Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Using singular value decomposition to analyze drug/β-cyclodextrin mixtures: insights from X-ray powder diffraction patterns†
Kanji
Hasegawa
,
Satoru
Goto
 *,
Chihiro
Tsunoda
,
Chihiro
Kuroda
,
Yuta
Okumura
,
Ryosuke
Hiroshige
,
Ayako
Wada-Hirai
,
Shota
Shimizu
,
Hideshi
Yokoyama
and
Tomohiro
Tsuchida
*,
Chihiro
Tsunoda
,
Chihiro
Kuroda
,
Yuta
Okumura
,
Ryosuke
Hiroshige
,
Ayako
Wada-Hirai
,
Shota
Shimizu
,
Hideshi
Yokoyama
and
Tomohiro
Tsuchida

Faculty of Pharmaceutical Sciences, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan. E-mail: s.510@rs.tus.ac.jp
First published on 20th September 2023
Abstract
The article discusses the use of mathematical models and linear algebra to understand the crystalline structures and interconversion pathways of drug complexes with β-cyclodextrin (β-CD). It involved the preparation and analysis of mixtures of indomethacin, diclofenac, famotidine, and cimetidine with β-CD using techniques such as differential scanning calorimetry (DSC), X-ray powder diffraction (XRPD), and proton nuclear magnetic resonance (1H-NMR). Singular value decomposition (SVD) analysis is used to identify the presence of different polymorphs in the mixtures of these drugs and β-CD, determine interconversion pathways, and distinguish between different forms. In general, linear algebra or artificial intelligence (AI) is used to approximate the contribution of distinguishable entities to various phenomena. We expected linear algebra to completely reveal all eight entities present in the diffractogram dataset. However, after performing the SVD procedure, we found that only six independent basis functions were extracted, and the entities of the INM α-form and the CIM B-form were not included. It is considered that this is due to that data processing is limited to revealing only six or seven independent factors, as it is a small world. The authors caution that these may not always reproduce or approach reality in complicated real-world situations.
1. Introduction
Several fields require investigation of polymorphism to regulate crystallization phenomena. The formation of polymorphs involves various factors, including physical conditions such as temperature, humidity, and pressure, chemical conditions (solvents, additives and others), and preparation/storage processes. In materials science, crystals' optical effects are applied to the development of optical communications and devices, while the study of the crystalline structure is useful for the development of new materials.1–3 In earth science, we can understand the origin of the earth from the formation and structure of crystals.4–6 The crystallizing phenomenon is particularly important in pharmaceutical, cosmetic, and food sciences.7–10 A compound has different crystal polymorphs, known or unknown, and there may be differences in their physicochemical properties and efficacy.11–13 It is necessary to control crystallization to maintain high quality at various steps in the manufacturing process.7,14 The crystal habit (i.e., crystal structure, such as cutting direction and crystallinity) can influence crystal formation and can be useful in analyzing the effects of adjustments such as heating, cooling, pressurization, the addition of semi-solidifying agents, and additives. A crystal habit control technology is used for this purpose.15 Since pharmaceuticals are manufactured and stored under various conditions, the crystal habit of active pharmaceutical ingredients (APIs) may change even if their crystal polymorph does not change. Differences in crystal habit, as well as polymorphism of APIs, can affect the physical and chemical properties and stability of pharmaceuticals.16,17 For example, different crystallizing behavior may result in different solubility of APIs. An unstable crystal may improve solubility, but a stable crystal may decrease solubility.18,19 Furthermore, differences in crystallizing behavior may affect APIs' pulverization-ability, compressibility, water retention, and emulsification properties.20 To maintain pharmaceutical quality, it is important to pay attention to changes in crystallizing behavior and create an appropriate manufacturing and storage environment. The present study's aim is quantitatively to distinguish and classify the crystalline states (polymorphism and habits) of pure or complex active APIs.Shimada and Tateuchi focused on the importance of the effects of basic drugs on the physical properties of indomethacin.21,22 A quantitative structure–activity relationship study of the solubility of indomethacin (INM) showed that the value of the partition coefficient of basic drugs and the decrease in melting point (ΔTm) due to physical mixing influence the solubility of INM.21 In a recent study, the radar chart analysis for Lipinski's drug-likeness was used to search for candidate compounds as useful drugs for sulfa drug-pyrrole complexes.23 Kasai and Shiono et al. found that famotidine (FAM) and cimetidine (CIM), which have similar structures to acidic drugs, show marked differences in their behavior.24 Tsunoda et al. have discussed the individual intermolecular interactions of these basic drugs with acidic drugs.25 Therefore, indomethacin and diclofenac were selected as low-solubility acidic drugs with the aim of clarifying the difference in the effects of FAM and CIM.
Explainable artificial intelligence (XAI) refers to the development of AI systems that can provide understandable and transparent explanations of their recognition and decision-making processes.26–32 IBM Watson uses various techniques to achieve XAI, such as explaining how a particular decision was made using a set of predefined rules, responding to user queries with relevant explanations through natural language processing, generating visualizations to help users understand the logic behind the decision, allowing interactive feedback during the decision-making process, and more.33 Incorporating transparency and interpretability aims to increase human trust in AI systems. However, it is noteworthy that providing explanations alone does not guarantee trust or acceptance of the system's decisions.34,35 To build trust in AI systems, it is crucial to consider a range of ethical and social considerations and tailor the explanations and interactions provided by the system to the specific needs and context of the users.36,37 While IBM Watson can provide a large amount of information, it is worth noting that the number of sources surveyed by the system does not prove the objective authenticity of scientific findings.38 The validity of scientific discoveries depends on the quality of evidence and the reproducibility of results, rather than popularity or the number of sources surveyed. In scientific research, scientists use inductive or deductive logic. Particularly, experimental chemists presuppose the deductive components consisting of chemically/physically pure entities (namely, molecular species, resonance structures, racemic compounds, and others) or analytically indistinguishable molecular assemblies to verify the validity of their findings.
In our previous work, we explored a novel approach to analytical chemistry that involved using linear algebra and basis functions derived from various spectroscopic techniques, such as Fourier-transform infrared (FTIR),39 circular dichroism (CD),40 ultraviolet/visible light (UV/vis),41,42 electron paramagnetic resonance (ESR),43,44 and fluorescence (FL).45,46 This approach enabled us to reconstruct observed data by applying a similar technique to analyze diffraction patterns obtained from X-ray powder diffractometry (XRPD).47,48
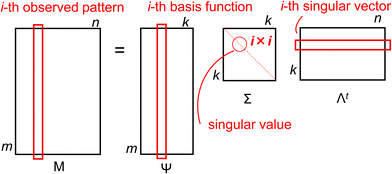
In the present study, we assumed that the observable diffractogram patterns were linear combinations of latent patterns for unknown molecular entities or distinguishable complexes (independently adding the newly synthesized or derived complex), which we arranged into a matrix M based on the experimental conditions. Using singular value decomposition (SVD) on the M matrix, we obtained a basis function matrix Ψ, a diagonal matrix Σ of singular values, and the transposed matrix Λt of singular vectors: M = ΨΣΛt, where the i-th vector in the Ψ matrix represents a latent pattern for an assumed entity, the i-th singular value in the Σ matrix indicates the statistical variance in the contribution of the corresponding entity, and the j-th singular vector in the Λt matrix corresponds to the vector descriptor used to represent each observed pattern using the linear combination coefficients for the basis functions and singular values (see Scheme 1). Thus, any arbitrary spectrum observed can be rationally reproduced by a linear combination of the confirmable basis functions. Refer to our previous publications for a more detailed explanation of the mathematical protocol used for the SVD procedure.39–46
Based on this reflection, we reported that an approach with a melting entropy-phase diagram indicated whether inclusion complexation with HP-β-CD succeeded for nifedipine or nicardipine hydrochloride.49 Different polymorphs were found in a mixture of urea and naphthalene, indicating that entropy plays an important role.48 We recently applied the melting entropy-phase diagram approach to determine the efficiencies of the inclusion complexation of INM with HP-β-CD for mixtures prepared using the physical mixture (PM) method and those prepared using the solution mixture (SM) method.50 We considered that these complexes contained independent molecules with single polymorphs. Therefore, for transformation among the polymorphs, we analyzed their DSC thermograms using rough approximations.
This linear algebra approach may not provide the same level of advanced prediction and decision-making capabilities as AI, while it does offer chemists a comprehensive way to identify the latent pure entities that contribute to the apparent patterns of various states.34–38 This approach offers a more explainable and traceable (but quite simple) alternative to deep learning procedures that typically rely on complicated mathematical approaches. The induced basis functions help us understand why an unknown combination of components is expected to derive its own XRPD pattern. Each basis function corresponds to a curated pure molecular entity or analytically distinguishable complex. The decision-making process involved in arriving at these expectations is similar to the deductive thinking used by chemists, with the logic built using a linear combination of observable basis functions. While the predicted patterns may appear complex, these methods are only slightly different from general analyses that rely on calibration and measurement (it means about procedures, not evaluation of data).
