Open Access Article
Open Access ArticleTowards more reproducible and FAIRer research data: documenting provenance during data acquisition using the Infofile format†
Bernd
Paulus
and
Till
Biskup‡
 *
*
Physikalische Chemie, Albert-Ludwigs-Universität Freiburg, Albertstr. 21, 79104 Freiburg, Germany. E-mail: research@till-biskup.de
First published on 23rd December 2022
Abstract
Information, i.e. data, is regarded as the new oil in the 21st century. The impact of this statement from economics for science and the research community is reflected in the hugely increasing number of machine-learning and artificial intelligence applications that were one driving force behind writing out the FAIR principles. However, any form of data (re)use requires the provenance of the data to be recorded. Hence, recording metadata during data acquisition is both an essential aspect of and as old as science itself. Here, we discuss the why, when, what, and how of research data documentation and present a simple textual file format termed Infofile developed for this purpose. This format allows researchers in the lab to record all relevant metadata during data acquisition in a user-friendly and obvious way while minimising any external dependencies. The resulting machine-actionable metadata in turn allow processing and analysis software to access relevant information, besides making the research data more reproducible and FAIRer. By demonstrating a simple, yet powerful and proven solution to the problem of metadata recording during data acquisition, we anticipate the Infofile format and its underlying principles to have great impact on the reproducibility and hence quality of science, particularly in the field of “little science” lacking established and well-developed software toolchains and standards.
1 Introduction
In archaeological excavations, reproducibility (in the sense of repeatability) is rarely an issue, as digging out artifacts is an irreversible and intrinsically irreproducible process. Therefore, archaeologists are trained early on to painstakingly document every step and the context of an artifact in great detail during excavations.1 Experimental scientists, in contrast, often seem much more relaxed in this regard, assuming that data missing the necessary documentation can easily be reacquired. While this was never true, it would at least be economically infeasible and largely inefficient. Furthermore, numerical data without accompanying metadata are like dug-out artifacts in archaeology: lacking relevant context and hard to interpret. In both cases, once the context is lost, getting it back is nearly impossible. Scientific record keeping, thus creating data about data, i.e. metadata,2,3 is therefore both an essential aspect of conducting science and crucial for knowledge creation and dissemination.4 Documenting data during acquisition is part of what has lately been called “research data management”5–7 and resides fully in the realm and responsibility of the individual scientist. Furthermore, it is an essential aspect of FAIR(er) data.The FAIR principles8 (Fig. 1), which themselves rest on earlier work,9 usually focus on reuse of data independent of the original data creators or collectors, often in the context of big (uniform) data and machine learning. Nevertheless, adhering to these principles clearly enhances reproducibility, even with “little” (and diverse) data, such as in spectroscopy. The difference between “big” and “little” science goes back to de Solla Price,10 with big science meaning large, collaborative scientific endeavours that can mainly be traced back to the Manhattan project. This distinction between big and little science led in turn to distinguishing “big data” from “little data”11 and the discussion of the “long tail of data”12,13 for those diverse kinds of data that are obtained by individual and independent researchers with highly specialised and sometimes unique techniques rather than the large amount of uniform and highly standardised data originating in big international research collaborations. Attempts to create the necessary research data infrastructure to allow for reuse of data,14–19 termed e-science or cyberinfrastructure depending on their geographic origin, predate the FAIR principles by far, but have recently gained interest again, as evidenced by initiatives such as the European Open Science Cloud (EOSC)20 or the German National Research Data Infrastructure (NFDI).21
 | ||
| Fig. 1 The FAIR Guiding Principles for scientific data management and stewardship,8 in short “the FAIR principles”, provide guidelines for improving the reuse of data, and hence rely intrinsically on reproducibility and sufficiently well documented data. | ||
While both the FAIR principles and large-scale research data infrastructure focus mostly on big (and uniform) data11,14 and data reuse, reproducibility requires relevant metadata to be collected close to the actual data acquisition, i.e., much earlier in the research data life cycle22 (Fig. 2). Furthermore, particularly with “little data” (sometimes termed the “long tail” of data12,13), reuse by people other than the “future me” of the scientist originally collecting the data is rare at best. Nevertheless, documenting data with rich metadata to make research as reproducible as possible is an imperative and connected to the professional ethics of researchers,23,24 though reuse (by others) is probably not the most convincing argument for preparing research data for sharing or publishing.
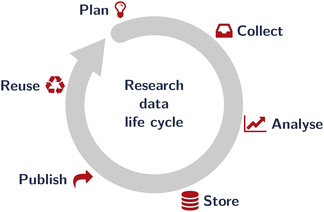 | ||
| Fig. 2 The research data life cycle as an idealised sequence of steps taken on research data. The actual life cycle may be much more complicated. While the FAIR principles8 (Fig. 1) focus pretty much on the last aspect (“reuse”) and particularly on big (and homogeneous) data, a necessary prerequisite is to document data provenance during data acquisition (the “collect” step), hence at the beginning of the research data life cycle. Here, we focus exclusively on documentation during data acquisition. | ||
Infrastructure relies on funding bodies and institutions, and while there are a few examples of successful long-term data repositories, e.g., the PDB,25–27 CCDC/CSD,28 and NCBI,29 there are enough documented examples of highly ambitious projects that stopped being funded and hence ceased.11,30,31 Documenting provenance during data acquisition, however, is both the responsibility of the individual scientist in the lab and entirely under their control. Therefore, we focus here on the early stages of the research data life cycle, namely data collection, and discuss the prerequisites for robust, resilient, and reliable metadata collection that is in itself a necessary prerequisite for FAIR data.
Probably everyone who is active in experimental science knows the phenomenon: you have performed an experiment, are evaluating it weeks or months later – and suddenly realize that a small, but now essential piece of information about the experiment has not been documented. This is all the more true if setups were used for the experiments that consist of many interchangeable components – as is the case with laboratory-built setups. Another scenario: not everyone keeps a clean laboratory notebook in which everything that was done is listed in a comprehensible manner at all times.32
In both cases, what helps is a structured filing of all important information in a kind of form containing all necessary “fields”. Since today there is a computer at almost every experimental setup, it is obvious to record this information before/during the experiment and to use the possibilities of modern electronic data processing for this purpose. Of course, one can think of web forms or something else for this purpose,33 but still the most flexible and robust solution, not depending on any additional infrastructure, is a plain text file with a clear and specified internal structure. This is the background of the development of the Infofile: to create a structured possibility to store all important information about an experiment in a key–value store with a small footprint and minimum technological dependencies. The idea behind the development of the basic file format was to create a solution that is both easy and convenient to write and read for humans and readable by computers.
