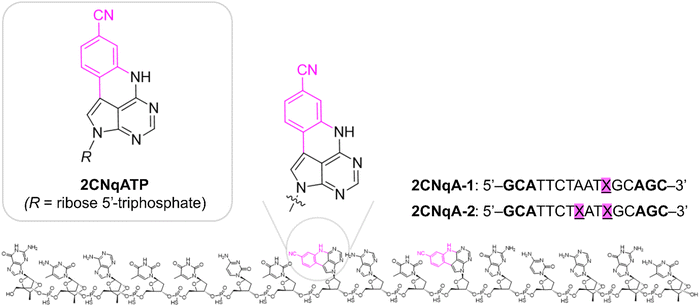
Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multiphoton characterization and live cell imaging using fluorescent adenine analogue 2CNqA†
Jesper R.
Nilsson
a,
Carlos
Benitez-Martin
 a,
Henry G.
Sansom
b,
Pauline
Pfeiffer
a,
Tom
Baladi
a,
Henry G.
Sansom
b,
Pauline
Pfeiffer
a,
Tom
Baladi
 c,
Hoang-Ngoan
Le
c,
Anders
Dahlén
c,
Steven W.
Magennis
c,
Hoang-Ngoan
Le
c,
Anders
Dahlén
c,
Steven W.
Magennis
 b and
L. Marcus
Wilhelmsson
b and
L. Marcus
Wilhelmsson
 *a
*a
aDepartment of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg SE-412 96, Sweden. E-mail: marcus.wilhelmsson@chalmers.se; Tel: +46-31-772 3051
bSchool of Chemistry, University of Glasgow, University Avenue, Glasgow, G12 8QQ, UK
cOligonucleotide Discovery, Discovery Sciences, R&D, AstraZeneca, Gothenburg, Sweden
First published on 17th July 2023
Abstract
Fluorescent nucleobase analogues (FBAs) are established tools for studying oligonucleotide structure, dynamics and interactions, and have recently also emerged as an attractive option for labeling RNA-based therapeutics. A recognized drawback of FBAs, however, is that they typically require excitation in the UV region, which for imaging in biological samples may have disadvantages related to phototoxicity, tissue penetration, and out-of-focus photobleaching. Multiphoton excitation has the potential to alleviate these issues and therefore, in this work, we characterize the multiphoton absorption properties and detectability of the highly fluorescent quadracyclic adenine analogue 2CNqA as a ribonucleotide monomer as well as incorporated, at one or two positions, into a 16mer antisense oligonucleotide (ASO). We found that 2CNqA has a two-photon absorption cross section that, among FBAs, is exceptionally high, with values of σ2PA(700 nm) = 5.8 GM, 6.8 GM, and 13 GM for the monomer, single-, and double-labelled oligonucleotide, respectively. Using fluorescence correlation spectroscopy, we show that the 2CNqA has a high 2P brightness as the monomer and when incorporated into the ASO, comparing favorably to other FBAs. We furthermore demonstrate the usefulness of the 2P imaging mode for improving detectability of 2CNqA-labelled ASOs in live cells.
Introduction
The unique ability of fluorescence techniques to study biomolecules, such as oligonucleotides, under physiological conditions, with excellent sensitivity and spatiotemporal resolution, has made them indispensable tools in various fields of biochemical and biophysical research. To enable efficient detection, however, oligonucleotides require fluorescence labelling, as the canonical nucleobases are virtually non-emissive.1 An interesting alternative to the established labelling method of grafting bulky amphiphilic fluorophores such as ATTO-, AlexaFluor-, or cyanine dyes to the oligonucleotide, is to substitute one or several canonical nucleobases in the sequence for fluorescent nucleobase analogues (FBAs). FBAs are structural and functional mimics of the canonical nucleobases, and therefore act as an integral part of the oligonucleotide by maintaining its native base-pairing pattern and secondary structure.2–4 FBAs have been successfully employed as in-sequence fluorescent reporters in various oligonucleotide investigations, providing information on for instance base pairing dynamics,5 rotational dynamics,6 ligand- and protein binding,7,8 duplex- and triplex formations,9,10 and duplex form transitions.11,12 FBAs have also been used to label and track oligonucleotides with therapeutic potential, such as antisense oligonucleotides (ASOs) and mRNA, in live cells.13,14A recognized drawback of FBAs, however, is that they typically absorb light primarily in the UV region, with comparably low molar absorptivity (ε < 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).15 This means that researchers looking to image FBAs in a biological matrix are left with the options of either exposing the sample to potentially harmful doses of UV light, or settling for suboptimal excitation in the blue end of the visible spectrum. One way to overcome this is by using multiphoton excitation, in which two or more low energy photons can combine to access high-energy excited states.16 In addition to better matching with the FBA absorption, multiphoton excitation through the so-called therapeutic optical window may also allow for deep tissue- and in vivo fluorescence