 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Crystal engineering with 1,3,4-oxadiazole derivatives: on the importance of CH⋯N and CH⋯π interactions†
Muhammad Naeem
Ahmed
 *a,
Maheen
Akhtar
a,
Hina
Andleeb
b,
Muhammad Adnan
Bashir
*a,
Maheen
Akhtar
a,
Hina
Andleeb
b,
Muhammad Adnan
Bashir
 c,
Mahmoud A. A.
Ibrahim
c,
Mahmoud A. A.
Ibrahim
 d,
Peter A.
Sidhom
d,
Peter A.
Sidhom
 e,
Ifzan
Arshad
e,
Ifzan
Arshad
 f,
Muhammad Nawaz
Tahir
g,
Diego M.
Gil
f,
Muhammad Nawaz
Tahir
g,
Diego M.
Gil
 h,
Rosa M.
Gomila
h,
Rosa M.
Gomila
 i and
Antonio
Frontera
i and
Antonio
Frontera
 *i
*i
aDepartment of Chemistry, The University of Azad Jammu and Kashmir, Muzaffarabad 13100, Pakistan. E-mail: drnaeem@ajku.edu.pk
bDepartment of Pharmaceutical Sciences, Bouvé College of Health Sciences, Northeastern University, Boston, Massachusetts 02115, USA
cThe State Key Laboratory of Chemical Oncogenomics, Guangdong Provincial Key Laboratory of Nano-Micro Materials Research, School of Chemical Biology and Biotechnology, Shenzhen Graduate School of Peking University, Shenzhen 518055, China
dComputational Chemistry Laboratory, Chemistry Department, Faculty of Science, Minia University, Minia, Egypt
eDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Tanta University, Tanta, Egypt
fInstitute for Advanced Study, Shenzhen University, Shenzhen 518060, Guangdong, China
gDepartment of Physics, University of Sargodha, Sargodha, Pakistan
hINBIOFAL (CONICET – UNT), Instituto de Química Orgánica, Facultad de Bioquímica, Química y Farmacia, Universidad Nacional de Tucumán, Ayacucho 471, T4000INI, San Miguel de Tucumán, Argentina
iDepartament de Química, Universitat de les Illes Balears, Crta. de Valldemossa km 7.5, 07122 Palma de Mallorca, Baleares, Spain. E-mail: toni.frontera@uib.es
First published on 10th November 2023
Abstract
Three new 1,3,4-oxadiazoles (1–3) have been synthesized. The crystal structure of two of them were solved by single crystal X-ray diffraction analysis and a detailed quantitative analysis of the weak noncovalent interactions have been performed by using DFT calculations. In both compounds, the formation of recurrent H-bonded motifs involving the 1,3,4-oxadiazole is observed. In addition, a variety of CH⋯π interactions are established, involving both aliphatic and aromatic C–H bonds and the π-system of the electron rich tert-butylphenyl ring. QTAIM analysis and NCI plots were used to study the nature and the extent of different intermolecular interactions observed in these structures, which were rationalized using MEP surface plots.
Introduction
For the development of new drugs, nitrogen containing heterocyclic compounds are considered to be the potential source of natural products.1 Oxadiazoles, triazoles, pyridines, indoles, and their derivatives, commonly used as medicines and pesticides,2 are examples of such compounds. Among these, 1,3,4-oxadiazole and its derivatives3,4 have gained attention not only for their significant biological abilities,5,6 including antiviral,7 antifungal,8 insecticidal,9 antibacterial,10–14 and antitumor activities,9 but also for their high activity, good selectivity, and low toxicity. In particular, compounds having 1,3,4-oxadiazole cores display broad-spectrum bioactivities in agriculture, with some related commercial pesticides successfully developed, such as oxadiargyl, oxadiazon, and insecticide metoxadiazone.15 Therefore, the 1,3,4-oxadiazole scaffold is an important pharmacophore in the development of agrochemical research. Due to their crucial role in supramolecular chemistry, molecular recognition and materials chemistry,16 noncovalent interactions are important for structural chemists.17–19 The hydrogen bonding (HB) interactions are extremely important in biological systems as can be shown in nucleic acids where the assembly is controlled by a combination of H-bonds and π⋯π stacking interactions.20 It is well-known that relatively weak intermolecular interactions such as C–H⋯X (X = halogens, O, S, N) hydrogen bonds play a vital role in the crystal packing of different kinds of molecules.21–27In light of this background, we report herein the synthesis and characterization of three new 1,3,4-oxadiazole derivatives 1–3 (Scheme 1). The crystal structures of compounds 1 and 2 were solved by single crystal X-ray diffractions and the computed molecular structure has been investigated by DFT calculations focusing on the energetic analysis of the CH⋯π and H-bonding interactions. The latter form H-bonded motifs where 1,3,4-oxadiazole acts as a double H-bond acceptor.
Moreover, in continuation of our ongoing research on structural properties and biological applications26 of different heterocyclic compounds,28 especially the azole derivative,13 herein we report the structural as well as LOX inhibition studies (ESI†) of three 1,3,4-oxadiazole derivatives having thioether connectivity,4 as shown in Scheme 1. The main advantage of these new derivatives over the already reported derivatives of 2-(benzylsulfanyl)-5-phenyl-1,3,4-oxadiazole is that compound 1 and 2 contain a tertiary butyl group while compound 3 contains 3-FPh which may behave structurally and biologically different.