Here, we present both the specification of the Infofile format and the general ideas behind it that can easily be adapted to other, more generic file formats, such as JSON34 or YAML.35 As is often the case, the key is not so much the file format itself as the idea and concepts that gave rise to its development, as well as the information stored within the files and the guidance the keys provide to collect the relevant bits of information.
2 The why, when, what, and how of research data documentation
The work presented here is rooted in experimental spectroscopy and hence “little science”, due to the experience and background of the authors. This clearly informs the answers to the questions raised: the why, when, what, and how of research data documentation. However, the questions as such need to be addressed in any case. Only the degree of automation may vary substantially between big and little science. Furthermore, while developed in a spectroscopic context, the concepts have been (partly) adapted to computational chemistry by the authors and can be easily adapted to other disciplines as well. For details of how to contribute and further develop the format, see the discussion section below. Due to the focus of the Infofile format, the following discussion is mostly restricted to the “collect” phase of the research data life cycle (Fig. 2). Of course, documenting research data is relevant in other phases as well, particularly during processing and analysis. This is the realm of scientific workflow systems such as the ASpecD framework.36 The FAIR principles8 (Fig. 1) can help in figuring out what to document along the entire research data life cycle, and guides and check lists are increasingly available,37,38 as well as a whole body of literature regarding research data management.5–72.1 Why document research data?
Nowadays, the usual narrative goes as follows: data need to be properly documented with metadata in order to allow others to reuse these data for their own purposes, often in the context of machine learning and artificial intelligence applications.39 This is one of the driving forces behind the FAIR principles,8 and there is some truth in mocking the FAIR acronym as “Finally AI-Ready”. Certainly, there is great value in the exponentially increasing amount of available data,19,40 making data-driven research—the “Fourth Paradigm” of Jim Gray14,41—possible. Therefore there is a great need for high-quality, i.e. properly documented, data for their reuse. However, reusing other people's data (outside one laboratory or group) is mostly an issue of “big science”.10 From our own experience, the average spectroscopist is not too concerned with data sharing and data publication, as they usually deal with their acquired data themselves, only sharing the final analysis and interpretation within collaborations and eventually in traditional text publications with static images. Of course, reproducibility and hence proper data documentation is no less of an issue here. However, it needs a different motivation. The best motivation is to think of the future self. Every scientist wants to be able to comprehend what they have done. This may be motivated either by egoism or by professional ethics. In addition, most group leaders will be familiar with the issue of trying to make sense of data acquired by a student who has long since left, but desperately needing the results for a publication. While this is technically speaking reuse of somebody else’s data, it is still within the context of a group and hence requires much less effort in documenting, as the overall context of original data acquisition and the actual meaning of the data is much better conserved or at least easier to reconstruct.Automating data processing and analysis using scientific workflow systems such as the ASpecD framework36 is another motivation to document research data. Given the documentation in the form of machine-actionable metadata, the processing and analysis routines can gain a “semantic understanding” of the data. Thus, careful documentation during data acquisition directly pays off later. For details see the discussion on how to document research data below.
Finally, it deserves mentioning that both funders9,21,42,43 and publishers increasingly require data to be documented and available, although it remains to be seen whether and how fast this will really lead to more reproducible and FAIRer data particularly in “little science”. As Briney7 correctly states, a data management plan (as required by a funder) and actual data management are two different things, and only the latter will make using our data easier for us (and others), and enhance reproducibility and thus the overall quality of our research.
2.2 When to document research data?
Documenting research data should be done in parallel to data acquisition. The reason is simple: the available information is maximal at this point in time, and deciding upon and extracting the relevant bits from all available information is the crucial part that usually requires a lot of experience and expertise. Here, it is most valuable to learn from those who spend the most time in the laboratory, often the technical staff. Documenting during data acquisition already constrains the way documentation can be sensibly done. On the one hand, it comes in quite handy to have some kind of form. Thus, you need not think about which parameters to record, and can concentrate on the important aspects (see ref. 44, p. 34). On the other hand, documentation should be as independent as possible from technical infrastructure: pen and paper are probably no longer adequate in a digital world. However, a simple but structured text file is clearly superior to a web form in this regard. Still, many setups, though they are controlled by computers, are not connected to a network, let alone the outside world, if only due to security concerns and outdated software that cannot be updated. In any case, leaving documentation until later is not an option, as it will only result in documentation never being done, or in the best case being incomplete and unreliable.2.3 What needs to be documented?
Having dealt with why and when to document, the next question is: what needs to be documented? The simple answer would be: everything that is relevant for reproducible research. Of course this is not helpful (due to being circular), and it is important to stress once again that here, we deal only with documenting the data acquisition and data provenance. Other aspects of reproducible research45 such as a complete audit trail of data processing and analysis are out of scope, but dealt with by, e.g. scientific workflow systems, such as the ASpecD framework.36There are, however, some minimal criteria for information to be documented: you should be able to create the complete materials and methods part of a publication for the given method and acquired data from the metadata recorded during data acquisition. But be aware that materials and methods parts are rarely sufficiently detailed. Hence a better question to answer would be which information you would need to perform a comparable experiment or even repeat the described experiment (ignoring availability of samples and the like). On a more abstract level, data documentation should provide answers to six questions: who has done what, with whom, when, how, and why? Asking for the reason (why?) is often highly important, as it helps to decide upon the quality and context of the data, but is often forgotten or neglected.
A general rule for setups that are laboratory-built or contain exchangeable components is to detail all components with kind, manufacturer, and exact type designation. An example from our own experience may serve as an illustration: if you replace the components of a setup all day long, hunting for the origin of spurious signals, but do not carefully and painstakingly document each step, you cannot assign the individual test measurements to a concrete incarnation of your setup, even by the evening of the very same day. This renders any analysis in the aftermath, which is often sensible and necessary, simply impossible.
To realise which available information is relevant is a matter of the power of observation. While you can train this skill, it is not evenly distributed among scientists. Giving an example of what information may be necessary to record and how important it is to even document things that seem too obvious: strong magnetic fields lead to orientation of chloroplasts in cells, and hence give rise to an orientation-dependent time-resolved electron paramagnetic resonance spectroscopy signal.46 This was only revealed due to the responsible scientist carefully documenting whether the sample had been frozen and hence immobilised within or outside the magnetic field.47
Last but not least, there is a difference between parameters that can be detected or measured and in some cases even controlled on the one hand and all other types of observation on the other hand. While the former should enter the documentation in a formalised way, the latter are typically entered as part of a comment, but can be of tremendous importance, as discussed above.
