Open Access Article
Open Access ArticleCollaborative methods to enhance reproducibility and accelerate discovery
Drew A.
Leins
 *a,
Steven B.
Haase
b,
Mohammed
Eslami
c,
Joshua
Schrier
*a,
Steven B.
Haase
b,
Mohammed
Eslami
c,
Joshua
Schrier
 d and
Jared T.
Freeman
a
d and
Jared T.
Freeman
a
aAptima, Inc., USA. E-mail: drewleins@gmail.com
bDuke University, Depts. of Biology and Medicine, USA
cNetrias, LLC., USA
dFordham University, Department of Chemistry, USA
First published on 29th November 2022
Abstract
Many domains across physical, life, and social sciences suffer from incomplete models of constructs (e.g., organisms, environments, behaviors), which hinders reproducibility and the pace of discovery. Critically, the prevailing research paradigm, of individuals or small groups working within the resource constraints of their own organization, does little to support model completion and discovery. It does not integrate capabilities, enable investigators to generate data at scale, or offer a path to sharing knowledge at the level of data (versus at the level of conclusions). To develop and deploy a new paradigm for conducting science, The Defense Advanced Research Projects Agency (DARPA) created the Synergistic Discovery and Design (SD2) program. The SD2 program proposed a novel method of conducting science that (1) integrates the capabilities of multiple organizations across multiple domains and (2) makes implicit knowledge explicit at all stages of the scientific process. It assembled and integrated a sociotechnical system that aimed to overcome the limitations of conventional scientific methods. In this paradigm, scientists and technologists collaborated to develop technologies, share data, and conduct science in ways that were faster, more efficient, more complete, and more productive than was possible outside of this program. This paper describes the SD2 approach to developing that sociotechnical system—of selectively applying conventional methods of science, embracing a more collaborative paradigm, and establishing an infrastructure—to drive discovery.
Many domains across physical, life, and social sciences suffer from a common problem: Incomplete models of constructs (e.g., organisms, environments, behaviors) hinder reproducibility and the pace of discovery. For example, in synthetic biology, until recently only a fraction of the genomic interactions and regulatory network of E. coli was documented, making it difficult to design compatible genetic components and systems that reliably persist in E. coli across environmental conditions. This necessarily constrained the ways E. coli can be used as a testbed to design organisms that perform useful functions. Critically, the way science is currently conducted in many fields does little to support model completion and discovery. It does not (1) facilitate integration of diverse capabilities, (2) position investigators to generate quantities of data needed to drive advanced analytics (e.g., machine learning), or (3) offer a path to sharing knowledge when it is most profitable to do so, at the level of data (vs. at the level of conclusions).
To develop and deploy a new paradigm for conducting science, The Defense Advanced Research Projects Agency (DARPA) created the Synergistic Discovery and Design (SD2) program. The SD2 program proposed a novel method of conducting science that (1) integrates the capabilities of multiple organizations across multiple domains and (2) makes implicit knowledge explicit at all stages of the scientific process. It assembled and integrated biologists, chemists, computer scientists, and social scientists to develop and embody a sociotechnical system that aimed to overcome the limitations of conventional scientific methods. This paper describes the SD2 approach to developing that sociotechnical system—of selectively applying conventional methods of science, embracing a more collaborative paradigm, and establishing an infrastructure—to drive discovery.
The prevailing paradigm for research
To better understand the methods developed and deployed by the SD2 program, it helps to understand the benefits and limitations of the prevailing, conventional paradigm of science, one we call the individualist paradigm.1 Individualist science is characterized by an individual or small group working within the resource constraints of their own organization to design, test, and analyze constructs.Benefits of individualist science
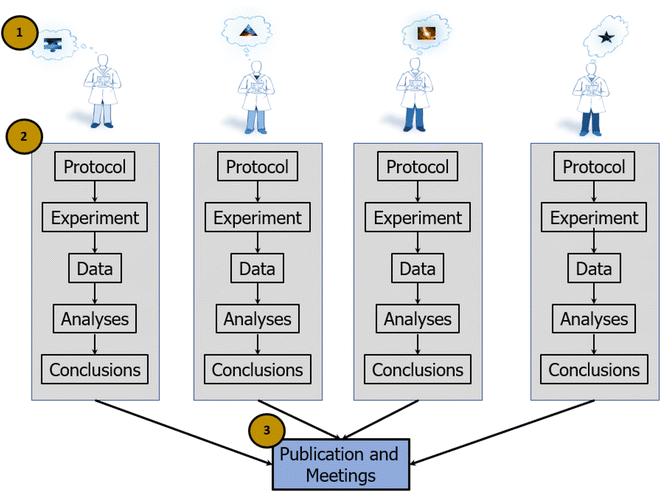
In the individualist paradigm, single investigators or small groups complete an entire research cycle on their own. Investigators generate focused questions within tractable regions of a research space, develop experimental and analytical approaches to addressing the questions, perform the experiments, analyze the data, and then evaluate whether the results support or refute a model or a theory (Fig. 1). It is largely a flexible paradigm, as it allows investigators to choose their own questions and search over a broad scientific space (vs. confining investigators to operating in a narrow space defined by a central authority). This method is well suited for scientific exploration that does not require extensive resources or interdisciplinary capabilities. From an organizational perspective, individualist science is efficient, as it necessarily co-locates planning, data generation, and analysis. If investigators work only with members of their own organization (or their own lab), it obviates the need for cross-organization communication (e.g., contractual negotiations), training (e.g., how to implement a protocol), and the extensive data processing and tracking required when sharing data outside of an organization.This paradigm can also be operationally efficient because it allows investigators to leverage their own strengths, knowledge, and previous methods and results. Such an approach allows for continuity in developing, conducting, and disseminating a line of research. For example, iterative or derivative methods that address the same research question may yield results that are comparable across experiments and thus easily aggregated and interpreted. The individualist paradigm also consolidates scientific knowledge so that the individuals who know the most about a dataset are those who generated it, thus accelerating the process of reporting results. In short, the individualist paradigm allows investigators to conduct principled, efficient, and sometimes impactful science. However, it imposes constraints that put grand discovery out of reach, slows the pace of progress, and causes confusion generated by a lack of reproducibility.
Limitations of individualist science
Individualist science is inadequate for solving complex problems that require multiple experts, diverse resources, or rapid generation of data at scale. Complex problems often require multiple experts from different disciplines to contribute to solutions. Examples include interventions for cancer or Alzheimer's disease, or rapid vaccine development in response to a pandemic. Of course, individual scientists can contribute to solving these problems; however, scientists operating within the confines of their own group are necessarily constrained by the capabilities and expertise of that group. Hence, these groups often investigate only part of a complex problem. For, example, in the study of cell division and cancer, individual labs tend to specialize in the regulation of a specific event or phase (e.g., DNA replication in S, or growth-factor signaling in G1). Alternatively, labs may focus on a specific mode of regulation (e.g., kinase/phosphorylation or transcriptional regulation). In the study of materials science, some groups specialize in the synthesis of novel materials, others in characterization, and others in processing and device fabrication. The implicit understanding is that somehow all these labs will congregate and determine how all of these activities and pieces fit together to inform a complete model (e.g., of cell division or a new functional material). Yet the data and models generated by these labs are unlikely to be compatible, so aggregating understanding at this level is unlikely to yield the necessary insights. As laboratories become more specialized, they may become even more siloed and less able to share protocols, data, and insights, let alone to do so in rapid, scalable ways. As problems become more complex, interdisciplinary, and urgent, the paradigm for solving them must enable rapid integration and scaling of data-collection and analysis capabilities. The individualist paradigm hinders labs from integrating capabilities and collaborating effectively.The prevailing approach to authoring protocols constrains scientists to designing experiments that feature and leverage their own organization's resources. As noted previously, capabilities may not be compatible across organizations; thus, even in response to a common problem, different labs often author protocols that call for different experimental materials, instruments, and resources. These differences necessarily render the datasets incompatible and ill-suited for integration. Instead, aggregation of relevant scientific knowledge must then occur at the level of conclusions. This may be acceptable when conclusions are compatible;10 however, it becomes problematic when conclusions are incompatible because they are based on different findings yielded by different experimental methods. Identifying the precise point in the research cycle where variance was introduced and influenced the results is critical, but very difficult to pinpoint across organizations working in siloed, parallel fashion.
Within the individualist paradigm, it has become common to collaborate in ways that separate data-collection groups and activities from data-analysis groups and activities. This style of collaboration is prevalent where the data collected are large (e.g., ‘omics data sets). As experimentalists advance from design to experiment to data collection, implicit knowledge (e.g., the intent of an experiment), if not made explicit, may fail to propagate to the data analysis groups (or if it propagates, it does so ambiguously, leaving room for confusion). The lack of integration between experimentalists and analysts in this scenario can result in conclusions that are misaligned with experimental intent and methods. Consequently, it becomes difficult to reconcile conclusions derived from analysis across experiments.