2. Experimental section
2.1. Materials
DCF acid and its sodium salt were purchased from Tokyo Chemical Industry (Tokyo, Japan). INM, FAM, CIM, and β-cyclodextrin (β-CD; a cyclic heptamer of glucosides) were obtained from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan). All other commercially available materials and solvents were of analytical reagent grade.Two mixing methods were used to produce the API/β-CD mixtures: physical mixing (PM) and solvent mixing (SM). The PM method involved kneading the mixed API and β-CD with an agate mortar and pestle. Neat API and plain β-CD samples were used after grinding, similar to the PM preparation. The PM-prepared mixtures were produced as an equimolar mixture of the API and β-CD. Meanwhile, the SM method involved mixing an ethanol solution of completely dissolved API and β-CD, removing the ethanol by evaporation, and then drying in a vacuum desiccator at 298 K overnight to obtain a powder. The SM-prepared mixtures were provided in molar ratios (mole fraction of API) of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (100%), 3
0 (100%), 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (75%), 2
1 (75%), 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (67%), 1
1 (67%), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (50%), 1
1 (50%), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (33%), 1
2 (33%), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (25%), and 0
3 (25%), and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (0%). Because the SM-prepared INM/β-CD mixture induced the α-form of INM,50 the neat INM dried in a vacuum was used as the γ-form and the α-form crystal was yielded by recrystallization with ethanol, evaporating overnight at a temperature of 278 K.
1 (0%). Because the SM-prepared INM/β-CD mixture induced the α-form of INM,50 the neat INM dried in a vacuum was used as the γ-form and the α-form crystal was yielded by recrystallization with ethanol, evaporating overnight at a temperature of 278 K.
2.2. Thermal analysis of neat API and API mixture with β-CD
Differential scanning calorimetry (DSC) was used to measure the thermal behavior of neat API and API mixture with β-CD. The DSC instrument (DSC8230, Rigaku Co., Tokyo, Japan) was used to scan the samples from 303 K to 453 K at a rate of 5.0 or 10.0 K min−1, under a nitrogen gas flow of 30 mL min−1. The scanning range for DCF was 303–463 K. Tm, which is the melting start temperature, was obtained using Thermo Plus 2 software (Rigaku Co., Tokyo, Japan) by measuring the intersection of the baseline extension and the maximum slope point of the peak. If the thermogram showed a simple endothermic peak, the area enclosed by the endothermic curve and the baseline was converted to the total melting enthalpy (ΔfusH) for a given mass amount of the component. The total melting entropy (ΔfusS) was simultaneously approximated using the quotient of ΔfusH divided by Tm, according to the classical definition of Clausius.2.3. Nuclear magnetic resonance spectroscopy
The 1H-NMR measurements were performed at 400 MHz using LA-400 (JEOL Ltd). The samples were dissolved in either deuterated DMSO (DMSO-d6) or deuterated water (D2O) at a temperature of 298 K. The chemical shift of the solvent is used as a reference point (DMSO-d6 2.500 ppm, D2O 4.800 ppm). The ppm of the solvent was calibrated with tetramethylsilane (TMS) set as the origin.2.4. XRPD diffraction of API and API mixture with β-CD
XRPD pattern measurements were used to identify the polymorph of neat API and API mixture with β-CD. RINT 2000 instrument (Rigaku Co., Tokyo, Japan) was used with a Cu Kα radiation source at a voltage of 40 kV and a current of 40 mA, filtered by the Ni filter to produce monochromatic radiation. The X-ray irradiation was performed using the parallel-beam method in a 2 by theta (2θ) range from 5 to 40 degrees at a scanning rate of 0.02 degree steps. 2θ from 5 to 40 degrees is customary in pharmaceutical analysis.51 Angles smaller than 5 degrees fall within the range of X-ray Small-Angle Scattering (SAXS) measurements, which offer distinct information from the molecular-level details obtained from XRPD.52 Whereas, the use of high theta (higher angle than 40 degrees) data is used to obtain resolution at a small distance and to obtain total scattering profiles (such as pair distribution functions, i.e., PDF, or similar Diff(r) functions), that are very useful in materials science. This could more easily be achieved with molybdenum, silver radiation, or synchrotron. In this study, by focusing on such commonly used measurement parameters, we have avoided unnecessary complexities in interpretation. The spectra were presented as the average of five scans, and the scanning sequences were conducted in triplicate or more times. The samples were crushed in an agate mortar and pestle, and then the powders were mixed. The diffractograms obtained by XRPD were published in the ESI,† TXT (406 K) and TXT (304 K), which were described in comma separated values (CSV).To identify the polymorph, we compared the observed diffractograms of single-crystal structures of the API with reference ones. They were obtained by converting 3D coordinates using the reflex module of powder diffraction on BIOVIA/ACCELRYS Materials Studio 2022 (Dassault Systems) and calculating the Miller indices of conspicuous peaks. The 3D crystalline coordinates were retrieved from the Cambridge Crystallographic Data Centre (CCDC).
For INM, we surveyed the most stable γ-form and metastable α-form from the CCDC reference codes INDMET (1972) and INDMET04 (2011), respectively. DCF was found in three polymorphs: the most stable HD2-form retrieved as SIKLIH (1990), metastable HD1-form as SIKLIH02 (1997), and metastable HD3-form as SIKLIH04 (2001).
CIM contained four forms: the A-form as CIMETD02 (1979), the B-form as CIMETD06 (2019), the C-form as CIMETD04 (2013), and the z-form as CIMETD01 (1984). FAM obtained two forms: its A-form as FOGVIG01 (1989) and B-form as FOGVIG02 (2002).
As previously reported, the CIM complexes with β-CD and γ-CD (the cyclic octamer of glucosides) formed the polymorph of channel form, while the non-complexed CDs and the CIM complex with α-CD (the cyclic hexamer of glucosides) induced the structure of the cage form, which was confirmed with the simulated pattern from BCDEXD03 (1994).51 The channel form was verified with the diffraction angles of the para-hydroxybiphenyl complex with β-CD from the reference code OFAXID (2007).
2.5. Singular value decomposition (SVD) procedure applied to diffractograms generated by XRPD analysis
The observed i-th diffractogram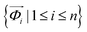 of the sample was represented as an m-dimensional vertical vector measured at the specific 2θ. Since 2θ is measured over a range of 5–40 degrees with an interval of 0.02 degrees, the value of m is 1751. Matrix M was composed of a horizontal sequence of vectors from the first diffractogram vector and the i-th diffractogram vectors, with an m × n rectangular matrix defined as eqn (1):
of the sample was represented as an m-dimensional vertical vector measured at the specific 2θ. Since 2θ is measured over a range of 5–40 degrees with an interval of 0.02 degrees, the value of m is 1751. Matrix M was composed of a horizontal sequence of vectors from the first diffractogram vector and the i-th diffractogram vectors, with an m × n rectangular matrix defined as eqn (1): | (1) |
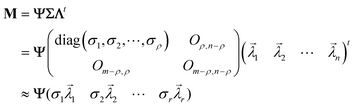 | (2) |
The diagonal matrix Σ contains the diagonal elements {σi|1 ≤ i ≤ ρ} that have positive real values ordered in a descending order. These elements represent the singular values, which indicate dispersion. The i-th column of the orthogonal matrix Λ is the coefficient vector corresponding to the singular value σi, and vector ![[small lambda, Greek, vector]](https://www.rsc.org/images/entities/i_char_e121.gif) i is called a specific singular vector. The rows of the matrix Ψ are denominated as basis function vectors.39–48 The principal component vector
i is called a specific singular vector. The rows of the matrix Ψ are denominated as basis function vectors.39–48 The principal component vector ![[small omega, Greek, vector]](https://www.rsc.org/images/entities/i_char_e13e.gif) i is the coefficient vector
i is the coefficient vector ![[small lambda, Greek, vector]](https://www.rsc.org/images/entities/i_char_e121.gif) i multiplied by the corresponding singular value σi.
i multiplied by the corresponding singular value σi.
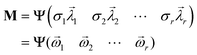 | (3) |
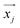 composed of the i-th elements xi,j:
composed of the i-th elements xi,j: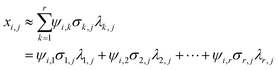 | (4) |
Principal component analysis (PCA)47 was performed on σiλi extracted by SVD. The 3D plots of principal component (PC) vectors are shown in the Supplementary Movie (ESI†).
3. Results
3.1. Preparation of acidic API complexes with β-CD and their diffractograms
The SM-prepared INM/β-CD mixtures at molar ratios of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and 1
2, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, the SM-treated INM (at a molar ratio of 1
3, the SM-treated INM (at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), the neat INM (γ-form), the α-form INM crystal, the SM-treated β-CD (0
0), the neat INM (γ-form), the α-form INM crystal, the SM-treated β-CD (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
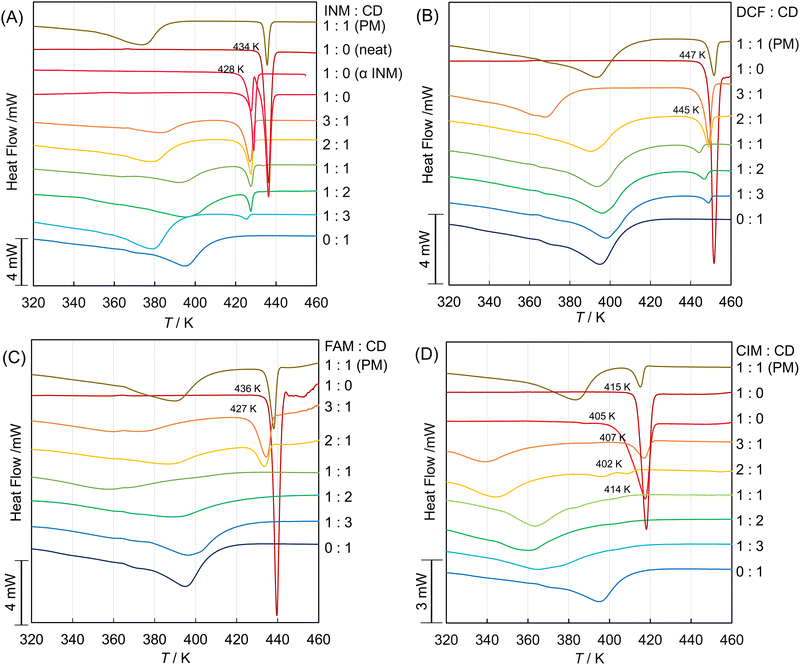
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the plain β-CD were prepared as described in the Experimental section. Their DSC thermograms are shown in Fig. 1A. The thermogram of the SM-prepared INM/β-CD equimolar mixture contained two endothermic troughs at the dropping temperature of 428 K and 434 K and their intermedial peak. The left and right troughs were corresponding to the signal of the neat INM and that of the α-form INM crystal, respectively. The intermedial peak indicated the phase transition from the metastable α-form to the most stable γ-form.50 The SM-prepared INM/β-CD equimolar mixture showed the broad signal of the cage form of β-CD at about 380–400 K and the sharp signal of the α-form INM at the dropping temperature of 428 K. Compared to the thermogram of the PM-prepared INM/β-CD equimolar mixture, the INM/β-CD inclusion complex was partially obtained, but the crystalline INM of α-form would remain.
