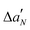Open Access Article
Open Access ArticleThe anticancer peptide LL-III alters the physico-chemical properties of a model tumor membrane promoting lipid bilayer permeabilization†
Marco
Campanile
,
Rosario
Oliva
,
Gerardino
D’Errico
,
Pompea
Del Vecchio
and
Luigi
Petraccone
 *
*
Department of Chemical Sciences, University of Naples Federico II, Via Cintia 4, 80126 Naples, Italy. E-mail: luigi.petraccone@unina.it
First published on 9th December 2022
Abstract
LL-III is an anticancer peptide and has the ability to translocate across tumor cell membranes, which indicates that its action mechanism could be non-membranolytic. However, the exact mechanism through which the peptide gains access into the cell cytoplasm is still unknown. Here, we use a plethora of physico-chemical techniques to characterize the interaction of LL-III with liposomes mimicking the lipid matrix of the tumor cell membrane and its effect on the microstructure and thermotropic properties of the membrane. Furthermore, the effect of the presence of Ca2+ cations at physiological concentration was also investigated. For comparison, the interaction of LL-III with liposomes mimicking the normal cell membrane was also studied. Our results show that the peptide selectively interacts with the model tumor cell membrane. This interaction does not disrupt the lipid bilayer but deeply alters its properties by promoting lipid lateral reorganization and increasing membrane permeability. Overall, our data provide a molecular level description of the interaction of the peptide with the model tumor membrane and are fully consistent with the non-membranolytic action mechanism.
1. Introduction
Cancer is a group of diseases characterized by excessive and uncontrolled proliferation of cells and represents one of the major causes of human death.1 The currently available treatments are based on radiotherapy, chemotherapy and surgery which can lead to a plethora of side effects, mainly due to the lack of specificity.2 In the search for more effective and selective therapeutics, interest has been developed towards host defense peptides (HDPs), a class of natural compounds identified in most living organisms as the first line of defense of the innate immune system against a broad range of pathogens such as bacteria, virus and fungi.3 HDPs were historically investigated mainly for their potential application as antimicrobial agents to address the problem of antibiotic resistance. However, after the discovery of the potent antitumor activity of rabbit macrophage peptides MCP-1 and MCP-2 in 1985, an increasing number of peptides were shown to selectively target tumor cells and thus classified as anticancer peptides (ACPs).4 Most of these peptides act by inducing cell membrane disruption but some of them can translocate into the cytoplasm, where they interact with intracellular targets. However, the exact action mechanism of ACPs is still under debate and several models have been proposed to describe it; nonetheless, a major role is known to be played by the tumor membrane lipid matrix.5 Indeed, different from healthy eukaryotic cells, malignant cells are characterized by a net negative charge on the external surface. Cell transformation involves, in fact, a loss of membrane asymmetry due to the exposure of the outer leaflet of the anionic lipid phopshoserine (PS), a lipid which is exclusively located in the inner leaflet under physiological conditions.6 The exposure of PS molecules allows the interaction of their head groups with substances contained in the extracellular matrix such as calcium ions. Previous studies have shown that Ca2+ is mainly attracted by negatively charged membranes containing PS.7,8 It has been shown that the preferential coordination of Ca2+ to this lipid head group alters the physicochemical properties of the lipid bilayer by increasing the rigidity and lipid packing and inducing membrane thickening.9The antimicrobial and anticancer peptide LL-III (VNWKKILGKIIKVVK) is a member of the family of Lasioglossins, three bioactive peptides extracted from the venom of the bee Lasioglossum laticeps. It is an amphipathic pentadecapeptide characterized by a naturally amidated N-terminus and a net charge of +6 below pH 7.4 (Fig. 1).
 | ||
| Fig. 1 The LL-III sequence in an extended conformation with amino acid side chains highlighted. The color code is as follows: yellow for hydrophobic, green for polar uncharged, blue for the polar positively charged residues. The peptide structure at pH 6.5 was generated using the online tool “PepDraw” developed by the Tulane University (Lousiana, USA) available at https://pepdraw.com/.10 | ||
Previous studies showed that LL-III exhibits a low toxicity towards healthy eukaryotic cells and is active in vitro against some cancer cell lines such as human lymphoblastic leukemia, human promyelocytic leukemia, human cervix carcinoma and human colon adenocarcinoma.11 Particularly, it has been reported that this peptide can translocate across tumor cell membranes in a non-disruptive manner, suggesting that its action mechanism involves the recognition of one or more intracellular targets.12 However, the exact mechanism through which the peptide reaches the cell cytoplasm is still unknown. Here, we have biophysically characterized LL-III interaction with model tumor membranes in order to clarify its membrane perturbing effects and to provide a reasonable explanation for the translocation mechanism. We have selected the lipid pair 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-rac-phosphoseryne (POPS) to mimic the basic characteristics of the tumor membrane lipid matrix. Particularly, we chose POPS as a representative of phosphatidylserines for two main reasons: it is characterized by a Tm of 14 °C, which guarantees that at room temperature it is in a liquid crystalline phase like POPC13 and it shows higher miscibility with phosphatidylcholines when compared, for example, to the saturated lipid DPPS.14,15 Moreover, as the extracellular matrix contains a Ca2+ concentration of ∼1–2 mM and no description of the effects of its coordination to PS lipids on ACP binding is currently available, we performed all our experiments both in the absence and in the presence of a physiological concentration of Ca2+.16 For comparison, the interaction of LL-III with the liposome of POPC/Chol mimicking the normal cell membrane was also studied. In this work, the ability of LL-III to selectively recognize and perturb the model tumor membrane was shown and the effect of Ca2+ on such interactions was revealed to be only marginal. Our results are compatible with a peptide surface binding mode as no deep penetration into the hydrophobic core has been observed. As shown in our calorimetric data, the main effect of the peptide is to induce lipid membrane lateral reorganization, likely due to the specific interaction with the anionic component PS, which involves the progressive substitution of the coordinated Ca2+. These membrane physicochemical property alterations allow LL-III to permeabilize the membrane (as shown in our leakage experiment), providing an explanation for its translocation mechanism across the tumor membrane. Overall, our data are consistent with the non-membranolytic action mechanism.