Moreover, a methodological paradigm that fails to support the aggregation of knowledge before the level of conclusions is necessarily slow and difficult to accelerate. When national- or global-level problems require rapid, large-scale responses, interested and capable organizations cannot simply join the cause and contribute their data unless those data are compatible. Instead, responding organizations typically operate in parallel, siloed fashion, executing full experimental cycles before sharing potentially incompatible conclusions. For example, in response to the COVID-19 pandemic, there was a large-scale globally distributed effort to generate data relevant to pathogen genomics, clinical outcomes, and epidemiological factors. Recognizing the value of rapid dissemination of information, many publications provided open access to COVID-19 research and released data prior to peer review.13 Useful as these measures were, the data that were made available were not necessarily easily aggregated for analysis, not least of all because data collection methods were not standardized or transparent and there was no globally accessible infrastructure for transforming and aggregating idiosyncratic data for analysis.14 Thus, it required extensive manual labor to reconcile and compile these data. Such outcomes are suboptimal. By contrast, a paradigm that aggregates knowledge at the level of data supports scaling and acceleration to address a wide range of problems more quickly and comprehensively.
In short, the individualist model of science fails to support reproducibility, data sharing, advances in experimentation and analytics, and rapid progress to common conclusions. Hurdles to generating, analyzing, and sharing data at scale prevent scientists from accelerating model development and discovery. A new paradigm is needed to overcome these hurdles.
An alternative to individualist science
To overcome the limitations of individualist science and accelerate the pace of discovery, scientists must engage in activities that promote collaboration across disciplines and enable production of larger, more completely described, shareable, and reproducible datasets. Rather than bound scientific activities by the physical and intellectual resource constraints of individual organizations, scientists should seek to enhance their organizational capabilities and leverage the complementary capabilities of other organizations. The SD2 program did this by integrating existing, distributed methods and capabilities into a sociotechnical system that allows scientists to collaborate to share protocols and data more easily, generate data more reliably, and leverage cross-organizational capabilities that support data generation and analysis at scale.One can look to a recent example in the study of protein structure—the success of AlphaFold—to illustrate the benefits of collaborative science and, in particular, standardized data formats and advances in data analytics.16 For decades, numerous researchers worked individually to document and collect many high-resolution structures of diverse proteins. To be publishable, these data had to be in a standard format and posted to a shared repository, the Worldwide Protein Data Bank (wwPDB).17 This data bank, which currently supports large-scale data sharing and reuse, took more than 20 years to gain traction with scientific, publishing, and funding communities. It was founded in 1971 by a relatively small community of scientists who were willing to share their data. At that time, the larger scientific community was less inclined to share data, at least in part because of unresolved questions regarding the scientific and commercial value of withholding data (e.g., to improve their accuracy or realize financial gains).18 It was not until 1989 that the International Union of Crystallography's (IUCr) Biological Macromolecule commission addressed these concerns and articulated its policy that structures be deposited in the PDB prior to publication and that their public release could be delayed for a limited time.19 Then, in 1992, more than 20 years after the PDB was founded, the NIH and other funding agencies began to adopt formal policies mandating deposition of research results in the PDB.20 Since the year 2000, over 209![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 data sets have been deposited to the PDB. These data are freely and publicly available. Consequently, they have been used to inform a vast array of prospective solutions associated with predicting protein structure. For example, the ten most cited structures in this database had been cited, collectively, over 31
000 data sets have been deposited to the PDB. These data are freely and publicly available. Consequently, they have been used to inform a vast array of prospective solutions associated with predicting protein structure. For example, the ten most cited structures in this database had been cited, collectively, over 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times through 2018.21 Most recently, wwPDB datasets fed novel data-driven neural-network models of machine learning to provide a computationally efficient method of predicting structures that is as accurate as experimental methods that are far more costly and less efficient.22 The SD2 program aimed to accomplish a similar outcome, though more quickly, in synthetic biology and materials chemistry, as these fields have suffered from a paucity of large, accessible datasets conducive to applications of advances in AI/ML (except see resources such as SynBioHub,23 the Cambridge Structural Database24 [CSD] and the Inorganic Crystal Structure Database25 [ICSD], which provide access to large repositories of structural data for some biological and chemical constructs). Here, we describe components of a framework for collaborative science that aggregates and integrates scientific insight at the level of data and generates datasets of sufficient quantity and quality to support better experimental decision-making, leverage advances in analytics, and accelerate discovery.
000 times through 2018.21 Most recently, wwPDB datasets fed novel data-driven neural-network models of machine learning to provide a computationally efficient method of predicting structures that is as accurate as experimental methods that are far more costly and less efficient.22 The SD2 program aimed to accomplish a similar outcome, though more quickly, in synthetic biology and materials chemistry, as these fields have suffered from a paucity of large, accessible datasets conducive to applications of advances in AI/ML (except see resources such as SynBioHub,23 the Cambridge Structural Database24 [CSD] and the Inorganic Crystal Structure Database25 [ICSD], which provide access to large repositories of structural data for some biological and chemical constructs). Here, we describe components of a framework for collaborative science that aggregates and integrates scientific insight at the level of data and generates datasets of sufficient quantity and quality to support better experimental decision-making, leverage advances in analytics, and accelerate discovery.
Solving complex problems that require multiple experts, a diversity of resources, or data at scale
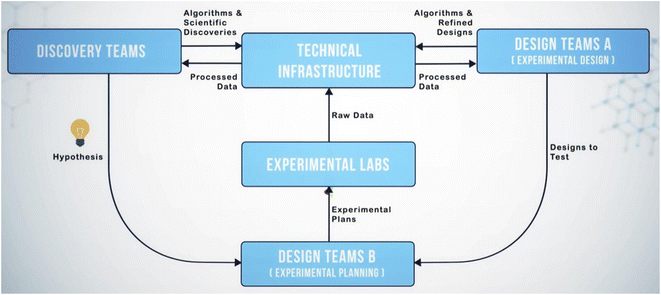
The SD2 program focused on three core scientific topics: (1) designing proteins for better binding and stability, (2) engineering genetic components and networks for sensing and signaling in yeast and bacteria, and (3) identifying materials and methods for synthesizing perovskite crystals. These are high-dimensional problems that require diverse expertise and capabilities and data at scale to fully investigate. Because these types of problems require more comprehensive investigation, it is necessary to bring a more complete set of resources to bear on them. This was a fundamental question for SD2: How does one integrate the diverse capabilities of distributed organizations to solve high-dimensional problems more quickly and effectively? The solution was to design a sociotechnical system that identified and integrated complementary expertise and capabilities across individual organizations. For example, consider a high-level workflow for investigating a model representing an organism with a synthetic circuit that renders the organism a sensor for a molecule in its environment. Investigators design the model, including the synthetic parts and network, write a protocol to test it, implement the protocol (e.g., build parts, run an experiment, generate data), analyze the data, and generate conclusions. It would require tremendous resources to complete this workflow at scale. A single organization likely could not do this efficiently, end-to-end, and generate the data required to fully interrogate the model. The SD2 program, however, divided the required scientific and engineering labor across more than 20 organizations spread over five technical areas. These technical areas included the following:• Discovery, responsible for applying machine learning and other sophisticated analytics to data, to inform model development.
• Design, responsible for designing model constructs and developing protocols for testing those constructs.
• Experimentation, responsible for executing protocols and generating raw data.
• Data Infrastructure, responsible for providing and maintaining the hardware and software for transferring, storing, curating, and processing data at Petabyte scale.
• Socio-technical Integration, responsible for facilitating, monitoring, and measuring collaboration in, and the evolution of, the sociotechnical system.