Experimental
Instrumentation and reagents
All solvents and reagents were purchased from commercial suppliers and used as received unless noted. All products were purified by column chromatography on silica gel and recrystallization. Melting points were determined on a Yanaco melting point apparatus and are not corrected. FT-IR spectra were recorded on a SHIMADZU FTIR-8400S spectrophotometer using the KBr disc method. 1H-NMR (400 MHz) spectra were measured on a Bruker spectrometer in CDCl3 and TMS as internal standard. Chemical shifts of 1H-NMR were reported in parts per million relative to the solvent residual peak (δ 7.26). Chemical shifts of 13C-NMR were reported relative to CDCl3 (δ 77.16). Reaction progress was monitored by thin layer chromatography (TLC). FT-IR spectra were recorded on a SHIMADZU FTIR-8400S spectrophotometer using the KBr disc method. Suitable crystals were coated with paratone 8772 oil and placed on a glass fiber analysed by using a Bruker Kappa APEX-IICCD diffractometer equipped with graphite monochromated Mo-Kα radiation. Good quality single crystals of compounds 1–2 suitable for XRD analysis were grown from a mixture of EtOH and EtOAc (dissolving 150 mg of each compound in 5–10 mL of solvent) by slow evaporation over a period of 48 hours at ambient temperature. The substrates were prepared according to the literature method.4,13General procedure for the synthesis of (1–3)
The compounds (1–3) were synthesized by an already reported procedure4,13 with slight modifications by treating 5-aryl-1,3,4-oxadiazole with aromatic alkyl halides in the presence of potassium carbonate in acetone with simple stirring (Scheme 2). Synthesized compounds were purified by thin layer chromatography (TLC), column chromatography (CC) and recrystallization.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc 7
EtOAc 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); solubility: CHCl3/DMSO; FTIR (KBr, cm−1): 1560 (C
3); solubility: CHCl3/DMSO; FTIR (KBr, cm−1): 1560 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Caro), 1563 (C
Caro), 1563 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1173 (C–O), 1045 (C–O–C oxadiazole), 1H-NMR (400 MHz, CDCl3): δ 8.04–7.95 (m, 2H), 7.55–7.45 (m, 3H), 7.42–7.35 (m, 4H), 4.52 (s, 2H), 1.32 (s, 9H); 13C-NMR (101 MHz, CDCl3): δ 165.81, 164.03, 151.19, 132.48, 131.59, 129.01, 128.84, 126.68, 125.76, 123,73, 36.64, 34.58, 31.26; analysis: for C19H20N2OS found (calculated): C, 70.34 (70.30); H, 6.21 (6.25); N, 8.63 (8.60)%.
N), 1173 (C–O), 1045 (C–O–C oxadiazole), 1H-NMR (400 MHz, CDCl3): δ 8.04–7.95 (m, 2H), 7.55–7.45 (m, 3H), 7.42–7.35 (m, 4H), 4.52 (s, 2H), 1.32 (s, 9H); 13C-NMR (101 MHz, CDCl3): δ 165.81, 164.03, 151.19, 132.48, 131.59, 129.01, 128.84, 126.68, 125.76, 123,73, 36.64, 34.58, 31.26; analysis: for C19H20N2OS found (calculated): C, 70.34 (70.30); H, 6.21 (6.25); N, 8.63 (8.60)%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc 7
EtOAc 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); solubility: CHCl3/DMSO; FTIR (KBr cm−1): 1572 (C
3); solubility: CHCl3/DMSO; FTIR (KBr cm−1): 1572 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Caro), 1521 (C
Caro), 1521 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1010 (C–O–C oxadiazole); 1H-NMR (400 MHz, CDCl3): δ 7.43–7.34 (m, 4H), 7.23 (s, 2H), 4.52 (s, 2H), 3.93 (s, 9H), 1.31 (s, 9H); 13C-NMR (101 MHz, CDCl3): δ 165.71, 163.83, 153.70, 151.22, 132.46, 128.83, 125.75, 118.73, 104.13, 99.99, 60.97, 56.40, 36.67, 34.58, 31.25: analysis: for C20H26N2O4S found (calculated): C, 63.75 (63.72); H, 6.32 (6.30); N, 6.76 (6.71)%.
N), 1010 (C–O–C oxadiazole); 1H-NMR (400 MHz, CDCl3): δ 7.43–7.34 (m, 4H), 7.23 (s, 2H), 4.52 (s, 2H), 3.93 (s, 9H), 1.31 (s, 9H); 13C-NMR (101 MHz, CDCl3): δ 165.71, 163.83, 153.70, 151.22, 132.46, 128.83, 125.75, 118.73, 104.13, 99.99, 60.97, 56.40, 36.67, 34.58, 31.25: analysis: for C20H26N2O4S found (calculated): C, 63.75 (63.72); H, 6.32 (6.30); N, 6.76 (6.71)%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc 7
EtOAc 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); solubility: CHCl3/DMSO; FTIR (KBr cm−1): 1534 (C
3); solubility: CHCl3/DMSO; FTIR (KBr cm−1): 1534 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Caro), 1511 (C
Caro), 1511 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1013 (C–O–C oxadiazole); 1H-NMR (400 MHz, CDCl3): δ 7.88 (d, J = 8.5 Hz, 1H), 7.60–7.53 (m, 2H), 7.38 (dd, J = 8.5, 2.0 Hz, 1H), 7.30 (tdd, J = 7.6, 5.3, 1.8 Hz, 1H), 7.15–7.00 (m, 2H), 4.57 (s, 2H); 13C-NMR (101 MHz, CDCl3): δ 164.64, 163.54, 162.02, 160.04, 138.08, 133.83, 131.54, 131.39 (d, J = 3.4 Hz), 131.21, 130.15 (d, J = 8.2 Hz), 127.59, 124.37 (d, J = 3.7 Hz), 123.03 (d, J = 14.5 Hz), 121.44, 115.65 (d, J = 21.1 Hz), 30.20; analysis: for C15H9Cl2FN2OS found (calculated): C, 50.72 (50.68); H, 2.55 (2.57); N, 7.89 (7.90)%.