2. Materials and methods
2.1 Materials
The peptide lasioglossin-III (LL-III) with the sequence NH2-VNWKKILGKIIKVVK-CONH2 was purchased from Primm Srl (Milano, Italy) with a purity of >95%. The lipids cholesterol (Chol), 1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine (DPPC), 1-palmitoyl-2oleoyl-sn-glycero-3-phosphatidylserine (POPS) and 1-palmitoyl-2-oleoyl-glycero-3-phosphatidylcholine (POPC) as well as spin-labeled phosphatidylcholines used as EPR probes (1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphatidylcholine, nPC-SL, n = 5, 7, 10, and 14) were provided by Avanti Polar Lipids (Alabaster, United States). The fluorescent probes, 1,6-diphenylhexatriene (DPH) and 6-dodecanoyl-2-dimethylaminonaphthalene (Laurdan), chloroform, methanol, dimethylformamide (DMF), cacodylic acid, calcium chloride (CaCl2), sodium chloride (NaCl), sodium hydroxide (NaOH) and carboxyfluorescein (CF) were acquired from Merck (Darmstadt, Germany). Sodium cacodylate buffer (10 mM) was prepared by solubilizing cacodylic acid in deionized water and adjusting the pH to 6.5 or 7.0 by adding an appropriate amount of NaOH. The pH values of 6.5 and 7.0 were selected to mimic tumor and healthy cell extracellular matrices, respectively.172.2 Liposome preparation
Lipids were weighed in a darkened glass vial and solubilized in a chloroform/methanol mixture (2/1 v/v). A thin layer of an appropriate lipid mixture was obtained by removing the organic solvent with a gentle nitrogen flux. The sample was kept drying under vacuum for at least 4 hours in order to promote the evaporation of the remaining traces of organic solvents. The lipid film was then hydrated by adding sodium cacodylate buffer at the desired pH with or without 1 mM CaCl2 at a temperature major than the lipid transition temperature. The sample was vigorously mixed to obtain multilamellar vesicles (MLVs). To incorporate DPH in the lipid vesicles, a solution of the probe in chloroform was added to the lipid mixture in the thin layer formation step at a lipid/DPH mole ratio of 150. The same procedure was employed to prepare vesicles containing Laurdan, by adding a probe solution in DMF to achieve a lipid/Laurdan mole ratio of 30. For electron paramagnetic resonance (EPR) measurements, appropriate aliquots of nPC-SL solution in ethanol (1% w/w) were added to the lipid mixture in organic solvents to obtain 2% mol mol−1 of spin-labelled phosphatidylcholines over total lipids. When required for the experimental technique, MLVs were extruded at least 27 times through a porous polycarbonate membrane to obtain ∼100 nm large unilamellar vesicles (LUVs) using a Mini-Extruder (Avanti Polar Lipids), equipped with two glass syringes of 1 mL. The LUV size was checked by determining the hydrodynamic diameter thanks to dynamic light scattering measurements performed on a Zetasizer Nano ZS from Malvern Instruments (Malvern, United States). LUVs containing carboxyfluorescein (CF) were prepared by hydrating the lipid film with a solution 40 mM CF in sodium cacodylate buffer and the obtained MLVs were extruded as previously described. The CF in solution was removed by means of gel-filtration, passing the suspension through a G25 Sephadex medium column (1.45 × 5.0 cm). As elution buffer sodium cacodylate 10 mM in the presence of 1 mM CaCl2 and 150 mM NaCl was employed to prevent the vesicles from disrupting due to osmotic stress. The fractions containing the vesicles were collected and unified. As the procedure involves the loss of lipids, their concentration was determined by performing Stewart assay.18 The selected compositions to mimic the tumor cell membranes were POPC/POPS 8/2 mol mol−1 and DPPC/POPS 8/2 mol mol−1 while POPC/Chol 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mol mol−1 was used for the healthy eukaryotic cell membrane. The appropriate amounts of a peptide solution were added to the lipid suspensions to obtain the desired lipid to peptide (L/P) mole ratios for the experiments.
2 mol mol−1 was used for the healthy eukaryotic cell membrane. The appropriate amounts of a peptide solution were added to the lipid suspensions to obtain the desired lipid to peptide (L/P) mole ratios for the experiments.
2.3 Steady-state fluorescence spectroscopy
Fluorescence experiments were performed on a Horiba Fluoromax-4 from Horiba Scientific (Edison, USA) at a controlled temperature of 25 °C and in 10 mM sodium cacodylate buffer at pH 6.5 or 7.0.2.4 Circular dichroism (CD)
To evaluate the LL-III secondary structure, the far-UV spectra of the peptide at a fixed concentration of 15 μM were recorded using a Jasco 1500 spectropolarimeter from Jasco Corporation (Tokyo, Japan) in the absence or in the presence of POPC/POPS or POPC/Chol LUVs at a L/P ratio of 100. All the samples were prepared in 10 mM sodium cacodylate buffer at pH 6.5 and 1 mM CaCl2. A quartz 0.1 cm path-length cuvette was employed, and the temperature was fixed at 25 °C. The spectra were recorded between 200 and 260 nm with steps of 0.5 nm, a scan rate of 50 nm min−1, a bandwidth of 4.0 nm and a response time of 2 s. Each spectrum was the average of 5 measurements.2.5 Electron paramagnetic resonance spectroscopy (EPR)
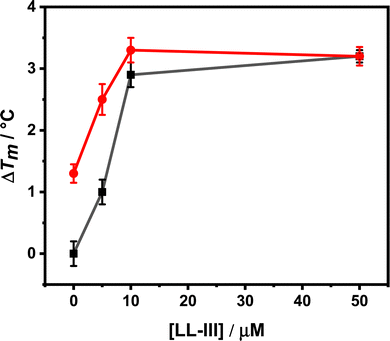
The EPR spectra of nPC-SL in POPC/POPS MLV suspensions were recorded in the absence or in the presence of LL-III at a L/P mole ratio of 10. The total lipid concentration in each sample was 10 mM. Samples were prepared in 10 mM sodium cacodylate buffer in the absence or in the presence of 1 mM CaCl2. EPR experiments were performed using a 9 GHz Bruker Elexys E-500 spectrometer (Bruker, Rheinstetten, Germany). Capillaries containing the samples (∼25 μL) were placed in a standard 4 mm quartz sample tube. The instrumental settings were as follows: sweep width, 100 G; resolution, 1024 points; modulation frequency, 100 kHz; modulation amplitude, 1.0 G; time constant, 20.5 ms; and incident power, 5.0 mW. Several scans, typically 8, were accumulated to improve the signal-to-noise ratio. A quantitative analysis of nPC-SL spectra was performed by determining the acyl chain order parameter, S, and the nitrogen isotropic hyperfine coupling constant, , as described in the literature.22 The differential order parameters, ΔS, and differential nitrogen isotropic hyperfine coupling constants,
, as described in the literature.22 The differential order parameters, ΔS, and differential nitrogen isotropic hyperfine coupling constants, 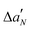 , were calculated subtracting the values determined for POPC/POPS MLVs alone from the values determined for each sample. The experiments were performed in triplicates.