imaging with improved capabilities for optical sectioning by avoiding out-of-focus bleaching.17 The propensity of a molecule to be excited via a two-photon (2P) process is characterized by its 2P absorption cross section (σ2PA, unit: 1 GM = 10−50 cm4 s photon−1), which for strong 2P absorbers that are comparable in size to FBAs is in the range of 100–500 GM.18 However, the required design criteria for FBAs, i.e. being highly fluorescent, having a hydrogen bonding pattern tailored to specific base-pairing, and not perturbing oligonucleotide secondary structures or interactions, result in σ2PA values which typically are 10–100-fold lower. A further challenge with FBAs is that their fluorescence quantum yield (ΦF) generally decreases upon incorporation into oligonucleotides, meaning that the 2P brightness (B2P = ΦF × σ2PA), which ultimately dictates the fluorophore's detectability, decreases proportionally. For these reasons, designing FBAs that are suitable for imaging nucleic acids using 2P excitation is a complex task and successful examples are rare. The uridine-inspired DMAthaU, reported by Nobis et al., was shown to have a brightness (B2PA = 18 GM) which was sufficient for single-molecule detection as a monomer; it was not, however, evaluated inside an oligonucleotide.19 Samaan et al. recently reported the highly fluorescent ABN, with a structure inspired by the pyrimidine nucleobase scaffold and push–pull motif of rhodamine B.20 Although detectable at a single-molecule level inside DNA using one-photon total internal reflection fluorescence (TIRF) microscopy, and thus constituting an excellent example of a bright, in-sequence fluorophore, ABN's ability to mimic the interactions of a specific nucleobase remains to be investigated. An interesting 2P-absorbing FBA that recently emerged from our lab is the pentacyclic adenine analogue pA, which has been characterized both as a monomer and inside DNA, where it replaces adenine with high fidelity.21,22 Its high 2P brightness inside single- and double-stranded DNA (1.3 GM and 0.35 GM, respectively) makes it useful as a base mimicking fluorescent reporter.22
000 M−1 cm−1).15 This means that researchers looking to image FBAs in a biological matrix are left with the options of either exposing the sample to potentially harmful doses of UV light, or settling for suboptimal excitation in the blue end of the visible spectrum. One way to overcome this is by using multiphoton excitation, in which two or more low energy photons can combine to access high-energy excited states.16 In addition to better matching with the FBA absorption, multiphoton excitation through the so-called therapeutic optical window may also allow for deep tissue- and in vivo fluorescence imaging with improved capabilities for optical sectioning by avoiding out-of-focus bleaching.17 The propensity of a molecule to be excited via a two-photon (2P) process is characterized by its 2P absorption cross section (σ2PA, unit: 1 GM = 10−50 cm4 s photon−1), which for strong 2P absorbers that are comparable in size to FBAs is in the range of 100–500 GM.18 However, the required design criteria for FBAs, i.e. being highly fluorescent, having a hydrogen bonding pattern tailored to specific base-pairing, and not perturbing oligonucleotide secondary structures or interactions, result in σ2PA values which typically are 10–100-fold lower. A further challenge with FBAs is that their fluorescence quantum yield (ΦF) generally decreases upon incorporation into oligonucleotides, meaning that the 2P brightness (B2P = ΦF × σ2PA), which ultimately dictates the fluorophore's detectability, decreases proportionally. For these reasons, designing FBAs that are suitable for imaging nucleic acids using 2P excitation is a complex task and successful examples are rare. The uridine-inspired DMAthaU, reported by Nobis et al., was shown to have a brightness (B2PA = 18 GM) which was sufficient for single-molecule detection as a monomer; it was not, however, evaluated inside an oligonucleotide.19 Samaan et al. recently reported the highly fluorescent ABN, with a structure inspired by the pyrimidine nucleobase scaffold and push–pull motif of rhodamine B.20 Although detectable at a single-molecule level inside DNA using one-photon total internal reflection fluorescence (TIRF) microscopy, and thus constituting an excellent example of a bright, in-sequence fluorophore, ABN's ability to mimic the interactions of a specific nucleobase remains to be investigated. An interesting 2P-absorbing FBA that recently emerged from our lab is the pentacyclic adenine analogue pA, which has been characterized both as a monomer and inside DNA, where it replaces adenine with high fidelity.21,22 Its high 2P brightness inside single- and double-stranded DNA (1.3 GM and 0.35 GM, respectively) makes it useful as a base mimicking fluorescent reporter.22
