Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A quaternary solid-form of ritonavir: an oxalate salt oxalic acid co-crystal acetone solvate†
Chang
Wang
ab,
Thomas D.
Turner‡
 *a,
Cai Y.
Ma
a,
Christopher M.
Pask
c,
Ian
Rosbottom
a,
Richard S.
Hong
d,
Ahmad Y.
Sheikh
*a,
Cai Y.
Ma
a,
Christopher M.
Pask
c,
Ian
Rosbottom
a,
Richard S.
Hong
d,
Ahmad Y.
Sheikh
 d,
Qiuxiang
Yin
d,
Qiuxiang
Yin
 b and
Kevin J.
Roberts
b and
Kevin J.
Roberts
 a
a
aCentre for the Digital design of Drug Products, School of Chemical and Process Engineering, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK. E-mail: t.d.turner@leeds.ac.uk
bSchool of Chemical Engineering and Technology, State Key Laboratory of Chemical Engineering, Tianjin University, Tianjin 300072, China
cSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK
dSolid State Chemistry, Process R&D, AbbVie Inc., North Chicago, IL 60064, USA
First published on 17th February 2023
Abstract
Ritonavir has been reported in seven crystal forms notably form I, II, IIIb and IV, as well as a hydrate, an L-tyrosine co-crystal and a formamide solvate. A new form is reported here with a novel quaternary structure with ritonavir as an oxalate salt, oxalic acid co-crystal and acetone solvate in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5
0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 stoichiometry. The new oxalate salt form (CCDC deposition number 2009282), was co-crystallised with oxalic acid from a supersaturated acetone solution, and exhibits a polar monoclinic crystal structure with a blocky needle-like crystal habit. The molecular conformation of ritonavir for the new form shows significant differences with respect to the well-characterised form I, II and IIIb polymorphs. Its crystallography is characterised by a 2-fold screw axis along the b axis, in a structure that exhibits multiple hydrogen bonds formed between amide–ureido, oxalate–oxalic acid and amide–amide groups in a layered intermolecular packing structure. Intermolecular stacking of the benzene and thiazole rings takes place along the crystallographic a axis with the needle axis growth of the crystals being likely to be governed by these interactions. The oxalate salt form co-crystal is found to have the characteristic conformations of the N-methyl urea and carbamate groups being in a trans and trans conformation respectively, as is the case for the recently discovered form IIIb, but in distinct contrast to cis and trans for form I, and trans and cis for form II. This may suggest that a cis and cis conformation for another new form may be feasible. The recovery of the form IV crystals at the lower concentrations of oxalic acid is perhaps indicative of its close similarity with the new oxalate salt co-crystal solvate form presented here.
0.5 stoichiometry. The new oxalate salt form (CCDC deposition number 2009282), was co-crystallised with oxalic acid from a supersaturated acetone solution, and exhibits a polar monoclinic crystal structure with a blocky needle-like crystal habit. The molecular conformation of ritonavir for the new form shows significant differences with respect to the well-characterised form I, II and IIIb polymorphs. Its crystallography is characterised by a 2-fold screw axis along the b axis, in a structure that exhibits multiple hydrogen bonds formed between amide–ureido, oxalate–oxalic acid and amide–amide groups in a layered intermolecular packing structure. Intermolecular stacking of the benzene and thiazole rings takes place along the crystallographic a axis with the needle axis growth of the crystals being likely to be governed by these interactions. The oxalate salt form co-crystal is found to have the characteristic conformations of the N-methyl urea and carbamate groups being in a trans and trans conformation respectively, as is the case for the recently discovered form IIIb, but in distinct contrast to cis and trans for form I, and trans and cis for form II. This may suggest that a cis and cis conformation for another new form may be feasible. The recovery of the form IV crystals at the lower concentrations of oxalic acid is perhaps indicative of its close similarity with the new oxalate salt co-crystal solvate form presented here.
