Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biological noncovalent N/O⋯V interactions: insights from theory and protein data bank analyses†
Sergi
Burguera
,
Antonio
Frontera
 and
Antonio
Bauzá
and
Antonio
Bauzá
 *
*
Department of Chemistry, Universitat de les Illes Balears, Ctra. de Valldemossa km. 7.5, 07122, Palma de Mallorca, Islas Baleares, Spain. E-mail: antonio.bauza@uib.es
First published on 23rd October 2023
Abstract
Computations at the PBE0-D3/def2-TZVP level of theory in conjunction with a Protein Data Bank (PDB) survey have provided first time evidence of favorable noncovalent interactions between ADP metavanadate (VO4) and ADP orthovanadate (VO5) and electron rich atoms. These involve a σ-hole present in the V atom and the lone pairs belonging to (i) protein residues (e.g., serine (SER), glutamate (GLU) or histidine (HIS)), (ii) backbone carbonyl groups and (iii) water molecules. A computational study has been carried out to rationalize the physical nature and directionality of the interaction in addition to its plausible biological role. The results reported herein are expected to have an impact in the fields of medicinal chemistry, bioinorganic chemistry and chemical biology.
Introduction
Vanadium plays an essential role in several biological processes,1 such as those involving nitrogenases and haloperoxidases. Concretely, vanadium nitrogenases2–5 convert atmospheric nitrogen into biologically useful forms, thus facilitating nitrogen fixation. Similarly, vanadium haloperoxidases6–9 are critical for various marine organisms, assisting in the synthesis of halogenated organic compounds that play crucial roles in their defense mechanisms.Vanadate (VO43−), a compound containing vanadium in its highest oxidation state (+5), exhibits a structure closely resembling that of phosphate (PO43−), which allows it to interact with and modulate various enzymes and protein targets, competitively inhibiting phosphate-dependent enzymes.10,11 This can be observed in Fig. 1, where a comparison between metavanadate and phosphate and between orthovanadate and the transition state (TS) corresponding to a phosphoryl group transfer reaction is illustrated.
In this regard, adenosine diphosphate (ADP)-vanadate12 is a subclass of vanadate derivative that has attracted considerable attention, owing to its potential value in a range of biological applications. These include mimicking the activity of adenosine triphosphate (ATP) and interacting with the binding site of ATP-dependent proteins. By acting as an ATP equivalent, ADP-vanadate can modulate the enzymatic activity of proteins involved in crucial cellular processes such as phosphorylation, signal transduction, and energy metabolism,13–17 thus opening the way to a myriad of applications in the fields of medicinal chemistry and rational drug design.
Despite its promising potential, the use of ADP-vanadate species is an evolving field that demands further exploration and understanding to unravel the precise mechanisms of action and optimize the therapeutic applications of these compounds in the treatment of specific diseases. To tackle this point, we have inspected the Protein Data Bank18 (PDB) to investigate the molecular forces responsible for the stabilization of the metavanadate/orthovanadate moiety present in ADP-vanadate species, in combination with a computational study at the PBE0-D3/def2-TZVP level of theory.
Using Quantum Mechanics tools, we have demonstrated the presence of an electrophilic region over the V atom (commonly known as a σ-hole19–21), in both tetrahedral (metavanadate, VO4) and trigonal bipyramidal (orthovanadate, VO5) molecular arrangements, both of them being identified during the PDB survey. A σ-hole (defined as a region of low electron density on the extension of a covalent bond)22 can typically be found along the P–F/C–I bonds in trifluorophosphane or iodoethane molecules. There are two factors that govern the σ-hole donor ability, which are the polarizability value of the element bearing the σ-hole (which increases on going from the lighter to heavier elements) and the EWGs (electron withdrawing groups) attached to it. This leads to the use of heavier elements and strong EWGs as the best combination to enhance the σ-hole donor ability of a given compound.
Recently, scientists have expanded the σ-hole concept from the p-block to the transition metal block elements (composed of groups 3 to 12). In this context, several new noncovalent forces have been theoretically proposed and experimentally exploited, such as regium/coinage bonding (RgB)23,24 and spodium bonding interactions (SpB),25 which refer to the attraction between electron-deficient sites on a coinage-metal atom (group 11) and a spodium atom (group 12) and an electron donor molecule, respectively. Also, σ-holes have been found in other groups from the d-block,26–29 and in a very recent report30 in group 5 (V, Nb and Ta).
The presence of a σ-hole allows the V atom to favorably interact with O/N rich species, such as the protein residues (e.g., serine (SER), glutamate (GLU) or histidine (HIS)), backbone carbonyl groups and water molecules studied herein. These contacts were computationally studied for the first time, suggesting a plausible biological role for this interaction. In addition, we have also explored the physical nature and directionality of the A⋯V (A = O, N, S and C) interaction through a theoretical study at the PBE0-D3/def2-TZVP level of theory to confirm their noncovalent nature. Owing to the potential applications and future perspectives regarding ADP-vanadate, we believe the results reported herein will contribute to the understanding of the biological roles of V as well as to the fields of medicinal chemistry, enzyme catalysis and chemical biology.