In this work, we continue our exploration of FBAs with multiphoton bioimaging potential by investigating the quadracyclic adenine 2CNqA (Fig. 1), which was previously established as an excellent adenine analogue that is highly emissive as a monomer23 as well as inside DNA,24 RNA,24 and antisense oligonucleotides (ASOs).13 Herein, the multiphoton absorption properties and detectability of 2CNqA as a ribonucleoside triphosphate (2CNqATP, Fig. 1), and incorporated at one or two positions in an ASO (2CNqA-1 and 2CNqA-2, respectively, Fig. 1) are investigated. We also explore the usefulness of multiphoton excitation for visualizing the FBA-labelled oligonucleotides inside live cells using non-linear fluorescence microscopy. ASOs are an emerging class of therapeutic oligonucleotides that induce their therapeutic effect by binding to RNA via Watson–Crick base-pairing and several drugs based on this modality have achieved regulatory approval.25 The motivation for using the ASO design (Fig. 1) in this work is that (1) it internalizes spontaneously into cells, thereby avoiding potentially emission-perturbing formulation protocols such as lipoplexation, (2) the constrained ethyl and phosphorothioate chemistries in the oligonucleotide backbone, which are common ASO modifications, make the oligonucleotide resistant to degradation,26,27i.e., intracellular fluorescence is likely to emanate from intact oligonucleotides rather than degradation products, and (3) it is nontoxic to cells up to at least 3 μM.13
This work contributes to the sparse catalogue of FBA studies carried out in a living environment and specifically explores, for the first time, the feasibility of multiphoton imaging of FBA-labelled oligonucleotides inside live cells.
Methods and materials
Synthesis; general methods and instrumentation
Commercially available reagents were used without further purification. The following chemicals were purchased as premixed oligonucleotide synthesis reagents from Sigma-Aldrich: DCA deblock for ÄKTA, CAP A for ÄKTA, CAP B for ÄKTA, and BTT Activator. 1H (500 MHz) NMR spectra were recorded at 300 K on a Bruker 500 MHz system equipped with a CryoProbe. 31P (202 MHz) NMR spectra were recorded at 300 K on a Bruker 500 MHz system. All shifts are recorded in ppm relative to the deuterated solvent (D2O). Further synthesis details are provided in ESI† Section 1.One-photon photophysical characterization
All measurements were performed at room temperature (ca. 22 °C) in aqueous buffer and concentrations were determined spectroscopically using the Lambert–Beer equation. Details on specific buffer compositions, molar absorptivities, and ASO:RNA duplex preparation are given in the ESI† Section 2. Absorption spectra were recorded on a Cary 50 Bio (Varian Technologies) spectrophotometer using a spectral bandpass of 1.5 nm, integration time of 0.1 s, and optical path length of 3.0 mm. Emission spectra were recorded on a SPEX Fluorolog (Jobin Yvon Horiba) fluorimeter. Samples were excited at 350 nm and emission was collected at a right angle using an integration time of 0.05–0.1 s and monochromator bandpass in the interval 1.0–3.0 nm on both the excitation and emission side. The fluorescence quantum yield and lifetime determinations of 2CNqATP are described in the ESI† Section 3.Fluorescence correlation spectroscopy (FCS)