Introduction
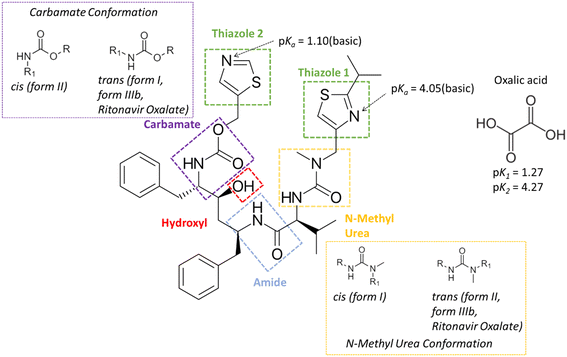
Ritonavir (Fig. 1) is a protease inhibitor for the treatment of acquired immunodeficiency syndrome (AIDS); the material's unexpected polymorphic transformation and resulting stability issues were well-documented two decades ago.1,2 Ritonavir was found to exhibit poor aqueous solubility and is consistent with a class 4 molecule under the biopharmaceutics classification system (BCS), and hence was marketed either as an oral liquid or as semi-solid capsules in ethanol/water based solutions. This formulation strategy and the propensity for polymorphism in organic materials3 provided an enhanced risk for the materials' transformation into a more stable and potentially more insoluble solid-form with time,4–6 and some two years after its release, ritonavir was found to fail its dissolution testing specification due to the presence of a new and more stable form (form II) being precipitated within the formulated product.1The structures of the form I and II polymorphs were determined and thoroughly characterised by Bauer et al.1 The conformational energy landscape of the ritonavir molecule was modelled using density functional theory (DFT) by Chakraborty et al.,7 highlighting that the identified molecular structure in its global minimum conformation exhibits stacking of its two thiazole rings. However, the conformational structures1 for molecules from both forms I and II were not found to adopt the global minimum molecular conformation when the molecules were packed within their crystallographic structures. This suggested that the unfavourable molecular conformational energy is offset by the formation of inter-molecular interactions once packed into the 3-D lattices for forms I and II. Additionally the form I conformation has been found to adopt a trans, cis conformation for its carbamate and n-methyl urea groups respectively whereas form II has been found to adopt a cis, trans conformation of the same groups.
High-throughput polymorph screening by Morissette et al.8 revealed (see Table 1) the existence of two new solvates (forms IIIa and V) as well as a further (metastable) phase (form IV), but up to now their full crystal structures have not been reported, albeit recent work9 through melt crystallisation has now resulted in a further solid form with its crystal structure (form IIIb,10 see Table 1). In this respect, due to the form III nomenclature being claimed for both the form identified from screening8 and that recently crystallised from the melt,9 for clarity we have referred to these two structures herein after as forms IIIa and IIIb, respectively. Surprisingly, due to its flexible molecular conformation and glass-forming tendency of the material, only a single co-crystal form of ritonavir co-crystallised with L-tyrosine have been reported.11–13 More recently, Wang et al.14 have characterised and compared, in significant detail, the crystal chemistry for both forms I and II in terms of their molecular conformations, polarizabilities, hydrogen bonding networks, intermolecular packing structures and crystal morphologies cross-correlating these to a wider assessment of their crystallisability behaviour and respective surface properties.
| Form | Crystal system | Solid-state structure | Melting point (°C) | Crystallisation method | Single crystal structure | Molecular Conformationa | References |
|---|---|---|---|---|---|---|---|
| a Refers to the molecular conformation with respect to the carbamate and N-methyl urea groups. | |||||||
| I | Monoclinic | Anhydrous pure form | 123 | Ethanol/H2O | Y | trans and cis | 1 |
| II | Orthorhombic | Anhydrous pure form | 126 | Ethanol/H2O | Y | cis and trans | 1 |
| IIIa | Monoclinic | Formamide solvate | 78–82 | Formamide/toluene or formamide/butyl acetate or formamide/acetone | N | Unknown | 8 |
| IIIb | Monoclinic | Anhydrous pure form | 114.6 | Melt | Y | trans and trans | 10 |
| IV | Unknown | Anhydrous pure form | 116 | Acetonitrile/butyl acetate or acetonitrile/isobutyl acetate or acetonitrile/isopropyl acetate | N | Unknown | 8 |
| IV | Unknown | Anhydrous pure form | 121.8 | Acetone/oxalic acid | N | Unknown | (This study, 2023) |
| V | Monoclinic | Trihydrate | 97 | Aqueous slurry conversion from III (readily transforms to I) | N | Unknown | 8 |
| Unknown | L-Tyrosine co-crystal | 112.9 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 liquid assisted grinding with methanol 1 liquid assisted grinding with methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1 water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