Separating design, discovery, experimentation, and data infrastructure created loops of data flow that required the scientists and technologists of SD2 (designated “program performers”) to collaborate to identify the most propitious ways to generate, share, and operate on data. Performers collaborated through several methods, including attending an in-person hackathon and quarterly program-wide integration meetings, participating in small groups to define and refine workflows and roles, interfacing on collaboration-focused platforms (e.g., Slack, Google Drive, GitHub, GitLab), and leveraging a shared infrastructure for housing data and software (the SD2 Environment or “SD2E”).26 Through these methods, performers negotiated the scientific questions they would address, the methods they would use to address them, and the division of labor across teams addressing a challenge. Simultaneously, performers identified friction points that slowed communication or the transfer or use of data and developed solutions to mitigate those friction points. For example, during a week-long program-wide hackathon at the start of the program, existing datasets relevant to each scientific topic were delivered to teams of performers clustered by each topic. As is typical of most research datasets, these were labeled and organized idiosyncratically, making it difficult for performers to understand the intent of the activities that generated the data and to identify methods for analyzing them. It became clear that if multiple teams were to operate on the same datasets, those teams needed a mechanism for understanding the data. This led to negotiating methods for describing activities, data, and metadata in a standardized way, so that any performer on the program could theoretically understand and operate on any SD2 dataset (akin to implementing FAIR data standards).27 Thus began development of the sociotechnical infrastructure that evolved into a system of people, technologies, and processes (the sociotechnical system, or STS) that guided the work within, and the collaboration across, each technical area. This system allowed design teams to send models to experiment planners who compiled and pushed protocols to experimental labs that generated and shared data with analysts who operated on it and fed results back to designers for model refinement (see Fig. 2). This became known as the design-build-test-learn (DBTL) loop. The general workflow of this loop has its roots in Walter Shewhart's specification–production–inspection cycle for quality control and iterative product development in manufacturing (1939). This was popularized by Edward Deming in the 1950s as the plan-do-check-act (PDCA) cycle. This is essentially a formalized version of the scientific method applied to manufacturing. The broad contours of iterative ligand synthesis and validation have existed for some time in pharmaceutical research, and certainly have been practiced without giving it an explicit name. Early invocations refer to this as cycles of design, synthesize, test, interpret28 or design, test, make, analyze,29 which consciously adopts quality management terminology for the discovery process. As implemented in SD2, it was an architecture for collaboratively and iteratively generating and analyzing data at scale and enhancing scientific models in ways not achievable through individualist science.
Supporting reproducibility
Protocol authoring
The experimental protocol is the core driver of data generation in the DBTL loop. Finding common ways to express protocols (and by extension their associated data and metadata) is the first step in solving reproducibility problems and enabling rapid data sharing.To facilitate protocol authoring and sharing across organizations, the SD2 program developed a suite of standardization tools (e.g., document templates and software applications) that integrated with commonly used document and spreadsheet formats. One of those tools, the Experiment Request, is a Google Docs-based template for organizing the prose of a protocol—the details that characterize a protocol—into a machine-readable format for driving an experiment.30 Critically, it is a dynamic document that records and timestamps changes to the experimental goal, rationale, execution plan (including a matrix of materials, parameters, and measurements), and analysis plan. It provides users with references to relevant artifacts, matrices of experimental materials and variables, and parameters for candidate analytics (e.g., expected data types and assertions of measurement dependencies). It also identifies metadata requirements, potential or expected challenges to running the experiment, and links to newly generated data. This template evolved with use over time, standardized the language of experimental protocols, and resulted in a more streamlined DBTL workflow. Consequently, performers spent fewer hours searching for clarification of critical details such as experimental parameters or data and metadata structures.
To further streamline the cross-organization workflow, program performers developed the Intent Parser,31 an application that processes the Experiment Request to further standardize the language in a protocol by mapping constructs in the request to canonical definitions in a shared database (e.g., a “Data Dictionary”)32 and linking these constructs to descriptions in a widely accessible resource (e.g., SynBioHub).23 When the Intent Parser encounters a novel term—for example, an idiosyncratic shorthand label for a material or procedure—it flags it for clarification. This allows scientists to either adopt others' (perhaps more common) terms and revise their protocol or to add new terms to the shared database. Thus, the Intent Parser helped standardize protocols while also allowing flexible protocol authoring. Critically, it reduced the need for all collaborators to know and use the same language during planning, as it discovers language discrepancies and incrementally nudges collaborators toward a shared lexicon.
This approach to protocol authoring defines the materials and methods of an experiment with sufficient precision to ensure reproducibility (or at least to raise the likelihood of reproducibility), yet it grants scientists license to use some local terminology by automating the standardization of terms. It also allowed a level of efficiency and productivity not realized prior to developing these applications: In a four-month period following development of this approach, 19 users from various organizations generated 34 experiment requests across three different protocols, yielding over 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 experimental samples generated across multiple sites. Moreover, the standardized language and formats of these protocols allowed simpler and faster analysis and more effective data sharing across the program31 It is important to note, however, that these applications were born of a struggle to develop a single, automated tool for ingesting the prose of any synthetic biology experiment protocol and translating it into a protocol that is executable by one of multiple laboratories with different infrastructures and capabilities. The work of developing such an automated tool—for example, of reconciling the different methods, materials, and lexicons of over a dozen organizations—proved to be too costly in time and resources to pursue. Instead, the SD2 community decided that the efficiency afforded by the combination of small modifications to the content and structure of written protocols and the relatively simple automation of the Experiment Request and Intent Parser tools outweighed the cost of developing a fully automated protocol translation tool.
000 experimental samples generated across multiple sites. Moreover, the standardized language and formats of these protocols allowed simpler and faster analysis and more effective data sharing across the program31 It is important to note, however, that these applications were born of a struggle to develop a single, automated tool for ingesting the prose of any synthetic biology experiment protocol and translating it into a protocol that is executable by one of multiple laboratories with different infrastructures and capabilities. The work of developing such an automated tool—for example, of reconciling the different methods, materials, and lexicons of over a dozen organizations—proved to be too costly in time and resources to pursue. Instead, the SD2 community decided that the efficiency afforded by the combination of small modifications to the content and structure of written protocols and the relatively simple automation of the Experiment Request and Intent Parser tools outweighed the cost of developing a fully automated protocol translation tool.
Whereas the synthetic biology working groups maintained and reconciled existing languages for representing new protocols, the materials chemistry working group had to invent new processes for representing protocols because the methods for testing their prescribed reactions had never before been automated. Hence, this group adopted a co-design model in which it simultaneously developed the underlying experimental laboratory protocol (designated as a “workflow” of reaction and characterization steps) and the software to support experimental specification and reporting. The experimental development was geographically distributed, with one team working on bench-scale development and the other team focused on automating those processes. A third team of software developers served as a conduit between the two experimental groups, capturing and documenting the necessary specifications and metadata required to perform the experiment. This informed both the software development as well as the evolution of the protocol and ensured that the protocol could be executed at different sites with different capabilities.
Transitioning protocols across organizations
To further support reproducibility, the SD2 program leveraged mechanisms for easily transitioning protocols to experimental labs. The program integrated software applications that compile and output human- or machine-readable protocols for guiding human or automated protocol execution. These applications allowed different organizations across technical areas to advance protocols while maintaining their own conventions for drafting and executing protocols. The synthetic biology groups largely used the following two applications to transition protocols to experimental labs: Aquarium33,34 and Autoprotocol.35 The materials chemistry group developed and deployed an application called Experiment Specification, Capture, and Laboratory Automation Technology (ESCALATE) to support protocol development and execution across labs.36Aquarium is a lab-management software application designed to support human-executed experimentation at the benchtop (though it can also drive automated experimentation). It is a user-friendly application that offers predefined, common experimental procedures and allows users to define their own protocol templates. Every defined, executable protocol step is represented as a unit that authors can select and chain to generate simple or complex protocols. This standardization generates protocols that are easy to repeat and share. By installing and using Aquarium at multiple performer sites, the SD2 program developed a network of laboratories that can leverage each other's capabilities to generate complementary datasets and test for reproducibility.
Autoprotocol is an experiment-building language designed to produce machine-executable protocols that enable automated experimentation. Similar to Aquarium, Autoprotocol allows an author to identify experimental parameters, compile a protocol of executable steps, and share the protocol with an appropriately equipped lab. Autoprotocol differs slightly from Aquarium in that it requires greater knowledge of coding; however, with modification, it can offer a user interface that allows scientists to select protocol elements and configure them in a workflow that can be easily read and interpreted by a human lab tech or an automated agent.
The software supporting materials chemistry experiments, ESCALATE, originated from cross-organization work and thus emphasizes cross-lab interactions. Because the labs participating in SD2 had different levels of automation, ESCALATE was developed to support both human and machine instructions. It was originally developed as a lightweight framework to prototype designs using a shared file system and spreadsheets. It was forged in the context of designing experiments to support metal halide perovskite crystallization. Using the lessons learned from the co-design process, a more general version was developed to allow for arbitrary experiments. Borrowing lessons learned from the development of Aquarium and Autoprotocol, the underlying model of experiments in ESCALATE allows for import and export of Autoprotocol experiment specifications. In principle, this can also be adapted for import and export of other emerging standards for describing reaction data, such as those found in the Open Reaction Database37 and those using the Universal Chemical Description Language (χDL).38
Aquarium, Autoprotocol, ESCALATE, and similar applications support collaborative science and reproducibility in several ways. First, these applications help standardize technical language, resulting in protocols that can be read and understood by users who share that language. Second, they allow scientists to author a protocol to be executed in any compatibly equipped lab. Collaborations can exist in which partner labs offer collaborators a menu of experimental capabilities from which to choose. Thus, for example, a scientist with no access to measurement devices could author a protocol to be run in a fully instrumented lab because both the author and the lab run instances of Aquarium. These applications can also be force multipliers. A scientific question requiring quick production of large datasets (e.g., data characterizing a novel virus at the start of a pandemic) could be addressed via multiple labs running the same protocol, thus consolidating an otherwise distributed workforce. A protocol that might have been executed by few can now be executed by many.