Analysis of biological O⋯V contacts
A PDB inspection revealed a total number of 32 structures involving metavanadate and orthovanadate moieties, with 25 of them exhibiting V⋯O/N noncovalent contacts (see the Methods section for the specific PDB search criteria below). Owing to its higher abundance, we have focused this section on studying the strength and directionality of V⋯O interactions present in four selected PDB structures (see Fig. 2 to 4) from a theoretical point of view, suggesting a potential role for these noncovalent contacts.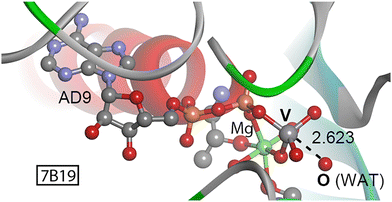 | ||
| Fig. 3 Partial view of the X-ray crystal structure of 7B19 with indication of the O⋯V contact. Distance in Å. | ||
The first two examples involve the Myosin protein family, which are motor proteins involved in distinct cellular pathways.31–33 Myosins contain actin- and ATP-binding sites, and conformational/chemical changes associated with ATP binding, hydrolysis and product release are crucial for their motility capacity. Since these proteins act in a reversible and fast way, small molecules can act as useful modulators of their functionality, leading to the development of novel therapeutic agents.34–36
X-ray structures of 2JJ937 and 2XEL38 (Fig. 2a and b) were obtained in the absence (2JJ9) and presence (2XEL) of an allosteric myosin inhibitor, named pentachloropseudilin (PClP). Interestingly, in 2JJ9 (Fig. 2a) two SER residues (181 and 236) located at the nucleotide binding site were noncovalently interacting with the metavanadate moiety, exhibiting O⋯V distances of 3.904 and 3.887 Å and OSER⋯V–O angles of 138.5 and 135.9°, respectively. The latter is involved in a noncovalent interaction (NCI) pathway (Fig. 2c) that communicates the allosteric site (where the inhibitor binds in the 2XEL structure) and the nucleotide binding site (where the ADP-metavanadate moiety is located).
This network of interactions, which extends up to 20 Å (mainly based on electrostatically enhanced hydrogen bonds (HBs)) is a crucial communication road between both domains and serves as a direct link between the inhibitor and the γ-phosphate/metavanadate moiety. Hence, the O⋯V interaction is implicated in the last step of this molecular relay, playing a key role in the allosteric communication pathway of myosin. A computational model of this O⋯V(+5) contact, including the ADP-vanadate moiety and the interacting SER residue revealed a strength of −3.0 kcal mol−1.
On the other hand, in the 2XEL structure (including the inhibitor bound to the allosteric site, Fig. 2b), the NCI pathway between both sites was altered, leading to an increase in the OSER236⋯V distance (from 3.887 Å in 2JJ9 to 3.968 Å in 2XEL), thus weakening the strength of interaction. In fact, the O⋯V contact observed in 2JJ9 was replaced by a O⋯HO HB involving a hydroxyl group from the vanadate moiety (not shown in Fig. 2b). Consequently, the O⋯V contact involved SER181 instead of SER236, at 3.708 Å.
To carry out ATP hydrolysis, a water molecule needs to be placed in position to perform a nucleophilic attack on the γ-phosphate of ATP.39 Structure 7B1940 (Fig. 3) corresponds to the motor domain of a mutant myosin M765GGG in complex with ADP-metavanadate, used to trap the enzyme in a pre-hydrolysis state. Interestingly, a water molecule is placed in the vicinity of the vanadate moiety (O⋯V distance of 2.623 Å and a OWAT⋯V–O angle of 177.3°) readily to undergo the nucleophilic attack on the metavanadate moiety. We theoretically modeled the interaction between the O from the water molecule and the vanadium center, resulting in a strength of −7.7 kcal mol−1. The presence of water molecules in a disposition prior to the nucleophilic attack was also observed in other structures retrieved from the search (2JJ9, 3MNQ, 3MJX37 and 7B1A40), exhibiting O⋯V(+5) distances ranging between 2.3 and 2.5 Å and interaction strengths of −2.6 (2JJ9), −3.1 (3MNQ) and −6.0 kcal mol−1 (3MJX). Thus, the formation of a pre-reactive O⋯V noncovalent complex is energetically favored and might assist in trapping the enzyme in a pre-catalytic stage by stabilization of the nucleophilic water molecule.
The last selected structure corresponds to the ATP-binding cassette transporter LptB2FG, which extracts lipopolysaccharides (LPS) out of the inner cellular membrane in Gram-negative bacteria in an ATP-dependent manner. A cryo-electron microscopy (Cryo-EM) study41 determined the structure of LptB2FGC in complex with ADP-vanadate trapped in the catalytic site, thus stabilizing the enzyme in an intermediate conformation after LPS exclusion. In Fig. 4, the V center exhibits a distorted trigonal bipyramidal shape, acting as a TS analog from a phosphoryl group transfer reaction (see also Fig. 1 above).
A close inspection of the O⋯V contacts present in 6MI8 revealed the interaction between a hydroxyl group from a neighboring SER residue (139) and the V atom from the orthovanadate moiety (O⋯V distance of 3.233 Å and OSER⋯V–O angle of 173.2°). Computations revealed a strength of −6.6 kcal mol−1 of the O⋯V(+5) interaction, thus contributing to the stabilization of ADP-orthovanadate in the catalytic cleft of the LptB2FG protein after the extrusion of the LPS molecule.
The selected structures involving different V coordination environments (tetrahedral in 2JJ9 and 2XEL and 7B19 and trigonal bipyramidal in 6MI8) and electron donor moieties (SER residues and water molecules) provided new insights into the molecular forces responsible for the stabilization of vanadate in proteins, serving as a brief example of the plausible biological significance of the O⋯V interaction.