FCS experiments were carried out on a home-built multiphoton setup.28 The excitation source was a broadband Ti:sapphire laser (Vitara UBB, Coherent). The pulse duration at the objective focus was 7 fs using a 145 nm bandwidth centered on 790 nm. The beam passed through a pulse shaper (MIIPS Box 640, Biophotonic Solutions Inc.) and was guided into an inverted microscope (IX-71, Olympus) and focused on to the sample by a 1.2 NA, 60× water immersion objective (UPlanSApo, Olympus). Samples were placed on a borosilicate glass coverslip (#1.5 thickness Menzel Gläser, Fisher Scientific). To minimise sample evaporation the temperature was maintained at 22 ± 1 °C by an incubator (CU-501, Live Cell Instrument). The fluorescence was captured by the same objective, transmitted through a dichroic (675dcspxr, Chroma), and the emission signal was split into its parallel and perpendicular components which were detected by two avalanche photodiodes (APDs) (PDM50c and $PD-050-CT, Micro Photon Devices). The signal from the APDs was detected by a hardware correlator (ALV-7002, ALV GmbH). All measurements were conducted at a concentration of 100 nM in aqueous buffer, containing 20 mM tris, 150 mM NaCl at pH 7.8. To remove fluorescent impurities the buffer was filtered through activated charcoal before use.Sample preparation for live cell microscopy
Wild-type Huh-7 cells were kindly provided by Prof. Samir El-Andaloussi at Karolinska Institute in Stockholm, Sweden, and verified to be mycoplasma free. The cells were cultured at 37 °C and 5% CO2 in complete medium composed of DMEM GlutaMax low glucose (Gibco, 21885025), with an addition of 10% fetal bovine serum (Gibco, 10270106). For sub-cultivation and seeding, the cells were washed twice with calcium/magnesium-free DPBS (Gibco, 14190250) and exposed to trypsin-0.25% EDTA to detach. For microscopy experiments, 500 μL cells in suspension (0.18 × 106 cells per mL, counted after trypsin neutralization), were seeded on sterile circular glass cover slips (16 mm in diameter, 0.16 mm thickness; Fischer Scientific No. 1 1/2) 24 h prior to compound exposure. The cells were then exposed to the ASOs at a final concentration of 3.0 μM in complete medium for 24 h at 37 °C and 5% CO2 prior to imaging. To facilitate imaging in the upright microscope setup, a cover slip sandwich was constructed using a custom-made cover slip holder, maintaining the glass-adhered cells in 160 μL ASO-containing complete medium, at 37 °C but without a CO2 atmosphere, for the duration of the imaging experiment (ca. 3–4 h).Multiphoton photophysical characterization and live cell fluorescence microscopy
Experiments were carried out on a Zeiss LSM 710 epi-fluorescence microscope equipped with an upright Axio Examiner.Z1 stage setup. For 1P excitation, the instrument-integrated 405 nm diode laser was used, while for 2P excitation, an InSight DeepSee (Spectra-Physics) tunable wavelength laser system, delivering sub-ps pulses at 80 MHz was used. Excitation and emission light were transmitted through a Plan-Apochromat 10×/0.45, ∞/0.17 objective with a working distance of 2.0 mm for the multiphoton absorption cross section and multiplicity determination, and a Plan-Apochromat 20×/1.0 DIC 75 mm, ∞/0.17 water immersion objective with a working distance of 1.9 mm for the cell imaging. The procedure for determining multiphoton absorption cross section and multiplicity is described in ESI† Section 4. For live cell microscopy, a stage with temperature control was used, maintaining 37 °C at the sample. Sample emission travelled through a 405 nm notch dichroic mirror (for 1P) or a 690 nm short pass dichroic mirror (for 2P), both of which transmit essentially all light in the selected emission range (410–680 nm). A pinhole of 1 airy unit was used for the 1P mode, so as to achieve a comparable spatial resolution as for the 2P mode, for which no pinhole was used. To allow for a comparison of 1P vs. 2P emission intensities under these conditions, identical detector gain was applied for both imaging modes. Images were acquired at 16-bit depth and the brightness’ were adjusted evenly across all images.Results and discussion
Synthesis
The details of the 2CNqATP synthesis are described in the ESI† Section 1. Briefly, the 2CNqA nucleoside (1, Fig. S1, ESI†) was prepared as described by Wypijewska et al.,24i.e. by a Silyl–Hilbert–Johnson glycosylation of 4-chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidine, followed by a Suzuki coupling to introduce a cyanophenyl moiety at position 5, and subsequent ring closure in basic conditions to furnish the quadracyclic nucleoside. Triphosphorylation of the 2CNqA nucleoside was then performed following a solid-phase procedure previously described by our group on a cytosine analogue, using the so-called cycloSal phosphoramidite as key reagent.14 2CNqATP was obtained in a good yield, similar to that observed for the former cytosine analogue, thus highlighting the robustness of the triphosphorylation process. The 2CNqA-labelled oligonucleotides were synthesized using a solid-phase oligonucleotide protocol, as described by Nilsson et al.13Photophysics