To support capability integration even further, at the time of this publication, an application—the Open Protocol Interface Language (OPIL)—is in development to translate protocols between Aquarium and Autoprotocol.39 With a common protocol language, the data and workflows from labs with different operating systems (e.g., human operated or semi-automated) will be interoperable. This will allow more efficient vetting of protocols and convergence on candidates for automated or high-throughput execution. As more labs integrate in this way, they can scale up responses to emergent problems without requiring investment in new infrastructure for automated workflows.
Supporting data sharing
To realize potential synergies across distributed organizations, the data generated by each organization must be shareable and interpretable. Standardizing protocols is a necessary early step, as experimental intent, input, and output must be interpretable. Similarly, standards for capturing and representing metadata are critical for sharing (propagating and tracking) data through a distributed workflow. In SD2, metadata took the form of contextual data (e.g., provenance of materials, descriptions of samples on plates) and the methods for generating, labeling, and processing those contextual data. One of the challenges of standardizing metadata across organizations is that different organizations have different conventions or schemas for representing metadata; for example, human-operated labs may record and convey metadata flexibly, in prose, whereas robotics labs may record and convey metadata rigidly, in machine-readable code. The conventions used by each are not easily abandoned. Thus, rather than develop a new convention and require all performers to adopt it, the SD2 program developed a post–hoc process for translating metadata from different organizations into a common lexicon. Initially, organizations with established conventions populated a table that mapped terms from convention X to convention Y to convention Z. This table allowed organizations to maintain their conventions and to translate their terms to and from other naming conventions. This eventually led to development of the previously referenced Data Dictionary32 and a stable schema that all performers could use to represent data and metadata for almost any experimental space in the program. It allowed data analysts to see the conditions of a given experiment, access a reference file, and query the data in a meaningful way. For example, when exploring the performance of genetic circuits in yeast, analysts needed to know what materials (e.g., yeast strain and reagent) were in each well of a sample plate. The Data Dictionary gave analysts access to a fully described experimental sample that allowed them to draw meaningful conclusions from the data. Critically, it provided a commonly accessible repository of complete data, which obviated the need to share data files repeatedly across communications platforms (e.g., email, Slack) and risk operating on incompatible data.Performers on SD2 also used metadata to describe analyses. Downstream, at the “learn” stage of the DBTL loop, many parameters were required to configure analytics. These parameters became metadata that described an analysis and allowed for easier sharing and interpretation of results. For example, when ensuring the quality of data and results, analysts could refer to metadata representing analytic choices (e.g., to use one control sample rather than another), to consider the validity of those choices and determine the quality of the results. Thus, by developing tools and processes for capturing metadata at various stages of the DBTL loop, SD2 performers could share data and compare apples to apples at each stage and be confident that they understood what the data and results represented.
The materials chemistry working group capitalized even further on metadata representations, eventually using them to refine their experimental search space. They used statistical analysis of metadata to identify anomalies associated with variations in laboratory conditions, which were in turn used as hypotheses for subsequent experiments.40 This type of “automated serendipity” enables flexible experimentation in which variations are permitted, documented, and analyzed for insights that inform model (or search space) refinement.
In addition to developing metadata tools and processes, SD2 performers leveraged a centralized, performer-maintained infrastructure to facilitate collaborative science and engineering, and in particular, data sharing. This infrastructure provided access to a research computing ecosystem with tremendous capacity to host software and to store and process data at scale.41 Importantly, it provided access to resources to allow a technically diverse user-base (from novice, point-and-click users to expert software developers) to participate in collaborative science and engineering. For example, it served as a platform for hosting the previously described data integration and metadata representation tools, as well as a diverse set of tools for analyzing data. Performers could access a dataset, a reference file for tracking metadata, an array of tools for analyzing the data, and tutorials or documentation for using the analysis tools. It was the core of development activities and a space where performers could contribute to the standards, methods, and tools that advanced a flexible and extensible approach to collaborative science.
Critically, the SD2 program leveraged a central authority to develop and impose mandates for sharing data. This was a key programmatic feature whose consequences—namely, quick negotiation and adoption of processes for sharing data—can be contrasted with the consequences of lacking a central authority. Recall that the PDB was conceived by practitioners in 1971,42 but was not enforced by some journals and funding institutions until 1992.20 In contrast, SD2 performers developed and adopted standards and a formal agreement for data sharing relatively quickly within the program. The program then promoted these standards in outreach to publishers, funders, and military research laboratories. However, it is important to note that the program reached consensus on some standards only after experiencing significant challenges to sharing and operating on data. Early in the program, the data scientists who were responsible for discovery through application of novel analytics, and who relied on the data to be FAIR (findable, accessible, interoperable, and reusable),27 were unable to apply their analytics across early datasets because those datasets were not necessarily interoperable or reusable. It took requests for better data “FAIRness” and quality to induce program performers to develop the processes and leverage the resources described in this section.
Engaging new experimental capabilities and generating data at scale
To generate data at scale, scientists must leverage synergies across labs and efficiencies in labs that enhance data generation through automation. Leveraging cross-organization synergies is relatively straightforward and takes advantage of the tools and applications previously discussed.With standardized, shareable protocols, distributed labs can run the same experiment and generate multiples of data. Collaborators who share an authoring and execution platform can leverage excess capacities for generating data in their partners' labs. Thus, if one lab is running experiments at full capacity, it can request to use spare capacity at a partner lab. In addition, distributed labs with complementary capabilities can collaborate to design and run experiments they would not be able to run alone. This enabled execution of different types of experiments—for example, single-crystal versus thin-film synthesis of perovskite crystals—across laboratories to study common phenomena.40
With automation, collaborators can scale their workflows to generate more data more efficiently. Leveraging automation to enhance data generation can take at least two forms. It can be as simple as adding automation to support human operations at the benchtop, or it can represent a fundamental departure from the benchtop approach, by replacing human operators with machinery. The SD2 program introduced automation to support data generation in both ways: (1) we paired software applications with human technicians (the semi-automated lab) and (2) we paired software applications with machinery (the fully automated lab).
In the semi-automated SD2 lab, applications such as Aquarium support competent, reliable execution and documentation of protocols. They provide human lab technicians explicit instructions for executing every step of a protocol. Lab techs can explore information associated with each step, preview future steps, and review past steps, to enhance their understanding of the protocol. During execution, the application prompts the lab tech to indicate when a step is complete and whether the step was modified in any way. Thus, the lab tech can annotate a step, for example, to record observations, changes to steps, durations of processes, or tips for implementing techniques. After a lab tech has completed a step and recorded any necessary metadata, the application advances to the next step and provides associated instructions. Thus, the lab tech will complete the protocol stepwise, receiving prompts to ensure that the actual conditions of execution are documented at each step. This approach allows the lab tech to exercise discretion in executing the protocol, to modify steps when appropriate, and to document the modifications. Of course, allowing lab techs discretion over how to execute a protocol can be problematic, as it requires competency. Applications such as Aquarium address this concern in two ways. First, they reduce the baseline level of competency required by lab techs by including contextual details (e.g., tips for techniques) and alternative operations. Consequently, lab techs do not have to rely on their own, potentially limited, knowledge of scientific principles or experience at the bench to determine whether an operation should be executed as written. Second, in cases in which lab tech competency is deficient or unknown, protocol authors can disable the features of the application that allow lab techs to exercise discretion (e.g., suggestions of alternative materials or procedures). In these cases, the application simply guides the lab tech rigidly through the protocol. This can alleviate some of the cognitive demand experienced by lab techs, for example in evaluating options or executing complex actions, and free up resources to focus on the important aspects of the protocol, thus reducing errors.13
Software applications that reduce the competence needed to execute experimental protocols should, however, be implemented strategically. Although they support efficient, reliable data generation, they represent an ambiguous good from an educator's perspective. On the one hand they may allow students or other workers with lower levels of training to participate in the research process; on the other hand, they reduce agency and the development of higher order skills associated with entry-level scientific apprenticeships. This tradeoff is not necessarily detrimental. As more scientific tasks become automated, the need for scientists to develop expertise relevant to formerly manual tasks decreases and is replaced by a need to develop expertise and knowledge relevant to automation. A discussion of the opportunities and training needs associated with automation in the chemical sciences can be found in a recent report by the US National Academy of Sciences.43 Incorporating automation technologies into pedagogical training is not inherently new,44,45 but recent efforts have focused on training students in the combination of experimental hardware and planning algorithms46,47 and on closely adjacent enabling technologies such as computer vision48,49 and speech recognition.50 In addition to producing a more technologically skilled workforce, this also provides an opportunity to create a more inclusive scientific workforce, as laboratory automation can remove barriers for students with visual or physical disabilities.51
In the fully automated lab, scientists design and submit protocols that can be read and executed by machines. In a lab with closed work cells, there is typically no human–machine interaction beyond uploading and selecting a protocol. The progress of an experiment is tracked through automated documentation of the movement of materials through a workflow. This is generally an efficient means of generating data, as it is not constrained by human resources (e.g., time, attention, availability). However, protocol flexibility and modification opportunities are limited, as changing hardware and software parameters can be resource intensive. Hence, automated workflows are ideal for protocols that have been pilot tested and vetted. This concern can be partially mitigated by an open, modular infrastructure in which workflows can move from module to module, or machine to machine, with humans transferring materials from one module to another. For example, one agent works on strain construction then a lab tech delivers the product to another agent that works on incubation and then on to another agent that draws and measures samples. Here, the progress of an experiment is tracked by human observers who can flexibly input metadata for each observation. In this modular workflow, small changes can be made at different points in a protocol (e.g., at strain construction or at sampling) if the downstream effects are acceptable and documented. A practical (but not insurmountable) technical challenge is developing automated modules that span the diverse range of activities (e.g., perturbation or materials handling) that are characteristic of biological and chemical protocols.