Results and discussion
Electrostatic potential study
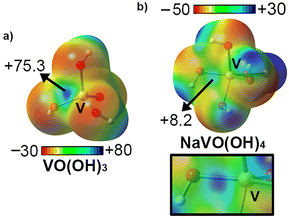
We started analyzing the V center from an electrostatic perspective by computing the molecular electrostatic potential (MEP) surfaces corresponding to metavanadate (VO(OH3)) and sodium orthovanadate (NaVO(OH)4) (see Fig. 5). As mentioned above, the V oxidation state in both compounds is +5; however, this is compensated by the presence of OH− and O2− groups. In fact, in the case of NaVO(OH)4, a sodium ion was used to mimic the Mg2+ ion present in biological media (see Fig. 1 and 2) as well as to keep the system neutral and comparable to VO(OH)3. As noted, in the case of VO(OH3), the MEP minima was located over the lone pairs belonging to the O atoms (−30 kcal mol−1) while the MEP maxima was located at the H atoms from the OH groups (+80 kcal mol−1). In addition, a region of very positive electrostatic potential was located on the prolongation of the V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, thus revealing a σ-hole, in a parallel way to that observed in the report of Calabrese and collaborators.30
O bond, thus revealing a σ-hole, in a parallel way to that observed in the report of Calabrese and collaborators.30
On the other hand, in the case of NaVO(OH)4, the MEP minima also corresponded to the lone pairs from the O atoms (−50 kcal mol−1) while the MEP maxima was located over the sodium ion, as expected. In this case, we also found an electrophilic region over the V atom placed along the vector of the V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, in line with the results obtained for metavanadate. Comparison of both σ-hole MEP values revealed a higher electrophilicity of VO(OH)3 (+75.3 kcal mol−1) and thus more favorable binding energy values are expected upon interacting with electron rich species. However, sodium orthovanadate also shows an electropositive σ-hole (+8.2 kcal mol−1) and thus, it can still attractively interact with electron rich species, such as SER139 residue in the 6MI8 structure.
O bond, in line with the results obtained for metavanadate. Comparison of both σ-hole MEP values revealed a higher electrophilicity of VO(OH)3 (+75.3 kcal mol−1) and thus more favorable binding energy values are expected upon interacting with electron rich species. However, sodium orthovanadate also shows an electropositive σ-hole (+8.2 kcal mol−1) and thus, it can still attractively interact with electron rich species, such as SER139 residue in the 6MI8 structure.
Energetic study
To rationalize the findings obtained from the PDB survey and investigate the σ-hole donor ability of both vanadate moieties, we designed a series of noncovalent complexes using the Lewis bases included in Fig. 6 (CO, OH2, O(CH3)2, SCH2, S(CH3)2, urea and pyridine), which resemble the electron donor molecules observed in the PDB structures. These were fully optimized at the PBE0-D3/def2-TZVP level of theory and the results are gathered in Table 1.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, in °) for complexes 1 to 16. VTd and VTp correspond to a tetrahedral and trigonal bipyramidal geometry, respectively
O, in °) for complexes 1 to 16. VTd and VTp correspond to a tetrahedral and trigonal bipyramidal geometry, respectively
| Complex | ΔE | d | ∠A⋯V![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
|---|---|---|---|
| 1 VTd⋯OC | −1.5 | 3.294 | 179.9 |
| 2 VTd⋯CO | −2.2 | 3.327 | 179.9 |
| 3 VTd⋯OH2 | −11.3 | 2.498 | 176.0 |
| 4 VTd⋯O(CH3)2 | −11.6 | 2.509 | 179.0 |
| 5 VTd⋯SCH2 | −7.0 | 3.170 | 179.5 |
| 6 VTd⋯S(CH3)2 | −11.1 | 2.952 | 179.2 |
| 7 VTd⋯O-urea | −18.7 | 2.334 | 179.6 |
| 8 VTd⋯N–pyridine | −16.8 | 2.456 | 179.9 |
| 9 VTp⋯OC | −1.1 | 3.498 | 179.9 |
| 10 VTp⋯CO | −0.9 | 3.758 | 179.9 |
| 11 VTp⋯OH2 | −3.0 | 3.028 | 180.0 |
| 12 VTp⋯O(CH3)2 | −5.5 | 3.078 | 179.6 |
| 13 VTp⋯SCH2 | −3.1 | 3.646 | 179.9 |
| 14 VTp⋯S(CH3)2 | −4.5 | 3.516 | 179.9 |
| 15 VTp⋯O-urea | −1.1 | 3.365 | 179.9 |
| 16 VTp⋯N-pyridine | −7.1 | 3.001 | 179.8 |
As noted, in all the cases attractive binding energy values were obtained, ranging between −18.7 and −0.9 kcal mol−1. In general, complexes involving metavanadate (1 to 8) achieved larger binding energy values compared to their corresponding orthovanadate (9 to 16) analogs, in line with the results derived from the electrostatic potential maps discussed above.