1), and the plain β-CD were prepared as described in the Experimental section. Their DSC thermograms are shown in Fig. 1A. The thermogram of the SM-prepared INM/β-CD equimolar mixture contained two endothermic troughs at the dropping temperature of 428 K and 434 K and their intermedial peak. The left and right troughs were corresponding to the signal of the neat INM and that of the α-form INM crystal, respectively. The intermedial peak indicated the phase transition from the metastable α-form to the most stable γ-form.50 The SM-prepared INM/β-CD equimolar mixture showed the broad signal of the cage form of β-CD at about 380–400 K and the sharp signal of the α-form INM at the dropping temperature of 428 K. Compared to the thermogram of the PM-prepared INM/β-CD equimolar mixture, the INM/β-CD inclusion complex was partially obtained, but the crystalline INM of α-form would remain.
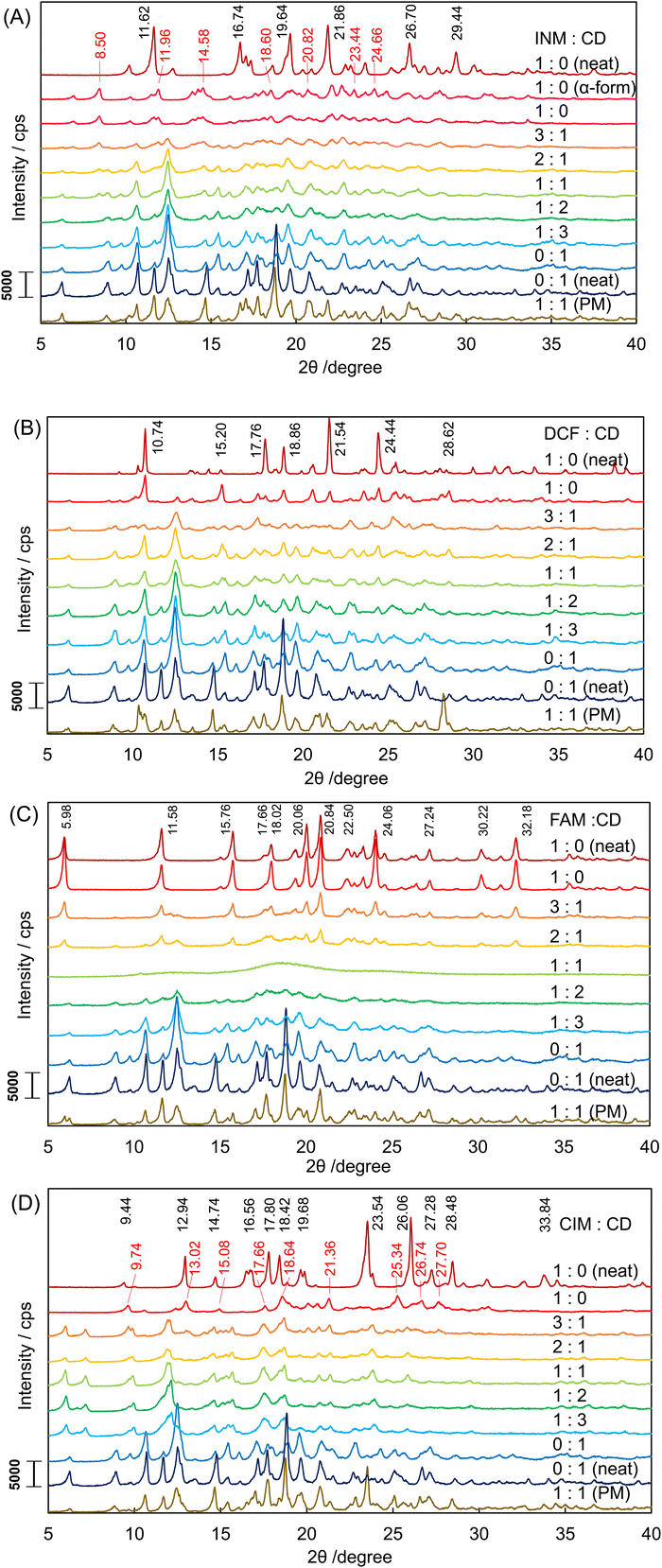
The corresponding results were obtained in the XRPD diffractograms in Fig. 2A. Signals from both the α-INM crystal and the plain β-CD were observed in the diffractogram of the SM-prepared INM/β-CD equimolar mixture. The diffractogram of neat INM was confirmed as the most stable γ-form with the simulated pattern in Fig. S2A (ESI†). Its signals did not appear any patterns of the SM-prepared mixture at various molar ratios and the PM-prepared equimolar mixture.
In Fig. 1B, the DSC thermograms of the DCF/β-CD mixtures show the endothermic signal at a dropping temperature of about 445–447 K except for that of the plain β-CD (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).46 Their XRPD diffractograms are shown in Fig. 2B. The diffractogram of neat DCF was verified as the most stable form HD2 with the simulated patterns in Fig. S2B (ESI†). Its signals were observed in the patterns of the DCF/β-CD mixtures. It indicated that the HD2 crystal was included in the SM-prepared mixtures at various molar ratios and the PM-prepared equimolar mixture.
1).46 Their XRPD diffractograms are shown in Fig. 2B. The diffractogram of neat DCF was verified as the most stable form HD2 with the simulated patterns in Fig. S2B (ESI†). Its signals were observed in the patterns of the DCF/β-CD mixtures. It indicated that the HD2 crystal was included in the SM-prepared mixtures at various molar ratios and the PM-prepared equimolar mixture.
The 1H-NMR spectra of plain β-CD, neat DCF, and their equimolar mixtures in DMSO-d6 and D2O were measured, as shown in Fig. S3A–C and S4A–C (ESI†). Although the signals of DCF showed few differences in the absence and presence of β-CD in deuterated DMSO, they shifted from neat DCF to its β-CD mixture in deuterated water. The doublet and double-triplet signals in the 2,6-dichloroaniline moiety and the double-triplet signal of the para-positioned proton in the phenylacetic acid moiety shifted to a lower magnetic field at about 0.01 ppm. This shift verified that the 2,6-dichloroaniline moiety partially intruded into the shielded interior cavity of β-CD. Since the H5 multiplet signal in the interior cavity of β-CD was superimposed onto the adjacent H6 double-doublet signal, its accurate chemical shift and coupling constants were unable to be determined. The H3 triplet signal in the interior cavity of β-CD provided only an insignificant difference in chemical shift because of the thermal perturbation of seven glucoside units attenuating the shift to 1/7 intensity as a time average.
3.2. Preparation of basic API complexes with β-CD and their diffractograms
The DSC thermograms of FAM and CIM are shown in Fig. 1C and D. Their equimolar mixtures with β-CD demonstrated no endothermic signals that appeared in the thermograms of the neat FAM and the neat CIM. The XRPD diffractograms of the SM-prepared FAM/β-CD mixture at various molar ratios, the PM-prepared FMA/β-CD equimolar mixture, the neat FAM, and the plain β-CD are shown in Fig. 2C. The SM-prepared DCF/β-CD equimolar mixture had the halo pattern. This is interpreted as indicating that the crystal structure was completely destroyed by the formation of an inclusion complex with cyclodextrin. The diffractograms of the neat DCF and the DCF mixtures at molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 3
0, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 2
1, and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were confirmed as containing the A-form of FAM with the patterns shown in Fig. S2C (ESI†). On the other hand, the diffractograms of the DCF mixtures at molar ratios of 1
1 were confirmed as containing the A-form of FAM with the patterns shown in Fig. S2C (ESI†). On the other hand, the diffractograms of the DCF mixtures at molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 0
3, and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 had the signals corresponding to those obtained from the plain β-CD (neat CD). It indicated that the DCF/β-CD mixtures were stoichiometrically formed in the inclusion complex at a molar ratio of 1
1 had the signals corresponding to those obtained from the plain β-CD (neat CD). It indicated that the DCF/β-CD mixtures were stoichiometrically formed in the inclusion complex at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
The 1H-NMR spectra of the neat FAM and the FAM/β-CD equimolar mixtures in deuterated DMSO and deuterated water were measured, as shown in Fig. S3D, E, S4D and E (ESI†). The signals of FAM shifted from neat FAM to the mixture in deuterated water. The singlet signal of 4-(2-guanydyl-1,3-thiazolyl)-methylene protons shifted to a lower magnetic field at about 0.10 ppm. The triplet signal of the 2-positioned aliphatic proton in the 1-aminosulphonylimino-propylamine moiety shifted to a lower magnetic field at about 0.03 ppm. As the signals of amino-protons vanished in deuterated water, the orientation of FAM intruding into β-CD was not specified. It suggested that even the sophisticated measurements (COSY and ROESY) are not expected to determine the interaction between FAM and β-CD in protic solvents. We speculate that the complexed FAM molecule penetrates the β-CD ring.
