calculate physicochemical properties of molecules for key hashing. Magenta (version 2.1.3) provided tools to manipulate MIDI files and train MusicVAE. Fluidsynth (version 2.2.3, installed via apt-get) was used to convert MIDI into wav format. Music21 (version 5.5.0) was used to create and read MIDI files. Matplotlib (version 3.2.2) was used to create plots and graphs. Sklearn was used to calculate the tSNE dimensionality reduction. Drug structures were collected from DrugBank Release Version 5.1.8 (2021-01-03).Data availability
Code to run the algorithm can be found here: https://github.com/cernaklab/samples. Trained VAE model can be downloaded from here: https://doi.org/10.5281/zenodo.6391612. Webapp of the algorithm can be used at: http://samples.cernaklab.com/Author contributions
All authors performed computational experiments and wrote the manuscript. T. C. supervised the work.Conflicts of interest
The Cernak Lab has received research funding or in-kind donations from MilliporeSigma, Relay Therapeutics, Janssen Therapeutics, Entos, Inc., SPT Labtech and Merck & Co., Inc. T. C. holds equity in Scorpion Therapeutics, and is a co-Founder and equity holder of Entos, Inc.Acknowledgements
The authors thank the University of Michigan College of Pharmacy for startup funds, as well as the NSF-Interdisciplinary REU Program (NSF-1851985) for funding J. F.Notes and references
- A. Kekule, Sur la constitution des substances aromatiques, Bull. Soc. Chim. Fr, 1865, 3, 98–110 Search PubMed.
- D. Weininger, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Comput. Sci., 1988, 28(1), 31–36 CrossRef CAS.
- Daylight Theory: SMARTS – A Language for Describing Molecular Patterns.
- S. Heller, A. McNaught, S. Stein, D. Tchekhovskoi and I. Pletnev, InChI - the worldwide chemical structure identifier standard, J. Cheminf., 2013, 5(1), 7 CAS.
- N. O'Boyle and A. Dalke, DeepSMILES: an adaptation of SMILES for use in machine-learning of chemical structures, ChemRxiv, 2018, preprint, DOI:10.26434/chemrxiv.7097960.v1.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Self-referencing embedded strings (SELFIES): A 100% robust molecular string representation, Mach. Learn.: Sci. Technol., 2020, 1(4), 045024 Search PubMed.
- D. Rogers and M. Hahn, Extended-Connectivity Fingerprints, J. Chem. Inf. Model., 2010, 50(5), 742–754 CrossRef CAS PubMed.
- H. L. Morgan, The Generation of a Unique Machine Description for Chemical Structures-A Technique Developed at Chemical Abstracts Service, J. Chem. Doc., 1965, 5(2), 107–113 CrossRef CAS.
- R. E. Carhart, D. H. Smith and R. Venkataraghavan, Atom pairs as molecular features in structure-activity studies: definition and applications, J. Chem. Inf. Comput. Sci., 1985, 25(2), 64–73 CrossRef CAS.
- S. Boobier, D. R. J. Hose, A. John Blacker and B. N. Nguyen, Machine learning with physicochemical relationships: solubility prediction in organic solvents and water, Nat. Commun., 2020, 11(1), 5753 CrossRef CAS PubMed.
- P. S. Kutchukian, J. F. Dropinski, K. D. Dykstra, B. Li, D. A. DiRocco and E. C. Streckfuss, et al., Chemistry informer libraries: a chemoinformatics enabled approach to evaluate and advance synthetic methods, Chem. Sci., 2016, 7(4), 2604–2613 RSC.
- F. Pereira and J. Aires-de-Sousa, Machine learning for the prediction of molecular dipole moments obtained by density functional theory, J. Cheminf., 2018, 10(1), 43 Search PubMed.
- T. Le, R. Winter, F. Noé and D.-A. Clevert, Neuraldecipher–reverse-engineering extended-connectivity fingerprints (ECFPs) to their molecular structures, Chem. Sci., 2020, 11(38), 10378–10389 RSC.
- M. Kumbar, Musical chemistry: Integrating chemistry and music, J. Chem. Educ., 2007, 84(12), 1933 CrossRef CAS.
- V. Tóth and L. Parkkonen, Autoencoding sensory substitution, arXiv, 2019, preprint, arXiv:1907.06286, DOI:10.48550/arXiv.1907.06286.
- Z. J. Baum, X. Yu, P. Y. Ayala, Y. Zhao, S. P. Watkins and Q. Zhou, Artificial intelligence in chemistry: current trends and future directions, J. Chem. Inf. Model., 2021, 61(7), 3197–3212 CrossRef CAS PubMed.
- J.-P. Briot, G. Hadjeres and F.-D. Pachet, Deep Learning Techniques for Music Generation. Computational Synthesis and Creative Systems, 2020 Search PubMed.
- T. Hermann, A. Hunt and J. G. Neuhoff. The sonification handbook: Logos Verlag Berlin; 2011 Search PubMed.