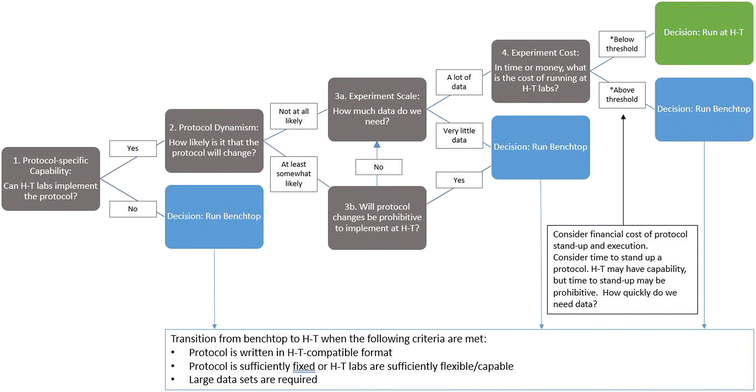
After the initial investment of writing and pilot testing a protocol, an automated, high-throughput approach offers significant gains in efficiency. Writing code to instruct an automated lab agent is more efficient than training a human lab tech to execute the same protocol. Moreover, an automated agent should, on average, offer greater reliability and availability than a human operator. Given a reliable source of power and regular maintenance, a machine's output should far surpass that of a human operator over increasingly longer intervals. Thus, optimized efficiency and maximized output are the primary benefits to this model. However, just as one must conduct a cost-benefit analysis before leveraging full automation, one must also conduct a cost-benefit analysis of exporting protocols to high-throughput labs. In SD2, scientists weighed the error rate of low-throughput, benchtop testing against the cost of that error rate amplified by high-throughput testing. In many instances, it was worth advancing a protocol to high-throughput testing; however, in some cases, it was justifiable to generate small datasets for emerging unvetted protocols or protocols that required extensive human operations. This was particularly the case when advancing a protocol to automated or high-throughput testing required extensive modifications either because the automated infrastructure lacked the machinery to perform a traditionally manual operation (e.g., mixing a sample by using a vortex device), thus requiring identification and description of a replacement procedure, or because the automated infrastructure had to be recoded to perform the required procedure. To guide decisions on whether to pursue high- versus low-throughput testing, performers developed a decision tree that prompted them to consider the capability needed to execute the protocol, how likely the protocol was to require modification, how much data were needed to answer the scientific question, and the cost of using each approach (e.g., benchtop vs. high throughput; see Fig. 3).
By and large, the approach toward automation on SD2—develop and deploy automation strategically and judiciously—is compatible with recent social critiques suggesting that aspirations toward full automation are suboptimal.52 To transform certain types of laboratory tasks through automation creates a need for highly technical development and maintenance (requiring specialized expertise) and precarious “piecework” to deal with errors in automation and interfaces between automated agents. Thus, full automation is likely to be unnecessary, potentially doomed to failure, and (if successful) exclusive of certain types of scientific investigation, thus landing scientists back in the confines of individualist science (albeit an automated individualist science). Instead, a better strategy may be to develop “islands of automation” in the laboratory, surrounded by manual tasks, thus enabling more flexible and inclusive science.
Engaging new analytic technologies
Bioinformatics has been the prevailing method for data processing and discovery in the biological domain. Recently, AI/ML techniques have seen more use given the growth of data and lack of well-defined models in the space. These new techniques present several opportunities and challenges. On one hand, they are purely data-driven, do not require expertise in the domain, and, with enough data, can discover non-linear, complex patterns in high-dimensional data. These patterns could be biomarkers or predictions of potential experimental failures that can inform experiment design. On the other hand, an incomplete understanding of the domain in which they are applied makes it difficult to transform the underlying objective of the model to be useful for downstream applications. This has an impact on how one defines “data at scale.” For example, a typical synthetic biology experiment uses a few (10–40) underlying genetic parts to create a circuit that will respond to a stimulus. The number of combinations of those parts grows combinatorially, and if one tests, for example, only three replicates of 5–10 designs, this represents a small fraction of the whole design space. Moreover, the variability of the response to a stimulus by a circuit can be large, and with only three replicates, such variability cannot be approximated. Thus, although such an experiment generates a lot of data, the amount is inadequate to train a design-space model using commonly available ML techniques. Unless researchers collect more data, they cannot avail themselves of emerging analytic techniques that have greater potential than bioinformatics to inform model completion.To address genetic-design challenge problems on SD2, experts in ML collaborated with experts in synthetic biology. In these collaborations, synthetic biologists explained the specific discovery objectives that could benefit from ML techniques, while ML experts explained the data requirements for training relevant ML algorithms. Together, they developed experimental designs and analytic workflows to comprehensively predict the outcome of an experiment, given data at scale. As a specific example, applying four inducers to an organism to stimulate a response requires data to be generated in five conditions (one control + 4 inducers) at the various time points of interest (e.g., log and stationary phase). At a sample level, this yields a total of 10 conditions for which “responses” can be observed, which is too few to train any ML model. By contrast, by scoping observations from the sample level to the level of genetic transcriptome—specifically, the response of a particular gene's expression given the features of the gene—the amount of data becomes the product of the gene's transcriptome (∼4000) across the conditions (×10), which yields a total of ∼40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 observations. When run across three replicates per condition, these conditions yield a total of 120
000 observations. When run across three replicates per condition, these conditions yield a total of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 observations. This is enough data to train a variety of models, given accurate identification of genetic features that can be linked to the gene's expression, such as its role in a network of genes. Using machine learning, the experimental conditions, and a vectorized representation of a gene's role in the network that represents the organism, SD2 performers were able to achieve greater than 90% accuracy in predicting whether the gene would be dysregulated, and an R2 ∼ 0.6 in quantifying the level of the gene's dysregulation.53
000 observations. This is enough data to train a variety of models, given accurate identification of genetic features that can be linked to the gene's expression, such as its role in a network of genes. Using machine learning, the experimental conditions, and a vectorized representation of a gene's role in the network that represents the organism, SD2 performers were able to achieve greater than 90% accuracy in predicting whether the gene would be dysregulated, and an R2 ∼ 0.6 in quantifying the level of the gene's dysregulation.53
In the materials chemistry thrust, work focused on accelerating the Edisonian trial-and-error process by using data at scale. A variety of experiment planning algorithms were tested for their ability to support interpolation of results,54 extrapolation to new chemical systems,36 combination of model predictions to identify anomalies,55 as well as active-learning54 and active-meta learning approaches56 for crystal growth control. These activities culminated in a competition between algorithms developed in the different problem domains.57 Automation was necessary to accumulate the initial datasets needed for algorithm development and testing, as well as to define statistically significant performance baselines. However, it should be emphasized that many of these methods are applicable to manual experimentation now that these initial data exist.56
The SD2 community applied collaboratively developed tools and methods—for example, automated data generation, pre-processing, normalization, and analysis—to several scientific questions. Among other advances, these efforts led to faster and more accurate predictions of protein stability,58–60 faster discovery of perovskite crystals,54,55,61 and more accurate predictions of the impact of synthetic biological circuits on host organisms.62
Enhancing discovery
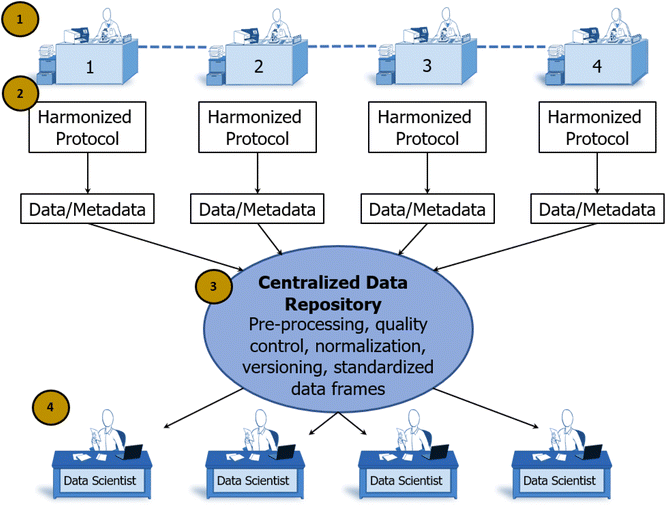
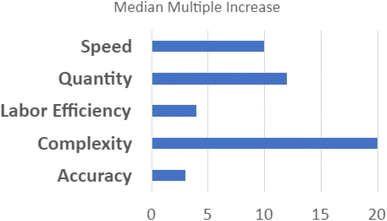