, were calculated subtracting the values determined for POPC/POPS MLVs alone from the values determined for each sample. The experiments were performed in triplicates.
2.6 Differential scanning calorimetry (DSC)
The heat capacity curves of DPPC/POPS vesicles in the absence and or in the presence of LL-III at L/P ratios of 100, 50 and 10 were recorded by means of a nano-DSC equipment from TA Instruments (New Castle, USA) equipped with two twin gold capillary cells of 300 μL. The total lipid concentration in each sample was 500 μM and MLVs were used in all the experiments as the associated calorimetric peaks have a higher resolution.23 Samples were prepared in 10 mM sodium cacodylate buffer in the absence or in the presence of 1 mM CaCl2. The temperature interval was 25–55 °C and the scan rate was set to 1 °C min−1. The capillary cells were subjected to a pressure of 3 atm to prevent bubble formation. A buffer only scan was recorded with the same settings and subtracted from the sample thermograms prior to the calorimetric peak analysis. For each sample, at least four heating and cooling scans were recorded to assess reproducibility and reversibility. The experiments were performed in triplicates and the average ΔHm and Tm and the standard deviations were calculated.3. Results
3.1 Binding of LL-III to model membranes
First, we evaluated the ability of LL-III to bind to the cancer cell model membrane by means of steady-state fluorescence spectroscopy. We monitored the variations in the emission of the LL-III Trp residue upon the addition of POPC/POPS LUVs. In addition, to explore the effect of Ca2+ on the binding constant, the experiments were performed both in the absence (Fig. 2, panel A) and in the presence (Fig. 2, panel B) of 1 mM of this cation. In the former case, the ionic strength was kept constant by adding 3 mM NaCl.As shown in Fig. 2, in both cases, an enhancement of fluorescence intensity and a blue shift of about 23 nm were observed upon the addition of POPC/POPS vesicles (at the highest concentration), revealing that the Trp residue experiences a more rigid and hydrophobic environment.24 These results are consistent with an interaction of the peptide with the POPC/POPS vesicles and further suggest a partial insertion of the peptide into the lipid bilayer. To estimate the effect of Ca2+ on binding, we evaluated the mole fraction partition constants (Kx) in the presence and in the absence of the cation by fitting the corresponding binding isotherms (insets of Fig. 1) as previously reported.19 Interestingly, the coordination of Ca2+ to the lipid head groups only marginally affects the partition equilibrium, with a reduction of Kx from 6.5 ± 1.4 × 105 to 4.8 ± 0.7 × 105. Overall, the coordination of Ca2+ to the POPC/POPS membrane does not hamper the LL-III membrane interaction.
We next explored the ability of LL-III to change its conformation upon binding with the POPC/POPS vesicle by means of circular dichroism measurements. Fig. 3 shows the CD spectra of the peptide in the absence and in the presence of lipid vesicles at a lipid to peptide mole ratio L/P of 100 (panel A) and the helical wheel projection of LL-III (panel B).
 | ||
| Fig. 3 Far-UV circular dichroism spectra of 15 μM LL-III alone (black curve) and in the presence of POPC/POPS LUVs at a lipid/peptide molar ratio of 100 (red curve). All the spectra were recorded at 25 °C in 10 mM sodium cacodylate buffer at pH 6.5 in the presence of 1 mM CaCl2. The approximate unfolded and folded LL-III structures were built using the software PyMol Version 2.5.2 available at https://pymol.org/2/(A).25 Helical wheel projection of LL-III with the hydrophobic amino acids colored in yellow, polar uncharged amino acids in green and polar positively charged amino acids in blue. The helical wheel projection was constructed using the online tool NetWheels available at https://netwheels.herokuapp.com/. The hydrophobic moment μH was calculated using the software MPEx available on https://blanco.biomol.uci.edu/mpex according to the Wimley-White interfacial hydrophobicity scale (B).26,27 | ||
The examination of the spectra reveals that in the presence of 1 mM CaCl2 the peptide is essentially unstructured as observed under neat buffer conditions (Fig. S1, ESI†), whereas in the presence of POPC/POPS vesicles a drastic change of the spectral features is observed. Particularly, upon vesicle addition, a shift of the minimum from 200 nm to about 208 nm is recorded, accompanied by the appearance of a second minimum around 222 nm. In addition, a positive band raises around 200 nm. These changes reveal that LL-III adopts an alpha helical structure upon the interaction with the model membrane. Furthermore, to elucidate the main characteristics of the peptide conformation, we drew the helical wheel projection and calculated the hydrophobic moment μH, a measure of the structure amphipathicity. Fig. 3 panel B shows that the LL-III polar and hydrophobic residues are well segregated in the helix (μH = 5.93), allowing for an effective interaction with phospholipids to occur at the membrane–water interface.