As shown in Fig. 2D, the diffractogram of the neat CIM was obtained, confirmed as the A-form from comparison to the simulated patterns in Fig. S2D (ESI†). But the SM-prepared CIM had its diffractogram inconsistent with that of the A-form, considering that it transformed to the B-form, which contains signals at 2θ angles of 9.60, 12.38, 14.88, 17.52, 18.38, 21.24, 25.12, 26.48, and 27.50 degrees in Fig. S2D (ESI†). The SM-prepared CIM/β-CD mixture at a molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 maintained the B-form signals. The greater the molar ratio of β-CD in the SM-prepared CIM mixtures, the more the B-form signals diminished. The diffractograms of the SM-prepared CIM/β-CD mixtures contained no halo pattern and were different from that of the plain β-CD (neat). As shown in Fig. 2A–C, the diffractograms of the plain β-CD (neat) and the SM-treated β-CD (0
1 maintained the B-form signals. The greater the molar ratio of β-CD in the SM-prepared CIM mixtures, the more the B-form signals diminished. The diffractograms of the SM-prepared CIM/β-CD mixtures contained no halo pattern and were different from that of the plain β-CD (neat). As shown in Fig. 2A–C, the diffractograms of the plain β-CD (neat) and the SM-treated β-CD (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) are similar to each other. Their signals accorded to the simulated pattern of the cage form from CCDC 3D coordinates, as shown in Fig. S2E (ESI†). Both of the plain β-CD (neat) and the SM-treated β-CD (0
1) are similar to each other. Their signals accorded to the simulated pattern of the cage form from CCDC 3D coordinates, as shown in Fig. S2E (ESI†). Both of the plain β-CD (neat) and the SM-treated β-CD (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were relationships of crystalline habits, which contained signals at the same angle but with different intensities.
1) were relationships of crystalline habits, which contained signals at the same angle but with different intensities.
The SM-prepared CIM/β-CD equimolar mixtures' diffractogram corresponded to the channel form's simulated pattern from CCDC 3D coordinates, as shown in Fig. S2F (ESI†). Namely, the CIM/β-CD complex formed the polymorph different from the plain β-CD.51 Such channel form was not found in the INM/β-CD, DCF/β-CD, and FAM/β-CD mixtures. The signal at the two by theta angle of 12.02 degrees would be of Millar's indices of hkl = 002 (corresponding to the signal at the two by theta angle of 12.18 degrees in the reference diffractogram of the channel form). This third axis C of crystalline coordinates was coincident with the rotational symmetry axis of the cylindrical β-CD molecule. The intensity of the diffraction at this angle increased, depending on the decrease of CIM or the increase of β-CD in the mixtures. Hence, it appeared that the crystal of cylindrical β-CD grew along the rotational symmetry axis with the increasing proportion of β-CD. We speculate that a small amount of the CIM molecule acts as a glue among the β-CD molecules to accumulate the channel cylinders.
Eventually, eight diffractograms of the distinguishable polymorphs were obtained as the γ-/α-forms of INM, the HD2 form of DCF, the A-form of FAM, the A-/B-forms of CIM, and the cage-/channel-forms of β-CD in the XRPD measurements of 35 as shown in Fig. 2A–D. In the present study, we dealt with these diffractograms to correspond to the information reproducing the individual chemical entities.
3.3. The basis functions of SVD analysis for diffractograms of the API/β-CD complexes
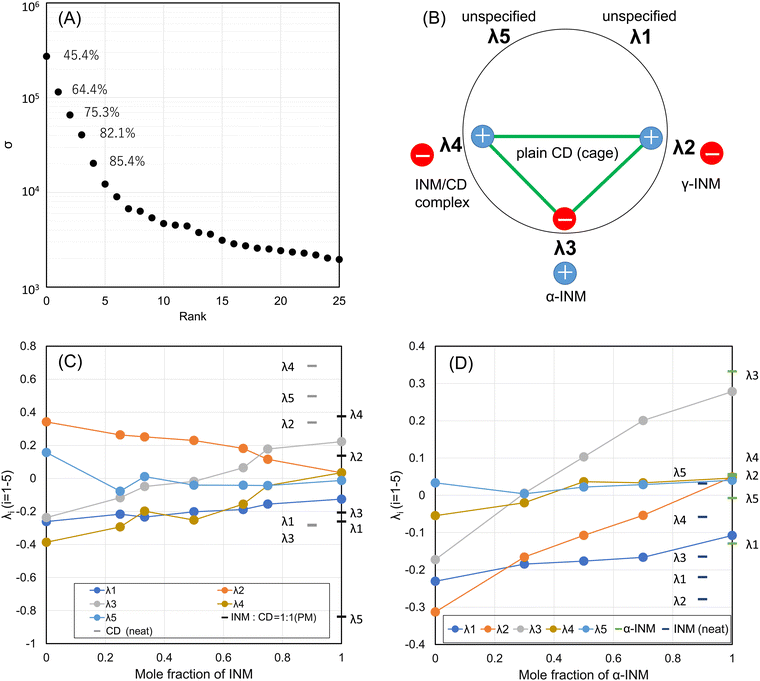
To distinguish the deductive components of chemical entities, i.e., eight polymorphs (and their crystalline habits, by chance), found in the API/β-CD mixtures of 35, their diffractograms combined as column vectors in the matrix M were treated by the procedure of the SVD method. The obtained singular values σi in the orthogonal matrix were plotted according to the rank on the common logarithm grid sorted in descending order, as shown in Fig. 3A. The first component showed the highest contribution of 33.7%. The cumulative contribution for the seventh component grew to 71.3%, and that for the eighth one to 75.1%. As the eighth σ plot seemed to be isolated from the adjacent plots of the previous and subsequent components, we retained not only the case of eight components but also that of seven components to be analyzed (r = 7, 8).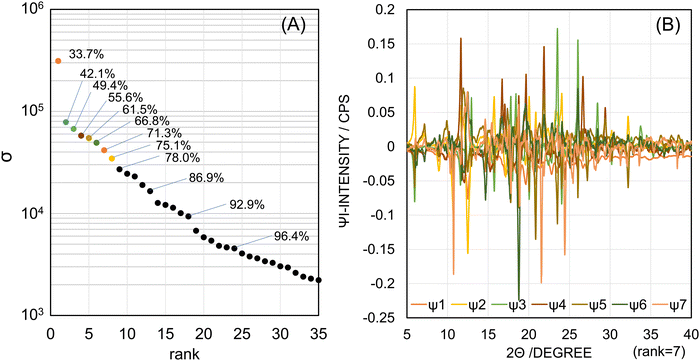 | ||
| Fig. 3 The SVD analysis for the matrix with 35 diffractograms of the API/β-CD mixtures: (A) the singular values and (B) the orthogonal basis functions of the obtained components. | ||
Fig. 3B shows their basis functions as the singular vectors {Ψi|1 ≤ i ≤ r}, in which distinctive peaks were observed. The individual basis functions are shown in Fig. S5 (ESI†). In Fig. S5A (ESI†), the first basis function Ψ1 has negative peaks and its signals were agreeable with the diffractogram of the cage form in the plain β-CD. In general, the first basis function corresponded to the average of all samples, so it often included little individual specificity. In Fig. S5B and C (ESI†), the second basis function Ψ2 has positive peaks, which corresponded to the diffractogram of the β-CD cage-form, and negative peaks, which corresponded to the XRPD pattern of FAM. Since the intensity of the negative peaks of the Ψ1 is almost half of that of the negative peaks of the Ψ2, the angle of the extracted pattern of the β-CD cage-form was about −120 degrees (cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) −120° = −0.5) to the plane of the Ψ1 and was about +150 degrees (cos
−120° = −0.5) to the plane of the Ψ1 and was about +150 degrees (cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150° = −0.866) to the perpendicular plane of the Ψ2. It meant that the plane of the pattern for the β-CD cage form made a dihedral angle of about 150 degrees to the plane of the pattern for FAM.
150° = −0.866) to the perpendicular plane of the Ψ2. It meant that the plane of the pattern for the β-CD cage form made a dihedral angle of about 150 degrees to the plane of the pattern for FAM.
In Fig. S5D and E (ESI†), the third basis function Ψ3 has positive peaks, which corresponded to the diffractogram of the CIM A-form, and negative peaks, which corresponded to the XRPD pattern of FAM. In Fig. S5F (ESI†), the fourth basis function Ψ4 has positive peaks, which corresponded to the diffractogram of the INM γ-form, and no negative peaks. In Fig. S5G and H (ESI†), the fifth basis function Ψ5 has positive peaks, which corresponded to the diffractogram of the β-CD channel form, and negative peaks, which corresponded to the XRPD pattern of FAM. In Fig. S5I (ESI†), the sixth basis function Ψ6 has negative peaks, which corresponded to the diffractogram of the CIM B-form. In Fig. S5J (ESI†), the seventh basis function Ψ7 has negative peaks, which corresponded to the diffractogram of the DCF HD2-form. In Fig. S5K (ESI†), the eighth basis function Ψ8 gave no distinctive peaks resembling the diffractograms of any polymorphs comparatively to other basis functions (Ψ1–Ψ7). This illustrated Ψ8 to be not essential to reproduce the observed patterns. Therefore, we determined r = 7.
The correspondence of the basis functions to the observed diffractograms of the polymorphs is summarized in Scheme 2. The diffractogram of the β-CD cage-form was divided into the negative peaks of Ψ1 and Ψ2 as described above. In this regard, the signals for the β-CD cage form were not yet observed in the positive peaks in Ψ6. The diffractogram of FAM seemed to be divided into the positive peaks of Ψ2 and the negative peaks of Ψ3 and Ψ5. From the above results, we found no basis functions indicating the INM α-form. As shown in Fig. S6 (ESI†), the signals depending on the INM α-form were not found in Ψ5 and other basis functions.