In the individualist paradigm, knowledge is shared only after an entire experimental cycle is complete and conclusions have been reached by the individual scientist. In SD2's collaborative paradigm, scientists worked together to share knowledge at all stages of the scientific method (see Fig. 4).This supports scalable, reproducible data generation and analysis by multiple groups. Protocol sharing and harmonization supports high-throughput experimentation. Data sharing at all levels supports experimental design conducive to AI/ML analytic approaches. Automated pipelines for pre-processing, normalization, and quality control increase the speed with which raw data become ready for analysis.30 Standardized data frames enable automated batch analysis of new data along with old data, substantially reducing the time and resources otherwise needed to complete an experimental cycle. In contrast to the scalability of the individualist approach, the speed of discovery scales with the addition of new laboratories in a collaborative paradigm because shared data accumulates more rapidly and accelerates all processes downstream of data production. At the end of the 4 year program, SD2 performers generated measurements of progress relative to an estimated pre-SD2 baseline.63 Analysts classified and normalized this diverse set of performer-generated, often domain-specific statistics. They reported that SD2 performers increased the number of constructs (e.g., perovskite crystals) designed or discovered by 12×; increased the complexity of model designs (e.g., genetic networks) by 20×; increased the speed of model design, build, test, or analysis by 10×; increased labor efficiency (e.g., output per individual) by nearly 4×; and increased the accuracy of model predictions (e.g., protein stability) by 3× (see Fig. 5).64 Critical to supporting rapid responses to emergent large-scale problems, the sociotechnical system developed in SD2 yielded notable gains in the speed to complete experimental cycles. Cycle speeds for designing proteins, engineering genetic components and networks, and synthesizing perovskite crystals increased dramatically (from 3–81×) over pre-SD2 rates.
Recommendations from the SD2 experience
SD2 performers learned many valuable lessons throughout the course of the program. The following are a few that stand out for their application to future collaborative enterprises that resemble the sociotechnical system (STS) developed and deployed in the SD2 program.In convening an STS, it will benefit the enterprise to engage in activities—for example, knowledge capture activities—that help identify the details of collaborators' methods that are critical to supporting collaboration and discovery. In the same way that making implicit knowledge explicit is critical to sharing knowledge and enabling discovery at the level of science, making methods and metamethods explicit is critical to sharing the knowledge (e.g., procedural, semantic, and institutional knowledge) that enables collaboration.
Throughout the collaboration, and especially early in the process, collaborators should be allowed to fail and encouraged to fail fast. Many professionals across industry and academia perceive failure as a negative outcome. This may obstruct their ability to view failure as an opportunity to improve or advance their work; consequently, rather than see and embrace failure, they push back and persist in an unrecognized failure mode. SD2 experienced both conditions. Not surprisingly, when SD2 performers recognized failure, accepted it, and sought opportunities to improve, they experienced success much more rapidly.
Collaborators must be flexible about retaining versus omitting metadata processes. Metadata collection, representation, and availability ended up being hallmark characteristics of the SD2 STS. However, it was challenging to identify what metadata to collect and how to organize them, because the metadata requirements depended on how the data would be used. Metadata uses are not always discernible early in research and they may rarely be anticipated for future research that will be conducted beyond the time and sociotechnical boundaries of the immediate STS. Moreover, some metadata requirements cannot be easily identified until a problem is discovered. Thus, flexibility is critical to managing metadata processes.
Regarding metadata and automation, if metadata processes—for example, collection and management—are candidates for automation, collaborators should consider automating them prior to running experiments. This may require the temporal staggering of work. Thus, it may not be possible to generate data while engineering data-management solutions, but this might avoid confusion and wasted work.
Regarding automation in general, collaborators should carefully consider the goals of introducing automation as a solution. It can enhance efficiency and free humans to engage in more fruitful activities, but it comes at a cost in set-up time, diagnosis, and flexibility. SD2 performers were initially focused on automating the full DBTL loop, which turned out to be too large and risky of an undertaking for processes that were still evolving, as there was high potential for wasting time automating the nascent, volatile processes. The balance struck by performers was to attempt to automate processes that were well defined and did not demand high flexibility. This purchased speedy execution for a large subset of data-production processes. In addition, performers worked to identify the processes that required the most human effort and to automate those, which resulted in reduced processing time and human errors. Ultimately, automation should support scientific outcomes, and it should support rather than fully replace the work of relevant human operators.
Finally, to engage potential users of emergent methods, technologies, and standards—and to avoid simply creating a new silo of information—collaborators should conduct outreach activities with relevant stakeholders. SD2 performers did so to promote the use of their scientific approach and the standards and tools that they developed. For example, the program sponsored a meeting with funders and publishers of science to address issues of open science, such as the publication of datasets. SD2 performers also engaged stakeholders, such as U.S. military research laboratories, in nearly all of their program-wide meetings, in part to elicit guidance that could shape the products of the program. In addition, SD2 performers published their research methods and findings extensively; and many of the tools and datasets developed in SD2 were made public at https://github.com/SD2E.65 However, like all DARPA programs, outreach activities were not formally funded after the end of the program. Thus, continued outreach, training, and maintenance of the SD2 infrastructure, tools, and data will likely require development of an entity (e.g., similar to U. S. Department of Energy laboratories or NIST) or a long-term program to fund research infrastructure. In the absence of such an entity or program, outreach beyond the lifespan of a government-funded program is left in the hands of individuals and groups who are invested in transitioning program products into adjacent research and development spaces.
Conclusion
The prevailing approach to conducting science—the individualist approach—is suboptimal, particularly in response to high-dimensional problems that call for a rapid response and data at scale. That paradigm often fails to support collaborative protocol authoring and data sharing; it confines scientists to infrastructures with limited capabilities; and it fails to support the integration of capabilities that would improve the pace of discovery and model development. A break from—or at least an enhancement to—the prevailing approach seems warranted. New approaches are needed that allow scientists to share protocols and data, and to leverage a fuller set of capabilities to generate, validate, and more deeply analyze larger datasets. The collaborative paradigm instantiated by the SD2 community in the form of a sociotechnical system is one such approach. It convened experts from diverse domains to develop a network of people, tools, and methods that could be applied to problems that have long suffered from the limitations of individualist science. No single organization was responsible for the work of all technical areas. Instead, organizations collaborated to identify synergies, select scientific challenges to address within a given topic, and divide the labor to generate and analyze data at scale.Although the work of SD2 was domain-specific, the collaborative paradigm it adopted can be generalized to support multidisciplinary work across several domains in life, physical, and social sciences. The SD2 sociotechnical system can serve as a model for developing similar systems that support a national infrastructure that is equipped and ready to respond to emergent, high dimensional problems that require diverse resources and capabilities to generate and analyze data at scale. To retain the status quo means failure to optimize resources, capabilities, and output at any stage of the scientific method, and hence missed opportunities to maximize discovery.
The discoveries that will advance science in profound ways will be made possible by collaborative, multidisciplinary efforts. These efforts require practices and incentives for sharing methods and data, and for identifying and leveraging complementary capabilities. This will allow for efficient generation and analysis of quality data at scale. This will lead to discovery.
