Open Access Article
Open Access ArticleARCHE-NOAH: NMR supersequence with five different CEST experiments for studying protein conformational dynamics†
Rodrigo
Cabrera Allpas
 a,
Alexandar L.
Hansen
a,
Alexandar L.
Hansen
 b and
Rafael
Brüschweiler
b and
Rafael
Brüschweiler
 *abc
*abc
aDepartment of Chemistry and Biochemistry, The Ohio State University, Columbus, Ohio 43210, USA. E-mail: bruschweiler.1@osu.edu
bCampus Chemical Instrument Center, The Ohio State University, Columbus, Ohio 43210, USA
cDepartment of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, Ohio 43210, USA
First published on 8th June 2023
Abstract
An NMR NOAH-supersequence is presented consisting of five CEST experiments for studying protein backbone and side-chain dynamics by 15N-CEST, carbonyl-13CO-CEST, aromatic-13Car-CEST, 13Cα-CEST, and methyl-13Cmet-CEST. The new sequence acquires the data for these experiments in a fraction of the time required for the individual experiments, saving over four days of NMR time per sample.
NMR is the method of choice for studying conformational exchange of biomolecules in solution providing atomic-resolution information of dynamics over timescales ranging from picoseconds to seconds.1,2 Various NMR experiments exist for the study of chemical exchange, including line shape analysis, R1p, Carr–Purcell–Meiboom–Gill (CPMG), and chemical exchange saturation transfer (CEST).3 CEST is a powerful experiment providing both kinetics and thermodynamics parameters of exchange processes (kex, pa, and pb) between the ground state a and the “invisible” excited state b, structural information in terms of their chemical shift differences (ΔΩ) and laboratory frame relaxation parameters R1, R2 of the ground state.4 For example, CEST-derived relaxation parameters can then be used to calculate S2 order parameters via a lean model-free approach (MFA) with good accuracy.4 Furthermore, the chemical shifts of the excited states can be used to model the structure of the excited state.5
The most common NMR spin probes for CEST in proteins are 15N and methyl-13C; however, other spin probes such as 13Cα, 13Cβ, and 13CO have been reported as well.5–10 These less commonly used NMR probes can give complementary information about the dynamic processes. For example, 13Cα and 13CO chemical shifts are very sensitive to protein backbone secondary structure.7,8 The most popular probe, 15N, does not report the dynamics of proline residues and is more susceptible to HN exchange with the water solvent, thereby losing dynamics information due to solvent exchange. For the study of aromatic residues, aromatic carbon probes have also been reported, but only for CPMG relaxation dispersion type experiments,11 despite their facility to form clusters and their important role in protein–protein interaction interfaces.12 Clearly, there is much conformational exchange information that can be gained for each residue of a protein using all available nuclei as probes. However, CEST experiments are rather time consuming to perform, since they are run in a pseudo-3D manner for the added resolution, taking a day to a week of experiment time for each type of spin. Hence, measuring the less commonly used spin probes can be prohibitively expensive in terms of spectrometer time, despite the complementary information one can obtain from each of them.
In recent years, NMR by ordered acquisition using 1H-detection (NOAH) supersequences has gained popularity for studying small molecules.13–16 The basic idea behind NOAH is to combine multiple separate pulse sequences (modules) into a supersequence, which employs only a single recovery delay (d1) throughout the pulse program. Because d1 is by far the longest time delay in an NMR experiment, a supersequence consisting of two separate pulse sequences with only a single recovery delay instead of two would effectively render the second experiment for “free” provided that other delays of the second experiment are minimal compared to d1 as is almost always the case. These supersequences are designed to use “leftover” magnetization from previous modules in subsequent modules without the need for lengthy d1 delays between modules to recover magnetization via spin-lattice relaxation. It is therefore critical to ensure that suitable pulse-sequence elements are employed that preserve unused magnetization for subsequent modules of the supersequence. Recently, small molecule applications of NOAH included as many as ten experiments, thereby drastically reducing the total experiment time17 and NOAH supersequences have been developed for complex mixture analysis in metabolomics.18–20
Despite much progress of NOAH for small molecule NMR, only a few applications exist for biomacromolecules. Early work of sequential NMR acquisition for proteins has been proposed for protein resonance assignment based on a non-standard assignment strategy.21,22 The NOAH approach was also applied to drug–protein interactions by combining 2D 1H-15N HSQC for the 15N-labeled protein with a 1H-13C-BIRD-HSQC for the ligand present at natural abundance.23 Recently, our group proposed a combination of two CEST pulse sequences using the NOAH approach for uniformly 15N, 13C-labeled proteins. The NOAH-(15N/13C)-CEST produced the same dynamics results as two independently acquired 15N-CEST and methyl-13C-CEST experiments, but required only a fraction of the time needed in the traditional approach.24 A key element of the approach is to use a 13C-start methyl CEST experiment that follows a regular 1HN start 15N-CEST. In this way, 1HN magnetization is exhausted during the first module, while retaining fresh 13C magnetization for the second module. While one would expect some major sensitivity loss when starting with thermal 13C instead of 1H magnetization, 1H composite-pulse decoupling during the extended saturation period Tex typically between 100–500 ms in the 15N-CEST leads to a heteronuclear nuclear Overhauser effect (hetNOE) resulting in a significant enhancement of the 13C spin magnetization at the start of the second module.