In the case of metavanadate complexes, complex 7 involving urea as the electron donor achieved the most favorable binding energy value of the set (−18.7 kcal mol−1), due to the simultaneous establishment of O⋯V interaction and NH⋯O HB (see the QTAIM analysis below). On the other hand, between complexes 1 and 2 involving CO as the Lewis base, the former obtained a poorer binding energy value (−1.5 kcal mol−1). The formation of ancillary HBs (involving the OH groups from metavanadate and the OH and CH bonds from the electron rich species) was also observed for complexes 3, 4 and 6 involving OH2, O(CH3)2 and S(CH3)2, also contributing to the interaction energies obtained (−11.3, −11.6 and −11.1 kcal mol−1, respectively). In addition to this, the directionality observed for the S/O⋯V interaction in these three complexes was noticeable, with the lone pairs of the O and S atoms clearly pointing to the V's σ-hole (S/O⋯V–O angle comprised between 176 and 180°). On the other hand, the geometry of complex 5 involving SCH2 was directed by a combination of a HB established between an OH group from metavanadate and a CH group from the electron donor molecule and the S⋯V interaction, resulting in a strength of −7.0 kcal mol−1. Lastly, complex 8 involving pyridine achieved an interaction energy of −16.8 kcal mol−1, with a contribution of two HBs involving the CH bonds of the electron donor molecule. Among the orthovanadate complexes (9 to 16), similar results were obtained for complexes 9 and 10 involving CO (−1.1 and −0.9 kcal mol−1, respectively).
On the other hand, in the case of complexes (11, 12 and 14) involving OH2, O(CH3)2 and S(CH3)2, the formation of a bifurcated HB (involving two OH groups from the orthovanadate moiety) was observed, thus lowering the directionality of the S/O⋯V interaction (the lone pairs of the O and S were not pointing to the V σ-hole), leading to less favorable binding energy values (−3.0, −5.5 and −4.5 kcal mol−1, respectively) compared to complexes 3, 4 and 6. Furthermore, complex 16 involving pyridine achieved the largest binding energy value in this set (−7.1 kcal mol−1), in line with the short equilibrium distance obtained (dN⋯V = 3.001 Å), while complex 15 involving urea obtained a very poor binding energy value (−1.1 kcal mol−1), contrary to that observed for complex 7, due to the absence of NH⋯O HB. A similar situation was observed for complex 14 involving CH2S as the electron donor moiety (−3.1 kcal mol−1).
Energy decomposition analysis
We have also performed an Energy decomposition analysis (EDA)42 on eight representative examples to understand the contribution of the different energy components (electrostatics, exchange-repulsion, orbital, dispersion, and electron correlation) in the stabilization of the noncovalent complexes studied herein (see Table 2). As noted, among the favourable energetic contributions, electrostatics is the dominating force in all the cases (exhibiting a contribution between 30% and 55%) except for complex 10 involving CO, where it represents 13% of the total attractive contributions.| Complex | Ele. | Ex-rep. | Orb. | Disp. | Corr. |
|---|---|---|---|---|---|
| 2 | −2.8 (41%) | 4.6 | −1.3 (19%) | −1.1 (16%) | −1.6 (24%) |
| 4 | −20.0 (50%) | 28.3 | −9.1 (23%) | −3.3 (8%) | −7.5 (19%) |
| 5 | −11.8 (46%) | 18.4 | −5.8 (23%) | −2.6 (10%) | –5.2 (21%) |
| 8 | –32.9 (55%) | 42.8 | –14.3 (24%) | –3.7 (6%) | –8.8 (15%) |
| 10 | –0.5 (13%) | 2.5 | –0.9 (27%) | –1.0 (30%) | –1.0 (30%) |
| 12 | –7.4 (39%) | 13.7 | –4.3 (23%) | –3.1 (16%) | –4.3 (22%) |
| 13 | −3.1 (30%) | 7.4 | −2.5 (24%) | −2.1 (20%) | −2.8 (26%) |
| 16 | −12.0 (43%) | 20.9 | −6.1 (22%) | −3.9 (14%) | −6.0 (21%) |
On the other hand, orbital and correlation terms possess a similar strength, exhibiting close values between them in the range of 15% to 30% of the total attractive contributions, except for complexes 4 and 8, where the orbital term is predominant (−9.1 and −14.3 kcal mol−1, respectively). Finally, the dispersion contribution seems to play a minor role in all the complexes studied (less than 10% of weight) except for complex 10, where all energy terms possess a similar contribution.
QTAIM and NCIplot analysis
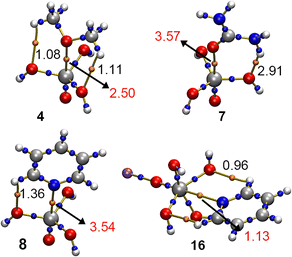
In Fig. 7, the Quantum Theory of Atoms in Molecules (QTAIM)43 is shown for four representative complexes (4, 7, 8 and 16). As noticed, a bond critical point (bcp) and a bond path connecting the O/N atoms from the electron donor molecule to the V atom from the vanadate moiety were found in these four examples, thus characterizing the N/O⋯V interaction. In addition, ancillary HBs were also denoted by the bcps and bond paths that connected the CH/NH groups from the Lewis base to the lone pairs of the OH groups belonging to the vanadate derivative.Also, in Fig. 8 the Non Covalent Interaction plot (NCIplot)44 analyses are shown regarding these four structures, exhibiting an NCI region that accounted for the presence of both the O/N⋯V interaction and the ancillary HBs observed in these complexes. Interestingly, for complexes 4 and 8 a marked difference in the isosurface color was observed by comparing the N/O⋯V interaction with the ancillary HBs, thus clearly indicating that the former directs the formation of the noncovalent complex. In the case of complex 7, both O⋯V and NH⋯O isosurfaces were blue, indicating a similar contribution of both noncovalent forces, in line with their respective ρ × 102 values (see Table 1 above). Lastly, for complex 16 the isosurface obtained was entirely green, thus also pointing to a similar contribution of the O/N⋯V interaction as well as the ancillary HBs.