Data availability
As this is a Perspective article, no primary research results, data, software or code have been included.Conflicts of interest
There are no conflicts to declare.Acknowledgements
For contributions to early versions of this paper: Enoch Yeung and Devin Strickland. For envisioning and architecting the SD2 program: Jennifer Roberts.Notes and references
- This includes the investigator-initiated and “lone-wolf” models of science.
- Open Science Collaboration, Estimating the reproducibility of psychological science, Science, 2015, 349(6251), aac4716, DOI:10.1126/science.aac4716.
- J. Ioannidis, Why most published research findings are false, PLoS Med, 2005, 2, e124, DOI:10.1371/journal.pmed.0020124.
- C. Begley and L. Ellis, Raise standards for preclinical cancer research, Nature, 2012, 483, 531, DOI:10.1038/483531a.
- M. McNutt, Reproducibility, Science, 2014, 343, 229 CrossRef CAS PubMed.
- S. Goodman, D. Fanelli and J. Ioannidis, What does research reproducibility mean?, Sci. Transl. Med., 2016, 8(341), 341ps12, DOI:10.1126/scitranslmed.aaf5027.
- C. Begley and J. Ioannidis, Reproducibility in science: improving the standard for basic and preclinical research, Circ. Res., 2015, 116(1), 116, DOI:10.1161/CIRCRESAHA.114.303819.
- T. Errington, E. Iorns, W. Gunn, F. Tan, J. Lomax and B. Nosek, Science forum: an open investigation of the reproducibility of cancer biology research, Elife, 2014, 3, e04333, DOI:10.7554/eLife.04333.
- J. Simmons, L. Nelson and U. Simonsohn, False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant, Psychol. Sci., 2011, 22(11), 1359, DOI:10.1177/0956797611417632.
- V. Danchev, A. Rzhetsky, and J. Evans, Centralized "big science" communities more likely generate non-replicable results, arXiv, 2018, preprint, arXiv:1801.05042, DOI:10.48550/arXiv.1801.05042.
- J. Wicherts, D. Borsboom, J. Kats and D. Molenaar, The poor availability of psychological research data for reanalysis, Am. Psychol., 2006, 61(7), 726 Search PubMed.
- However, proposals currently exist for filling this gap, for example, the compendium of discrete authenticable observations (CODA-O); Y. Hannun, Build a registry of results that students can replicate. To speed research, express conclusions as testable statements, and incorporate testing into training, Nature, 2021, 600, 571, DOI:10.1038/d41586.
- Editorial (4 Feb 2020), Calling all coronavirus researchers: keep sharing, stay open, Nature, 2020, 578(7), DOI:10.1038/d41586-020-00307-x.
- M. Kraemer, S. Scarpino, V. Marivate, B. Gutierrez, B. Xu and G. Lee, et al., Data curation during a pandemic and lessons learned from COVID-19, Nat. Comput. Sci., 2021, 1(1), 9–10, DOI:10.1038/s43588-020-00015-6.
- B. Xie and G. Salvendy, Review and reappraisal of modeling and predicting mental workload in single-and multi-task environments, Work Stress, 2000, 14(1), 74, DOI:10.1080/026783700417249.
- E. Callaway, ‘It will change everything’: DeepMind's AI makes gigantic leap in solving protein structures, Nature, 2020, 588, 203, DOI:10.1038/d41586-020-03348-4.
- H. Berman, K. Henrick and H. Nakamura, Announcing the worldwide protein data bank, Nat. Struct. Mol. Biol. , 2003, 10, 980, DOI:10.1038/nsb1203-980.
- M. Barinaga, The missing crystallography data: some disgruntled researchers are mounting a campaign to force crystallographers to make available key data when they publish the structure of complex molecules, Science, 1989, 245(4923), 1179–1181 CrossRef CAS PubMed.
- International Union of Crystallography, Policy on publication and the deposition of data from crystallographic studies of biological macromolecules, Acta Crystallogr., 1989, 45, 658 CrossRef.
- Public Health Service Policy Relating to Distribution of Unique Research Resources Produced with PHS Funding, NIH Guide, 1992, vol. 21, Number 33, September 11, https://grants.nih.gov/grants/guide/notice-files/not92-163.html.
- Z. Feng, N. Verdiguel, L. Di Costanzo, D. Goodsell, J. Westbrook, S. Burley and C. Zardecki, Impact of the Protein Data Bank Across Scientific Disciplines, Data Science Journal, 2020, 19(1), 25, DOI:10.5334/dsj-2020-025.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek and A. Potapenko, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583, DOI:10.1038/s41586-021-03819-2.
- J. McLaughlin, C. Myers, Z. Zundel, G. Misirli, M. Zhang, I. Ofiteru, A. Goñi-Moreno and A. Wipat, SynBioHub: A standards-enabled design repository for synthetic biology, ACS Synth. Biol. , 2018, 7(2), 682, DOI:10.1021/acssynbio.7b00403.
- C. Groom, I. Bruno, M. Lightfoot and S. Ward, The Cambridge Structural Database, Acta Crystallogr. , 2016, B72, 171–179, DOI:10.1107/S2052520616003954.
- G. Bergerhoff, I. Brown and F. Allen, Crystallographic Databases, International Union of Crystallography, Chester, 1987, vol. 360, pp. 77–95 Search PubMed.
- The tools and data developed in SD2 are largely available to the international community. This is consistent with DARPA’s intent and DARPA’s requirements for delivery of DARPA-funded intellectual property for unlimited use or government use. Some SD2 products are available publicly at https://github.com/SD2E, with liberal licenses (e.g., “Redistribution and use in source and binary forms, with or without modification, are permitted”). Others are available through program performers who developed the tools. In particular, the SD2 infrastructure, tools central to its workflows, and much of the SD2 data are archived at the Texas Advanced Computing Center for use in future research.
- M. Wilkinson, M. Dumontier, I. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J. Boiten, L. da Silva Santos and P. Bourne, et al., The FAIR Guiding Principles for scientific data management and stewardship, Sci. Data, 2016, 3, 160018, DOI:10.1038/sdata.2016.18.
- F. Ullman and R. Boutellier, A case study of lean drug discovery: from project driven research to innovation studios and process factories, Drug Discovery Today, 2008, 13(11–12), 543–550 CrossRef CAS PubMed.
- S. Andersson, A. Armstrong, A. Björe, S. Bowker, S. Chapman and R. Davies, et al., Making medicinal chemistry more effective—application of Lean Sigma to improve processes, speed and quality, Drug Discovery Today, 2009, 14(11–12), 598–604 CrossRef CAS PubMed.
- D. Bryce, R. Goldman, M. DeHaven, J. Beal, T. Nguyen, N. Walczak, M. Weston, G. Zheng, J. Nowak and J. Stubbs, et al., Round-Trip: An Automated Pipeline for Experimental Design, Execution, and Analysis, ACS Synth. Biol., 2022, 11(2), 608, DOI:10.1021/acssynbio.1c00305.
- T. Nguyen, N. Walczak, J. Beal, D. Sumorok, and M. Weston, Intent Parser: a tool for codifying experiment design, Proceedings of the International Workshop on Biodesign Automation, 2020, 66. https://www.iwbdaconf.org/2020/docs/IWBDA2020Proceedings.pdf Search PubMed.
- J. Beal, D. Sumorok, B. Bartley, and T. Nguyen, Collaborative terminology: SBOL project dictionary, Proceedings of the International Workshop on Biodesign Automation, 2020, https://jakebeal.github.io/Publications/IWBDA2020-SBOLProjectDictionary.pdf Search PubMed.
- B. Keller, A. Miller, G. Newman, J. Vrana, and E. Klavins, Aquarium: The Laboratory Operating System version 2.6.0, 2019, DOI:10.5281/zenodo.2583232.
- J. Vrana, O. de Lange, Y. Yang, G. Newman, A. Saleem, A. Miller, C. Cordray, S. Halabiya, M. Parks and E. Lopez, et al., Aquarium: open-source laboratory software for design, execution and data management, Synth. Biol., 2021, 6(1), ysab006 CrossRef CAS.
- Autoprotocol, https://autoprotocol.org/.
- I. Pendleton, G. Cattabriga, Z. Li, M. Najeeb, S. Friedler, A. Norquist, E. Chan and J. Schrier, Experiment Specification, Capture and Laboratory Automation Technology (ESCALATE): a software pipeline for automated chemical experimentation and data management, MRS Commun. , 2019, 9(3), 846, DOI:10.1557/mrc.2019.72.
- S. Kearnes, M. Maser, M. Wleklinski, A. Kast, A. Doyle, S. Dreher, J. Hawkins, K. Jensen and C. Coley, The Open Reaction Database, J. Am. Chem. Soc., 2021, 143(45), 18820, DOI:10.1021/jacs.1c09820.