N), 1013 (C–O–C oxadiazole); 1H-NMR (400 MHz, CDCl3): δ 7.88 (d, J = 8.5 Hz, 1H), 7.60–7.53 (m, 2H), 7.38 (dd, J = 8.5, 2.0 Hz, 1H), 7.30 (tdd, J = 7.6, 5.3, 1.8 Hz, 1H), 7.15–7.00 (m, 2H), 4.57 (s, 2H); 13C-NMR (101 MHz, CDCl3): δ 164.64, 163.54, 162.02, 160.04, 138.08, 133.83, 131.54, 131.39 (d, J = 3.4 Hz), 131.21, 130.15 (d, J = 8.2 Hz), 127.59, 124.37 (d, J = 3.7 Hz), 123.03 (d, J = 14.5 Hz), 121.44, 115.65 (d, J = 21.1 Hz), 30.20; analysis: for C15H9Cl2FN2OS found (calculated): C, 50.72 (50.68); H, 2.55 (2.57); N, 7.89 (7.90)%.
X-ray data collection and structure refinement
Suitable single crystals of compounds 1–2 were selected for X-ray analyses and diffraction data were collected on a Bruker Kappa APEX-II CCD detector with MoKα radiation at 100 K. Using the SADABS program, semiempirical correction was applied.29 The SHELX program was also used to solve all structures by direct method.30 Positions and anisotropic parameters of all non-H atoms were refined on F2 using the full matrix least-squares technique. The H-atoms were added at geometrically calculated positions and refined using the riding model. The details of crystallographic data and crystal refinement parameters for the compounds 1–2 are given in Table 1.| Crystal parameters | 1 | 2 |
|---|---|---|
| CCDC | 2241333 | 2241334 |
| Chemical formula | C19H20N2OS | C22H26N2O4S |
| M r | 324.43 | 414.51 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 296 | 296 |
| a, b, c (Å) | 10.5007(1), 19.5547(2), 8.6349(1) | 10.4066(1), 24.9033(2), 8.8792(1) |
| β (°) | 97.941(1) | 97.936(1) |
| V (Å3) | 1756.07(3) | 2279.08(4) |
| Z | 4 | 4 |
| Radiation type | Cu Kα | Cu Kα |
| μ (mm−1) | 1.67 | 1.50 |
| Crystal size (mm) | 0.36 × 0.24 × 0.22 | 0.36 × 0.28 × 0.24 |
| Diffractometer | Bruker Kappa APEXII CCD | Bruker Kappa APEXII CCD |
| Absorption correction | Multi-scan (SADABS) | Multi-scan (SADABS) |
| T min, Tmax | 0.545, 0.615 | 0.545, 0.615 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 21438, 3589, 3204 | 31041, 4391, 4023 |
| R int | 0.023 | 0.028 |
(sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ/λ)max (Å−1) θ/λ)max (Å−1) |
0.630 | 0.613 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.041, 0.123, 1.09 | 0.045, 0.137, 1.05 |
| No. of reflections | 3589 | 4391 |
| No. of parameters | 219 | 286 |
| No. of restraints | 12 | 14 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δ〉max, Δ〉min (e Å−3) | 0.19, −0.20 | 0.31, −0.26 |
Theoretical methods
The energies and topological analyses of the supramolecular assemblies investigated herein were computed at the PBE0-D3/def2-TZVP level of theory31–33 using the crystallographic coordinates and the program Turbomole 7.2.34 Grimme's D3 dispersion32 correction has been used since it is convenient for the correct evaluation of noncovalent interactions and especially those involving π-systems. The quantum theory of atoms in molecules (QTAIM)35 and noncovalent interaction (NCI) plot36 reduced density gradient (RGD) isosurfaces have been used to characterize non-covalent interactions. Both methods combined are useful to reveal noncovalent interactions in real space. The wavefunctions needed to generate the NCIPlot surfaces have been computed at the same level of theory using the Turbomole 7.2 program. The NCIPlot index RDG isosurfaces correspond to both favourable and unfavourable interactions, as differentiated by the sign of the second density Hessian eigenvalue and defined by the isosurface color. The color code used in this manuscript is blue and green for attractive interactions (strong and weak, respectively) and yellow and red for weakly and strongly repulsive, respectively. The NCIPlot cubes needed to construct the isosurfaces have been computed by means of the MULTIWFN program.37Hirshfeld surfaces and energetic calculations
The Hirshfeld surface analysis is a very useful method for studying intermolecular interactions which are responsible for the crystal packing of organic and inorganic compounds.38–41 The visualization of the intermolecular contacts in the crystal structure of compounds 1 and 2 was carried out by using the CrystalExplorer21.5 program.42 For the calculations, the input file was obtained from the CIF file obtained from the crystal structure determination. The normalized contact distance (dnorm) combines both de (distance taken from the nearest nucleus outside the surface), di (distance taken from the nearest nucleus inside the surface) and the vdW radii of the atoms involved in the intermolecular contact. The dnorm surfaces are mapped over a fixed colour scale of −0.025 a.u. (red) and +0.55 a.u. (blue). The scale colours used in analysing the main intermolecular contacts in the dnorm map are red (distances shorter than the sum of vdW radii of atoms), blue (distances longer than the sum of vdW radii) and white (distances equal to the sum of vdW radii of atoms involved in the intermolecular interactions). The combination of de and di in the form of two-dimensional fingerprint (FP) plots provides a summary of the contribution to the main intermolecular contacts to the total Hirshfeld surface area.40 We have also analysed an additional coloured property, namely the shape index, based on the local curvature of the surface. This Hirshfeld surface has been performed to explore the existence of C–H⋯π interactions.The energies of intermolecular interactions (Etot) for dimers formed in the crystal structure of both compounds were calculated using the CrystalExplorer 21.5 program.42 The electron density of the molecules for the intermolecular energy calculations has been obtained at the B3LYP/6-31G(d,p) level of theory. The total interaction energy (Etot) is partitioned into Coulombic (Ecoul), polarization (Epol), dispersión (Edis), and repulsión (Erep) energy contributions.