2.4 How to document research data?
The last question regarding the documentation of research data that needs to be discussed is how to document research data. Whatever way you decide upon, it should have a minimum entry hurdle: only systems that are easy to use and offer obvious advantages will be used. Furthermore, the advantages of using the system should be as obvious as possible, as this motivates researchers to accept the (initial) additional effort. As we are talking about file formats in the given context, as a very minimum they need to be human-writable and machine-readable. On this abstract level, four criteria can be named: (i) simple to write by the user, (ii) uniquely parsable, (iii) robust in the face of user errors, and (iv) easily extendable. The latter criterion is an example of the open–closed principle well-known from software engineering: a system should be open for extensions, but closed for changes that make it incompatible with existing external systems depending on it (see ref. 48, p. 57).Next, research data should be documented in a structured manner, i.e. using key–value pairs wherever possible. For some authors this is the defining aspect of the term “metadata”: structured and hence eventually machine-actionable data about data, not only data about data.49 To be of use, we need a fixed set of keys, although this will mostly be conventions agreed upon in a limited group, not official standards internationally agreed upon. In any case, these key–value pairs allow for a “semantic understanding” by the routines for data processing and analysis. Combined with a scientific workflow system such as the ASpecD framework,36 these machine-actionable metadata reveal their full potential, allowing automation of the processing and analysis pipelines to a great extent.
Furthermore, the metadata should be long-lasting and as permanent as possible. This imposes severe constraints on the available file formats. Long-term storage of digital artifacts is a problem not solved yet, but at least there are some clear lessons from experience as to what does not work (i.e., proprietary formats). Additionally, there is no agreement on what long-term storage of research data (and hence the accompanying metadata) really means, but ten years seems to be a sensible minimum time span.
Documenting research data should be resilient and independent. No man-made system is perfect, hence we need to build resilience right into the systems we use. And documentation is no exception to this rule. This means, e.g., not relying on a single code table connecting different parts, such as a single list connecting sample numbers with sample descriptions. Resilience often comes with duplication that is hard to keep consistent without automatic cross-checking and additional measures. A prime example of resilience (of a certain kind) is the Rosetta Stone: the same information is coded in three different languages. To be independent, on the other hand, echoes having minimum technical requirements and making things as simple and obvious as possible.50
When choosing appropriate file formats for storing (data and) metadata, both criteria, long-lasting and permanent, and resilient and independent, should be carefully considered, as both are related to long-term accessibility of the information: file formats should be robust and long-lasting, with a good chance of accessing them after decades. Besides that, they should be open and well-documented, and ideally internationally standardised, in contrast to proprietary, vendor-specific formats found frequently in spectroscopy. On the time scale of science, the development of computers is a rather short period, not to mention the usual lifetime of file formats. The only robust file format that is fully platform-independent and accessible basically without any special program appears to be bare text, ideally restricted to ASCII 7-bit characters, but nowadays probably UTF-8.51 This is the reason why data exchange in Unix operating systems and their descendants uses text files near-exclusively.52 Using bare text files for storing information may deserve a comment. Insisting on their use implies in no way that these files should be unstructured. On the contrary, structuring these files, and thus creating specific formats, is an important aspect of retrieving information in a fully automated way.
Taken together, metadata should be stored in plain (but structured) text files that reside directly next to the measured data. A useful convention is to name these metadata files identically to the data files (except for the file extension).
2.5 Prerequisites
Having discussed the questions of why, when, what, and how to document research data, a series of prerequisites emerge that need to be fulfilled in order to allow for appropriate metadata acquisition.First of all, a structure or data model is needed for the metadata to be recorded. This requires a thorough understanding of the processes, resulting in a mental model of a dataset that can only be achieved and created by means of sufficient experience. One possible implementation of this model, consisting of the recorded (mostly numeric) data and the accompanying, machine-actionable metadata, is the dataset in the context of the ASpecD framework.36 The data model needs to be modular and expandable, another application of the open–closed principle (see ref. 48, p. 57) mentioned above. Furthermore, the data model and the resulting metadata structure should be a consense, at least within the group of people using the actual format.
Next is the need for simple tools for metadata acquisition. Metadata should be recorded preferably by digital means, and definitely be platform-independent. Furthermore, they need to be machine-readable and human-writable, with a clear emphasis on the latter: as usual, only systems that are sufficiently easy to operate will be used. Additionally, the tools used to record metadata should have minimal external dependencies. Definitely, network connection and internet access are dependencies that, from our own experience, cannot be taken for granted and should hence be avoided. Finally, the tools should be robust against human error, i.e., resilient.
Last but not least, metadata and parameters should be acquired automatically wherever possible. Many parameters can be read out from sensors or by software connected to the measurement device or setup. This does not mean that these parameters should not end up in a metadata file, but rather that the values of those parameters recorded by sensors or software take precedence over the values of the same parameters recorded manually.53 However, this is less a matter of the format the metadata are stored in than the software stack used to record and harmonise the eventual metadata file. The same is true for (automatic) checks for consistency which are highly valuable.