- A. Hammer, A. Leonov, N. Bell and L. Cronin, Chemputation and the Standardization of Chemical Informatics, JACS Au, 2021, 1(10), 1572, DOI:10.1021/jacsau.1c00303.
- B. Bartley, J. Beal, D. Bryce, R. Goldman, B. Keller, J. Ladwig, P. Lee, R. Markeloff, T. Nguyen, J. Nowak, and M. Weston, Open Protocol Interface Language, 2021, https://github.com/SD2E/OPIL-specification Search PubMed.
- P. Nega, Z. Li, V. Ghosh, J. Thapa, S. Sun, N. Hartono, M. Najeeb Nellikkal, A. Norquist, T. Buonassisi, E. Chan and J. Schrier, Using automated serendipity to discover how trace water promotes and inhibits lead halide perovskite crystal formation, Appl. Phys. Lett., 2021, 119(4), 041903, DOI:10.1063/5.0059767.
- A subset of performers who assumed responsibility of the technical infrastructure created several pieces of technology, each designed and implemented in collaboration with program performers. The software foundation for this was Tapis, a web services platform, developed by the Texas Advanced Computing Center, which supports containerized high-performance computing (HPC) applications, data management, and reactive functions. This group also created a network of data processing pipelines to support management (e.g., upload, processing, and sharing) of raw and transformed data, reference data, and analytic output. To chain pipelines and other containerized applications, the group developed a common software framework, called Reactors, which enabled integrated, automated data workflows. Data scientists could then visualize, explore, and interpret results using Jupyter Notebooks shared in a centralized code repository.
- https://www.cshl.edu/labdish/to-celebrate-40th-anniversary-the-protein-data-bank-returns-toits-birthplace/ .
- National Academies of Sciences, Engineering, and Medicine, Empowering Tomorrow's Chemist: Laboratory Automation and Accelerated Synthesis: Proceedings of a Workshop in Brief, The National Academies Press, 2022, Washington, DC, DOI:10.17226/26497.
- J. Strimatis, Robots in the laboratory- an overview, J. Chem. Educ., 1989, 66(1), A8, DOI:10.1021/ed066pA8.
- C. Nichols and L. Hanne, Automated Combinatorial Chemistry in the Organic Chemistry Majors Laboratory, J. Chem. Educ., 2010, 87(1), 87–90, DOI:10.1021/ed800013g.
- S. Vargas, S. Zamirpour, S. Menon, A. Rothman, F. Häse, T. Tamayo-Mendoza, J. Romero, S. Sim, T. Menke and A. Aspuru-Guzik, Team-Based Learning for Scientific Computing and Automated Experimentation: Visualization of Colored Reactions, J. Chem. Educ., 2020, 97(3), 689–694, DOI:10.1021/acs.jchemed.9b00603.
- L. Saar, H. Liang, A. Wang, A. McDannald, E. Rodriguez, I. Takeuchi, and A. Kusne, A Low Cost Robot Science Kit for Education with Symbolic Regression for Hypothesis Discovery and Validation, arXiv, 2022, preprint, arXiv:2204.04187.
- Y. Kosenkov and D. Kosenkov, Computer Vision in Chemistry: Automatic Titration, J. Chem. Educ., 2021, 98(12), 4067–4073, DOI:10.1021/acs.jchemed.1c00810.
- A. Sharma, Laboratory Glassware Identification: Supervised Machine Learning Example for Science Students, J. Comput. Sci. Educ., 2021, 12(1), 8–15, DOI:10.22369/issn.2153-4136/12/1/2.
- F. Yang, V. Lai, K. Legard, S. Kozdras, P. Prieto, S. Grunert and J. Hein, Augmented Titration Setup for Future Teaching Laboratories, J. Chem. Educ., 2021, 98(3), 876–881, DOI:10.1021/acs.jchemed.0c01394.
- R. Soong, K. Agmata, T. Doyle, A. Jenne, A. Adamo and A. Simpson, Rethinking a Timeless Titration Experimental Setup through Automation and Open-Source Robotic Technology: Making Titration Accessible for Students of All Abilities, J. Chem. Educ., 2019, 96(7), 1497–1501, DOI:10.1021/acs.jchemed.9b00025.
- L. Munn, Automation is a Myth, Stanford University Press, 2022 Search PubMed.
- M. Eslami, A. Borujeni, H. Eramian, M. Weston, G. Zheng, J. Urrutia, C. Corbet, D. Becker, P. Maschhoff and K. Clowers, et al., Prediction of whole-cell transcriptional response with machine learning, Bioinformatics, 2022, 38(2), 404, DOI:10.1093/bioinformatics/btab676.
- Z. Li, M. Najeeb, L. Alves, A. Sherman, V. Shekar, P. Parrilla, I. Pendleton, W. Wang, P. Nega, M. Zeller, J. Schrier, A. Norquist and E. Chan, Robot-accelerated perovskite investigation and discovery, Chem. Mater., 2020, 32(13), 5650, DOI:10.1021/acs.chemmater.0c01153.
- Y. Tang, Z. Li, M. Nellikkal, H. Eramian, E. Chan, A. Norquist, D. Hsu and J. Schrier, Improving data and prediction quality of high-throughput perovskite synthesis with model fusion, J. Chem. Inf. Model., 2021, 61(4), 1593, DOI:10.1021/acs.jcim.0c01307.
- V. Shekar, G. Nicholas, M. Najeeb, M. Zeile, V. Yu, X. Wang, D. Slack, Z. Li, P. Nega, E. Chan and A. Norquist, Active meta-learning for predicting and selecting perovskite crystallization experiments, J. Chem. Phys., 2022, 156(6), 064108, DOI:10.1063/5.0076636.
- V. Shekar, V. Yu, B. Garcia, D. Gordon, G. Moran, D. Blei, L. Roch, A. García-Durán, M. Ani Najeeb and M. Zeile, et al., Serendipity based recommender system for perovskites material discovery: balancing exploration and exploitation across multiple models, ChemRxiv, 2022, DOI:10.26434/chemrxiv-2022-l1wpf.
- J. Singer, S. Novotney, D. Strickland, H. Haddox, N. Leiby, G. Rocklin, C. Chow, A. Roy, A. Bera and F. Motta, et al., Large-scale design and refinement of stable proteins using sequence-only models, bioRxiv, 2021, DOI:10.1371/journal.pone.0265020.
- A. Zaitzeff, N. Leiby, F. Motta, S. Haase and J. Singer, Improved datasets and evaluation methods for the automatic prediction of DNA-binding proteins, Bioinformatics, 2022, 38(1), 44, DOI:10.1093/bioinformatics/btab603.
- J. Estrada Pabón, H. Haddox, G. Van Aken, I. Pendleton, H. Eramian, J. Singer and J. Schrier, The Role of Configurational Entropy in Miniprotein Stability, J. Phys. Chem. B , 2021, 125(12), 3057, DOI:10.1021/acs.jpcb.0c09888.
- M. Najeeb Nellikkal, R. Keesey, M. Zeile, S. Venkateswaran, Z. Li, N. Leiby, M. Zeller, E. Chan, J. Schrier and A. Norquist, A spatiotemporal route to understanding metal halide perovskite crystallization. “A spatiotemporal route to understanding metal halide perovskitoid crystallization”, Chem. Mater., 2022, 34, 5386–5396, DOI:10.1021/acs.chemmater.2c00247.
- A. Hasnain, S. Sinha, Y. Dorfan, A. Borujeni, Y. Park, P. Maschhoff, U. Saxena, J. Urrutia, N. Gaffney and D. Becker, et al., A data-driven method for quantifying the impact of a genetic circuit on its host, Biomedical Circuits and Systems Conference (BioCAS), IEEE, 2019, 1, DOI:10.1109/BIOCAS.2019.8919140.
- SD2 performers estimated baselines for these metrics—speed, quantity, labor efficiency, complexity, and accuracy—based on the state of the science at the start of the program (i.e., without the benefit of the methods and tools developed on the program). For example, in the perovskite experimental effort, two post-doctoral researchers and two bachelor-level technicians were able to build the automated system and produce new single-crystal diffraction quality samples for novel perovskites at 3.7× the rate of unaided chemists doing manual experimentation. That is, four workers with robots could produce the equivalent of 15 workers without robots, when averaged over the first 3 years of the project.
- These increases are all median values.
- Like most, if not all, military-funded research programs, SD2 engaged only the funded performers in developing its technologies and standards. This was in part due to constraints and traditions of military research, in which vetted individuals and institutions engage under contracts that specify intellectual property rights, data-sharing obligations, and publication review requirements; it was in part also due to the challenge of coordinating activity among just the program’s roughly 300 performing scientists and technologists, much less additional external participants.
| This journal is © The Royal Society of Chemistry 2023 |