Results and discussion
Synthesis and characterization
The compounds (1–3) under study were synthesized by a simple and faster procedure. Equimolar quantities of the respective 1,3,4-oxadiazole and aryl halides in acetone as solvent in the presence of potassium carbonate as base react by stirring during 2 hours at 45–50 °C, leading to the corresponding 1,3,4-oxadiazole derivatives (1–3). Column chromatography and re-crystallization from ethanol afford the products 1 and 2 as crystalline solids in good yields. Only very small and twinned crystals with low X-ray scattering ability were obtained for compound 3.The 1H NMR (400 MHz, CDCl3) data for all compounds shows a multiplet in the range of 8.00–7.05, corresponding to the nine, six and seven aromatic protons in the molecule. A singlet at 4.56–4.51 indicates the presence of two methylene protons in all the three compounds. Similarly, nine tertiary butyl protons resonate at 1.31 ppm. A singlet at 1.31 also corresponding to the nine tertiary butyl protons in compound 1 while two singlet signals at 3.93 and 1.30 confirms the presence of nine methoxy protons and nine tertiary-butyl protons in compound 2. All carbon resonances fall between 165.81–30.19 ppm.
Description of crystal structures of compounds 1–2 and DFT study
In 2-[(4-tertbutylphenylmethyl)sulfanyl]-5-(3,4,5-trimethoxy-phenyl)-1,3,4-oxadiazole, one of the C-atoms of the methoxy group and the terminal C-atoms of the tertbutyl group are disordered over two set of sites with an occupancy ratio of 0.699(3)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.301(3). In this molecule, the benzene ring of the trimethoxy group A (C14–C19), the 1,3,4-oxadiazole-2-thiol group B (C1/C2/N1/N2/O1/S1) and part of the toluene group C (C3–C9) are planar having r.m.s. deviations of 0.0036, 0.0099 and 0.0028, respectively. The dihedral angles between A/B, B/C and C/A are 5.22(7)°, 76.39(6)° and 81.11(6)°, respectively (Fig. 1).
0.301(3). In this molecule, the benzene ring of the trimethoxy group A (C14–C19), the 1,3,4-oxadiazole-2-thiol group B (C1/C2/N1/N2/O1/S1) and part of the toluene group C (C3–C9) are planar having r.m.s. deviations of 0.0036, 0.0099 and 0.0028, respectively. The dihedral angles between A/B, B/C and C/A are 5.22(7)°, 76.39(6)° and 81.11(6)°, respectively (Fig. 1).
In 2-[(4-tertbutylphenylmethyl)sulfanyl]-5-(phenyl)-1,3,4-oxadiazole, the terminal C-atoms of the tertbutyl group are disordered over two set of cites with an occupancy ratio of 0.869(3)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.131(3). In this molecule, the benzene ring A (C14–C19), the 1,3,4-oxadiazole-2-thiol group B (C1/C2/N1/N2/O1/S1) and part of the toluene group C (C3–C9) are planar having r. m. s. deviations of 0.0022, 0.0126 and 0.0366, respectively. The dihedral angles between A/B, B/C and C/A are 10.59(5)°, 66.66(5)° and 75.98(6)°, respectively. Molecules form dimers via C–H⋯N bonding and these dimers are stacked due to C–H⋯π interactions (Fig. 2).
0.131(3). In this molecule, the benzene ring A (C14–C19), the 1,3,4-oxadiazole-2-thiol group B (C1/C2/N1/N2/O1/S1) and part of the toluene group C (C3–C9) are planar having r. m. s. deviations of 0.0022, 0.0126 and 0.0366, respectively. The dihedral angles between A/B, B/C and C/A are 10.59(5)°, 66.66(5)° and 75.98(6)°, respectively. Molecules form dimers via C–H⋯N bonding and these dimers are stacked due to C–H⋯π interactions (Fig. 2).