3 The Infofile format
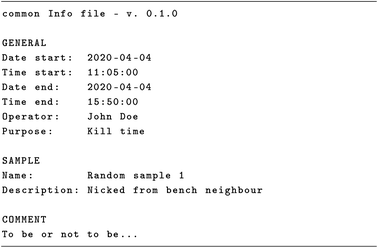
Having set the stage, we will now present an actual file format, the Infofile format, for recording metadata during data acquisition and storing all relevant and important metadata belonging to a measurement. Originally, the format was developed for use with a series of MATLAB® toolboxes54–59 used for processing and analysing different kinds of spectroscopic data, i.e. the predecessors of what later became the ASpecD framework for reproducible spectroscopic data processing and analysis36,60 and the packages based on it.61–64 The information contained in these files is transferred to the associated dataset, one of the key concepts not only of the ASpecD framework, the inseparable unit of data and its accompanying metadata. For a first impression, Scheme 1 shows the most generic version of an Infofile, lacking any specific details for an actual method.3.1 Criteria for the file format
Three criteria were crucial for developing the Infofile format: it should be human-writable (and readable) and machine-readable, with an emphasis on the first part, due to being used by the scientist in the lab on a daily basis. Next, it needed to be plain text,52 and due to being used with MATLAB®, there was a hard requirement to restrict characters to ASCII 7-bit. At the time of development of the format (about 2011) and until recently, MATLAB® had no support for UTF-8. However, the restriction to the ASCII 7-bit character set is relieved now and an Infofile may use the full UTF-8 character set. Additionally, the file format needed to be unambiguously identified, using an identifier in the first or second line that includes the version number. The latter is crucial not only to check whether the file read by a piece of software is an actual Infofile, but also to handle different versions, as the metadata schema will always evolve with time. And last but not least, the file should be self-contained, i.e. understandable without external documentation. All these criteria are nicely reflected in the example of the most general case presented in Scheme 1.3.2 Characteristics of the Infofile format
Generally, the Infofile format is a key–value store with (only) two hierarchy levels: keys are grouped in blocks. This is similar to configuration files such as the INI files popular with the Windows operating system for a long time. The format comes with minimum formatting overhead, as is obvious from Scheme 1: the only formatting users need to be aware of and to pay attention to is block names that appear in all-caps, and the use of colons as key–value separators. This is pretty much self-explanatory, and the format explicitly refrains from using all kinds of brackets, tags, or whitespace characters (indentation) in a syntactically meaningful way. This makes the format pretty robust and resilient. Spaces are allowed nearly everywhere (and are mostly ignored). While the values have been vertically aligned in Scheme 1, this is only for enhanced readability and hence a matter of convenience, but is not at all a requirement of the format itself.Another characteristic of the Infofile that adds to its user-friendliness: keys can contain spaces, making them more human-readable and familiar. Usually, in the software reading the Infofile contents, the spaces will be converted, as depending on the programming language, keys in key–value stores (associative arrays) may not be allowed to contain spaces due to being handled like variable names. However, this is entirely a matter of the importer routines and the processing and analysis framework used, not of the file format as such.
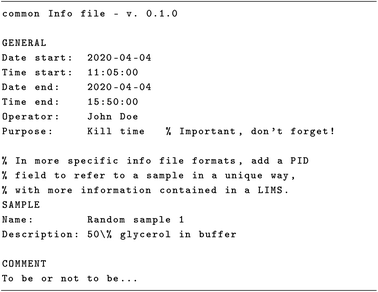
Sometimes it comes in quite handy to add comments to the Infofile that are not meant to be processed by the software reading the file. Hence, comments can be added everywhere, on a line by themselves or at the end of a line, using a special comment character (by default the percent character), cf.Scheme 2. Sometimes, the comment character needs to be used with its actual meaning as character, not in its special function. This is possible by escaping it in the standard UNIX way (prefixing it with a backslash), following the principle of least surprise (see ref. 52, p. 42). As general advice, use comments sparingly, as comments can do more harm than good if not done correctly, and they generally tend to distract from the important aspects (see ref. 65, ch. 4 and ref. 66, ch. 32).
The last characteristic of the Infofile format worth mentioning here is the comment block at the end of the file. The comment block always comes last, allowing for maximum flexibility in formatting and character use, as all the remaining content of the file can be parsed as a comment. While this is mostly a matter of convenience for the parser to be implemented, the comment block as such is an integral part of the Infofile format and the underlying concept of metadata recording. While all recurring observations and parameters of a measurement should (and eventually will) be coded in key–value pairs, there is often the need to note additional observations. Furthermore, this adds to the flexibility of the format, allowing detection of recurring pieces of information in the comment block that lead to extending the metadata model and in turn the introduction of additional keys.
3.3 Heuristics for choosing keys and entering information
Many criteria have been listed above regarding how to choose appropriate keys and decide on which pieces of information to record in the first place, namely being able to create a materials and methods part from the information recorded and to be able to perform a comparable experiment or even repeat the original experiment. However, there is a series of heuristics for how to identify appropriate, useful keys that has more to do with practical aspects.Already from the most generic incarnation of an Infofile presented in Scheme 1, two blocks and a series of keys can be inferred that, while being pretty obvious, deserve some comments. The GENERAL block answers the three questions of who has done something when and why, while the second block, SAMPLE, answers the question “with whom” something has been done. Date and time should be recorded for both the start and end of measurements, as a measurement easily runs either overnight or even for longer than 24 hours. While one could think of using other formats for date and time such as ISO 860167 or RFC 333968 combining both in one string, separating date and time into two fields is much more readable (and writable) for people not familiar with programming. The operator field is also quite important, not least to document who has been involved in recording data. For several operators, use a comma-separated list of names. In the examples shown here, the names are given. One could think of adding persistent identifiers (PIDs) such as the ORCID number. However, not everybody involved in data acquisition in a lab will necessarily have such an ID. Just to repeat, never underestimate the usefulness of explicitly stating the purpose of a measurement or data acquisition. Often, we perform series of experiments, varying one parameter for optimisation, or to get a first overview. Having this piece of information stored in the metadata can be tremendously helpful for deciding which data to process and analyse further. Regarding the details for the sample—whatever a sample may be in a given context—, name and description as shown in the generic example are probably the minimum information necessary. Often, adding a reference by means of a (persistent) identifier is very helpful. This allows for looking up further information. However, for enhanced robustness and independence of the Infofile from external infrastructure, the minimum necessary information regarding the sample should be stored within the file as well. If applicable, chemical identifiers of the sample like InChI69 or SMILES70,71 could be added to the sample section.
Of course, the generic version of the Infofile format (Scheme 1) lacks any details of the method and its specific experimental parameters. This is the realm of dedicated versions of Infofiles for the actual method, and examples are provided in the ESI† and online.72 Nevertheless, here as well, some heuristics can be given. Scheme 3 lists two blocks frequently encountered with spectroscopic methods, namely details about the spectrometer and the temperature. The spectrometer block may be generalised for setups outside spectroscopy. In any case, it should at least contain information on the model (and manufacturer) and as far as possible an exact type designation. Furthermore, as most setups nowadays are software-controlled, details on the software used are necessary as well. The information on the measurement software should include version numbers, ideally with a PID, and the manufacturer if applicable. If your setup is controlled by lab-written software, make sure all the best practices of (scientific) software development apply, as a bare minimum using a version control system and unique version numbers. For an overview see ref. 73 and 36 and references therein.