In Table 3, the value of the density (ρ × 100) at the bcp/NCI region that characterizes the A⋯V interaction as well as the ancillary HBs present in complexes 1 to 16 are shown. As noted, the values of rho gathered for the A⋯V (A = O, N, S and C) interaction are of larger magnitude than those referring to the ancillary HBs (except for complex 12 involving O(CH3)2), thus highlighting the directing role of the V σ-hole in the formation of the noncovalent complexes studied herein.
| Complex | ρ A⋯V | ρ HB | ∇2ρ | −G/V | H |
|---|---|---|---|---|---|
| a No ancillary HBs were found. | |||||
| 1 | 0.36 | —a | 1.57 | 1.43 | 0.09 |
| 2 | 0.55 | —a | 1.88 | 1.26 | 0.08 |
| 3 | 2.63 | —a | 10.56 | 1.02 | 0.05 |
| 4 | 2.50 | 1.08/1.11 | 10.04 | 1.03 | 0.07 |
| 5 | 1.38 | 1.18 | 3.37 | 0.98 | −0.01 |
| 6 | 2.21 | 1.71 | 4.96 | 0.89 | −0.16 |
| 7 | 3.57 | 2.91 | 15.96 | 1.02 | 0.08 |
| 8 | 3.54 | 1.36 | 11.19 | 0.93 | −0.22 |
| 9 | 0.36 | —a | 1.27 | 1.26 | 0.06 |
| 10 | 0.37 | —a | 1.00 | 1.31 | 0.04 |
| 11 | 0.87 | 0.79 | 3.24 | 1.14 | 0.09 |
| 12 | 0.85 | 0.93/0.93 | 2.87 | 1.14 | 0.08 |
| 13 | 0.60 | —a | 1.64 | 1.21 | 0.06 |
| 14 | 0.76 | —a | 1.98 | 1.13 | 0.05 |
| 15 | 0.45 | —a | 1.58 | 1.27 | 0.07 |
| 16 | 1.13 | 0.96/0.96 | 3.70 | 1.03 | 0.03 |
In addition, we also indicated the values of the Laplacian at the bcp/NCI region that characterizes the A⋯V interaction (∇2ρ × 100), resulting in positive values in all cases, as it is common in closed shell calculations. Furthermore, the values of the potential and kinetic energy densities lie within the same range in all the cases, thus confirming the noncovalent nature of the A⋯V interaction (−G/V) ≈ 1.
NBO analysis
We were also curious about the orbital interactions responsible for the stabilization of the A⋯V complexes studied herein. Therefore, we used the Natural Bonding Orbital (NBO)45 approach with particular emphasis on the second-order perturbation analysis for complexes 1 to 16, owing to its usefulness when studying donor–acceptor interactions (see Table 4).| Complex | Donor | Acceptor | E 2 |
|---|---|---|---|
| 1 | LP O | BD* V–O | 0.65 |
| 2 | LP C | BD* V–O | 2.89 |
| 3 | LP O | BD* V–O | 22.69 |
| 4 | LP O | BD* V–O | 14.93 |
| LP O | BD* C–H | 0.69 | |
| 5 | LP S | BD* V–O | 12.55 |
| LP O | BD* C–H | 0.65 | |
| 6 | LP S | BD* V–O | 22.50 |
| LP O | BD* C–H | 0.44 | |
| 7 | LP O | BD* V–O | 42.82 |
| LP O | BD* N–H | 8.52 | |
| 8 | LP N | BD* V–O | 32.60 |
| LP O | BD* C–H | 0.73 | |
| 9 | LP O | BD* V–O | 0.48 |
| 10 | LP C | BD* V–O | 0.90 |
| 11 | LP O | BD* V–O | 4.57 |
| LP O | BD* H–O | 0.40 | |
| 12 | LP O | BD* V–O | 3.56 |
| LP O | BD* C–H | 1.18 | |
| 13 | LP S | BD* V–O | 0.81 |
| 14 | LP S | BD* V–O | 2.24 |
| 15 | LP O | BD* V–O | 1.13 |
| 16 | LP N | BD* V–O | 8.37 |
| LP O | BD* C–H | 1.08 |
Interestingly, in all the cases an orbital contribution consisting of the donation from the lone pairs (LP) of either an O (complexes 1, 3, 4, 7, 9, 11, 12 and 15), C (complexes 2 and 10), S (complexes 5, 6, 13 and 14) or N atom (complexes 8 and 16) to an antibonding V–O (BD* V–O) orbital was observed, spawning from 0.48 to 42.82 kcal mol−1. This broad range of energies is in line with the short equilibrium distances obtained in some complexes (e.g., 3, 4, 7 and 8), which are those that exhibited the largest orbital contributions. Also, the high directionality observed for the computed complexes (see Table 1), with interaction angle values between 176° and 180° also influenced these results, particularly in ensuring a better overlap between the lone pairs and antibonding orbitals of both electron donor and acceptor counterparts.