Molecular dimers and intermolecular interaction energies
The calculations performed with the CrystalExplorer 21.5 (ref. 42) program reveals three molecular dimers for compounds 1 and 2 which are energetically significant in the stabilization of the crystal packing. Molecules of 1 interact through C–H⋯N and C–H⋯π intermolecular interactions, while the crystal packing of 2 is mainly stabilized by a combination of C–H⋯N, C–H⋯O and C–H⋯π interactions. Table 2 shows pair-wise interaction energies associated to intermolecular hydrogen bonds and C–H⋯π contacts related to three molecular dimers for 1 and 2, along with the geometrical parameters. For 1, the most stabilized dimer (D1, Fig. 3) is formed by two different C–H⋯N hydrogen bonds, one involves the H3B of the methylene group and the N1 atom of the oxadiazole moiety as acceptor and other involving the H5 atom of the phenyl ring and the N2 atom. Dimer D1 is also stabilized by C–H⋯π interactions involving the H17B atom of the tert-butyl group and the centroid of the C4–C9 ring. The contribution of electrostatic and dispersion energies is 37 and 63% towards the stabilization of this dimer.| Compound 2 | |||||||
|---|---|---|---|---|---|---|---|
| a Cg1 and Cg2 are the centroids of the N1/N2/C1/C2/O1 and C4–C9 rings, respectively for 1 and Cg1 and Cg2 are the centroids of the C14–C19 and C4–C9 rings, respectively for 2. b Geometry of intermolecular interactions (Å, °). Scale factors used to determine Etot at the B3LYP/6-31G(d,p) level of theory: Ecoul = 1.019, Epol = 0.651, Edis = 0.901, Erep = 0.811. | |||||||
| D1 | C20–H20A⋯Cg2 | 2.61 | −5.41 | −1.09 | −13.7 | 6.53 | −13.7 |
| D2 | C22–H22C⋯N1 | 2.72/116 | −4.83 | −1.23 | −10.2 | 3.73 | −12.5 |
| C5–H5⋯N2 | 2.74/136 | ||||||
| D3 | C21–H21A1⋯O4 | 2.78/147 | −2.85 | −0.31 | −10.1 | 3.76 | −9.50 |
| C20–H20B⋯Cg1 | 2.98 | ||||||
 | ||
| Fig. 3 Partial view of the crystal packing of compound 1 showing the main dimers (D1–D3) involved in the intermolecular interactions. The intermolecular contacts are shown as dashed lines. | ||
The next most stabilized dimer D2 (Etot = −7.23 kcal mol−1, Fig. 3) is stabilized by intermolecular C–H⋯π interactions. The H18 atom of the phenyl group and the C4–C9 phenyl ring participate in C–H⋯π interactions as a donor and as an acceptor, respectively. The contribution of electrostatic energy is significantly reduced (≈16%) compared to that on dimer D1. This difference could be mainly attributed to the absence of hydrogen bonds in the stabilization of dimer D2. The H6 atom of the phenyl ring is involved in an intermolecular C–H⋯π interaction with the centroid of the oxadiazole ring (Fig. 3). It is important to note that the dispersion energy is dominant (82%) towards the stabilization of this dimer.
In the solid-state structure of 2, we have identified three structural dimers (D1–D3) with significant intermolecular interactions energies (Etot) as shown in Table 2 and Fig. 4. These energies range from −13.7 to 9.50 kcal mol−1. The basic structural dimer D1 (Etot = −13.7 kcal mol−1, Fig. 4) consists of inversion related molecules stabilized by C–H⋯π interactions between the H20A atom of one methoxy group and the centroid of the C4–C9 ring. The dispersion energy contributes about 68% towards the stabilization of dimer D1 in this structure. The second strong dimer D2 (Etot = 12.5 kcal mol−1, Fig. 4) is formed by a combination of C22–H22C⋯N1 and C5–H5⋯N2 hydrogen bonds, involving the N1 and N2 of the oxadiazole ring as acceptor. The contribution of electrostatic and dispersion energy for the stabilization of dimer D2 is 37 and 63%, respectively. Dimer D3 (Etot = −9.50 kcal mol−1) is stabilized by a combination of C21A–H21A1⋯O1 and C20–H20B⋯π intermolecular interactions. The C–H⋯π contact involves the H20B atom of one methoxy moiety and the centroid of the C14–C19 phenyl ring. For the stabilization of this dimer, the dispersion energy contributes 76% (Fig. 4).
 | ||
| Fig. 4 Partial view of the crystal packing of compound 2 showing the main dimers (D1–D3) involved in the intermolecular interactions. The intermolecular contacts are shown as dashed lines. | ||
MEP, QTAIM and NCIPlot study of dimers D1–D3
The dimers described above have been further analyzed using three computational tools: MEP surface plots of the monomers and QTAIM/NCIPlot analysis of the dimers. The utilization of QTAIM and NCIPlot methods combined is useful to reveal interactions in real space.First, the molecular electrostatic potential (MEP) surfaces of compounds 1 and 2 have been computed to investigate the most electrophilic and nucleophilic parts of the molecules. They are represented in Fig. 5, evidencing that the MEP minimum is located at the N-atoms of the 1,3,4-oxadiazole ring (−37.0 kcal mol−1) in 1 and at the O-atom of the methoxy group (−37.7 kcal mol−1) in 2. The MEP at the O-atom of the five membered ring is significantly more positive (−7.5 kcal mol−1 in 1 and −6.3 kcal mol−1 in 2).
 | ||
| Fig. 5 MEP surfaces of compounds 1 (a) and 2 (b). The energies at selected points are given in kcal mol−1. Isosurface 0.001 a.u. | ||
The MEP surfaces also show that the MEP values are positive at the aromatic and aliphatic C–H bonds, ranging from +11 to +17 kcal mol−1 on the dihydropyrazole ring in both complexes. The MEP is negative over the six-membered rings, in both compounds, ranging from −9.4 to −13.8 kcal mol−1, thus explaining their ability to participate in C–H⋯π interactions. Moreover, the MEP surface analysis and the location of the MEP maximum and minimum agrees well with the important C–H⋯N interactions described above in both compounds, which are relevant in their crystal packing. Finally, it is worth mentioning the existence of two weak σ-holes at the S-atoms, opposite to the C–S bonds with values ranging from 1.3 to 5.0 kcal mol−1.
The analysis of the three dimers of compound 1 using QTAIM and NCIPlot indices is depicted in Fig. 6, focusing solely on intermolecular interactions for clarity. In dimer D1, the QTAIM/NCIPlot analysis identifies three CH⋯N bonds. Each bond is characterized by a bond critical point (CP, marked as a red sphere) and a bond path (highlighted with an orange line) linking the H-atom to the N-atom of the five-membered ring. Notably, a green reduced density gradient (RDG) isosurface, coinciding with the bond CP, further characterizes each hydrogen bond (HB). The unified QTAIM/NCIPlot examination also unveils additional contacts between C–H groups and the π-systems of tert-butylphenyl rings, marked by bond CPs, bond paths, and green isosurfaces.