Measured values should have both a numeric value and a unit. Usually, a number without an accompanying unit is rather useless, and the units are ambiguous much more often than not. While value and unit may well be separated into different fields in the metadata model implementation used in processing and analysis software,36 having them together in one field in the Infofile adds to the convenience of writing such files.74
Another important aspect regarding the recording of metadata is the information on whether parameters have been properly recorded or controlled or whether they are only approximate values. A prime example here is temperature which can be “room temperature” (a highly variable value, often abbreviated as RT), a value properly measured by a sensor, or even a value controlled by a temperature control unit (be it a cryostat or a simpler device). Hence, while the block “temperature” in Scheme 3 provides fields for the actual temperature reading, and the controller, cryostat, and cryogen used, in practice some of these values may not be relevant and hence can be replaced with a string such as “N/A” to explicitly mark them as not available/not applicable.
Setups that are either lab-built or consist of exchangeable or replaceable parts deserve special attention. In this case, it is crucial to document all relevant information for each individual component, such as the probehead in the case of an EPR spectrometer, as exemplified in Scheme 4. In an optical spectroscopy experiment, this would similarly apply to the optical cell used. Beware that explicitly documenting each potentially replaceable part is crucial, even though you never intend to replace it. For each variable component, the manufacturer, kind, and exact type designation should be recorded.
Finally, an excellent heuristic for identifying additional metadata keys is to carefully monitor the information entered into the comment block at the end of an Infofile. First of all, it is important to make use of the comment block to record all additional information that seems relevant but does not (yet) fit to any key–value pair. However, as soon as the same piece of information is repeatedly entered in the comment block, think of making it a proper key.
3.4 Examples
Since its development in 2011, the Infofile format has been successfully used on a daily basis for a series of different spectroscopic methods and experiments, such as time-resolved EPR (trepr) spectroscopy,63 optical transient absorption (TA) spectroscopy (including a variant for detecting magnetic field effects), and continuous-wave EPR (cwepr) spectroscopy.61,62 Due to their verbosity, examples of Infofiles for different spectroscopic methods are given in the ESI,† but are available via GitHub as well.72The software supporting the Infofile format was originally the different MATLAB® toolboxes written by the authors to process and analyse trepr,57 cwepr,56 and TA59 data. The current reference implementation for a parser for the Infofile format is part of the ASpecD framework,36,60 and support for special formats (and mappings) is contained in derived packages, namely the trepr63 and cwepr61,62 Python packages. The source code of the ASpecD framework and hence the reference implementation of the Infofile parser is available via GitHub.75
4 Discussion
The Infofile format is not the first and it will definitely not be the last format for recording metadata during data acquisition. Hence, how does it compare to similar developments, and how does it relate to electronic laboratory notebooks (ELNs) and laboratory information systems (LIMSs), to scientific workflow systems, and to other, more standardised formats?4.1 Comparable developments
One format that comes immediately to mind due to its overall similarity is the FMF format.76 So what makes the Infofile format different, and in some sense probably superior? The FMF format contains both data and metadata, and depends on the actual setup to directly write data (and metadata) in FMF format, while the Infofile is a completely independent and additional file. That means that the FMF file can only be (sensibly) used with setups where the experimenter has full control over the software used to record the data. The Infofile, in contrast, works well with any kind of setup and control software, be it commercial or laboratory-written.Another recent development with similarities to the Infofile format is Adamant, a JSON schema-based metadata editor for research data management workflows.33 According to its authors, Adamant has been developed to systematically collect research metadata as early as the conception of the experiment, and it aims at supporting the whole research data management process. Relying on JSON as an internal format is definitely a sound decision, and making use of a web frontend for metadata recording by the experimenter results in a highly platform-independent solution. However, quite in contrast to the Infofile concept, this requires at least network access and a web server running the Adamant backend somewhere at least in the local network. As mentioned previously, from our own multiple experiences there is a good chance that the ubiquitous computers controlling experimental setups are not connected to the local network, let alone the internet. Hence, a solution such as the Infofile without any external technical dependency besides a simple text editor available on probably every relevant operating system is a clear advantage.
4.2 Relation to data formats
Most scientific data formats except for the most primitive ones will always contain metadata besides the actual numerical data. This is true for both vendor formats and open formats used regularly for data exchange, such as HDF5,77 NetCDF,78 FITS,79 JCAMP-DX,80 or NMReData,81 to name but a few. As these formats all store data and metadata together, similar restrictions to those discussed above for the FMF format apply.76 While instrument control software will usually record parameters as structured metadata, there will always be some information that is not collected this way. Hence the need for a solution to store additional metadata during data acquisition, as provided by the Infofile format. Depending on the data exchange format used, it will be possible to include the metadata recorded using the Infofile afterwards in a separate step, though. In any case, the Infofile format cannot and will not replace formats for storing both data and metadata, but complement these formats at least during data acquisition.4.3 Relation to electronic laboratory notebooks (ELNs)
Electronic laboratory notebooks (ELNs) are pretty en vogue currently, and there are a number of open-source developments gaining momentum and adoption, such as openBIS,82 elabFTW,83 and Chemotion.84 The Infofile can be seen as a lightweight ELN, and it has been used as such in the authors' lab. Furthermore, the Infofile could be included in an ELN record and even in a paper lab book as a printout, the latter having been done in practice. Hence, in some way, the Infofile format can be regarded as a predecessor of an ELN, but it is probably much more an independent aspect of reproducible research, taking care of recording all relevant metadata during data acquisition.Furthermore, an ELN is not necessarily structured, clearly not only consisting of machine-actionable metadata, unlike the Infofile. Additionally, a text file provides much higher flexibility than an online form, e.g. in an ELN. Eventually, however, the information contained in an Infofile could be automatically imported into an ELN, given an API of the latter. Therefore, the Infofile and an (electronic) laboratory notebook complement each other favourably, rather than being competitors. Besides its natural connection to ELNs, the Infofile format does integrate well with a LIMS, mostly by means of references (PIDs) to samples and the like. In the authors' lab, the Infofile is used in the larger context of the LabInform LIMS.85
4.4 Provenance of data analysis and scientific workflow systems
The focus of the Infofile format is on recording metadata during data acquisition, and this is the only aspect of reproducible research and the research data life cycle (cf.Fig. 2) it deals with. Nevertheless, data provenance and recording metadata during data acquisition are only one step towards FAIRer research data. Therefore, the Infofile format does not exist in isolation, but directly connects to scientific workflow systems such as the ASpecD framework36 which provides a gap-less record of each individual processing and analysis step performed on data. However, the ASpecD framework and packages built on top of it61–64 rely on the metadata stored within the Infofile and imported during data import.4.5 Standard formats