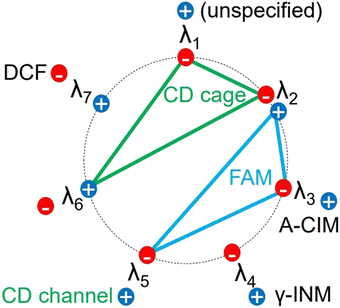 | ||
| Scheme 2 Summarized scheme of the obtained components by the SVD analysis for the matrix with 35 diffractograms of the API/β-CD mixtures. | ||
3.4. The coefficient vectors for diffractograms of the API/β-CD complexes
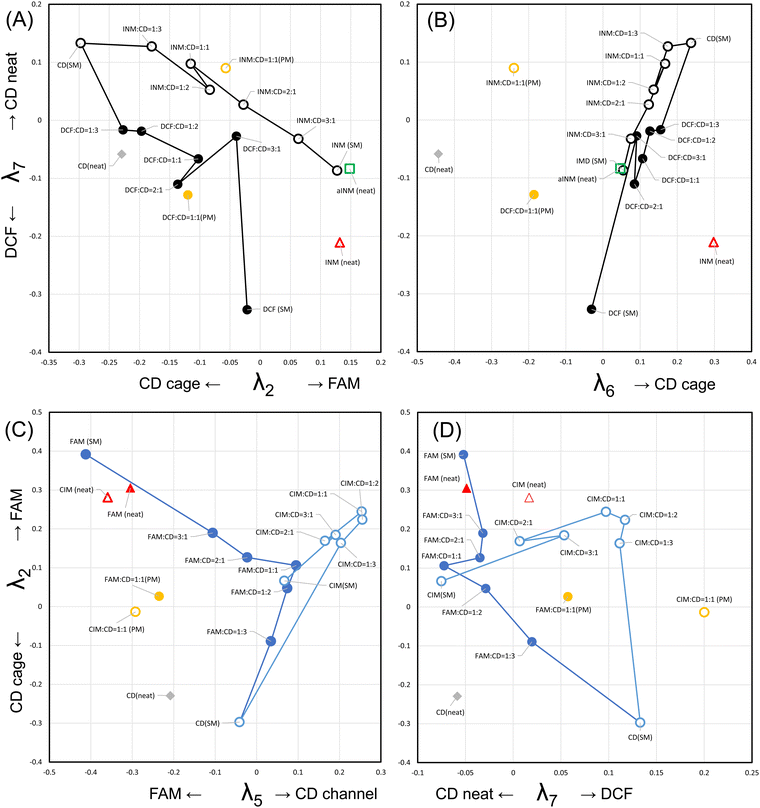
The basis functions presented the hyperdimensional “rulers” to translate the observed diffractograms to the linear combination of the pattern elements that correspond to the consisting entities. Subsequently, to verify the actual representations with the specific coefficient vectors in the right singular matrix, Fig. 4 shows the components {λij|1 ≤ i ≤ r, 1 ≤ j ≤ n} of the j-th specific coefficient vector plotted as a function of the mole fractions of the APIs for the SM-prepared API/β-CD mixtures. The first component λ1 (hereafter, we omit subscript j) monotonically increased in all the API/β-CD mixtures. The second component λ2 changed parallel to λ1, but that in the FAM/β-CD mixture increased beyond zero of the ordinate in Fig. 4C. It reflected that the positive peaks in the second basis function Ψ2 represented the diffractogram of FAM. In Fig. 4D, λ2 increased in the CIM/β-CD mixture, which has the channel form of β-CD. It indicated that the cage form of β-CD vanished in the CIM/β-CD mixtures.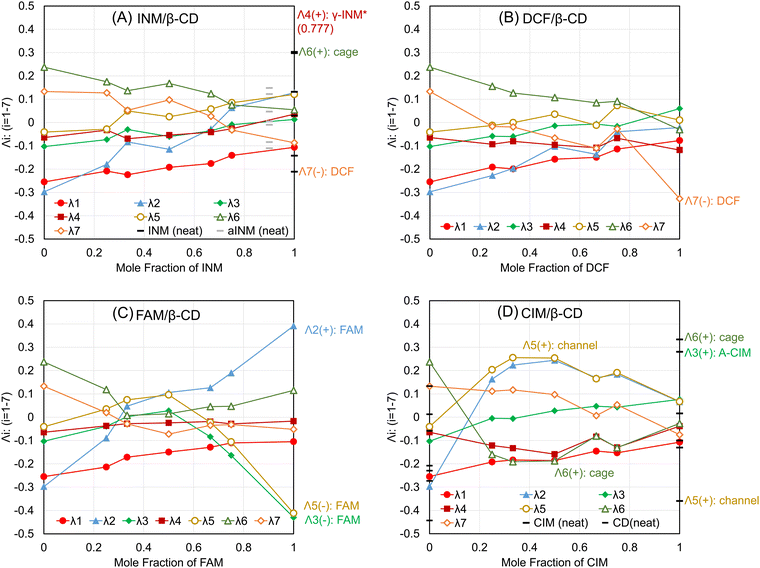 | ||
| Fig. 4 The specific coefficient vectors for the (A) INM/β-CD mixtures, (B) DCF/β-CD mixtures, (C) FAM/β-CD mixtures, and (D) CIM/β-CD mixtures. | ||
In association, the fifth component λ5 showed a positive plateau just for the CIM/β-CD mixture in Fig. 4D. It was reasonable because the positive peaks of the fifth basis function Ψ5 represented the diffractogram of the channel form of β-CD. As the negative peaks of Ψ5 corresponded to the pattern of FAM, λ5 decreased in the high mole fraction of FAM in Fig. 4C. Suppose that the pattern of FAM was considered to be divided into the basis functions Ψ5 (negative peaks), Ψ3 (negative peaks), and Ψ2 (positive peaks), it was comprehensible that the components λ3 and λ2 simultaneously decreased and increased, respectively.
The third component λ3 might have positive peaks indicating the contribution of the CIM A-form, concurrently with its negative peaks reflecting the diffractogram of FAM. In Fig. 4D, the specific coefficient vector of the neat CIM (A-form) was expressed as bars at a mole fraction of 100%, overlapped with the plots of the vector components of the SM-treated CIM (B-form). The λ3 of the neat CIM was a positively large contribution. The fifth component λ5 and the sixth component λ6 of the neat CIM were negatively and positively large, respectively. Assume the positive peaks of λ5 corresponded to the channel form of β-CD and the positive peaks of λ6 demonstrated the cage form of β-CD; these two components λ5 and λ6 would represent that the neat CIM included neither channel nor cage forms of β-CD. On the other hand, the vector components of the SM-treated CIM (B-form) resulted in small values under ±0.1 on the ordinary axis. In Fig. S5I (ESI†), the sixth basis function Ψ6 had the negative peaks of four corresponding to the diffractogram of the CIM B-form, but the intensities of its matched peaks were all small. It was reviewed to contain little evidence that λ6 correlated to the diffractogram of the CIM B-form (denied in Scheme 2). The results of the SM-treated CIM in Fig. 5D suggested the positive peaks of Ψ6 to partially represent the contribution of the cage form of β-CD. The positive peaks at the angles of 12.54 and 19.64 degrees were considered to share the contribution of the diffractogram of the cage form of β-CD, as shown in Fig. S5I (ESI†).
 | ||
| Fig. 5 The trajectories of the interconversion pathways for the (A) DCF/β-CD and INM/β-CD mixtures and (B) FAM/β-CD and CIM/β-CD mixtures. See Supplementary Movies, MP4 (7273 K) and MP4 (8521 K) (ESI†) of the three dimensional matrix. | ||
The fourth and seventh components λ4 and λ7 were characterized as representing the contributions of the INM γ-form and DCF, respectively. λ4 nearly unchanged depending on the mole fraction of APIs in the SM-prepared mixtures, as shown in Fig. 4A–D. Its large value appeared in the neat INM (γ-form), the specific components of which overlapped with those of the SM-treated INM plotted at a mole fraction of 100% in Fig. 4A. As its value of the λ4 of the neat INM was 0.777, its plot bar was beyond the ordinate axis region. It was concluded that the basis function Ψ4 and the specific coefficient component λ4 independently expressed the diffractogram and its contribution to the INM γ-form. λ7 showed a little positive value at the lower mole faction of API in the SM-prepared mixtures. The large value of λ7 was obtained in the neat DCF positioned at a mole fraction of 100% in Fig. 4B. As the negative peaks of Ψ7 corresponded to the diffractogram of DCF, the neat DCF showed a negatively large value. As shown in Fig. S5J (ESI†), the positive peaks at angles of 12.48 and 18.72 degrees were obtained, contributing very slightly to representing the cage form of β-CD.
The results are summarized in Scheme 2, but we were unable to obtain the basis function corresponding to the diffractograms of the INM α-form and the CIM B-form in the eight independent polymorphs. In Fig. 4A, the specific coefficient components of the recrystallized α-form of INM are expressed with gray bars at a mole fraction of 90%. All absolute intensities of the components were less than ±0.15, so no basis functions reflected the diffractogram of the INM α-form. Furthermore, no basis functions were obtained also as a corresponding pattern to the diffractogram of the CIM B-form. Similar analyses were performed for r = 8, but the full set of basis functions corresponding to the diffractograms of eight polymorphs could not be obtained. In Fig. 3A, the components with the higher σ in the rank of 13 (r = 13) showed a cumulative contribution of 86.99%, and those in the rank of 18 (r = 18) showed a cumulative contribution of 92.9%. By assembling such components, the represented patterns were refined to the corresponding diffractograms. However, the obtained variations of basis functions were not improved. The use of more basis functions to represent the experimental diffractograms could increase the reproducibility of the generated XRPD patterns. However, chemists do not expect the observation of experimental data to be refined. The aim should be to clarify the factors that independently make up the phenomena. In the present work, the number of independent entities was six, which includes the INM γ-form, DCF, FAM, the CIM A-form, the β-CD cage form, and the β-CD channel form.