The photophysical properties of the two ASOs 2CNqA-1 and 2CNqA-2, and the monomeric 2CNqATP were investigated in aqueous buffer by determining their absorption spectra and cross sections, and emission spectra following 1P- and multiphoton excitation (Fig. 2a).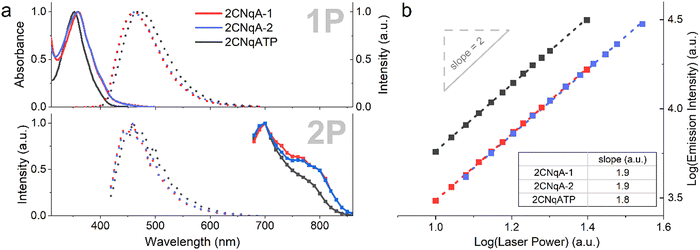 | ||
| Fig. 2 Photophysical properties of 2CNqA-1 (red), 2CNqA-2 (blue), and 2CNqATP (black). (a) Top panel: Normalized 1P absorption spectra (solid lines) and emission spectra (dotted lines, λexc = 350 nm). Bottom panel: normalized 2P emission spectra (dotted lines, λexc = 700 nm) and 2P cross section spectra (boxed solid lines, λem = 460 nm). (b) Absorption multiplicity plot, where integrated emission intensities were determined at various excitation intensities (λexc = 700 nm). Linear regressions (dashed lines, see ESI† Table S1 for details on fitting) resulted in the slopes depicted in the table inset. | ||
The 1P spectra of 2CNqA-1 and 2CNqA-2 were reported in previous work13 and the 2CNqATP exhibits similar spectral characteristics, albeit with a slight blue-shift in absorption (ca. 5 nm, λmax = 352 nm) and red-shift in emission (ca. 10 nm, λmax = 471 nm) (Fig. 2a). The fluorescence quantum yield and lifetime of 2CNqATP were herein determined to 0.48 and 9.9 ns, respectively (see ESI† Section 3), which is in line with reported values for a similar 2CNqA monomer.23 The 2-photon excitation maxima were observed at 700 nm for all compounds, with a pronounced shoulder for the ASOs appearing at ca. 780 nm (Fig. 2a, bottom panel). As expected, excitation into the 2P band using 700 nm pulsed excitation produces the same emission spectra as the corresponding 1P process (compare 1P vs. 2P dashed lines in Fig. 2a); note that the lower signal-to-noise ratio observed in the 2P emission spectra reflects the less-than-optimal performance of the acousto-optic filter monochromator applied in the microscope. The absorption cross sections at 700 nm were determined to be 6.9 GM, 13 GM, and 5.8 GM for 2CNqA-1, 2CNqA-2, and 2CNqATP, respectively, which among reported FBAs is exceptionally high, and the doubled absorption cross section of 2CNqA-2 compared to 2CNqA-1 agrees with the number of 2CNqA molecules in the oligonucleotides. The multiplicities of the absorption events were evaluated by recording the emission at various excitation intensities, which in a log–log plot results in a linear dependency with slopes of 1.9, 1.8, and 1.9 for 2CNqA-1, 2CNqA-2, and 2CNqATP, respectively (Fig. 2b), confirming a two-photon process. Since the therapeutic mechanism of ASOs inside cells involves the ASO binding to its complementary target RNA,29 we decided to also characterize the photophysical properties of 2CNqA-1 and 2CNqA-2 as RNA-bound duplexes and found that the absorption cross sections and multiplicities were largely retained (ESI† Fig S8). This, along with the nearly unchanged fluorescent quantum yield for these ASOs upon RNA binding,13 could make 2CNqA a useful probe for multiphoton techniques aimed at quantifying oligonucleotide concentration via fluorescence readout.
To further explore the non-linear photophysics of the 2CNqA fluorophore as a monomer compared to in an oligonucleotide, we measured fluorescence correlation spectra of the 2CNqATP and 2CNqA-1 using broadband multiphoton excitation (Fig. 3).
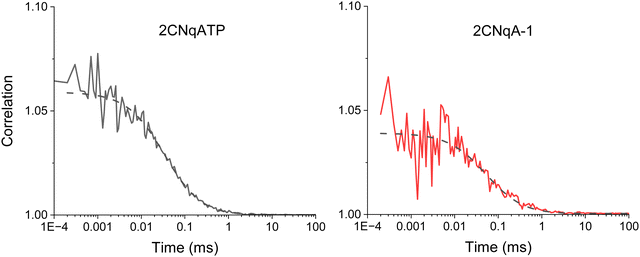 | ||
| Fig. 3 Fluorescence correlation spectroscopy of 100 nM samples of 2CNqA as a monomer (2CNqATP, left) and oligomer (2CNqA-1, right) in aqueous buffer with 11.6 mW excitation power (the dashed lines show the fits; see ESI† Table S2 for details on fitting). For 2CNqATP the CPM was 2.7 ± 0.2 kHz with an average of 7.6 ± 1.3 molecules in the focus; the diffusion time was 36 ± 2.4 μs. For the 2CNqA-1 the CPM was 1.0 ± 0.2 kHz with of 7.8 ± 0.8 molecules in the excitation volume; the diffusion time was 50 ± 5.2 μs. The errors are the sample standard deviation from four 30-minute measurements for 2CNqATP and three 30-minute measurements for 2CNqA-1. | ||
The 2P brightness, expressed as count rate per molecule (CPM), of 2CNqA-1 is 1.0 kHz, which is twice that of the previously reported pA when incorporated into a similar length oligomer.21 The 2CNqATP monomer, with a CPM of 2.7 kHz, is also one of the brightest FBAs yet measured, approaching that of ABN at 7 kHz.20