Finally, for comparison, the interaction of LL-III with POPC/Chol liposomes mimicking the normal cell membrane was also verified. In contrast with POPC/POPS, the addition of POPC/Chol vesicles results in a very small decrease of the fluorescence intensity and no significant wavelength shift (Fig. S2 panel A, ESI†). Furthermore, no conformational changes were observed, suggesting a negligible interaction of LL-III with the eukaryotic-like vesicle (Fig. S2 panel B, ESI†); thus, we did not perform additional experiments with this lipid composition.
3.2 Effect of LL-III on the lipid bilayer microstructure and hydration properties
To explore in detail the effect of peptide binding on the microstructure of the membrane polar head group region and the hydrophobic core, appropriate fluorescent probes were incorporated in the POPC/POPS LUVs. Particularly, the fluorescent probes Laurdan and DPH were selected as it is known that they partition at the height of glycerol (just below the head group region) and in the hydrophobic core of the bilayer, respectively. Fluorescence anisotropy provides information about the membrane order. Indeed, anisotropy depends on the rotational mobility of the probes and increases if the motion is hindered as an effect of membrane local stiffening. Fig. 4 shows the approximate location of the probes (panel A) and the millianisotropy (〈r〉) values of DPH (panel B) and Laurdan (panel C) as a function of the peptide concentration both in the absence and in the presence of Ca2+.Interestingly, upon increasing the LL-III concentration in the absence of Ca2+, the anisotropy of both DPH and Laurdan increases, indicating a reduction of the two probes rotational mobility and revealing that the peptide rigidifies the model membrane both in the head group and the hydrophobic core region. Moreover, the increase of DPH anisotropy also suggests that the peptide, most likely, does not penetrate deep into the hydrophobic core of the membrane (otherwise, a reduction of anisotropy should occur, as observed for the membrane penetrating P9Nal(SS) peptide28). Similar observations were already reported for the peptide trematocine (which is supposed to act through a carpet mechanism), where an increase of DPH anisotropy embedded in PC/PG liposomes was observed.29 Thus, the reported results are compatible with a surface binding mode of LL-III to PC/PS liposomes. Fig. 4, panel C, shows that the trend of Laurdan anisotropy in the presence of Ca2+ is similar to that observed in the absence of the cation, revealing that the LL-III surface binding mode is not significantly affected by Ca2+ coordination in the head group region. Remarkably, the increase of the anisotropy for DPH (Fig. 4 panel B) is quite modest in the presence of Ca2+, suggesting that the presence of the cation partially counterbalances the rigidification effect of LL-III in the membrane hydrophobic core. Indeed, the millianisotropy values of DPH in the absence and in the presence of Ca2+ are 113 and 112, respectively, suggesting a slight fluidization effect of calcium of the inner hydrophobic core of the membrane (this effect was also inferred from the EPR data, see Fig. 6, panel A). These results indicate that the cation binding to the head group region increases the probe rotational mobility. This could be due to the induction of a local negative curvature that can increase the spacing among lipid chains. Thus, the almost constant r of DPH reported in Fig. 4, panel B, can only be due to a counterbalance effect between the peptide (increase of anisotropy) and calcium (decrease of anisotropy).
We next monitored the hydration changes of the lipid head group region taking advantages of the Laurdan emission properties. Indeed, the Laurdan spectra are the result of the emission from two distinct electronic states: a locally excited state, which prevails in hydrophobic environments and is characterized by a higher energy transition and a solvent-relaxed state, which is shifted to lower energies as a consequence of stabilizing solvent interactions.30 The relative contribution from these two emission states can be quantitatively described by the parameter GP (see Materials and methods section) which provides information on the membrane head group region hydration state. Commonly, the GP values depend on the particular analyzed lipid system in terms of composition and phase and are typically comprised between about 0.6 (less hydrated gel phase) and −0.2 (more hydrated liquid phase).31Fig. 5 shows the GP values recorded for Laurdan embedded in POPC/POPS vesicles, upon increasing the LL-III concentration both in the absence and in the presence of Ca2+.
In the absence of Ca2+, the negative GP value reveals that the model membrane is partially hydrated as expected for a lipid bilayer in the fluid phase. The addition of Ca2+ leads to a significant dehydration of the membrane surface in agreement with previous studies.7,9 Upon LL-III addition, the membrane surface hydration state is altered under both conditions but in an opposite way: starting from the Ca2+ containing sample, LL-III binding results in a partial re-hydration whereas in the absence of the cation it leads to a reduction of the water content. Interestingly, the two curves obtained converge at the highest LL-III concentration suggesting that in the presence of Ca2+ the peptide progressively replaces the coordinated cation from its binding sites mainly represented by phosphatidylserine head groups. This observation is consistent with a preferential interaction of LL-III with the PS anionic lipid.
To further investigate the effect of LL-III on the lipid bilayer microstructure, an EPR investigation was undertaken. Four spin-labelled phosphatidylcholines bearing the cyclic nitroxide label at various positions along the sn-2 acyl chain (nPC-SL, n = 5, 7, 10, 14) were alternatively incorporated in the membrane (2% mol mol−1 over total lipid). This approach allows the lipid organization to be investigated in detail as a function of the distance from the bilayer surface. The nPC-SL spectra (reported in Fig. S3 and S4, ESI†) were quantitatively analyzed as described in the literature in order to obtain the nitrogen hyperfine coupling constant  , which is related to the polarity of the microenvironment in which the nitroxide label is embedded, and the order parameter S, a measure of the orientational ordering of the labelled segment of the acyl chain with respect to the normal to the bilayer surface.22 The POPC/POPS bilayers present S and
, which is related to the polarity of the microenvironment in which the nitroxide label is embedded, and the order parameter S, a measure of the orientational ordering of the labelled segment of the acyl chain with respect to the normal to the bilayer surface.22 The POPC/POPS bilayers present S and 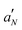 profiles typical of membranes in the Ld state.32 Both parameters decrease with n, the more dramatic variation being observed for 14PC-SL. Thus, the bilayer inner core is rather disordered and hydrophobic, see Table S1 (ESI†).
profiles typical of membranes in the Ld state.32 Both parameters decrease with n, the more dramatic variation being observed for 14PC-SL. Thus, the bilayer inner core is rather disordered and hydrophobic, see Table S1 (ESI†).