N | Unknown | 13 | |
| Monoclinic | Salt co-crystal solvate | 125.3 | Acetone/oxalic acid | Y | trans and trans | (This study, 2023) | |
Generally, a selected co-former can be incorporated into the crystal lattice with molecules of charged or uncharged active pharmaceutical ingredients (API) when a salt or co-crystal, respectively, is formed.15–17 To distinguish between whether a salt or a co-crystal might form, the difference in pKα between the API and the co-former usually provides a useful guide to predict the possibility of proton transfer during the co-crystallisation process (ΔpKα = pKα(API) − pKα(co-former)). It is generally accepted18,19 that a non-ionised co-crystal complex would more likely to be formed when the magnitude of ΔpKα ≤ 0, although this has been recently refined <−1 through examination of >6000 molecules within the Cambridge Structural Database (CSD).20 In contrast, when the ΔpKα > 4 it would be expected that any resultant solid-form would most likely form a salt.20 For the borderline region with ΔpKα (0–3), it is not generally regarded that predictive methods can accurately evaluate whether a co-crystal or salt form will be produced. Ritonavir has reported pKα1 and pKα2 values of 1.9 and 2.5 for the two thiazole groups,21 whilst the pKα values of oxalic acid have been found to be 1.27 and 4.27 for pKα1 and pKα2, respectively.22
Reflecting its small molecular size and large number of hydrogen bond accepter/donor options, oxalic acid (Fig. 1) is a good co-former to access potentially novel co-crystal solid forms. In the case of its use as a co-former for ritonavir, the ΔpKα values are 0.63 and 1.23 with the two thiazole groups, respectively. Given the high propensity of potential H-bond donors and acceptors in ritonavir,14,23 its NH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups offer the most likely options for the formation of intermolecular H-bonds with co-formers.
O functional groups offer the most likely options for the formation of intermolecular H-bonds with co-formers.
In this paper, the crystallisation of ritonavir in the presence of oxalic acid has been explored with the aim to further examine its solid-form landscape. The work also builds upon our recent in-depth examination of the solid-state and surface properties of forms I and II,14 their intermolecular hydrogen bonding networks and how these various solid forms relate to the molecular conformation in the solid-state.
Experimental section
Materials
Ritonavir form II was obtained from AbbVie Inc. and used as received without further purification. Anhydrous oxalic acid (>99.0%) was purchased from Sigma Aldrich, acetone HPLC grade (>99%) was used as supplied by VWR International.Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 was chosen due to the di-acid nature of oxalic acid, however the impact of oxalic acid concentration was not extensively investigated here). The composition was heated to ensure complete dissolution of solids and agitated with a magnetic stirrer at 700 rpm. 10 ml aliquots of solution were then filtered into 20 ml vials through a 0.45 μm filter membrane.
5 was chosen due to the di-acid nature of oxalic acid, however the impact of oxalic acid concentration was not extensively investigated here). The composition was heated to ensure complete dissolution of solids and agitated with a magnetic stirrer at 700 rpm. 10 ml aliquots of solution were then filtered into 20 ml vials through a 0.45 μm filter membrane.
The solutions were subsequently used for both slow evaporative and fast cooling crystallisation experiments. For the former case the vials were sealed with parafilm which were perforated with several small holes in order to ensure slow evaporation (not to dryness) at room temperature (∼25 °C). For the latter case, the vials were sealed and quench cooled to 5 °C to achieve high supersaturations.
In both the above cases, the resulting crystals were filtered and dried after solvent evaporation with the solid phase being analysed using a differential scanning calorimeter with samples contained in a loosely covered aluminium pan using a sample weight of 2–5 mg and a heating rate of 10 °C per minute and a nitrogen flow of 40 mL min−1 (DSC) (Mettler-Toledo DSC-1), Olympus optical microscope and powder X-ray diffraction (PXRD, Bruker D8 diffractometer, Cu Kα 1.54 Å).