3.5. The trajectory of the interconversion pathway in diffractograms of the API/β-CD complexes
Although we cannot directly recognize the hyperdimensional vectors and their polytope, projecting them onto a three-dimensional space can help us imagine their geometrical properties.52–54 To achieve this, we decided on the combination of three axes in the projected space based on the wide distributions (standard deviations) of the plots along the axes (see Fig. 5A and B). Also, the 3D plots of PC vectors obtained by performing PCA on σiλi extracted by SVD are shown in the Supplementary Movie, MP4 (7273 K) (ESI†). We visualized the linear polygonal lines of the crystalline states in the projected Cartesian space and traced the interconversion pathway from the state of SM-treated β-CD to that of the α-form of INM (open circles).54However, as strains of the coefficient vectors were conspicuous in the projected diagrams (see Fig. 4A and B), the observed linearity might be hard to distinguish, and the pathways seemed to meander. Therefore, in lower-dimensional projections, the strains would be often emphasized, just as a 3D perfect circle looks like an ellipse on a planner projection. The coordinates for the DCF mixture were λ2 = −0.062, λ6 = −0.059, and λ7 = −0.799, and its plot was located 0.46 units under the plot of the SM-treated DCF on the λ7 coordinate. This distance between the plots for the SM-treated DCF and the neat DCF was considered to reflect their crystalline habits, which were confirmed in Fig. 2B. The interconversion pathway of DCF/β-CD mixtures (closed circles) would sprawl from the plot for the plain β-CD (as a beginning) to that for the SM-treated DCF (as a destination).
In Fig. 5C and D, the characteristic bending lines of pathways for the FAM and CIM mixtures were observed in the Cartesian space consisting of the λ5, λ7, and λ2 axes. The 3D plots of PC vectors obtained by performing PCA are shown in the Supplementary Movie, MP4 (8521 K) (ESI†). The interconversion pathway of the FAM/β-CD mixtures prepared with SM (closed circles) lay on the λ5–λ2 diagram plane in Fig. 5C. The plot for the plain CD was located at the λ2 coordinate of −0.3, and that for the equimolar mixture of FAM was located at the λ2 coordinate of +0.1. The interconversion pathway progressed directly to the region of high λ2 and low λ5, depending on the molar ratio of FAM, and then progressed to the plot of the SM-treated FAM. Although the pathway from the equimolar mixture to the SM-treated FAM appeared to curve towards zero on the λ7 axis in Fig. 5D, the bent at the plot for the equimolar mixture was more prominent. This bending trajectory in the hyperdimensional space is referred to as the V-shape.
Meanwhile, the interconversion pathway of the CIM/β-CD mixture prepared with SM (open circles) lay on the λ2–λ7 diagram plane, with the equimolar mixture acting as a corner. The plot of plain β-CD was shared with the pathway for the FAM mixtures, and the pathway lay along the rough trajectory from the plot of the plain β-CD to the corner. The continuous trajectory bent from the corner and spanned a border of λ7 = 0, then meandered, reaching the plot for the SM-treated CIM. The trajectory of the CIM/β-CD mixtures can also be considered to take the V-shape if details are not taken into account. The verified trajectories in Fig. 5, i.e., the linear shapes for the INM and DCF mixtures and the V-shapes for the FAM and CIM mixtures, reveal the apparent variations of the diffractograms in Fig. 2.
In Fig. 5C, the V-shaped trajectory of the interconversion pathway from the plain β-CD to the SM-treated FAM via their equimolar mixture spanned the λ2–λ5 diagram plane. In Fig. 5D, the trajectory from the plain β-CD to the SM-treated CIM via their equimolar mixture spanned the λ2–λ7 diagram plane, being perpendicular to that for the β-CD to FAM. In Fig. 5C and D, the distance between the equimolar FAM/β-CD mixture and the SM-treated CIM is significantly close. The differences from the seven-dimensional vector of the equimolar FAM/β-CD mixture (at the center in Fig. 4C) to that of the SM-treated CIM (at the right edge in Fig. 4D) included the λ1 of +0.05 and the λ3 of −0.05. However, the similarity between the diffractograms of the equimolar FAM/β-CD mixture in Fig. 2C (halo pattern) and the SM-treated CIM in Fig. 2D (assigned to the B-form of CIM in Fig. S2D, ESI†) cannot be recognized. The basis functions of Ψ1, Ψ2, and Ψ3 seemed to involve the peaks related to neither the halo pattern nor the CIM B-form. The configuration space of the interconversion pathways of API/β-CD mixtures was reproduced in the seven-dimensional vector space, but we obtained no diffractograms of the CIM B-form as one of the entities.
3.6. SVD analysis for diffractograms of the INM mixtures of its polymorphs and those with β-CD
Based on our analysis of the diffractogram dataset, we found that the analyzed samples contain both γ- and α-forms of INM and A- and B-forms of CIM. However, using SVD analysis, we were able to distinguish between the presence of the γ-form of INM and the A-form of CIM. Our analysis identified six basis functions as independent factors (chemical entities) that can be used to identify these forms. However, we were unable to identify other states, such as the α-form of INM and the B-form of CIM. If any machine learning procedure was used, any unidentified states, such as the INM α-form and the CIM B-form, could be labeled with identification. Whether using linear algebra or machine learning (with any functional or any multiple-layered function), the mathematical mapping of the states and the interconversion can be processed, and there would be no essential difference. The remaining states can be reproduced because alternative factors (opposite poles) can be transformed by each other using linear algebra. While this interpretation seems reasonable, it may not be universal. For example, the channel forms of β-CD obtained in the CIM/β-CD mixtures were also alternatives to the cage form of β-CD. However, we can recognize the basis function for the channel form and the combination of those for the cage form.To determine whether a single basis function or dual functions are provided by such an alternative pair (opposite poles) of chemical entities, we conducted an SVD analysis for both the combination of the α- and γ-forms of INM and that of the plain β-CD and the INM/β-CD complex. We measured the mixtures of α- and γ-crystals of INM at various molar ratios and merged them with those of the SM-prepared INM/β-CD mixtures, resulting in 26 samples. The SVD analysis was conducted, and the results are summarized in Fig. 6. The first five higher ranks showed a cumulative contribution of 85.4%.
In Fig. S7 (ESI†), the first basis function Ψ1 represented the diffractogram of the INM γ-form. The second basis function Ψ2 corresponded to the diffractogram of the plain β-CD and the INM γ-form. The third basis function Ψ3 matched the diffractogram of the plain β-CD and the INM α-form. The fourth basis function Ψ4 matched the diffractogram of the plain β-CD and the INM/β-CD complex with a crystalline habit different from the plain β-CD. The fifth and sixth basis functions were not consistent with either the diffractograms of INM forms or those of β-CDs.
In Fig. 6, the specific coefficient components λi were plotted as a function of the mole fraction of INM in the SM-prepared INM/β-CD mixtures. λ2 and λ5 decreased, while λ4 increased depending on the molar ratio of β-CD. These changes were attributed to the positive peaks of Ψ2 and negative peaks of Ψ4 correlated to the diffractogram of the plain β-CD (cage form). The assignment for Ψ5 was ambiguous, but λ5 seemed to be responsible for the amount of plain β-CD. λ3 increased depending on the molar ratio of INM, reflecting that the diffractogram of the INM α-form corresponds to the positive peaks of Ψ3. As always, λ1 was probably proportional to the average intensity of all peaks. The peaks of neat β-CD were shown with gray bars at a mole fraction of 0%. λ4 for neat β-CD was the highest because the positive peaks of Ψ4 corresponded to neat β-CD. The peaks of the PM-prepared equimolar mixture were expressed with bars at a mole fraction of 50%. The reason why λ5 for the PM-treated equimolar mixture was the lowest might be that Ψ5 was responsible for the amount of plain β-CD. Fig. 6D shows the coefficient components for the α- and γ-forms of INM mixtures. It suggested that λ2 and λ3 corresponded to γ- and α-forms, respectively. λ4 and λ5 did not respond to the molar ratio of INM polymorphs.