There are a number of formats that may be used instead of the Infofile format proposed here, such as XML,86 JSON,34 and YAML.35 Nevertheless, there are good reasons to prefer the Infofile format over each of the named formats that will be detailed below. Eventually, deciding on a particular format is to a certain extent a matter of taste and personal familiarity, and it cannot be overstated that having an appropriate structured metadata model containing all necessary parameters is much more important than the actual file format used to store this information. The reason not to use XML is simple: XML has a far too verbose markup and will most probably never be used by a scientist in the lab, although other disciplines have much better experience with using and manually writing XML, e.g. to annotate text.87,88 In a similar vein, the reason for preferring YAML over JSON is the cleaner structure, basically omitting any brackets, and overall less markup of the former. Eventually, the reason for using the Infofile format rather than JSON or YAML is that the Infofile format is more robust towards users’ mistakes and has a cleaner and more obvious structure with less hierarchic levels, although the latter can become a disadvantage, too. While JSON requires using brackets that add “unnecessary” markup from the perspective of a non-technical person, YAML relies on consistent indentation. While both JSON and YAML can easily and automatically be validated, the Infofile format requires much less formatting than JSON and is more forgiving regarding whitespace as compared to YAML.4.6 Extending and further developing the Infofile format
The current reference implementation for a parser for the Infofile format is part of the ASpecD framework,36,60 with the source code available via GitHub.75 Additionally, examples of Infofiles for different spectroscopic formats are available via GitHub as well,72 together with information on how to contribute to their further development. Due to the permissive license, everybody is welcome to use and further develop the Infofile format for their own purposes. Development of the templates for specific methods will always be closely connected to the respective data model, e.g. in the context of the trepr63 and cwepr61,62 Python packages.5 Outlook
The Infofile format has been used successfully in the authors' lab for more than a decade and for a series of different spectroscopic methods. Although the individual formats for the methods have evolved, the pace of new versions has been slowed down over time, as is to be expected. Two directions for further developments can be anticipated. One is to minimise the contents of the Infofile that need to be entered manually by the operator. This could be achieved by files containing only those parameters not collected automatically by the setup, and automatically adding all the other parameters afterwards with the software used to process and analyse the data. Beware, however, that the “truth” is not necessarily always in the parameter values collected by the setup—for a more detailed discussion see the ESI.† The ASpecD framework36,60 not only allows parsing and importing Infofiles, but also to write reports based on templates. The latter could be used to automatically create Infofiles containing both manually user-recorded and automatically collected information on a measurement. This would ease the manual metadata acquisition by the experimenter, while retaining the full information necessary to reproduce an experiment in a textual file easily accessible and independent of any additional infrastructure and dedicated software. The other direction is to eventually transfer to the YAML format which simply did not exist yet when the Infofile format was developed. This direction is pursued within the UVVisPy package,64 but builds upon experience with the Infofile format and follows the same general principles laid out here. Furthermore, the arguments in favour of the Infofile format put forward above still hold.6 Conclusions
Taken together, we have discussed in quite some detail why and how metadata documenting the provenance of research data need to be recorded in order to enable and enhance the FAIRness of these data. The five key takeaways from this discussion are: (i) data without metadata are useless. Both form an inseparable unit. (ii) The amount of information available is maximal during data acquisition. The crucial task is to reduce it to the relevant facts. (iii) Metadata should allow for a “semantic understanding” by the routines for data processing and analysis. (iv) Metadata should be stored in a structured manner readable by both humans and machines, i.e. computers. (v) A format for metadata should be platform-independent and as simple as possible to use. Furthermore, we have presented a simple, yet powerful file format, the Infofile format, allowing all relevant metadata to be recorded during data acquisition. The Infofile not only provides the ASpecD framework and derived packages with the information necessary for automated data processing and analysis, but it makes research more reproducible and the data documented this way overall FAIRer. Given the lack of recording sufficient metadata during data acquisition, particularly in spectroscopy, and the user-friendliness of the Infofile format, we anticipate the solution described here to have high potential towards making research, particularly in the field of “little science” lacking the established and well-developed software toolchain and standards, more reproducible.Data availability
The code, analysis scripts, and datasets supporting this article have been uploaded as part of the ESI.† The Infofile templates are available via GitHub: https://github.com/tillbiskup/infofile and viahttps://doi.org/10.5281/zenodo.7452780.Author contributions
B. P. motivated and co-developed the format and was key to its practical adoption, as he made its use obligatory for all students he supervised; T. B. developed the format, formalised its specification, implemented the importers, and wrote the paper. All authors read and approved the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Th. Berthold for introducing them to the general idea of collecting all relevant information about data acquisition in a text file sitting next to the actual data on the file system, as well as for being a person with excellent powers of observation, making him a superior scientist. Furthermore, they acknowledge U. Heinen for insightful discussions and all those using the Infofile and realising its relevance, in order of their involvement: D. Nohr, K. Serrer, D. Meyer, J. Popp, C. Matt, K. Stry, P. Jung, M. Schröder. Last but not least, K. Schmitt is acknowledged for fruitful discussions regarding reproducibility and documentation in archaeology and for providing related references.Notes and references
- T. Hölscher, Klassische Archäologie Grundwissen, Wissenschaftliche Buchgesellschaft, Darmstadt, 2nd edn, 2006 Search PubMed.
- M. L. Zeng, Metadata, Facet Publishing, London, 3rd edn, 2022 Search PubMed.
- J. Riley, Understanding Metadata, National Information Standards Organization (NISO), Baltimore, MD, 2017 Search PubMed.
- K. Shankar, J. Am. Soc. Inf. Sci. Technol., 2007, 58, 1457–1466 CrossRef.
- C. Strasser, Research Data Management, National Information Standards Organization (NISO), Baltimore, MD, 2015 Search PubMed.
- L. Corti, V. Van den Eynden, L. Bishop and M. Woollard, Managing and Sharing Research Data: A Guide to Good Practice, SAGE Publications, Thousand Oaks, CA, 2020 Search PubMed.
- K. Briney, Data Management for Researchers: Organize, Maintain and Share your Data for Research Success, Pelagic Publishing, Exeter, UK, 2015 Search PubMed.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- OECD, Recommendation of the Council concerning Access to Research Data from Public Funding, OECD Technical Report OECD/LEGAL/0347, 2006 Search PubMed.
- D. J. de Solla Price, Little science, big science, Columbia University Press, New York, 1963 Search PubMed.