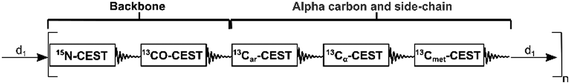
Here, we report a new NOAH supersequence, named ARCHE-NOAH for All-Residue-CHemical-Exchange-NOAH, that measures 5 protein CEST modules in a single experiment with a single recovery delay d1 (Fig. 1). This supersequence acquires data for CEST experiments of nearly all usable probes of amino-acid residues in proteins, namely backbone 15N-CEST, 13Cα-CEST, and 13CO-CEST along with side-chain 13C-aromatic-CEST and 13C-methyl-CEST in a substantially accelerated manner. ARCHE-NOAH uses band-selective pulses for all carbons, thereby carefully preserving the starting 13C magnetization for the subsequent CEST module (see below). NMR experiments were performed on an 850 MHz Bruker Ascend magnet equipped with an Avance III HD console and a triple resonance inverse cryoprobe. In order to assess the performance of the ARCHE-NOAH supersequence, it was compared with the individual modules as standalone 1H-start sequences recorded with the same parameters applied to 1 mM colicin E7 immunity protein Im7 in 97%/3% H2O/D2O at 298 K. The resulting spectra were processed using NMRPipe, followed by peak-picking, peak assignment, and cross-peak quantification. CEST profiles were analyzed using the ChemEx software (https://www.github.com/gbouvignies/chemex). More details of the methods and NMR acquisition parameters can be found in the ESI.†
The comparison of the sensitivity of the supersequence and NMR time with its standalone 1H-start counterparts is summarized in Table 1. As should be expected from the aforementioned hetNOE enhancement, the 13C-CEST module experiences the most enhancement from 1H-decoupling blocks in the preceding sequences. We find differential hetNOEs for the different types of 13C atoms, namely 13Cmet > 13Cα ≅ 13Car > CO, which can be explained by the number of protons attached to the carbon consistent with the literature.25
| NMR experiment | S/Na | Experiment time (h min) | Scaled experiment timeb (h min) |
|---|---|---|---|
a Average signal-to-noise of all non-overlapped signals of the respective sequence. Noise was calculated by taking the median absolute deviation (MAD) of the processed NMRPipe files in an empty region with no signals.
b Scaled experiment time (h min) for the standalone NMR experiments obtained by the formula 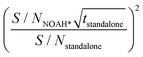 .
c Due to minimal sampling requirements for these pseudo-3D CEST experiments, this projected measurement time is not feasible in practice. .
c Due to minimal sampling requirements for these pseudo-3D CEST experiments, this projected measurement time is not feasible in practice.
|
|||
| 15N-CEST | 1086 | 32 h 25 min | 27 h 52 min |
| 13CO-CEST | 299 | 29 h 17 min | 2 h 41 minc |
| 13Car-CEST | 331 | 28 h 35 min | 5 h 42 minc |
| 13Cα-CEST | 454 | 29 h 59 min | 5 h 34 minc |
| 13Cmet-CEST | 980 | 29 h 57 min | 32 h 46 min |
| Total experiment time | 150 h 13 min | 74 h 35 min | |
| NOAH-15N-CEST | 1007 | 52 h 59 min | — |
| NOAH-13CO-CEST | 90 | ||
| NOAH-13Car-CEST | 148 | ||
| NOAH-13Cα-CEST | 196 | ||
| NOAH-13Cmet-CEST | 1025 | ||
The total experiment time saving of the ARCHE-NOAH sequence is 65% corresponding to almost 100 hours (>4 days) of instrument time. Shorter experiment times are especially critical for samples with limited lifetime for which ARCHE-NOAH provides an efficient way to acquire five different CEST types. In fact, the acquisition time of the new supersequence is faster than acquiring the two most often used CEST sequences, 15N-CEST and 13Cmet-CEST, as standalone experiments, while providing three extra CEST experiments that carry unique information, albeit at a lower sensitivity.