Fluorescence microscopy
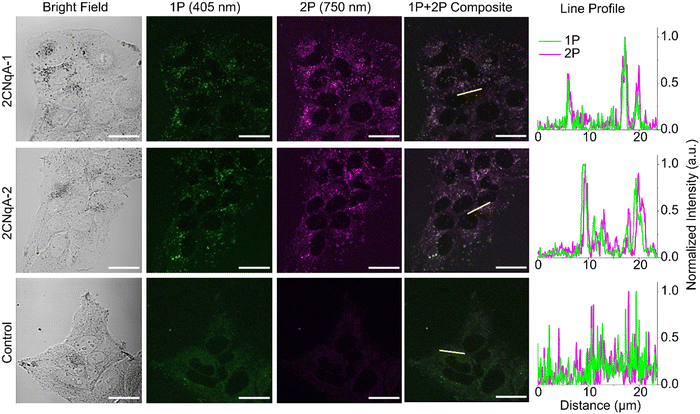
The potential for oligonucleotide-incorporated 2CNqA to be used in 2P live-cell imaging was investigated using a fluorescence microscope capable of confocal operation combined with a pulsed laser for non-linear excitation. Live human liver cells (Huh7) were exposed to 3.0 μM 2CNqA-1 or 2CNqA-2 for 24 h, whereafter bright field, 1P, and 2P images of the same field of view were acquired (Fig. 4).The 2CNqA-labeled ASOs are clearly detectable inside the cells using either 1P or 2P excitation, with a punctuated uptake pattern consistent with endo/lysosomal localization observed in the ASO-exposed samples (Fig. 4). The excellent overlap in the composite images, further illustrated by the alignment of the intensity profiles (Fig. 4, yellow lines and line profiles), shows that the 1P and 2P fluorescence, as expected, are colocalized in the images of the cells. In contrast, the emission from the control cells (autofluorescence only) is comparably low and evenly distributed throughout the cytosol. The poor colocalization in the control samples is likely due to different chemical species in the cells contributing to 1P vs. 2P autofluorescence.
Fluorophore detectability in biological samples depends on the intensity ratio of fluorophore emission to cell autofluorescence. To investigate this for the 2CNqA-labeled ASOs using 1P vs. 2P excitation, we imaged the cells by optical sectioning (z-stacks). Basing the analysis on z-stacks, rather than on single images, cancels out potential perturbing effects caused by small misalignments of the 1P and 2P beam paths in the microscope. Scanning through the cell monolayer and masking out extracellular regions in the ensuing image analysis (see ESI† Section 7 for details), allowed for quantification of the total intracellular emission per area unit for the different compound exposures and imaging modes (Fig. S11, ESI†). The ratio of the total intracellular emission from the 2CNqA ASO-exposed cells to that from the corresponding control cells gives a measure of detectability (Fig. 5).
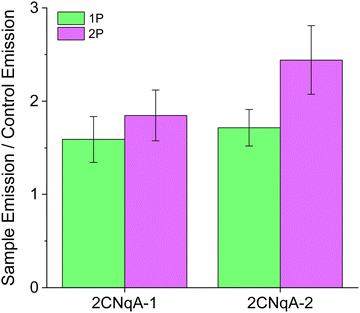 | ||
| Fig. 5 Quantification of 2CNqA detectability in live Huh7 cells using 1P and 2P fluorescence microscopy. Instrument settings and cell exposure conditions were identical to those used in Fig. 4. Sample Emission refers to the total intracellular emission from 2CNqA-ASO exposed cells while Control Emission is the total intracellular emission from non-exposed cells (see ESI† Section 7 for further details). | ||
Although the excitation intensity dependence for the two imaging modes is different (Iem ∝ Iexc for 1P and Iem ∝ Iexc2 for 2P), the detectability ratios in Fig. 5 are independent of the applied laser powers, as they are divided by the control emission acquired with the same settings. In addition to fluorophore brightness, this ratio also depends on experimental parameters such as compound concentration, uptake efficiency, and autofluorescence levels. A plausible explanation for the modest detectability ratios observed here (for both excitation modes) could be elevated autofluorescence levels caused by a lack of CO2 during image acquisition. Nevertheless, since these factors are identical for 1P and 2P excitation, the relative detectability for the two imaging modes can be compared directly. Here, it is seen that 2P excitation performs better (ca. 40% higher for 2CNqA-2) compared to 1P (Fig. 5). Knowing that prolonged imaging can result in fluorophore bleaching, we also included a comparison of the 1P vs. 2P photobleaching kinetics of 2CNqATP using the microscope setup. It was seen that at excitation powers that result in similar 1P and 2P emission intensities, the 2P emission remains stable over time, while the 1P signal decreases significantly (Fig. S12, ESI†). This illustrates the benefit of the 2P imaging mode's ability to avoid out-of-focus excitation. Finally, it deserves mentioning that a so-called descanned type detection was used for the imaging in this work, which produces representative images for samples that exhibit low scattering (e.g. the herein used cell monolayer) using either 1P or 2P excitation. For samples with higher optical density, however, the option of non-descanned detection – uniquely available to 2P operation, will further improve the FBA's detection.