3.7. The trajectory of the interconversion pathway in diffractograms of the INM mixtures
Fig. 7A and B show views from both sides in a Cartesian space made up of the λ2, λ3, and λ4 axes of the polytope of the interconversion pathway in an INM mixture of its polymorphs and those with β-CD at various molar ratios. The Supplementary Movie, MP4 (7146 K) (ESI†) shows views from both sides with the PC1, PC2, and PC3 axes, which correspond to the four-dimensionally rotating body of the projected three-dimensional space in Fig. 7A and B. The interconversion pathways of INM/β-CD mixtures and mixtures of α- and γ-polymorphs move in meandering but single-direction trajectories when viewed from any direction in the hyperspace. In Fig. 7B, these trajectories overlap. In other figures, it can be seen that both trajectories share an intersection at the plot corresponding to the INM α-form (around the zero λ2 coordinate). From this starting point, the trajectory for INM/β-CD mixtures moves in the direction of the plot for plain β-CD (positive PC1 (λ2) coordinate and PC3 (λ4) coordinate with a large distribution range in the movie file), while the trajectory for INM mixtures of polymorphs traces the plot for the INM γ-form (negative PC1 (λ2) coordinate and PC3 (λ4) coordinate with a small distribution range in the movie file). The degree of λ4 is thought to reflect the transition between the crystalline habits of β-CD, regardless of the amount and polymorphs of INM. Therefore, the current SVD analysis can distinguish between two opposing concepts: the crystalline habits of β-CD in the cage form and the α- or γ-polymorphs of INM.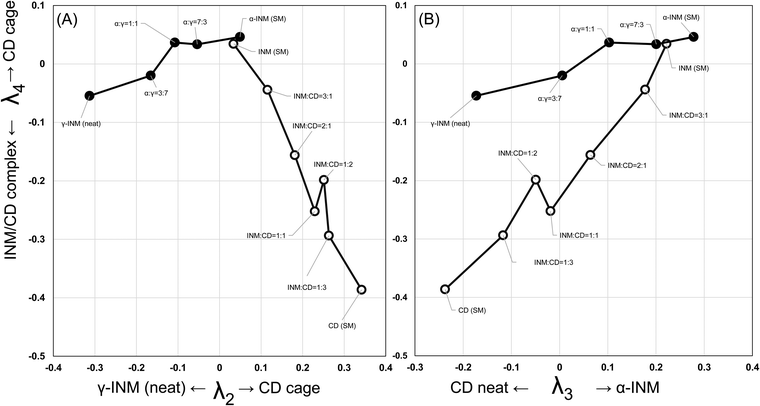 | ||
| Fig. 7 The trajectories of the interconversion pathways for the INM mixtures of the α- and γ-crystals and the INM/β-CD mixtures, projected on the λ2–λ4 plane (A) and the λ3–λ4 plane (B). See Supplementary Movie, MP4 (7146 K) (ESI†) of the three dimensional matrix. | ||
4. Discussion
In 1967, social psychologist Stanley Milgram claimed ‘six degrees of separation’, corresponding to his well-known and many times criticized experiments about a social network structure of human relationships. His hypothesis was subsequently called a small-world phenomenon.55 In 1998, sociologist Watts and mathematician Strogatz found that the addition of a small number of random links shortened the distance between any two vertices, compared with a regular lattice in the network.56 They accounted for it as a small-world phenomenon.In our linear algebra study, the relationships among the X-ray powder diffractograms of the mixtures of INM and β-CD showed the state of elements linked with direct tie lines. Meanwhile, the addition of DCF, FAM, and CIM caused to loss of the basis function for the diffractions of the INM α-form. To explain the observed diffractogram, those of the individual states (polymorphs and crystalline habits) are required, although the results of linear algebra analysis can reproduce the diffractograms constituted a combination of selected components (Fig. S8, ESI†). Tie lines in the network of the interconversion pathways would be shortened (reduction of dimensionality) and the reproduction was achieved with a small number of basis functions. We speculate it occurred due to one of the small-world phenomena.
The interconversion pathway (φi) in the observable instrumental quantity is essentially defined as a linear combination of differential equations and basis functions, as shown in eqn (5):
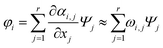 | (5) |
Configuration space is a mathematical space that describes the nodes (states) and edges (interconversion pathways) of all parts of a system.54,57–59 It is typically represented by a hyperdimensional space, where each dimension corresponds to a degree of freedom of the system. By analyzing it, analysts can determine the range of possible diversity and regulate the pathway that the assembled materials should follow to reach a particular state (existence).40–42,54 One of the challenges of working with configuration space is that it can be very complex and high-dimensional, especially for systems with many degrees of freedom. In such cases, it may be necessary to use techniques to analyze the configuration space and regulate state interconversion. It is important to note that alternative factors can be transformed into each other using linear algebra. For example, if entities x1 and x2 interact to produce entities x3 and x4, respectively, the allowed configuration space would have four dimensions, consisting of two independent axes for the interconversion from x1 to x3 and from x2 to x4, and two independent axes for the sum of x1 and x3 and that of x2 and x4. However, since the sum of x1 and x3 and that of x2 and x4 can be represented by their molar ratio under stoichiometric interaction, the dimensionality reduces to three. This experimental setup corresponds to the analysis of diffractograms of mixtures such as the α- and γ-polymorphs and the INM/β-CD mixtures. In the SVD analysis, their particular states and interconversion resulted in two degrees of freedom, meaning that the configuration space was two-dimensional. This configuration space, including the V-shaped trajectory shown in Fig. 6, is a two-dimensional manifold in a three-degree-of-freedom space. The reduction in dimensionality (from 3 to 2) is due to the dependence of INM on the α-polymorph as long as it is present in the INM/β-CD complex. As a result, it was not possible to obtain the state of the γ-form of INM included in β-CD, indicating that not all entities present can be extracted from experiments or experiences.
Regarding our analysis of the dataset of diffractograms of API/β-CD mixtures, we found that the samples contain both γ- and α-forms of INM and A- and B-forms of CIM. However, the SVD analysis was able to distinguish between the presence of the γ-form of INM and the A-form of CIM, identifying six basis functions as independent factors (chemical entities). Nevertheless, some unidentified states, such as the α-form of INM and the B-form of CIM, remain. If a machine learning procedure was applied, any unidentified states, such as the INM α-form and the CIM B-form, could be labeled with identification. The mathematical mapping of both the states and interconversion can be processed using linear algebra or machine learning with any multiple-layered functions or functionals, so there would be no essential difference. The remaining states can be reproduced because alternative factors (opposite poles) can be transformed into each other using linear algebra. The addition of DCF, FAM, and CIM resulted in highly clustered entities; yet the interconversion pathways between them were shortened. This interpretation seems reasonable, but it may not be universal. For example, the channel forms of β-CD obtained in the CIM/β-CD mixtures were also alternatives to the cage form of β-CD.
We expected linear algebra to completely reveal all eight entities present in the diffractogram dataset. However, after performing the SVD procedure, we found that only six independent basis functions were extracted, and the entities of the INM α-form and the CIM B-form were not included. This could be explained by information about these hidden entities being shared by other basis functions. On the other hand, this could also indicate that obtaining the full set of basis functions consisting of all diffractograms is not possible, and there is a limit to the amount of information or knowledge that can be processed. It is considered that this is due to the fact that data processing is limited to revealing only six or seven independent factors, as it is a small world.
The diffractogram of the NM α-form as an independent entity was hidden in the SVD analysis of the API/β-CD mixtures, while it was recovered in that of the mixtures of α-/γ-polymorphs and the INM/β-CD mixtures. Suppose the upper limit of the number of dependent parameters was seven (or the number of connections six). It suggested that linear algebra should be limited to applying the dataset which involves seven or fewer independent factors. The X-ray diffractograms depend on the crystalline periodic features of the mixed powdered solids, so they are inappropriate to the continuous figures as the response in the scientific experiments. Because the change observed in diffractograms is only dependent on the quantities of component entities, the interpretation of data is simple and independent. However, to study the relationship between the complexity of data and recognizability in linear algebra approaches, other observations (FTIR, for example) as new subjects should be considered.
5. Conclusion
In the present study, we measured and analyzed the X-ray powder diffractograms of APIs, β-CD, and their mixtures. APIs and β-CD transformed their polymorphs and crystalline habits depending on their compositions, but their transformations cannot randomly occur. The practical observations of solid states were unattainable to the number of the capable components (chemical entities and undistinguishable complexes) although we expected to extract their individual diffractograms (basis functions) using linear algebra. Nevertheless, the INM α-form and the CIM B-form were able to be reproduced as the linear combinations of these basis functions. The projection of the interconversion pathway in the hyperdimensional space allows us to visualize the relationship between the polymorphs or crystalline habits. We highlighted that the alternative factors can be transformed into each other using linear algebra but cannot be extracted as deductive entities from experiments or experiences. The authors further discussed the limitations of experimental chemists in verifying the validity of their findings without presupposing the deductive components consisting of chemical/physical pure entities, while AI may recognize such entities with the assistance of machine learning. They concluded by discussing the limitations of XAI in providing a deductive explanation and obtaining independent deductive entities from experiments and experiences.Author contributions
Kanji Hasegawa: investigation, visualization, writing – original draft. Satoru Goto: investigation and supervision. Chihiro Tsunoda: review and editing. Chihiro Kuroda: review and editing. Yuta Okumura: review and editing. Ryosuke Hiroshige: review and editing. Ayako Wada-Hirai: review and editing. Shota Shimizu: review and editing. Hideshi Yokoyama: review and editing. Tomohiro Tsuchida: review and editing.Conflicts of interest
There are no conflicts to declare.References
- K. Sakoda, Optical Properties of Photonic Crystals, Springer Series in Optical Sciences, Springer, Berlin, 2nd edn, 2005, vol. 80 Search PubMed.
- B. T. Hoga, E. Kovalska, M. F. Craciun and A. Baldycheva, J. Mater. Chem. C, 2017, 5, 11185–11195 RSC.
- T. Kato, M. Gupta, D. Yamaguchi, K.-P. Gan and M. Nakayama, Bull. Chem. Soc. Jpn., 2021, 94, 357–376 CrossRef CAS.
- K. D. Putirka and F. J. Tepley, Minerals, Inclusions and Volcanic Processes, Reviews in Mineralogy and Geochemistry, Mineralogical Society of America, Virginia USA, 2008, vol. 69 Search PubMed.
- N. L. Bowen, J. Geol., 1919, 27, 393–430 CrossRef CAS.
- T. Mikouchi, M. Komatsu, K. Hagiya, K. Ohsumi, M. E. Zolensky, V. Hoffmann, J. Martinez, R. Hochleitner, M. Kaliwoda, Y. Terada, N. Yagi, M. Takata, W. Satake, Y. Aoyagi, A. Takenouchi, Y. Karouji, M. Uesugi and T. Yada, Earth, Planets Space, 2014, 66, 82 CrossRef.