The QTAIM/NCIPlot analysis of dimer D2 (Fig. 6b) reveals several bond CPs connecting both monomers. Beyond the bond CPs and RDG isosurfaces signifying the CH⋯π interactions, the QTAIM analysis indicates connections between each S-atom and the O-atom of the oxadiazole ring, and to an H-atom from the phenyl ring. The diminutive size of these green RDG isosurfaces implies the relative weakness of these contacts.
For dimer D3 (Fig. 6c), the QTAIM/NCIPlot assessment identifies two CH⋯N interactions. Each is marked by a bond CP and bond path linking the H-atom to the N-atom of the five-membered ring. Two more bond CPs and RDG isosurfaces highlight the CH⋯π interactions. The QTAIM evaluation further detects a CH⋯S bond involving a CH bond from the t-butyl group.
A comparative analysis was conducted on the three dimers of compound 2, as depicted in Fig. 7. In dimer D1, each symmetrically identical and notably short CH⋯π contact is distinguished by a bond CP and bond path, linking the H-atom to a C-atom of the ring. The π-character of this contact is more distinctly highlighted by the RDG isosurface, which envelops a significant portion of the π-cloud of the aromatic ring.
For dimer D2 (Fig. 7b), the QTAIM/NCIPlot evaluation verifies the presence of three CH⋯N hydrogen bonds, marked by their respective bond CPs, bond paths, and green isosurfaces. This analysis also uncovers the involvement of two CH⋯π contacts associated with the p-tert-butylbenzyl groups.
Lastly, the QTAIM/NCIPlot assessment of the centrosymmetric dimer D3, aside from identifying the CH⋯π contacts, reveals two CH⋯O and CH⋯S hydrogen bonds. These bonds engage with the H-atoms of the methoxy group, aligning well with the MEP analysis results for compound 2, as illustrated in Fig. 7b.
In summary, the six dimers showcase a complex array of interactions, providing context for the moderately strong interaction energies calculated for these assemblies (see Table 2).
Hirshfeld surface analysis
The Hirshfeld surfaces mapped over the dnorm function for compounds 1 and 2 are shown in Fig. 8. The dominant intermolecular contacts in both structures are shown as bright red areas. These representations reveal that the crystal packing of both compounds is mainly stabilized by non-classical hydrogen bonds and additional non-conventional intermolecular contacts observed in the crystal structure. The full two-dimensional fingerprint plots and those delineated into H⋯S/S⋯H and O⋯S/S⋯O for 1 and H⋯H and H⋯N/N⋯H for 2 are shown in Fig. 9.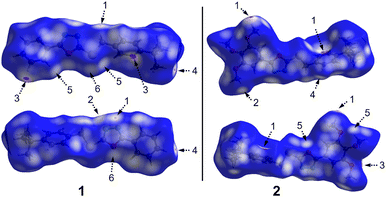 | ||
| Fig. 8 Hirshfeld surfaces mapped over dnorm property for compounds 1 and 2. The second molecule is 180° rotated around the horizontal axis of the plot. The labels are discussed in the text. | ||
In compound 1, the small red spots labelled 1 and 2 are attributed to C3–H3B⋯N1 and C5–H5⋯N2 hydrogen bonds, respectively, with 11.3% contribution to the total Hirshfeld surface area. The red bright red spots labelled 3 on the dnorm map are assigned to C–H⋯π interactions, involving the H5 atom and the centroid of the C4–C9 phenyl ring. These contacts are visible as a pair of wings in the top left and bottom right region of the fingerprint plot (Fig. 9), which comprise 25.8% of the total Hirshfeld surface area.
The white regions labelled 5 are attributed to weak C6–H6⋯S1 hydrogen bonds. These contacts are visible in the fingerprint plots as a pair of spikes at (de + di) ≈ 3.0 Å, in accordance with the H6⋯S1 distance of 3.165 Å. These H⋯S/S⋯H contacts comprise 5.9% of the Hirshfeld surface. The appearance of red regions around the S1 and O1 atoms are indicative of S1⋯O1 chalcogen bonding interactions. These contacts contribute 2.0% to the total Hirshfeld surface area. The presence of H⋯H interactions in the crystal structure of 1 is evidenced by the presence of bright red areas labelled 4 in the dnorm surface, attributed to H17⋯H12B dihydrogen interactions [d(H17⋯H12B) = 2.39 Å], with a distance shorter than the sum of vdW radii of H-atoms. These contacts are highlighted in the middle of scattered points of the fingerprint plot, with a minimum value of (de + di) ≈ 2.4 Å, and the highest contribution of 49% of the total Hirshfeld surface area.
In 2, the bright red spots labelled 1 in the dnorm surface are mainly attributed to C20–H20⋯π interactions involving the H20 atom of the methyl group from the methoxy moiety and the centroid of the C4–C9 ring. These interactions are also visible on the fingerprint plot (Fig. 9) as a pair of wings, with a contribution of 19.9% to the total Hirshfeld surface. The red spots labelled 2 and 3 are attributed to C22–H22C⋯N1 and C13–H13B⋯O4 hydrogen bonds, respectively, while the red regions labelled 4 and 5 are attributed to C5–H5⋯N2 and C21–H21A⋯O1 interactions, respectively. The contribution of H⋯N/N⋯H and H⋯O/O⋯H contacts to the total Hirshfeld surface area is 8.8 and 14.2%, respectively.