- M. Muller-Eberstein and N. van NoordTranslating Visual Art Into Music, 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), 2019 Search PubMed.
- Making music from spider webs.
- N. Garrido, A. Pitto-Barry, J. J. Soldevila-Barreda, A. Lupan, L. C. Boyes and W. H. C. Martin, et al., The Sound of Chemistry: Translating Infrared Wavenumbers into Musical Notes, J. Chem. Educ., 2020, 97(3), 703–709 CrossRef CAS.
- A Self-Consistent Sonification Method to Translate Amino Acid Sequences into Musical Compositions and Application in Protein Design Using Artificial Intelligence.
- otnemrasordep. GitHub - otnemrasordep/sonification-bela: Final Assignment for ECS7012P: Music and Audio Programming.
- M. Milazzo and M. J. Buehler, Designing and fabricating materials from fire using sonification and deep learning, Iscience, 2021, 24(8), 102873 CrossRef PubMed.
- N. W. Tay, F. Liu, C. Wang, H. Zhang, P. Zhang and Y. Z. Chen, Protein music of enhanced musicality by music style guided exploration of diverse amino acid properties, Heliyon, 2021, 7(9), e07933 CrossRef CAS PubMed.
- I. Su, I. Hattwick, C. Southworth, E. Ziporyn, A. Bisshop and R. Mühlethaler, et al., Interactive exploration of a hierarchical spider web structure with sound, J. Multimodal User Interfaces, 2022, 1–15 Search PubMed.
- D. K. F. Meijer, I. Jerman, A. V. Melkikh and V. I. Sbitnev. Biophysics of consciousness: A scale-invariant acoustic information code of a superfluid quantum space guides the mental attribute of the universe. Rhythmic Oscillations in Proteins to Human Cognition. 2021, pp. 213–361 Search PubMed.
- Towards molecular musical instruments: interactive sonification of 17-alanine, graphene and carbon nanotubes, ed. T. J. Mitchell, A. J. Jones, M. B. O'Connor, M. D. Wonnacott, D. R. Glowacki and J. Hyde, 2020, pp. 214–221 Search PubMed.
- M. Groß, Die Musik der Proteine, Nachr. Chem., 2019, 67(10), 98 CrossRef.
- S. L. Franjou, M. Milazzo, C.-H. Yu and M. J. Buehler, Sounds interesting: Can sonification help us design new proteins?, Expert Rev. Proteomics, 2019, 16(11–12), 875–879 CrossRef CAS PubMed.
- A. Borgonovo and G. Haus, Sound synthesis by means of two-variable functions: experimental criteria and results, Comput. Music. J., 1986, 10(3), 57–71 CrossRef.
- M. A. Garcia-Ruiz and J. R. Gutierrez-Pulido, An overview of auditory display to assist comprehension of molecular information, Interact. Comput., 2006, 18(4), 853–868 CrossRef.
- C. A. Lipinski, Lead- and drug-like compounds: the rule-of-five revolution, Drug Discovery Today: Technol., 2004, 1(4), 337–341 CrossRef CAS PubMed.
- K. Ning. Most used keys on Spotify, 2020, Available from: https://forum.bassbuzz.com/t/most-used-keys-on-spotify/5886.
- N. C. Chung, B. Miasojedow, M. Startek and A. Gambin, Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data, BMC Bioinform., 2019, 20(Suppl 15), 644 CrossRef PubMed.
- G. Maggiora, M. Vogt, D. Stumpfe and J. Bajorath, Molecular similarity in medicinal chemistry: miniperspective, J. Med. Chem., 2014, 57(8), 3186–3204 CrossRef CAS PubMed.
- A. Roberts, J. Engel, C. Raffel, C. Hawthorne and D. Eck, A Hierarchical Latent Vector Model for Learning Long-Term Structure in Music, 2018.
- J. Besnard, G. F. Ruda, V. Setola, K. Abecassis, R. M. Rodriguiz and X.-P. Huang, et al., Automated design of ligands to polypharmacological profiles, Nature, 2012, 492(7428), 215–220 CrossRef CAS PubMed.
- M. H. S. Segler, T. Kogej, C. Tyrchan and M. P. Waller, Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks, ACS Cent. Sci., 2018, 4(1), 120–131 CrossRef CAS PubMed.
- T. Mikolov, I. Sutskever, K. Chen, G. Corrado and J. Dean, Distributed Representations of Words and Phrases and their Compositionality, arXIv, 2013, preprint, arXiv:1310.4546, DOI:10.48550/arXiv.1310.4546.
- Global Vectors for Word Representation, ed. J. Pennington, R. Socher and C. Manning, 2014, pp. 1532–1543, https://aclanthology.org/D14-1162/ Search PubMed.
- R. Gómez-Bombarelli, J. N. Wei, D. Duvenaud, J. M. Hernández-Lobato, B. Sánchez-Lengeling and D. Sheberla, et al., Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules, ACS Cent. Sci., 2018, 4(2), 268–276 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00008g |
| This journal is © The Royal Society of Chemistry 2023 |