Fig. 6 shows the variation of these two parameters due to the presence of LL-III or/and Ca2+. The peptide causes an increase of the acyl chain order at all label positions.33 Particularly, the highest ΔS is observed for 5PC-SL, the value becoming lower as the label is stepped down along the acyl chain (Fig. 5, panel A). This evidence clearly points to a peptide positioning at the bilayer interface. The concurrent increase of the local polarity (Fig. 5, panel B) supports this hypothesis and can be ascribed to the polarity of the peptide, which is highly charged, and/or to the peptide hydration which is not completely lost upon membrane interaction.
The adsorption of Ca2+ ions on the bilayer surface causes opposite effects on lipid ordering depending on the distance from the head group: the positive ΔS values are observed for 5 and 7PC-SL, while ΔS turns to slightly negative values for 10 and 14PC-SL (Fig. 5, panel A). These evidences suggest that the divalent cations lead to a lipid crowding which results in the stiffening of the more external bilayer section. This perturbation of the lipid organization leaves the tail termini free to move and to assume disordered conformations. No Ca2+ effect is observed on the 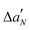 profile (Fig. 6, panel B). The expected polarity increase due to the ion interaction could be proposed to be balanced by the bilayer de-hydration observed in the fluorescence experiments.34
profile (Fig. 6, panel B). The expected polarity increase due to the ion interaction could be proposed to be balanced by the bilayer de-hydration observed in the fluorescence experiments.34
The effect of LL-III and Ca2+ seems to be additive in that their simultaneous presence causes the highest increase of the lipid ordering observed in this work. Even in this case, the stronger variations are observed for 5PC-SL, indicating the peptide interaction to occur at the bilayer interface. The peptide determines the  profile, independent of the Ca2+ presence.
profile, independent of the Ca2+ presence.
3.3 Effect of LL-III binding on the lipid bilayer thermotropic properties
To further explore the effects of peptide binding on the model membrane physicochemical properties, we performed DSC experiments both in the absence and in the presence of Ca2+. As POPC is characterized by a transition temperature below 0 °C, this lipid was replaced by DPPC to obtain a system with a transition temperature suitable for the instrument.13 The resulting thermograms are shown in Fig. 7. The transition temperatures Tm and the transition enthalpies ΔHm are listed in Table 1.| L/P ratio | Buffer | +1 mM CaCl2 | ||
|---|---|---|---|---|
| ΔHm (kJ mol−1) | T m (°C) | ΔHm (kJ mol−1) | T m (°C) | |
| / | 28.8 ± 0.9 | 37.1 ± 0.2 | 30.2 ± 2.6 | 38.5 ± 0.1 |
| 100 | 25.1 ± 1.6 | 38.1 ± 0.2 | 26.0 ± 0.6 | 39.5 ± 0.3 |
| 50 | 21.8 ± 1.4 | 39.9 ± 0.2 | 22.4 ± 1.1 | 40.4 ± 0.2 |
| 10 | 16.7 ± 0.5 | 40.5 ± 0.1 | 18.2 ± 2.3 | 40.4 ± 0.1 |
The DSC thermogram of DPPC/POPS vesicles under neat buffer conditions is characterized by a broad transition centered at 37.1 °C which is due to the melting of the lipid acyl chain and the associated increase in conformational disorder (Fig. 7, panel A). The addition of Ca2+ determines the shift of the main transition to higher temperatures and an increase of the transition enthalpy (Table 1 and Fig. 7, panel B). Furthermore, the calorimetric peak is sharper indicating that the lipids melt more cooperatively. Collectively, these data show that Ca2+ stabilizes the gel phase accordingly with previous studies.9 Upon LL-III addition, a similar trend was observed both in the presence and in the absence of Ca2+: on increasing the peptide concentration, the transition peak is shifted at higher temperatures and the enthalpy progressively decreases (Table 1). At the maximum LL-III concentration employed (L/P 10), the peak shows a double component DSC profile, with a shoulder extending towards lower temperatures (Fig. 7). The main peak is centered at about 40.4 °C, a temperature which is similar to that of DPPC alone under similar conditions (40.9 °C), suggesting a loss of membrane lateral organization homogeneity due to the formation of lipid domains.35 This observation is consistent with a preferential interaction of the peptide with the negatively charged PS and the formation of regions enriched in this component which melt at lower temperatures and regions enriched of DPPC which melt at higher temperatures. We further compared the observed ΔTm on increasing the peptide concentration in the absence and in the presence of Ca2+ (Fig. 8). The trends are very similar and at the maximum LL-III concentration explored the ΔTm values ultimately converge. This result supports the hypothesis that LL-III competes with Ca2+ for the binding to the PS head group and, at higher concentrations, replaces it.
3.4 LL-III effects on membrane permeability
Next, as our data suggest that LL-III does not disrupt the model membrane, we have evaluated its ability to permeabilize the lipid bilayer in the presence of Ca2+. To this end, we have performed leakage assay by encapsulating 30 mM carboxyfluorescein (CF) into POPC/POPS LUVs. At a high local concentration, the CF emission is self-quenched.36 This allows its release in solution to be monitored by following the fluorescence enhancement as a function of time both in the absence and in the presence of an increasing peptide concentration. The kinetic curves and the leakage percentage at 1500 s are reported in Fig. 9. In the absence of LL-III, the fluorescence signal is steady and low due to the self-quenching of the entrapped CF (Fig. 9, panel A). The profile of the recorded kinetic curves is significantly dependent on the peptide concentration added, a feature more consistent with a transient permeabilization rather than equilibrium pore formation.37 To compare the data at the various peptide concentrations employed, the leakage percentage at 1500 s was reported (Fig. 9, panel B). 50% leakage was observed at a LL-III concentration of 126 nM, corresponding to a bound peptide to lipid mole ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. Therefore, LL-III is able to exert a permeabilizing effect on the model membrane even at a low bound concentration.