One way to compare measurement times and sensitivity is to scale down the signal-to-noise of the 5 standalone experiments to the signal-to-noise of the ARCHE-NOAH experiment and calculate how much experiment time would be needed to match their signal-to-noise. In this case, the time saving is 29%. While in theory this is an accurate way of calculating the time saved by our method, in practice one cannot reduce the experiment time of the standalone experiments to this extent, since each experiment requires a minimal number of total scans. For example, it is not possible to lower the number of scans of the 13CO-CEST by a factor of 10 to obtain the desired experiment time to match the signal-to-noise even with non-uniform-sampling methods. Therefore, in practice the time saving is much closer to 65%.
Additionally, it is worth nothing that if we were to leave out the least sensitive experiment 13CO-CEST, our time savings after applying the correction done above would be 33% for a 15N-13Car-13Cα-13Cmet-CEST sequence. If a second CEST block is left out, the corrected time savings would be 33% for 15N-13Cα-13Cmet-CEST and 36% for a 15N-13Car-13Cmet-CEST. If both 13CO- and 13Car-CEST are left out, the resulting time saving for 15N-13Cmet-CEST is 37% (Table 2). Thus, leaving out one or several CEST blocks that have lower sensitivity only results in a modest time gain, while losing the protein dynamics information contained in those experiments. Nevertheless, our sequence is sufficiently flexible that if one is not interested in a particular CEST module, the module can be left out and one still benefits from an overall time savings with the remaining modules. In practice, this means that if one has a uniformly 15N,13C-labeled protein and is planning to acquire a 15N-CEST and 13C-methyl-CEST, there is no actual time gain by not acquiring all five CEST experiments with ARCHE-NOAH with the added benefit of obtaining complementary protein dynamics information.
| ARCHE-NOAH supersequencea | Time savings (%) | Scaled time savings (%) |
|---|---|---|
| a Each CEST module on the NOAH supersequence is defined in the following way: N = 15N-CEST, CO = 13CO-CEST, Car = 13Car-CEST, Cα = Cα-CEST, and Cmet = Cmet-CEST. | ||
| NOAH-(N-CO-Car-Cα-Cmet) | 65 | 29 |
| NOAH-(N-Car-Cα-Cmet) | 60 | 33 |
| NOAH-(N-Cα-Cmet) | 53 | 34 |
| NOAH-(N-Car-Cmet) | 53 | 36 |
| NOAH-(N-Cmet) | 39 | 37 |
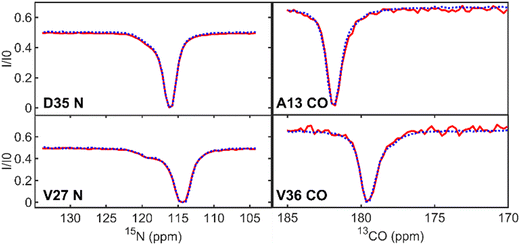
Next, we demonstrate that the CEST profiles that we obtained using the NOAH supersequence are essentially identical to the profiles obtained by the corresponding standalone 1H-start sequences. For backbone CEST profiles (Fig. 2), we have the 15N-CEST and 13CO-CEST experiments, which form the first and second block of the supersequence. Unlike 13CO-CEST, 15N-CEST does not suffer from signal-to-noise losses as is best visible in the baseline of the profile. Despite the increased noise level of the 13CO-CEST profiles, they accurately match the reference profiles and the asymmetry of the main dip reflecting the presence of exchange can still be clearly discerned. A comparison between standalone and NOAH of these two experiments can be found in the ESI† (Fig. S1 and S2).
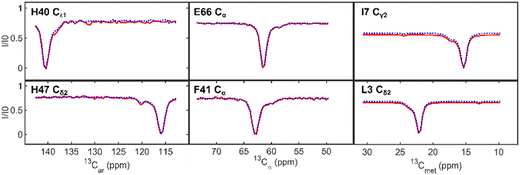
The α-carbons, aromatic carbons, and methyl-CEST profiles are shown in Fig. 3. Their slightly lower signal-to-noise is less of an issue. However, the 13Cα-CEST, both when part of ARCHE-NOAH and as a standalone experiment, has a significant residual water signal that perturbs the baseline of nearby peaks; therefore, we limited the comparison to residues further away from the water resonance with 1H chemical shifts outside the 4.7–4.9 ppm region. The largest difference in our profiles is seen in a slight baseline offset of the methyl carbons. This is because of two 180° pulses that perturb them in the preceding 13Cα-CEST module during the INEPT transfers, tipping the aliphatic carbon magnetization to −z. Despite using selective 180° pulses to get the methyl carbon magnetization back to +z shortly after the INEPT transfers, they result in minor 13C-R1 relaxation rate differences. This difference is about ∼0.1 s−1 and only occurs if the Cα-module is used. The shoulder peaks in all of these profiles, even if minor, are however accurately reproduced. Lastly, although not visible in the profiles themselves, a small systematic chemical shift variation of about 0.005 ppm (4.2 Hz) can be seen in the NOAH versus the standalone spectra. This effect, which is due to extra heating during the decoupling in each CEST block in the supersequence, is inconsequential for the analysis. A comparison between standalone and NOAH is given in the ESI† (Fig. S3–S5).