- C. L. Borgman, Big Data, Little Data, No Data: Scholarship in the Networked World, MIT Press, Cambridge, MA, 2015 Search PubMed.
- P. B. Heidorn, Libr. Trends, 2008, 57, 280–299 CrossRef.
- C. L. Borgman, J. Am. Soc. Inf. Sci. Technol., 2012, 63, 1059–1078 CrossRef.
- The Fourth Paradigm, ed. T. Hey, S. Tansley and K. Tolle, Microsoft Research, Redmont, Washington, 2009 Search PubMed.
- Open, Social and Virtual Technology for Research Collaboration, e-Science, ed. C. Koschtial, T. Köhler and C. Felden, Springer, Cham, 2021 Search PubMed.
- J. Gray, A. S. Szalay, A. R. Thakar, C. Stoughton and J. vandenBerg, Virtual Observatories, 2002, pp. 103–107 Search PubMed.
- J. Gray, D. T. Liu, M. Nieto-Santisteban, A. Szalay, D. J. DeWitt and G. Heber, SIGMOD Rec., 2004, 34, 35–41 Search PubMed.
- G. Bell, J. Gray and A. Szalay, Computer, 2006, 39, 110–112 CrossRef.
- A. Szalay and J. Gray, Nature, 2006, 440, 413–414 CrossRef CAS PubMed.
- European Commission and Directorate-General for Research and Innovation, Realising the European open science cloud : first report and recommendations of the Commission high level expert group on the European open science cloud, Publications Office, 2016 Search PubMed.
- RfII – German Council for Scientific Information Infrastructures, Enhancing Research DataManagement: Performance through Diversity. Recommendations regarding structures, Processes, and Financing for Research Data management in Germany, 2016, http://nbn-resolving.de/urn:nbn:de:101:1-20161214992 Search PubMed.
- A. M. Cox and W. W. T. Tam, Aslib J. Inf. Manag., 2018, 70, 142–157 CrossRef.
- National Academy of Sciences and National Academy of Engineering and Institute of Medicine, On Being a Scientist: A Guide to Responsible Conduct in Research, The National Academies Press, Washington, DC, 3rd edn, 2009 Search PubMed.
- National Academies of Sciences, Engineering, and Medicine, Reproducibility and Replicability in Science, The National Academies Press, Washington, DC, 2019 Search PubMed.
- H. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. Bhat, H. Weissig, I. Shindyalov and P. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS.
- H. Berman, K. Henrick and H. Nakamura, Nat. Struct. Biol., 2003, 10, 980 CrossRef CAS PubMed.
- wwPDB consortium, Nucleic Acids Res., 2019, 47, D520–D528 CrossRef.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS PubMed.
- E. W. Sayers, E. E. Bolton, J. R. Brister, K. Canese, J. Chan, D. C. Comeau, R. Connor, K. Funk, C. Kelly, S. Kim, T. Madej, A. Marchler-Bauer, C. Lanczycki, S. Lathrop, Z. Lu, F. Thibaud-Nissen, T. Murphy, L. Phan, Y. Skripchenko, T. Tse, J. Wang, R. Williams, B. W. Trawick, K. D. Pruitt and S. T. Sherry, Nucleic Acids Res., 2022, 50, D20–D26 CrossRef CAS PubMed.
- T. Hey and A. E. Trefethen, Future Gener. Comput. Syst., 2002, 18, 1017–1031 CrossRef.
- T. Hey and A. E. Trefethen, Int. J. High Perform. Comput. Appl., 2004, 18, 285–291 CrossRef.
- Note that for the sake of argument, it doesn’t matter whether the lab book is a physical paper book or electronic. While electronic lab books clearly offer advantages, such as easy cross-referencing and access from multiple locations, understanding both the process to be documented and what information to record is a necessary prerequisite that is overlooked far too often. In short: having a tool doesn’t spare you from learning how to make proper use of it—and from understanding whether it is the right tool for the task at hand.
- I. C. Siffa, J. Schäfer and M. M. Becker, F1000Research, 2022, 11, 475 Search PubMed.
- T. Bray, The JavaScript Object Notation (JSON) Data Interchange Format, RFC Editor STD 90, RFC Editor, 2017 Search PubMed.
- YAML: YAML Ain't Markup Language, 2022, https://yaml.org/ Search PubMed.
- J. Popp and T. Biskup, Chem.: Methods, 2022, 2, e202100097 CAS.
- Y. Li and G. P. Ahlqvist, Preparing Your Chemical Data for Publishing and FAIR Sharing, 2021, DOI:10.17605/OSF.IO/VCSNP.
- ETH-Bibliothek and E. P. F. L. Bibliothèque, Data Management Checklist, 2016, DOI:10.5281/zenodo.633701.
- E. Alaydın, Introduction to Machine Learning, The MIT Press, Cambridge, MA, 3rd edn, 2014 Search PubMed.
- G. Bell, T. Hey and A. Szalay, Science, 2009, 323, 1297–1298 CrossRef CAS PubMed.
- T. Hey and A. Trefethen, Inform. -Spektrum., 2020, 42, 441–447 CrossRef.
- Allianz der deutschen Wissenschaftsorganisationen, Grundsätze zum Umgang mit Forschungsdaten, 2010, DOI:10.2312/ALLIANZOA.019.
- Deutsche Forschungsgemeinschaft, Guidelines for Safeguarding Good Research Practice. Code of Conduct, 2022, DOI:10.5281/zenodo.6472827.
- A. N. Whitehead, An Introduction to Mathematics, Dover Publications, Mineola, 2017 Search PubMed.
- Implementing Reproducible Research, ed. V. Stodden, F. Leisch and R. D. Peng, CRC Press, Boca Raton, 2014 Search PubMed.
- T. Berthold, M. Bechtold, U. Heinen, G. Link, O. Poluektov, L. Utschig, J. Tang, M. C. Thurnauer and G. Kothe, J. Phys. Chem. B, 1999, 103, 10733–10736 CrossRef CAS.
- Personal communication G. Kothe.
- B. Meyer, Object-Oriented Software Construction, Prentice Hall PTR, Upper Saddle River, New Jersey, 1997 Search PubMed.
- A. Brand, F. Daly and B. Meyers, Metadata Demystified, The Sheridan Press & NISO Press, Hanover, PA, 2003 Search PubMed.
- One prime example of storing information in a way that has minimum technical requirements to recover—besides the ability to read—is the microfilm archive in the German Barbarastollen, a disused mine, intended to preserve Germany’s cultural heritage from man-made or natural disaster.