1000. Therefore, LL-III is able to exert a permeabilizing effect on the model membrane even at a low bound concentration.
4. Discussion
LL-III is an antimicrobial and anticancer peptide and has the ability to translocate across tumor cell membranes, probably according to a non-membranolytic action mechanism. However, the molecular basis of the interaction of LL-III with pathogenic membranes and the mechanism through which it gains access to the cell cytoplasm are still unknown. In this paper, a series of physico-chemical techniques have been used to fully characterize the interaction of the anticancer LL-III peptide with the model tumor membrane and the effect of the peptide on the membrane microstructure and thermotropic properties. To mimic the negatively charged surface of tumor cell membranes, we modelled the lipid matrix using POPC/POPS liposomes.6 Moreover, as the extracellular matrix contains a millimolar concentration of Ca2+,16 which is known to preferentially interact with phosphatidylserines altering lipid bilayer properties,7,9 the effect of this ion on the LL-III interaction was explored by performing the experiments in the absence and in the presence of Ca2+. In addition, the effects of LL-III on POPC/Chol liposomes mimicking healthy cell membrane were also investigated for comparison. The results of the steady-state fluorescence experiments clearly show that the peptide selectively interacts with the POPC/POPS membrane and the presence of Ca2+ has a negligible effect on the partition constant. Furthermore, Trp emission (Fig. 2) is enhanced and blue shifted upon lipid addition, revealing that this residue is partially inserted into the membrane and suggesting that LL-III partitions at the water–membrane interface. Furthermore, CD experiments show that upon vesicle binding the peptide partially adopts an α-helical structure upon membrane interactions which involves the segregation of polar and hydrophobic residues on different faces of the structure (μH = 5.93). This feature is common with other active anticancer peptides in which the adoption of an amphipathic alpha helical structure favors the interaction with the target membrane.38 To obtain additional information on the peptide binding mode, we monitored the effect of the peptide on different regions of the membrane by following the changes in the fluorescence anisotropy of the two probes Laurdan and DPH embedded in the lipid bilayer. Specifically, Laurdan provides information on the membrane surface dynamical properties whereas DPH mainly reflects the hydrophobic core of the membrane. Our results clearly show that peptide binding induces a rigidification of the membrane both in the absence and in the presence of Ca2+, suggesting that the peptide is, most likely, localized on the membrane surface at the water–membrane interface and does not penetrate deeply in the membrane hydrophobic core. This hypothesis was further supported by EPR measurements showing that the magnitude of peptide-induced rigidification reduces moving from the surface to the lipid acyl chain termini. Interestingly, DPH anisotropy appears to be less sensitive to the addition of LL-III in the presence of Ca2+ (Fig. 4, panel B), suggesting that the presence of the cation partially counterbalances the rigidification effect of LL-III in the hydrophobic region of the membrane. The increase of the probe rotational mobility could be due to a local negative curvature induced by cation coordination which results in an increase of spacing between the lipid acyl chain and in a reduction of the order parameter as sensed by the EPR probe 14-PCSL. Indeed, previous studies have shown that the presence of Ca2+ at sub-millimolar and millimolar bulk concentrations can induce the formation of invaginations on the surface of membranes containing anionic lipids (e.g. DOPE/DOPS bilayers)40 as a consequence of lipid clustering and of a reduction of the spontaneous curvature.39 The Laurdan GP measurement in the presence and absence of Ca2+ allowed us to monitor the hydration state of the membrane surface on increasing the peptide concentration. We found that in the absence of LL-III the hydration of the membrane is very different after the addition of Ca2+, being much more dehydrated compared to the neat buffer. Despite this difference, the addition of the peptide leads to a bound state under both conditions showing a very similar GP value, suggesting that LL-III preferentially interacts with PS and progressively substitutes the coordinated Ca2+ from its binding sites. To verify the ability of the peptide to preferentially bind the anionic lipids, we monitored the effect of peptide addition on the thermotropic properties of DPPC/POPS vesicles by means of DSC measurements. The examination of the DSC profiles shows that LL-III turns the single component DSC profile of DPPC/POPS into a multicomponent DSC profile, which consists of two overlapping peaks, indicating phase segregation. This observation is fully consistent with a preferential interaction of the peptide with the lipid anionic component resulting in a lateral redistribution of membrane lipids and domain formation. Overall, the calorimetric, EPR and fluorescence data clearly reveal that LL-III binds on the membrane surface and does not significantly perturb the membrane integrity, indicating a non-membranolytic action mechanism (i.e., a mechanism that does not involve permanent damages in the membrane as a cause of cell death),41 in agreement with the previously reported data showing that the tumor cell membrane is still intact after peptide translocation into the cytoplasm.12 In addition, we can speculate about the peptide binding mode. Starting from the peptide helical projection (Fig. 3 panel B) and assuming that the helix is oriented parallel on the membrane surface with its hydrophobic side (where Trp occupies a central position) pointing to the membrane hydrophobic core, we suggest that Lys4, Lys9 and Lys15 are the amino acids most likely involved in an electrostatic interaction with PS head groups whereas Lys5 and Lys12 probably interact with water molecules.Then, we evaluated the ability of the peptide to permeabilize the membrane by performing a leakage assay after encapsulating carboxyfluorescein in the lipid vesicles. Our data (Fig. 9) indicate that LL-III favors the release of the vesicle content with a concentration-dependent rate and plateau level, which is compatible with membrane permeabilization due to a transient and local bilayer destabilization,37 rather than a true equilibrium pore formation which would lead to the complete loss of the vesicles content even at the lowest peptide concentration employed.42 We propose that this effect is a consequence of the clustering of anionic lipids promoted by peptide binding. Indeed, the presence of packing irregularities at the lipid domain boundaries can locally lower the permeability barrier allowing LL-III translocation into the cancer cell cytoplasm.43
In summary, we performed detailed biophysical characterization of the interaction of LL-III with a lipid bilayer containing the anionic lipid phosphatidylserine and in the presence of a physiological concentration of Ca2+. Overall, our data are consistent with the observed non-membranolytic activity of LL-III. In addition, we showed that the presence of calcium (at a physiologically relevant concentration) has only minor effects on the LL-III mechanism of action. Furthermore, the obtained results provide a molecular level description of the interaction of the peptide with a lipid bilayer mimicking the lipid matrix of a tumor cell membrane, and such information can be useful in the development of new peptides that can act as anticancer agents.