The majority of the NMR acquisition parameters used to run the 1H-start standalone sequences are the same as those used in the ARCHE-NOAH supersequence. One unavoidable difference between standalone and the NOAH-CEST is the mandatory use of band-selective pulses in the 13CO-, 13Car-, and 13Cα-CEST modules. The replacement of regular broadband pulses by selective pulses is necessary to preserve carbon magnetization for the next modules of the supersequence. We used common selective pulses available in Topspin and their implementation can be automated using the WaveMaker app in Topspin 3.6 and later versions. Additionally, module-specific delays have unique identifiers to improve readability and ease of use and, as suggested in Table 2, the user can specify which of the inner-most CEST modules, i.e.13CO-, 13Car-, or 13Cα-CEST, should be included (or excluded). More information about the selective pulses employed in the ARCHE-NOAH supersequence can be found in Table S1 of the ESI.†
The ARCHE-NOAH can be applied to obtain chemical shifts of an “invisible” excited state of five NMR probes in one single supersequence for a uniformly 15N,13C-labeled sample. Three of these probes (13CO, 13Car, or 13Cα) have been seldom used in the past due to the additional spectrometer time requirements, whereas the new NOAH sequence collects them for “free”. Moreover, the total time of the ARCHE-NOAH with all five modules (53 h) is less than acquiring the 15N- and 13Cmet-CEST experiments as standalone 1H-start experiments (62 h). The chemical shifts from the probes obtained from our sequence provide a unique window into the structure of excited states of proteins.26 Although our method focused on covering most NMR probes encountered in amino-acid residues, a variation of our sequence with the four CEST modules (15N, 13CO, 13Car, and 13Cα) can be directly applied to uniformly 15N,13C-labeled RNA/DNA samples. In this case, the 15N, 13CO, and 13Car modules report on the nitrogenous base and the 13Cα module covers the ribose moiety offering a comprehensive view of their dynamics.27,28
In summary, the ARCHE-NOAH approach introduced here merges five different CEST modules (15N-CEST, 13CO-CEST, 13C-aromatic-CEST, 13Cα-CEST, and 13C-methyl-CEST) into a single supersequence with only a single recovery delay. The reported time savings are about 65%, which amounts to a saving of about 100 hours of high-field NMR instrument time. Hence, it is most advantageous to use ARCHE-NOAH when experiment time is of the essence; for example, due to protein stability, for the screening of protein dynamics in active enzyme-ligand systems, or when studying a protein under different experimental conditions, such as variable temperature or pressure. The only requirement is a uniformly 15N,13C-labeled protein sample, which can be the same sample used for backbone and side-chain resonance assignments and can be produced using a well-established and most economic labelling strategy. Moreover, the band-selective pulses used in the supersequence are readily available on standard Bruker spectrometers. The resulting spectra and CEST profiles compare well with the standalone experiments and are void of artifacts. The ARCHE-NOAH sequence facilitates the exploration also of less commonly studied atomic sites in amino-acid residues, providing novel complementary information on protein dynamics never studied before. It offers the comprehensive analysis of biomolecular dynamics and exchange processes that one would otherwise miss or simply choose not to pursue due to instrument time constraints.
We thank Ms Xinyao Xiang for providing the Im7 sample used in this work. This work was supported by the U.S. National Science Foundation (grant MCB-2103637 to R. B.). All the NMR experiments were performed at the Campus Chemical Instrument Center NMR facility at Ohio State University. The ARCHE-NOAH pulse sequence and AU programs used for processing can be found at https://github.com/RCabreraAllpas.
Conflicts of interest
There are no conflicts to declare.Notes and references
- I. R. Kleckner and M. P. Foster, Biochim. Biophys. Acta, Proteins Proteomics, 2011, 1814, 942–968 CrossRef CAS PubMed.