The existence of the C–H⋯π interactions has been evidenced by the appearance of patches with a large red depression above the π-system, and a blue region surrounding the C–H donor on the Hirshfeld surfaces mapped over the shape index property (Fig. 10). These results are in accordance with the dnorm, fingerprint plots and with the crystallographic results.
 | ||
| Fig. 10 Hirshfeld surfaces of 1 (top) and 2 (bottom) mapped over shape index property in the colour range of −1.0 a.u. (concave) to 1.0 a.u. (convex). | ||
Conclusions
Three new oxadiazole derivatives were synthesized and two of them were X-ray characterized. The research explored molecular dimers and their intermolecular interaction energies in compounds 1 and 2. Both compounds have three energetically significant molecular dimers crucial for crystal packing stabilization. Compound 1 interacts through C–H⋯N and C–H⋯π, whereas compound 2 is stabilized primarily by C–H⋯N, C–H⋯O, and C–H⋯π interactions. Dimer D1 in both compounds emerged as the most stable, with interaction energies heavily influenced by electrostatic and dispersion contributions.Several computational tools, including MEP surface plots, QTAIM, and NCIPlot, provided deeper insights into these interactions confirming the significance of C–H⋯π and C–H⋯N/O interactions in crystal packing. Hirshfeld surface analysis further highlighted that the crystal packing in both compounds is primarily stabilized by non-classical hydrogen bonds and additional unconventional intermolecular contacts.
Author contributions
MNA supervised the synthesis, write up and MA synthesised the compounds; HA performed molecular docking studies and LOX, AB performed 1H-NMR and 13C-NMR characterization; MAAI and PAS performed the conceptualization and theoretical studies; IA and MNT performed XRD, DMG performed the Hirshfeld surface analysis and the energetic study of dimers; RMG and AF performed DFT studies and write up.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are thankful to HEC-NRPU grant no.17373 and the MICIU/AEI (project PID2020-115637GB-I00, FEDER funds) for financial support. We thank the CTI (UIB) for computational facilities. A. F. thanks the Alexander von Humboldt fundation for the J. C. Mutis award.References
- M. J. Benedik, P. R. Gibbs, R. R. Riddle and R. C. Willson, Trends Biotechnol., 1998, 16, 390–395 CrossRef CAS PubMed.
- X. Gan, D. Hu, Z. Chen, Y. Wang and B. Song, Bioorg. Med. Chem. Lett., 2017, 27, 4298–4301 CrossRef CAS PubMed.
- F. Wang, B.-X. Yang, T.-H. Zhang, Q.-Q. Tao, X. Zhou, P.-Y. Wang and S. Yang, Arabian J. Chem., 2023, 16, 104479 CrossRef CAS.
- M. N. Ahmed, K. A. Yasin, S. Hameed, K. Ayub, I.-U. Haq, M. N. Tahir and T. Mahmood, J. Mol. Struct., 2017, 1129, 50–59 CrossRef CAS.
- S. S. Hamdani, B. A. Khan, M. N. Ahmed, S. Hameed, K. Akhter, K. Ayub and T. Mahmood, J. Mol. Struct., 2020, 1200, 127085 CrossRef.
- X.-M. Zhang, M. Qiu, J. Sun, Y.-B. Zhang, Y.-S. Yang, X.-L. Wang, J.-F. Tang and H.-L. Zhu, Bioorg. Med. Chem. Lett., 2011, 19, 6518–6524 CrossRef CAS PubMed.
- P.-Y. Wang, L. Zhou, J. Zhou, Z.-B. Wu, W. Xue, B.-A. Song and S. Yang, Bioorg. Med. Chem. Lett., 2016, 26, 1214–1217 CrossRef CAS PubMed.
- C.-J. Chen, B.-A. Song, S. Yang, G.-F. Xu, P. S. Bhadury, L.-H. Jin, D.-Y. Hu, Q.-Z. Li, F. Liu and W. Xue, Bioorg. Med. Chem., 2007, 15, 3981–3989 CrossRef CAS PubMed.
- A. S. N. Formagio, L. T. D. Tonin, M. A. Foglio, C. Madjarof, J. E. de Carvalho, W. F. da Costa, F. P. Cardoso and M. H. Sarragiotto, Bioorg. Med. Chem., 2008, 16, 9660–9667 CrossRef CAS PubMed.
- P. Li, J. Yin, W. Xu, J. Wu, M. He, D. Hu, S. Yang and B. Song, Chem. Biol. Drug Des., 2013, 82, 546–556 CrossRef CAS PubMed.
- W. Xu, S. Yang, P. Bhadury, J. He, M. He, L. Gao, D. Hu and B. Song, Pestic. Biochem. Physiol., 2011, 101, 6–15 CrossRef CAS.
- Y. Wang, X. Lu, J. Shi, J. Xu, F. Wang, X. Yang, G. Yu, Z. Liu, C. Li and A. Dai, Monatsh. Chem., 2018, 149, 611–623 CrossRef CAS.
- M. N. Ahmed, B. Sadiq, N. A. Al-Masoudi, K. A. Yasin, S. Hameed, T. Mahmood, K. Ayub and M. N. Tahir, J. Mol. Struct., 2018, 1155, 403–413 CrossRef CAS.
- M. N. Ahmed, I. Ashraf and K. A. Yasin, J. Chem. Soc. Pak., 2018, 40, 773–781 CAS.
- Q.-Q. Tao, L.-W. Liu, P.-Y. Wang, Q.-S. Long, Y.-L. Zhao, L.-H. Jin, W.-M. Xu, Y. Chen, Z. Li and S. Yang, J. Agric. Food Chem., 2019, 67, 7626–7639 CrossRef CAS PubMed.
- M. Mirzaei, H. Eshtiagh-Hosseini, N. Lotfian, A. Salimi, A. Bauzá, R. Van Deun, R. Decadt, M. Barceló-Oliver and A. Frontera, Dalton Trans., 2014, 43, 1906–1916 RSC.