Abbreviations
| ACPs | Anticancer peptides |
| CD | Circular dichroism |
| CF | Carboxyfluorescein |
| Chol | Cholesterol |
| DMF | Dimethylformamide |
| DPH | 1,6-Diphenylhexatriene |
| DPPC | 1,2-Dipalmitoyl-sn-glycero-3-phosphatidylcholine |
| DSC | Differential scanning calorimetry |
| EPR | Electron paramagnetic resonance |
| GP | General polarization |
| HDPs | Host defense peptides |
| Laurdan | 6-Dodecanoyl-2-dimethylaminonaphthalene |
| LL-III | Lasioglossin-III |
| LUVs | Large unilamellar vesicles |
| MLVs | Multilamellar vesicles |
| nPC-SL | 1-Palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphatidylcholine |
| POPC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine |
| POPS | 1-Palmitoyl-2-oleoyl-sn-glycero-3-rac-phosphoseryne |
| PS | Phosphatidylserine |
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors are grateful to the Italian MUR for granting Rosario Oliva with a research associated position (PON R&I 2014-2020, CUP: E65F21003250003).References
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Cancer statistics, Ca-Cancer J. Clin., 2022, 72, 7–33 CrossRef PubMed.
- F. Harris, S. R. Dennison, J. Singh and D. A. Phoenix, On the selectivity and efficacy of defense peptides with respect to cancer cells, Med. Res. Rev., 2013, 33, 190–234 CrossRef CAS PubMed.
- R. E. Hancock and G. Diamond, The role of cationic antimicrobial peptides in innate host defences, Trends Microbiol., 2000, 8, 402–410 CrossRef CAS PubMed.
- M. Xie, D. Liu and Y. Yang, Anti-cancer peptides: classification, mechanism of action, reconstruction and modification, Open Biol., 2020, 10, 200004 CrossRef CAS PubMed.
- D. Gaspar, A. S. Veiga and M. A. R. B. Castanho, From antimicrobial to anticancer peptides. A review, Front. Microbiol., 2013, 4, 1–16 Search PubMed.
- W. Chiangjong, S. Chutipongtanate and S. Hongeng, Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application (Review), Int. J. Oncol., 2020, 57, 678–696 CrossRef CAS PubMed.
- A. Melcrová, S. Pokorna, S. Pullanchery, M. Kohagen, P. Jurkiewicz, M. Hof, P. Jungwirth, P. S. Cremer and L. Cwiklik, The complex nature of calcium cation interactions with phospholipid bilayers, Sci. Rep., 2016, 6, 38035 CrossRef PubMed.
- M. L. Valentine, A. E. Cardenas, R. Elber and C. R. Baiz, Calcium-Lipid Interactions Observed with Isotope-Edited Infrared Spectroscopy, Biophys. J., 2020, 118, 2694–2702 CrossRef CAS PubMed.
- A. Melcrová, S. Pokorna, M. Vošahlíková, J. Sýkora, P. Svoboda, M. Hof, L. Cwiklik and P. Jurkiewicz, Concurrent Compression of Phospholipid Membranes by Calcium and Cholesterol, Langmuir, 2019, 35, 11358–11368 CrossRef PubMed.
- PepDraw: a tool to draw peptide primary structure and calculate theoretical properties, accessed November 2, 2022, https://pepdraw.com/.
- V. Cerovský, M. Budesínský, O. Hovorka, J. Cvacka, Z. Voburka, J. Slaninová, L. Borovicková, V. Fucík, L. Bednárová, I. Votruba and J. Straka, Lasioglossins: three novel antimicrobial peptides from the venom of the eusocial bee Lasioglossum laticeps (Hymenoptera: Halictidae), ChemBioChem, 2009, 10, 2089–2099 CrossRef PubMed.
- J. Slaninová, V. Mlsová, H. Kroupová, L. Alán, T. Tůmová, L. Monincová, L. Borovičková, V. Fučík and V. Ceřovský, Toxicity study of antimicrobial peptides from wild bee venom and their analogs toward mammalian normal and cancer cells, Peptides, 2012, 33, 18–26 CrossRef PubMed.
- Phase Transition Temperatures for Glycerophospholipids, https://avantilipids.com/tech-support/physical-properties/phase-transition-temps, accessed November 4, 2022.
- Miscibility of Phospholipid Binary Mixtures, https://avantilipids.com/tech-support/physical-properties/miscibility, (accessed November 4, 2022).
- A. Hinderliter, R. L. Biltonen and P. F. F. Almeida, Lipid Modulation of Protein-Induced Membrane Domains as a Mechanism for Controlling Signal Transduction, Biochemistry, 2004, 43, 7102–7110 CrossRef CAS PubMed.
- B. Alberts, Molecular biology of the cell, W. W. Norton & Company, New York, 7th edn, 2022 Search PubMed.
- V. Estrella, T. Chen, M. Lloyd, J. Wojtkowiak, H. H. Cornnell, A. Ibrahim-Hashim, K. Bailey, Y. Balagurunathan, J. M. Rothberg, B. F. Sloane, J. Johnson, R. A. Gatenby and R. J. Gillies, Acidity Generated by the Tumor Microenvironment Drives Local Invasion, Cancer Res., 2013, 73, 1524–1535 CrossRef CAS PubMed.
- J. C. M. Stewart, Colorimetric determination of phospholipids with ammonium ferrothiocyanate, Anal. Biochem., 1980, 104, 10–14 CrossRef CAS PubMed.
- A. S. Ladokhin, S. Jayasinghe and S. H. White, How to measure and analyze tryptophan fluorescence in membranes properly, and why bother?, Anal. Biochem., 2000, 285, 235–245 CrossRef CAS PubMed.