Conclusions
In this work, the quadracyclic adenine analogue 2CNqA was characterized as a multiphoton fluorescent probe and applied as an antisense oligonucleotide-incorporated fluorescent reporter for live cell imaging. We conclude that 2CNqA absorbs via a two-photon process with absorption cross section maxima at 700 nm of 5.8 GM, 6.9 GM, and 13 GM for the monomer 2CNqATP, singly, and doubly labelled ASOs, respectively, which for an FBA are among the highest values reported. These values are also largely retained in the corresponding ASO:RNA duplex. An excellent 2P brightness of 2CNqA was further demonstrated using fluorescence correlation spectroscopy, where it exhibits a CPM of 2.7 kHz as a monomer and 1.0 kHz when incorporated inside the ASO. Regarding imaging applicability, we show that the in-cell detectability increases by up to 40%, and that excessive 2CNqA photobleaching can be avoided by using 2P excitation instead of 1P, thus improving on a major drawback typically associated with FBA bioimaging. We anticipate that 2CNqA will be an attractive option for researchers looking to study oligonucleotides with maintained native behavior in optically dense biological matrices, such as in tissue or in vivo. Having herein established the multiphoton characteristics of 2CNqA, current efforts in our lab focus on investigating 2P imaging of this base analogue inside other types of oligonucleotides in combination with more challenging sample matrices.Author contributions
J. R. N. conceived and coordinated the study, performed the live cell microscopy, and wrote the paper. C.B.-M. characterized the two-photon properties of the compounds. H. G. S., with guidance from S. W. M., designed, performed, and evaluated the FCS measurements. P. P. carried out the cell culture work and the 1P characterization of 2CNqATP. Synthesis, purification, and purity analysis of the ASOs and 2CNqATP was performed by T. B. and H.-N. L., respectively, with guidance from A. D.. L. M. W. supervised the project as a whole. All authors provided feedback and input on the manuscript.Conflicts of interest
L.M.W., J.R.N., T.B., and P.P. own shares in Stealth Labels Biotech AB (LanteRNA); J.R.N. is also an employee there. A.D. are T.B. may own shares and are employees at AstraZeneca.Acknowledgements
The authors gratefully acknowledge the support from FoRmulaEx – Nucleotide Functional Drug Delivery Research Consortium, the Swedish Foundation for Strategic Research [IRC15-0065], and the support of the EPSRC for funding a studentship (H.G.S.). We furthermore thank the Centre for Cellular Imaging (CCI) at the University of Gothenburg and the National Microscopy Infrastructure, NMI (VR-RFI 2019-00217) for providing assistance in microscopy. Professor Elin K. Esbjörner is also acknowledged for providing materials and infrastructure for cell culture. Lastly, Mr Torbjörn Jönsson at the in-house workshop facility at Chalmers is recognized for his skilled contribution in creating the sample plates for the microscopy measurements.References
- C. E. Crespo-Hernández, B. Cohen, P. M. Hare and B. Kohler, Chem. Rev., 2004, 104, 1977–2020 CrossRef PubMed.
- W. Xu, K. M. Chan and E. T. Kool, Nat. Chem., 2017, 9, 1043–1055 CrossRef CAS PubMed.
- L. M. Wilhelmsson, Q. Rev. Biophys., 2010, 43, 159–183 CrossRef CAS PubMed.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579–2619 CrossRef CAS PubMed.
- A. Karimi, R. Börner, G. Mata and N. W. Luedtke, J. Am. Chem. Soc., 2020, 142, 14422–14426 CrossRef CAS PubMed.
- N. Grytsyk, L. Richert, P. Didier, D. Dziuba, S. Ciaco, V. Mazzoleni, T. Lequeu, M. Mori, Y. Tor, L. Martinez-Fernandez, R. Improta and Y. Mély, Int. J. Biol. Macromol., 2022, 213, 210–225 CrossRef CAS PubMed.
- T. Kumagai, B. Kinoshita, S. Hirashima, H. Sugiyama and S. Park, ACS Sens., 2023, 8, 923–932 CrossRef CAS PubMed.
- C. Steinmetzger, C. Bäuerlein and C. Höbartner, Angew. Chem., Int. Ed., 2020, 59, 6760–6764 CrossRef CAS PubMed.
- A. A. Martí, S. Jockusch, Z. Li, J. Ju and N. J. Turro, Nucleic Acids Res., 2006, 34, e50 CrossRef PubMed.