In addition, the orbital contributions regarding the ancillary HBs observed in the QTAIM and NCIplot analyses discussed above were also obtained. These mainly involved an LP from an O atom and a BD* involving (i) a C–H bond (in complexes 4 to 6, 8, 12 and 16) and (ii) a N–H bond (for complex 7). Also, complex 11 exhibited a HB interaction that involved an LP from an O atom belonging to a water molecule and a BD* H–O from the vanadate moiety.
Conclusions
In conclusion, the biological implications of N/O⋯V contacts involving ADP-vanadate species and protein residues/water molecules have been investigated for the first time through a combination of structural and computational analyses. Concretely, the PDB search complemented with a DFT-D3 study (PBE0-D3/def2-TZVP level of theory) provided structural and energetic evidence of the existence of noncovalent N/O⋯V interactions. This was complemented with a set of noncovalent complexes (involving both tetrahedral and trigonal bipyramid vanadate species and several Lewis bases) analyzed at the same level of theory. Lastly, EDA, QTAIM, NCIplot and NBO techniques were used to further characterize the noncovalent complexes studied herein, providing new insights into the physical nature and extension in real space of the interaction. Owing to the unique structural and chemical properties of vanadate, we expect that the results reported herein will be useful in the fields of medicinal chemistry, bioinorganic chemistry and chemical biology.Methods
Protein data bank search
An inspection of the PDB was carried out in June 2023 to investigate the formation of noncovalent complexes between vanadate derivatives and electron rich species. During our search, we considered both 4 (tetrahedral) and 5 (trigonal bipyramidal) coordination indexes. More in detail, the geometrical criteria considered during the search are shown in Fig. 9. | ||
| Fig. 9 Schematic representation of the V⋯A contacts searched in the PDB. All the A⋯V–O angles were considered during the search (only one is shown in the figure for the sake of clarity). The vdW radii of the V (2.42 Å), O (1.50 Å), N (1.66 Å) and S (1.89 Å) atoms were taken from the study of Alvarez.46 | ||
Using these criteria, a total number of 24 structures were found (see Tables 5 and 6).
| PDBID | Ligand ID | V Ci | ED | d | ∠ |
|---|---|---|---|---|---|
| 2JJ9 | AD9 | 4 | SER236 | 3.887 | 138.5 |
| WAT | 2.504 | 157.9 | |||
| 2JHR | AD9 | 4 | SER181 | 3.895 | 133.4 |
| 6Z2S | AD9 | 4 | SER236 | 3.785 | 144.4 |
| 3MNQ | AD9 | 4 | SER236 | 3.848 | 136.4 |
| WAT | 2.379 | 162.3 | |||
| 2XEL | AD9 | 4 | SER181 | 3.708 | 135.9 |
| 3MJX | AD9 | 4 | SER236 | 3.773 | 136.5 |
| WAT | 2.446 | 163.1 | |||
| 7B19 | AD9 | 4 | SER181 | 3.745 | 140.0 |
| WAT | 2.623 | 177.3 | |||
| 7B1A | AD9 | 4 | WAT | 2.317 | 163.6 |
| 2XO8 | AD9 | 4 | SER181 | 3.758 | 150.6 |
| 2X9H | AD9 | 4 | SER181 | 3.905 | 151.9 |
| 4AE3 | AOV | 5 | SER181 | 3.763 | 120.1 |
| 6MI8 | AOV | 5 | SER139 | 3.233 | 173.2 |
| 4RH7 | AOV | 5 | GLU1742 | 3.108 | 131.5 |
| 4BRL | GMV | 5 | THR118 | 3.633 | 136.6 |
| 4BRE | 50T | 5 | THR118 | 3.713 | 140.9 |
| PDBID | Ligand ID | V Ci | ED | d | ∠ |
|---|---|---|---|---|---|
| 7K1L | UVC | 5 | HIS235 | 3.657 | 139.4 |
| 6YO1 | UVC | 5 | HIS119 | 3.654 | 147.5 |
| 1JH7 | UVC | 5 | THR44 | 3.716 | 135.7 |
| 1RUV | UVC | 5 | HIS12 | 3.706 | 140.1 |
| 6RSA | UVC | 5 | HIS12 | 3.521 | 136.0 |
| 1M5O | AVC | 5 | A57 | 3.678 | 130.7 |
| 2P7E | AVC | 5 | A38 | 3.785 | 132.8 |
| 6RVZ | KL2 | 5 | ASN353 | 3.772 | 134.4 |
| 4NDG | V5A | 4 | HIS262 | 3.687 | 172.3 |
Computational methods
The MEP (molecular electrostatic potential) surfaces were computed at the PBE0-D3/def2-TZVP level of theory by means of the TURBOMOLE 7.7 program, analyzed using the Multiwfn software55 and visualized using Gaussview 5.0 program.56 The calculations for the QTAIM analysis were carried out at the PBE0-D3/def2-TZVP level of theory also using Multiwfn software. In addition, the energy decomposition analysis (EDA)57,58 scheme was used to understand the role of electrostatics, exchange-repulsion, orbital, dispersion and electron correlation contributions in the stabilization of the noncovalent complexes studied herein at the PBE0-D3/def2-TZVP level of theory, also using TURBOMOLE 7.7 software. The NBO calculations were carried out using the NBO 7.0 program59 at the HF/def2-TZVP level of theory.