- ISO Central Secretary, Information technology – Universal coded character set (UCS), International Organization for Standardization Standard ISO/IEC 10646:2020, 2020 Search PubMed.
- E. S. Raymond, The Art of UNIX Programming, Addison Wesley, Boston, 2004 Search PubMed.
- The parameter values stored by the control software of the setup need not necessarily reflect the “truth”, though. For details, see the ESI.†.
- T. Biskup and D. Meyer, common toolbox, 2022, DOI:10.5281/zenodo.7396144.
- T. Biskup and D. Meyer, EPR toolbox, 2022, DOI:10.5281/zenodo.7401982.
- T. Biskup and D. Meyer, cwEPR toolbox, 2022, DOI:10.5281/zenodo.7396037.
- T. Biskup, B. Paulus and D. Meyer, trEPR toolbox, 2022, DOI:10.5281/zenodo.7395548.
- D. Meyer and T. Biskup, Tsim toolbox, 2022, https://tsim.docs.till-biskup.de/, DOI:10.5281/zenodo.7395749.
- T. Biskup, TA toolbox, 2022, DOI:10.5281/zenodo.7395925.
- T. Biskup, ASpecD framework, 2022, https://docs.aspecd.de/, DOI:10.5281/zenodo.4717937.
- M. Schröder and T. Biskup, J. Magn. Reson., 2022, 335, 107140 CrossRef PubMed.
- M. Schröder and T. Biskup, cwepr Python package, 2021, https://docs.cwepr.de/, DOI:10.5281/zenodo.4896687.
- J. Popp, M. Schröder and T. Biskup, trEPR Python package, 2021, https://docs.trepr.de/, DOI:10.5281/zenodo.4897112.
- T. Biskup, UVVisPy Python package, 2021, https://docs.uvvispy.de/, DOI:10.5281/zenodo.5106817.
- R. C. Martin, Clean Code. A Handbook of Agile Software Craftmanship, Prentice Hall, Upper Saddle River, 2008 Search PubMed.
- S. McConnell, Code Complete. A practical handbook of software construction, Microsoft Press, Redmond, 2004 Search PubMed.
- ISO Central Secretary, Date and time – Representations for information interchange – Part 1: Basic rules, International Organization for Standardization Standard ISO 8601-1:2019, 2019 Search PubMed.
- G. Klyne and C. Newman, Date and Time on the Internet: Timestamps, RFC Editor RFC 3339, RFC Editor, 2002 Search PubMed.
- S. R. Heller, A. McNaught, I. Pletnev, S. Stein and D. Tchekhovskoi, J. Cheminf., 2015, 7, 23 Search PubMed.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- D. Weininger, A. Weininger and J. L. Weininger, J. Chem. Inf. Comput. Sci., 1989, 29, 97–101 CrossRef CAS.
- B. Paulus and T. Biskup, Infofile, 2022, https://github.com/tillbiskup/infofile, DOI:10.5281/zenodo.7452780.
- A. Prlić and J. B. Procter, PLoS Comput. Biol., 2012, 8, e1002802 CrossRef PubMed.
- Of course, having value and unit separated in different fields is helpful for machine-readability, as the parser needs not rely on the user adding at least one space between number and unit. However, the Infofile format values human-writability higher in this case.
- T. Biskup, ASpecD framework, 2022, https://github.com/tillbiskup/aspecd, DOI:10.5281/zenodo.4717937.
- M. Riede, R. Schueppel, K. O. Sylvester-Hvid, M. K. M. C. Röttger, K. Zimmermann and A. W. Liehr, Comput. Phys. Commun., 2010, 181, 651–662 CrossRef CAS.
- The HDF Group, Hierarchical Data Format, version 5, 1997–2022, https://www.hdfgroup.org/HDF5/ Search PubMed.
- Unidata, NetCDF, 2022, DOI:10.5065/D6H70CW6.
- D. C. Wells, E. W. Greisen and R. H. Harten, Astron. Astrophys., Suppl. Ser., 1981, 44, 363–370 Search PubMed.
- A. N. Davies, R. M. Hanson, P. Lampen and R. J. Lancashire, Pure Appl. Chem., 2022, 94, 705–723 CrossRef CAS.
- M. Pupier, J.-M. Nuzillard, J. Wist, N. E. Schlörer, S. Kuhn, M. Erdelyi, C. Steinbeck, A. J. Williams, C. Butts, T. D. Claridge, B. Mikhova, W. Robien, H. Dashti, H. R. Eghbalnia, C. Farès, C. Adam, P. Kessler, F. Moriaud, M. Elyashberg, D. Argyropoulos, M. Pérez, P. Giraudeau, R. R. Gil, P. Trevorrow and D. Jeannerat, Magn. Reson. Chem., 2018, 56, 703–715 CrossRef CAS PubMed.
- C. Barillari, D. S. M. Ottoz, J. M. Fuentes-Serna, C. Ramakrishnan, B. Rinn and F. Rudolf, Bioinformatics, 2016, 32, 638–640 CrossRef CAS PubMed.
- N. CARPi, A. Minges and M. Piel, J. Open Source Softw., 2017, 2, 146 CrossRef.
- P. Tremouilhac, A. Nguyen, Y. Huang, S. Kotov, D. S. Lütjohann, F. Hübsch, N. Jung and S. Bräse, J. Cheminf., 2017, 9, 54 Search PubMed.
- T. Biskup, LabInform: A modular laboratory information system built from open source components, ChemRxiv, 2022, preprint, DOI:10.26434/chemrxiv-2022-vz360.
- J. Paoli, E. Maler, T. Bray, F. Yergeau and M. Sperberg-McQueen, Extensible Markup Language (XML) 1.0, W3C W3C recommendation, (5th edn), 2008 Search PubMed.
- TEI Consortium, Guidelines for Electronic Text Encoding and Interchange, Version 4.5.0, 2022, Last updated on 25th October 2022, DOI:10.5281/zenodo.7382490.
- Personal communication C. Odebrecht.
Footnotes |
| † Electronic supplementary information (ESI) available: Specification of the Infofile format; examples for different spectroscopic methods; additional discussion. See DOI: https://doi.org/10.1039/d2dd00131d |
| ‡ Present address: Bundesinstitut für Risikobewertung, Max-Dohrn-Straße 8–10, 10589 Berlin, Germany. |
| This journal is © The Royal Society of Chemistry 2023 |