- S. H. White, W. C. Wimley, A. S. Ladokhin and K. Hristova, Protein folding in membranes: determining energetics of peptide-bilayer interactions, Methods Enzymol., 1998, 295, 62–87 CAS.
- T. Parasassi and E. Gratton, Membrane lipid domains and dynamics as detected by Laurdan fluorescence, J. Fluoresc., 1995, 5, 59–69 CrossRef CAS PubMed.
- S. Galdiero, A. Falanga, G. Vitiello, M. Vitiello, C. Pedone, G. D’Errico and M. Galdiero, Role of membranotropic sequences from herpes simplex virus type I glycoproteins B and H in the fusion process, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 579–591 CrossRef CAS PubMed.
- R. L. Biltonen and D. Lichtenberg, The use of differential scanning calorimetry as a tool to characterize liposome preparations, Chem. Phys. Lipids, 1993, 64, 129–142 CrossRef CAS.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Springer, New York, 3rd edn, 2006 Search PubMed.
- L. Schrödinger and W. DeLano, The PyMOL Molecular Graphics System (Version 2.5.2).
- A. R. Mól, M. S. Castro and W. Fontes, NetWheels: A web application to create high quality peptide helical wheel and net projections, Bioinformatics, 2018, 416347 Search PubMed.
- C. Snider, S. Jayasinghe, K. Hristova and S. H. White, MPEx: A tool for exploring membrane proteins: MPEx: A Tool for Exploring Membrane Proteins, Protein Sci., 2009, 18, 2624–2628 CrossRef CAS PubMed.
- R. Oliva, M. Chino, K. Pane, V. Pistorio, A. De Santis, E. Pizzo, G. D’Errico, V. Pavone, A. Lombardi, P. Del Vecchio, E. Notomista, F. Nastri and L. Petraccone, Exploring the role of unnatural amino acids in antimicrobial peptides, Sci. Rep., 2018, 8, 8888 CrossRef PubMed.
- G. Della Pelle, G. Perà, M. C. Belardinelli, M. Gerdol, M. Felli, S. Crognale, G. Scapigliati, F. Ceccacci, F. Buonocore and F. Porcelli, Trematocine, a Novel Antimicrobial Peptide from the Antarctic Fish Trematomus bernacchii: Identification and Biological Activity, Antibiotics, 2020, 9, 66 CrossRef CAS PubMed.
- K. Gaus, T. Zech and T. Harder, Visualizing membrane microdomains by Laurdan 2-photon microscopy (Review), Mol. Membr. Biol., 2006, 23, 41–48 CrossRef CAS PubMed.
- T. Parasassi, G. De Stasio, G. Ravagnan, R. M. Rusch and E. Gratton, Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence, Biophys. J., 1991, 60, 179–189 CrossRef CAS PubMed.
- G. Vitiello, A. Falanga, A. Alcides Petruk, A. Merlino, G. Fragneto, L. Paduano, S. Galdiero and G. D’Errico, Fusion of raft-like lipid bilayers operated by a membranotropic domain of the HSV-type I glycoprotein gH occurs through a cholesterol-dependent mechanism, Soft Matter, 2015, 11, 3003–3016 RSC.
- R. Oliva, A. Emendato, G. Vitiello, A. De Santis, M. Grimaldi, A. M. D’Ursi, E. Busi, P. Del Vecchio, L. Petraccone and G. D’Errico, On the microscopic and mesoscopic perturbations of lipid bilayers upon interaction with the MPER domain of the HIV glycoprotein gp41, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 1904–1913 CrossRef CAS PubMed.
- H. A. Hussain, The effect of cation concentration on the nitrogen splitting constant of nitroxide free radicals, Collect. Czech. Chem. Commun., 1990, 55, 2377–2380 CrossRef CAS.
- F. Battista, R. Oliva, P. Del Vecchio, R. Winter and L. Petraccone, Insights into the Action Mechanism of the Antimicrobial Peptide Lasioglossin III, Int. J. Mol. Sci., 2021, 22, 2857 CrossRef CAS PubMed.
- Y. N. Antonenko, G. S. Gluhov, A. M. Firsov, I. D. Pogozheva, S. I. Kovalchuk, E. V. Pechnikova, E. A. Kotova and O. S. Sokolova, Gramicidin A disassembles large conductive clusters of its lysine-substituted derivatives in lipid membranes, Phys. Chem. Chem. Phys., 2015, 17, 17461–17470 RSC.
- W. C. Wimley and K. Hristova, The Mechanism of Membrane Permeabilization by Peptides: Still an Enigma, Aust. J. Chem., 2020, 73, 96–103 CrossRef CAS PubMed.
- S. R. Dennison, M. Whittaker, F. Harris and D. A. Phoenix, Anticancer alpha-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes, Curr. Protein Pept. Sci., 2006, 7, 487–499 CrossRef CAS PubMed.
- C. Allolio and D. Harries, Calcium Ions Promote Membrane Fusion by Forming Negative-Curvature Inducing Clusters on Specific Anionic Lipids, ACS Nano, 2021, 15, 12880–12887 CrossRef CAS PubMed.
- Z. T. Graber, Z. Shi and T. Baumgart, Cations induce shape remodeling of negatively charged phospholipid membranes, Phys. Chem. Chem. Phys., 2017, 19, 15285–15295 RSC.
- V. Teixeira, M. J. Feio and M. Bastos, Role of lipids in the interaction of antimicrobial peptides with membranes, Prog. Lipid Res., 2012, 51, 149–177 CrossRef CAS PubMed.
- W. C. Wimley, Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model, ACS Chem. Biol., 2010, 5, 905–917 CrossRef CAS PubMed.
- R. F. Epand, G. Wang, B. Berno and R. M. Epand, Lipid Segregation Explains Selective Toxicity of a Series of Fragments Derived from the Human Cathelicidin LL-37, Antimicrob. Agents Chemother., 2009, 53, 3705–3714 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cp03528f |
| This journal is © the Owner Societies 2023 |