Lastly, the NCIplot60 isosurfaces correspond to both favorable and unfavorable interactions, as differentiated by the sign of the second density Hessian eigenvalue and defined by the isosurface color. The color scheme is a red–yellow–green–blue scale with red for repulsive (ρ+cut) and blue for attractive (ρ−cut) NCI interaction densities. Yellow and green surfaces correspond to weak repulsive and weak attractive interactions, respectively. The Visual Molecular Dynamics (VMD)61 program was used in the visualization of the results from the QTAIM and NCIplot analyses.
Author contributions
Most of the computational studies were conducted by S. B., and A. F. A. B. wrote the article and directed the study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank DGICYT of Spain (project PID2020-115637GB-I00 FEDER funds) for financial support. We thank the “Centre de les Tecnologies de la Informació” CTI (UIB) for computational facilities.Notes and references
- D. Rehder, Metallomics, 2015, 7, 730 CrossRef CAS PubMed.
- R. R. Eady, Coord. Chem. Rev., 2003, 237, 23 CrossRef CAS.
- Y. Zhao, S.-M. Bian, H.-N. Zhou and J.-F. J. Huang, Integr. Plant Biol., 2006, 48, 745 CrossRef CAS.
- Y. Hu, C. Chung Lee and M. W. Ribbe, Dalton Trans., 2012, 41, 1118 RSC.
- Y. Tanabe and Y. Nishibayashi, Coord. Chem. Rev., 2019, 381, 135 CrossRef CAS.
- A. Butler, Curr. Opin. Chem. Biol., 1998, 2, 279 CrossRef CAS PubMed.
- A. G. J. Ligtenbarg, R. Hage and B. L. Feringa, Coord. Chem. Rev., 2003, 237, 89 CrossRef CAS.
- A. Butler and J. N. Carter-Franklin, Nat. Prod. Rep., 2004, 21, 180 RSC.
- C. Leblanc, H. Vilter, J.-B. Fournier, L. Delage, P. Potin, E. Rebuffet, G. Michel, P. L. Solari, M. C. Feiters and M. Czjzek, Coord. Chem. Rev., 2015, 301–302, 134 CrossRef CAS.
- G. Huyer, S. Liu, J. Kelly, J. Moffat, P. Payette, B. Kennedy, G. Tsaprailis, M. J. Gresser and C. Ramachandran, J. Biol. Chem., 1997, 272, 843 CrossRef CAS PubMed.
- J. E. Benabe, L. A. Echegoyen, B. Pastrana and M. Martínez-Maldonado, J. Biol. Chem., 1987, 262, 9555 CrossRef CAS PubMed.
- A. S. Tracey, M. J. Gresser and S. Liu, J. Am. Chem. Soc., 1988, 110, 5869 CrossRef CAS.
- J. Chen, S. Sharma, F. A. Quiocho and A. L. Davidson, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 1525 CrossRef CAS PubMed.
- K. M. Kerr, Z. E. Sauna and S. V. Ambudkar, J. Biol. Chem., 2001, 276, 8657–8664 CrossRef CAS PubMed.
- D. R. Davies and W. G. J. Hol, FEBS Lett., 2004, 315–321 CrossRef CAS PubMed.
- I. L. Urbatsch, G. A. Tyndall, G. Tombline and A. E. Senior, J. Biol. Chem., 2003, 278, 23171 CrossRef CAS PubMed.
- D. Lacabanne, T. Wiegand, N. Wili, M. I. Kozlova, R. Cadalbert, D. Klose, A. Y. Mulkidjanian, B. H. Meier and A. Böckmann, Molecules, 2020, 22, 5268 CrossRef PubMed.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gillilland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235 CrossRef CAS PubMed.
- T. Clark, M. Hennemann, J. S. Murray and P. Politzer, J. Mol. Model., 2007, 2, 291 CrossRef PubMed.
- T. Brinck, J. S. Murray and P. Politzer, Int. J. Quant. Chem., 1992, 44, 57 CrossRef.
- P. Politzer, J. S. Murray and T. Clark, Phys. Chem. Chem. Phys., 2013, 14, 11178 RSC.
- P. Politzer and J. S. Murray, Crystals, 2017, 7, 212 CrossRef.
- J. H. Stenlid and T. Brinck, J. Am. Chem. Soc., 2017, 139, 11012 CrossRef CAS PubMed.
- J. H. Stenlid, A. J. Johansson and T. Brinck, Phys. Chem. Chem. Phys., 2018, 20, 2676 RSC.
- A. Bauzá, I. Alkorta, J. Elguero, T. J. Mooibroek and A. Frontera, Angew. Chem., Int. Ed., 2020, 59, 17482 CrossRef PubMed.
- G. Cavallo, P. Metrangolo, T. Pilati, G. Resnati and G. Terraneo, Cryst. Growth Des., 2014, 14, 2697 CrossRef CAS.
- A. Daolio, A. Pizzi, M. Calabrese, G. Terraneo, S. Bordignon, A. Frontera and G. Resnati, Angew. Chem., Int. Ed., 2021, 60, 20723 CrossRef CAS PubMed.
- A. Daolio, A. Pizzi, G. Terraneo, A. Frontera and G. Resnati, Chem. Phys. Chem., 2021, 22, 2281 CrossRef CAS PubMed.
- A. Frontera and A. Bauzá, Chem. – Eur. J., 2022, 28, e202201660 CrossRef PubMed.
- M. Calabrese, R. M. Gomila, A. Pizzi, A. Frontera and G. Resnati, Chem. – Eur. J., 2023, 29, e202302176 CrossRef CAS PubMed.