- T. R. Alderson and L. E. Kay, Cell, 2021, 184, 577–595 CrossRef CAS PubMed.
- P. Vallurupalli, A. Sekhar, T. Yuwen and L. E. Kay, J. Biomol. NMR, 2017, 67, 243–271 CrossRef PubMed.
- Y. Gu, A. L. Hansen, Y. Peng and R. Brüschweiler, Angew. Chem., Int. Ed., 2016, 55, 3117–3119 CrossRef CAS PubMed.
- P. Vallurupalli, G. Bouvignies and L. E. Kay, J. Am. Chem. Soc., 2012, 134, 8148–8161 CrossRef CAS PubMed.
- G. Bouvignies and L. E. Kay, J. Biomol. NMR, 2012, 53, 303–310 CrossRef CAS PubMed.
- A. L. Hansen, G. Bouvignies and L. E. Kay, J. Biomol. NMR, 2013, 55, 279–289 CrossRef CAS PubMed.
- P. Vallurupalli and L. E. Kay, Angew. Chem., 2013, 125, 4250–4253 CrossRef.
- Y. Zhou and D. Yang, J. Biomol. NMR, 2015, 61, 89–94 CrossRef CAS PubMed.
- D. Long, A. Sekhar and L. E. Kay, J. Biomol. NMR, 2014, 60, 203–208 CrossRef CAS PubMed.
- U. Weininger, M. Respondek and M. Akke, J. Biomol. NMR, 2012, 54, 9–14 CrossRef CAS PubMed.
- E. Lanzarotti, L. A. Defelipe, M. A. Marti and A. G. Turjanski, J. Cheminf., 2020, 12, 30 CAS.
- Ē. Kupče and T. D. W. Claridge, Angew. Chem., Int. Ed., 2017, 56, 11779–11783 CrossRef PubMed.
- Ē. Kupče and T. D. W. Claridge, J. Magn. Reson., 2019, 307, 106568 CrossRef PubMed.
- T. D. W. Claridge, M. Mayzel and Ē. Kupče, Magn. Reson. Chem., 2019, 57, 946–952 CrossRef CAS PubMed.
- J. R. J. Yong, A. L. Hansen, Ē. Kupče and T. D. W. Claridge, J. Magn. Reson., 2021, 329, 107027 CrossRef CAS PubMed.
- Ē. Kupče, J. R. J. Yong, G. Widmalm and T. D. W. Claridge, JACS Au, 2021, 1, 1892–1897 CrossRef PubMed.
- A. L. Hansen, E. Kupče, D.-W. Li, L. Bruschweiler-Li, C. Wang and R. Brüschweiler, Anal. Chem., 2021, 93, 6112–6119 CrossRef CAS PubMed.
- F. Tang and E. Hatzakis, Anal. Chem., 2020, 92, 11177–11185 CrossRef CAS PubMed.
- F. Tang, H. S. Green, S. C. Wang and E. Hatzakis, Molecules, 2021, 26, 310 CrossRef CAS PubMed.
- P. Bellstedt, Y. Ihle, C. Wiedemann, A. Kirschstein, C. Herbst, M. Görlach and R. Ramachandran, Sci. Rep., 2014, 4, 4490 CrossRef PubMed.
- C. Wiedemann, P. Bellstedt, A. Kirschstein, S. Häfner, C. Herbst, M. Görlach and R. Ramachandran, J. Magn. Reson., 2014, 239, 23–28 CrossRef CAS PubMed.
- V. M. R. Kakita, K. Rachineni, M. Bopardikar and R. V. Hosur, J. Magn. Reson., 2018, 297, 108–112 CrossRef CAS PubMed.
- R. C. Allpas, A. L. Hansen and R. Brüschweiler, Chem. Commun., 2022, 58, 9258–9261 RSC.
- P. M. Macdonald and R. Soong, J. Magn. Reson., 2007, 188, 1–9 CrossRef CAS PubMed.
- A. J. Baldwin and L. E. Kay, Nat. Chem. Biol., 2009, 5, 808–814 CrossRef CAS PubMed.
- B. Zhao, S. L. Guffy, B. Williams and Q. Zhang, Nat. Chem. Biol., 2017, 13, 968–974 CrossRef CAS PubMed.
- B. Zhao, J. T. Baisden and Q. Zhang, J. Magn. Reson., 2020, 310, 106642 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the NMR methodology used, details of the pulse sequence parameters (standalone and ARCHE-NOAH), side-by-side comparison of standalone vs. ARCHE-NOAH modular spectra, and selective pulse parameters. See DOI: https://doi.org/10.1039/d3cp01580g |
| This journal is © the Owner Societies 2023 |