- J. Orehek, D. Teslić and B. Likozar, Org. Process Res., 2021, 25, 16–42 CrossRef CAS.
- R. W. Hartel, Annu. Rev. Food Sci. Technol., 2013, 4, 277–292 CrossRef CAS PubMed.
- K. Sato, Crystallization of Lipids: Fundamentals and Applications in Food, Cosmetics, and Pharmaceuticals, John Wiley & Sons, 2018 Search PubMed.
- F. Artusio and R. Pisano, Int. J. Pharm., 2018, 547, 190–208 CrossRef CAS PubMed.
- B. Rodríguez-Spong, C. P. Price, A. Jayasankar, A. J. Matzger and N. Rodríguez-Hornedo, Adv. Drug Delivery Rev., 2004, 56, 241–274 CrossRef PubMed.
- K. Greco and R. Bogner, Mol. Pharmaceutics, 2010, 7, 1406–1418 CrossRef CAS PubMed.
- T. Van Duong, D. Lüdeker, P.-J. Van Bockstal, T. De Beer, J. Van Humbeeck and G. Van den Mooter, Mol. Pharmaceutics, 2018, 15, 1037–1051 CrossRef CAS PubMed.
- S. K. Jha, S. Karthika and T. K. Radhakrishnan, Resour.-Effic. Technol., 2017, 3, 94–100 Search PubMed.
- C.-S. Su, C.-Y. Liao and W.-D. Jheng, Chem. Eng. Technol., 2014, 38, 181–186 CrossRef.
- A. Heinz, C. J. Strachan, K. C. Gordon and T. Rades, J. Pharm. Pharmacol., 2009, 61, 971–988 CrossRef CAS PubMed.
- A. M. Healy, Z. A. Worku, D. Kumar and A. M. Madi, Adv. Drug Delivery Rev., 2017, 117, 25–46 CrossRef CAS PubMed.
- S. Sareen, G. Mathew and L. Joseph, Int. J. Pharm. Invest., 2012, 212–217 Search PubMed.
- D. J. Good and N. Rodríguez-Hornedo, Cryst. Growth Des., 2010, 10, 1028–1032 CrossRef CAS.
- K. Imamura, M. Nomura, K. Tanaka, N. Kataoka, J. Oshitani, H. Imanaka and K. Nakanishi, J. Pharm. Sci., 2010, 99, 1452–1463 CrossRef CAS PubMed.
- Y. Shimada, R. Tateuchi, H. Chatani and S. Goto, J. Mol. Struct., 2018, 1155, 165–170 CrossRef CAS.
- R. Tateuchi, N. Sagawa, Y. Shimada and S. Goto, J. Phys. Chem. B, 2015, 119, 9868–9873 CrossRef CAS PubMed.
- M. Gümüş, Ş. N. Babacan, Y. Demir, Y. Sert, İ. Koca and İ. Gülçin, Arch. Pharm., 2022, 355, e2100242 CrossRef PubMed.
- T. Kasai, K. Shiono, Y. Otsuka, Y. Shimada, H. Terada, K. Komatsu and S. Goto, Int. J. Pharm., 2020, 590, 119841 CrossRef CAS PubMed.
- C. Tsunoda, S. Goto, R. Hiroshige, T. Kasai, Y. Okumura and H. Yokoyama, Int. J. Pharm., 2023, 638, 122913 CrossRef CAS PubMed.
- M. Ridley, Inf. Technol. Librar., 2022, 41 DOI:10.6017/ital.v41i2.14683.
- A. B. Arrieta, N. Díaz-Rodríguez, J. Del Ser, A. Bennetot, S. Tabik, A. Barbado, S. Garcia, S. Gil-Lopez, D. Molina, R. Benjamins, R. Chatila and F. Herrera, Information Fusion, 2020, 58, 82–115 CrossRef.
- S. K. Jagatheesaperumal, Q.-V. Pham, R. Ruby, Z. Yang, C. Xu and Z. Zhang, IEEE Open J. Commun. Soc., 2022, 3, 2106–2136 Search PubMed.
- W. J. Murdoch, C. Singh, K. Kumbier, R. Abbasi-Asl and B. Yu, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(44), 22071–22080 CrossRef CAS PubMed.
- W. J. Murdoch, C. Singh, K. Kumbier and B. Yu, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22071–22080 CrossRef CAS PubMed.
- W. Samek and K.-R. Müller, Towards Explainable Artificial Intelligence, Explainable AI: Interpreting, Explaining and Visualizing Deep Learning, 2019, pp. 5–22 Search PubMed.
- W. Samek, T. Wiegand and K.-R. Müller, Explainable Artificial Intelligence: Understanding, Visualizing and Interpreting Deep Learning Models, arXiv, 2017, preprint, arXiv:1708.08296v1, DOI:10.48550/arXiv.1708.08296.
- N. Burkart and M. F. Huber, J. Artif. Intelligence Res., 2021, 70, 245–317 CrossRef.
- S. Lockey, N. Gillespie, D. Holm and I. A. Someh, Conference: Hawaii International Conference on System Sciences, 2021 Search PubMed.
- D. Shin, Int. J. Human-Comput. Stud., 2021, 146, 102551 CrossRef.
- R. Riedl, Electron. Mark., 2022, 32, 2021–2051 CrossRef.
- D. Leslie, Understanding artificial intelligence ethics and safety, 2019, Zenodo DOI:10.5281/zenodo.3240529.
- P. Tagde, S. Tagde, T. Bhattacharya, P. Tagde, H. Chopra, R. Akter, D. Kaushik and H. Rahman, Environ. Sci. Pollut. Res. Int., 2021, 28, 52810–52831 CrossRef PubMed.
- Y. Otsuka, W. Kuwashima, Y. Tanaka, Y. Yamaki, Y. Shimada and S. Goto, J. Pharm. Sci., 2021, 110, 1142–1147 CrossRef CAS PubMed.
- T. Shiratori, S. Goto, T. Sakaguchi, T. Kasai, Y. Otsuka, K. Higashi, K. Makino, H. Takahashi and K. Komatsu, Biochem. Biophys. Rep., 2021, 28, 101153 CAS.
- R. Hiroshige, S. Goto, R. Ichii, S. Shimizu, A. Wada-Hirai, Y.-P. Li, Y. Shimada, Y. Otsuka, K. Makino and H. Takahashi, J. Inclusion Phenom. Macrocyclic Chem., 2022, 102, 327–338 CrossRef CAS.
- R. Hiroshige, S. Goto, C. Tsunoda, R. Ichii, S. Shimizu, Y. Otsuka, K. Makino, H. Takahashi and H. Yokoyama, J. Inclusion Phenom. Macrocyclic Chem., 2022, 102, 791–800 CrossRef CAS.
- M. Takatsuka, S. Goto, K. Kobayashi, Y. Otsuka and Y. Shimada, BBA Adv., 2021, 1, 100030 CrossRef CAS PubMed.
- M. Takatsuka, S. Goto, K. Kobayashi, Y. Otsuka and Y. Shimada, Food Biosci., 2022, 48, 101714 CrossRef CAS.
- Y. Kurosawa, Y. Otsuka and S. Goto, Colloids Surf., B, 2022, 212, 112344 CrossRef CAS PubMed.
- Y. Kurosawa, S. Goto, K. Mitsuya, Y. Otsuka and H. Yokoyama, Phys. Chem. Chem. Phys., 2023, 25, 6203–6213 RSC.
- E. R. Henry and J. Hofrichter, Methods Enzymol., 1992, 210, 129–192 CAS.
- R. J. DeSa and I. B. C. Matheson, Methods Enzymol., 2004, 384, 1–8 CAS.
- Y. P. Lee, S. Goto, Y. Shimada, K. Komatsu, Y. Yokoyama, H. Terada and K. Makino, Phys. Chem. Biophys., 2015, 5, 1000187 Search PubMed.
- A. Wada-Hirai, S. Shimizu, R. Ichii, C. Tsunoda, R. Hiroshige, M. Fujita, Y.-P. Li, Y. Shimada, Y. Otsuka and S. Goto, J. Pharm. Sci., 2021, 110, 3623–3630 CrossRef CAS PubMed.
- S. Shimizu, A. Wada-Hirai, Y.-P. Li, Y. Shimada, Y. Otsuka and S. Goto, J. Pharm. Sci., 2020, 109, 2206–2212 CrossRef CAS PubMed.
- E. Flapan, When Topology Meets Chemistry, A Topological Look at Molecular Chemistry, Cambridge University Press, New York, 2000 Search PubMed.
- P. G. Mezey, Saphe in Chemistry, An Introduction to Molecular Shape and Topology, VCH Publisers, Inc., New York, 1993 Search PubMed.
- S. Goto, K. Komatsu and H. Terada, Bull. Chem. Soc. Jpn., 2013, 86, 230–242 CrossRef CAS.
- S. Milgram, Phyychol. Today, 1967, 2, 60–67 Search PubMed.
- D. J. Watts and S. H. Strogatz, Nature, 1998, 393, 440–442 CrossRef CAS PubMed.
- S. Goto and K. Komatsu, Hiroshima Math. J., 2012, 42, 115–126 Search PubMed.
- S. Goto, Y. Hemmi, K. Komatsu and J. Yagi, Hiroshima Math. J., 2012, 42, 253–266 Search PubMed.
- S. Goto, K. Komatsu and J. Yagi, Hiroshima Math. J., 2020, 50, 185–197 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cp02737f |
| This journal is © the Owner Societies 2023 |