- M. Singh, V. Aggarwal, U. Singh and N. Singh, Polyhedron, 2009, 28, 195–199 CrossRef CAS.
- A. Il'ya, D. Escudero, A. Frontera, P. V. Solntsev, E. B. Rusanov, A. N. Chernega, H. Krautscheid and K. V. Domasevitch, Dalton Trans., 2009, 2856–2864 Search PubMed.
- W. Wang, Y. Gao, M. Ji, H.-G. Yao and H. Qiu, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2010, 66, o2835 CrossRef CAS PubMed.
- C. F. Matta, N. Castillo and R. J. Boyd, J. Phys. Chem. B, 2006, 110, 563–578 CrossRef CAS PubMed.
- (a) M. Rocha, D. M. Gil, G. A. Echeverría, O. E. Piro, J. L. Jios and S. E. Ulic, J. Org. Chem., 2019, 84, 11042–11053 CrossRef CAS PubMed; (b) D. Sadhukhan, M. Maiti, G. Pilet, A. Bauzá, A. Frontera and S. Mitra, Eur. J. Inorg. Chem., 2015, 1958–1972 CrossRef CAS.
- (a) M. Rocha, G. A. Echeverría, O. E. Piro, J. J. Jios, R. D. Molina, M. E. Arena, S. E. Ulic and D. M. Gil, Aust. J. Chem., 2019, 73, 49–60 CrossRef; (b) S. Roy, M. G. B. Drew, A. Bauzá, A. Frontera and S. Chattopadhyay, Dalton Trans., 2017, 46, 5384–5397 RSC.
- (a) D. Dutta, S. M. Nashre-ul-Islam, U. Saha, A. Frontera and M. K. Bhattacharyya, J. Mol. Struct., 2019, 1195, 733–743 CrossRef CAS; (b) T. Maity, H. Mandal, A. Bauzá, B. C. Samanta, A. Frontera and S. K. Seth, New J. Chem., 2018, 42, 10202–10213 RSC.
- D. M. Gil, H. Pérez, G. A. Echeverría, O. E. Piro and A. Frontera, ChemistrySelect, 2020, 5, 6331–6338 CrossRef CAS.
- D. M. Gil, F. F. Salomón, G. A. Echeverría, O. E. Piro, H. Pérez and A. B. Altabef, Spectrochim. Acta, Part A, 2017, 185, 286–297 CrossRef CAS PubMed.
- (a) M. N. Ahmed, M. Arif, H. Andleeb, S. W. A. Shah, I. Arshad, M. N. Tahir, M. Rocha and D. M. Gil, CrystEngComm, 2021, 23, 955–971 RSC; (b) A. Bibi, I. Khan, H. Andleeb, J. Simpson, M. N. Tahir, S. Hameed and A. Frontera, J. Mol. Struct., 2021, 1227, 129425 CrossRef CAS; (c) H. Andleeb, I. Khan, A. Bauzá, M. N. Tahir, J. Simpson, S. Hameed and A. Frontera, Acta Crystallogr., Sect. C: Struct. Chem., 2018, 74, 816–829 CrossRef CAS PubMed.
- D. Quiñonero, A. Frontera, D. Escudero, P. Ballester, A. Costa and P. M. Deyà, Theor. Chem. Acc., 2008, 120, 385–393 Search PubMed.
- M. N. Ahmed, K. A. Yasin, K. Ayub, T. Mahmood, M. N. Tahir, B. A. Khan, M. Hafeez and M. Ahmed, J. Mol. Struct., 2016, 1106, 430–439 CrossRef CAS.
- G. Sheldrick, Bruker-Nonius, Madison, WI, 1990 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- C. Adamo and V. Barone, J. Chem. Phys., 1999, 110, 6158–6170 CrossRef CAS.
- S. Grimme, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- F. Weigend, Phys. Chem. Chem. Phys., 2006, 8, 1057–1065 RSC.
- R. Ahlrichs, M. Bär, M. Häser, H. Horn and C. Kölmel, Chem. Phys. Lett., 1989, 162, 165–169 CrossRef CAS.
- R. F. Bader, Chem. Rev., 1991, 91, 893–928 CrossRef CAS.
- J. Contreras-García, E. R. Johnson, S. Keinan, R. Chaudret, J.-P. Piquemal, D. N. Beratan and W. Yang, J. Chem. Theory Comput.., 2011, 7, 625–632 CrossRef PubMed.
- T. Lu and F. Chen, J. Comput. Chem., 2012, 33, 580–592 CrossRef CAS PubMed.
- M. A. Spackman and P. G. Byrom, Chem. Phys. Lett., 1997, 267, 215–220 CrossRef CAS.
- A. Parkin, G. Barr, W. Dong, C. J. Gilmore, D. Jayatilaka, J. J. McKinnon, M. A. Spackman and C. C. Wilson, CrystEngComm, 2007, 9, 648–652 RSC.
- M. A. Spackman and D. Jayatilaka, CrystEngComm, 2009, 11, 19–32 RSC.
- S. M. Nashre-ul-Islam, K. K. Borah, F. E. Öztürkkan, M. A. Raza, A. Frontera and D. M. Gil, J. Mol. Struct., 2022, 1268, 133686 CrossRef CAS.
- P. R. Spackman, M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, D. Jayatilaka and M. A. Spackman, J. Appl. Crystallogr., 2021, 54, 1006–1011 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: NMR spectra of all compounds and docking/LOX inhibition studies. CCDC 2241333 and 2241334. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3ce00944k |
| This journal is © The Royal Society of Chemistry 2023 |