- A. K. Shchyolkina, D. N. Kaluzhny, O. F. Borisova, M. E. Hawkins, R. L. Jernigan, T. M. Jovin, D. J. Arndt-Jovin and V. B. Zhurkin, Nucleic Acids Res., 2004, 32, 432–440 CrossRef CAS PubMed.
- A. F. Füchtbauer, M. S. Wranne, M. Bood, E. Weis, P. Pfeiffer, J. R. Nilsson, A. Dahlén, M. Grøtli and L. M. Wilhelmsson, Nucleic Acids Res., 2019, 47, 9990–9997 CrossRef PubMed.
- B. Dumat, A. F. Larsen and L. M. Wilhelmsson, Nucleic Acids Res., 2016, 44, e101 CrossRef PubMed.
- J. R. Nilsson, T. Baladi, A. Gallud, D. Baždarević, M. Lemurell, E. K. Esbjörner, L. M. Wilhelmsson and A. Dahlén, Sci. Rep., 2021, 11, 11365 CrossRef CAS PubMed.
- T. Baladi, J. R. Nilsson, A. Gallud, E. Celauro, C. Gasse, F. Levi-Acobas, I. Sarac, M. R. Hollenstein, A. Dahlén, E. K. Esbjörner and L. M. Wilhelmsson, J. Am. Chem. Soc., 2021, 143, 5413–5424 CrossRef CAS PubMed.
- D. Dziuba, P. Didier, S. Ciaco, A. Barth, C. A. M. Seidel and Y. Mély, Chem. Soc. Rev., 2021, 50, 7062–7107 RSC.
- R. K. P. Benninger and D. W. Piston, Curr. Protoc. Cell Biol., 2013, 59, 11–24 Search PubMed.
- E. Hemmer, N. Venkatachalam, H. Hyodo, A. Hattori, Y. Ebina, H. Kishimoto and K. Soga, Nanoscale, 2013, 5, 11339–11361 RSC.
- S. de Reguardati, J. Pahapill, A. Mikhailov, Y. Stepanenko and A. Rebane, Opt. Express, 2016, 24, 9053–9066 CrossRef CAS PubMed.
- D. Nobis, R. S. Fisher, M. Simmermacher, P. A. Hopkins, Y. Tor, A. C. Jones and S. W. Magennis, J. Phys. Chem. Lett., 2019, 10, 5008–5012 CrossRef CAS PubMed.
- G. N. Samaan, M. K. Wyllie, J. M. Cizmic, L.-M. Needham, D. Nobis, K. Ngo, S. Andersen, S. W. Magennis, S. F. Lee and B. W. Purse, Chem. Sci., 2021, 12, 2623–2628 RSC.
- R. S. Fisher, D. Nobis, A. F. Füchtbauer, M. Bood, M. Grøtli, L. M. Wilhelmsson, A. C. Jones and S. W. Magennis, Phys. Chem. Chem. Phys., 2018, 20, 28487–28498 RSC.
- M. Bood, A. F. Füchtbauer, M. S. Wranne, J. J. Ro, S. Sarangamath, A. H. El-Sagheer, D. L. M. Rupert, R. S. Fisher, S. W. Magennis, A. C. Jones, F. Höök, T. Brown, B. H. Kim, A. Dahlén, L. M. Wilhelmsson and M. Grøtli, Chem. Sci., 2018, 9, 3494–3502 RSC.
- A. Foller Larsen, B. Dumat, M. S. Wranne, C. P. Lawson, S. Preus, M. Bood, H. Gradén, L. M. Wilhelmsson and M. Grøtli, Sci. Rep., 2015, 5, 12653 CrossRef CAS PubMed.
- A. Wypijewska del Nogal, A. F. Füchtbauer, M. Bood, J. R. Nilsson, M. S. Wranne, S. Sarangamath, P. Pfeiffer, V. S. Rajan, A. H. El-Sagheer, A. Dahlén, T. Brown, M. Grøtli and L. M. Wilhelmsson, Nucleic Acids Res., 2020, 48, 7640–7652 CrossRef PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS PubMed.
- P. S. Pallan, C. R. Allerson, A. Berdeja, P. P. Seth, E. E. Swayze, T. P. Prakash and M. Egli, Chem. Commun., 2012, 48, 8195–8197 RSC.
- C. A. Stein, C. Subasinghe, K. Shinozuka and J. S. Cohen, Nucleic Acids Res., 1988, 16, 3209–3221 CrossRef CAS PubMed.
- D. Nobis, H. G. Sansom and S. W. Magennis, Methods Appl. Fluoresc., 2023, 11, 017001 CrossRef PubMed.
- A. Kilanowska and S. Studzińska, RSC Adv., 2020, 10, 34501–34516 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cp01147j |
| This journal is © the Owner Societies 2023 |