- J. L. Ross, M. Y. Ali and D. M. Warshaw, Curr. Opin. Cell Biol., 2008, 20, 41 CrossRef CAS PubMed.
- M. A. Hartman, D. Finan, S. Sivaramakrishnan and J. A. Spudich, Annu. Rev. Cell Dev. Biol., 2011, 27, 133 CrossRef CAS PubMed.
- M. A. Hartman and J. A. Spudich, J. Cell Sci., 2012, 125, 1627 CrossRef CAS PubMed.
- S. Sirigu, J. J. Hartman, V. J. Planelles-Herrero, V. Ropars, S. Clancy, X. Wang, G. Chuang, X. Qian, P.-P. Lu, E. Barrett, K. Rudolph, C. Royer, B. P. Morgan, E. A. Stura, F. I. Malik and A. M. Houdusse, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7448 CrossRef CAS PubMed.
- D. Lekaditi and S. Sakellaropoulos, Cardiol. Res., 2021, 12, 146 CrossRef PubMed.
- S. M. Day, J. C. Tardiff and E. M. Ostap, J. Clin. Invest., 2022, 132, e148557 CrossRef CAS PubMed.
- R. Fedorov, M. Böhl, G. Tsiavaliaris, F. K. Hartmann, M. H. Taft, P. Baruch, B. Brenner, R. Martin, H.-J. Knölker, H. O. Gutzeit and D. J. Manstein, Nat. Struct. Mol. Biol., 2009, 16, 80 CrossRef CAS PubMed.
- K. Chinthalapudi, M. H. Taft, R. Martin, S. M. Heissler, M. Preller, F. K. Hartmann, H. Brandstaetter, J. Kendrick-Jones, G. Tsiavaliaris, H. O. Gutzeit, R. Fedorov, F. Buss, H.-J. Knölker, L. M. Coluccio and D. J. Manstein, J. Biol. Chem., 2011, 286, 29700 CrossRef CAS PubMed.
- M. Priess, H. Göddeke, G. Groenhof and L. V. Schäfer, ACS Cent. Sci., 2018, 4, 1334 CrossRef CAS PubMed.
- P. Franz, W. Ewert, M. Preller and G. Tsiavaliaris, Int. J. Mol. Sci., 2021, 12, 104 Search PubMed.
- Y. Li, B. J. Orlando and M. Liao, Nature, 2019, 567, 486 CrossRef CAS PubMed.
- K. Kitaura and K. Morokuma, Int. J. Quantum Chem., 1976, 10, 325 CrossRef CAS.
- R. F. W. Bader, Acc. Chem. Res., 1985, 18, 9 CrossRef CAS.
- R. A. Boto, F. Peccati, R. Laplaza, C. Quan, A. Carbone, J.-P. Piquemal, Y. Maday and J. Contreras-García, J. Chem. Theory Comput., 2020, 16, 4150 CrossRef CAS PubMed.
- F. Weinhold, C. R. Landis and E. D. Glendening, Int. Rev. Phys. Chem., 2016, 35, 399 Search PubMed.
- S. Alvarez, Dalton Trans., 2013, 42, 8617 RSC.
- C. Adamo and V. Barone, J. Chem. Phys., 1999, 110, 6158 CrossRef CAS.
- M. Ernzerhof and G. E. Scuseria, J. Chem. Phys., 1999, 110, 5029 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- A. Schaefer, H. Horn and R. Ahlrichs, J. Chem. Phys., 1992, 97, 2571 CrossRef CAS.
- A. Daolio, A. Pizzi, M. Calabrese, G. Terraneo, S. Bordignon, A. Frontera and G. Resnati, Angew. Chem., Int. Ed., 2021, 60, 20723 CrossRef CAS PubMed.
- A. Daolio, A. Pizzi, G. Terraneo, A. Frontera and G. Resnati, Chem. Phys. Chem., 2021, 22, 2281 CrossRef CAS PubMed.
- B. Mallada, A. Gallardo, M. Lamanec, B. de la Torre, V. Špirko, P. Hobza and P. Jelinek, Science, 2021, 374, 863 CrossRef CAS PubMed.
- G. Balasubramani, et al. , J. Chem. Phys., 2020, 152, 184107 CrossRef PubMed.
- T. Lu and F. Chen, J. Comp. Chem., 2011, 33, 580 CrossRef PubMed.
- R. Dennington, T. A. Keith and J. M. Millam, GaussView, Version 5, Semichem Inc., Shawnee Mission, KS, 2016.
- L. Zhao and M. von Hopffgarten, WIREs Comp. Rev., 2018, 8, e1345 Search PubMed.
- K. Kitaura and K. Morokuma, Int. J. Quantum Chem., 1976, 10, 325 CrossRef CAS.
- E. D. Glendening, J. K. Badenhoop, A. E. Reed, J. E. Carpenter, J. A. Bohmann, C. M. Morales, P. Karafiloglou, C. R. Landis and F. Weinhold, Theoretical Chemistry Institute, University of Wisconsin, Madison, 2018.
- J. Contreras-García, E. R. Johnson, S. Keinan, R. Chaudret, J.-P. Piquemal, D. N. Beratan and W. Yang, J. Chem. Theory Comput., 2011, 7, 625 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14, 33 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Cartesian coordinates of complexes 1 to 16 as well as those corresponding to the H-optimized PDB models. See DOI: https://doi.org/10.1039/d3cp04571d |
| This journal is © the Owner Societies 2023 |