Results and discussion
Solid form characterisation
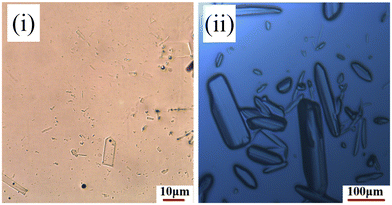
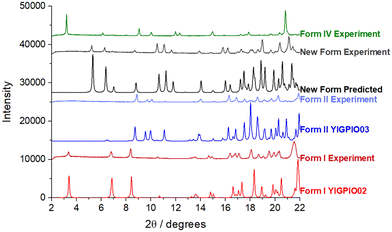
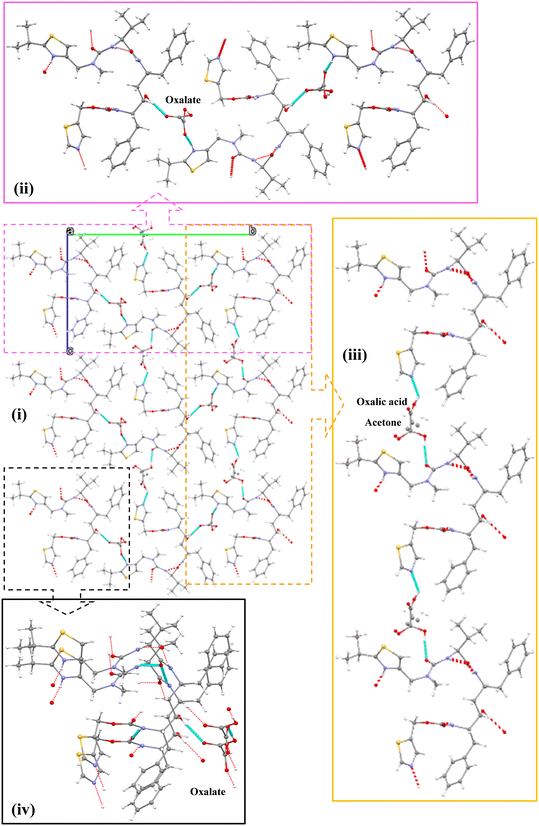
Form I was found to consistently crystallise when no oxalic acid was added to the supersaturated solutions, independent of the crystallisation method. However, microscopy analysis of the crystals harvested from the slow-solvent evaporation experiments in the presence of 5% oxalic acid, revealed a distinctly plate-like morphology (Fig. 2(i)) which was rather similar to that previously reported for form IV.8 However, with an increasing concentration of oxalic acid, a more block-like or tabular morphology (Fig. 2(ii)) was obtained from both the evaporative and rapid cooling crystallisation experiments.34Analysis of the PXRD patterns recorded from powdered samples of the crystals, as shown in Fig. 3, reveals distinct differences. The plate-like crystals exhibited characteristic diffraction peaks at 2θ values of 3.21°, 6.13°, 6.87°, 7.98° and 10.04° consistent with those of form IV.8,35 In contrast, the tabular crystals were found to exhibit characteristic peaks at 5.24°, 6.32°, 6.93°, 8.67° and 11.66°, which were not consistent with any of the previously observed solid forms as shown in Table 1 and hence were found to be consistent with the existence of a new form, which was found to be a ritonavir oxalate oxalic acid co-crystal acetone solvate, hereinafter for brevity “oxalate salt form”.
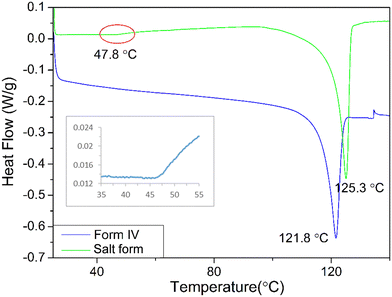
DSC analysis of the crystals of form IV and oxalate salt form (Fig. 4) revealed that the form IV data only exhibited one endothermic melting peak at about 121.8 °C which is close to that of 116 °C reported in the literature.8,35 The difference perhaps reflects variations in heating rate, crystal size and the DSC instrument precision between the data reported here and previous work. Analysis of the oxalate salt form crystals revealed two endothermic peaks at 47.8 and 125.3 °C, respectively. The first very small endothermic peak at 47.8 °C, being close to the boiling point of acetone (boiling point 56 °C), would be consistent with the removal of weakly bound surface solvent, most likely to be acetone due to the crystallisation conditions, and not crystallographically bound due to the very small peak area. The second endotherm at 125.3 °C would be more indicative of melting of the material.
Crystal structural determination
The crystal size of the isolated form IV crystals were unfortunately not found to be sufficiently large enough for a full single crystal diffraction analysis, and hence only the much larger single crystals of the salt form crystals were examined.34 Examination of optical micrographs of the single crystal selected for crystallographic analysis (see Fig. S2 in ESI†) revealed a crystal habit elongated along its crystallographic a axis.A more detailed analysis of the crystal structure of the new oxalate salt form revealed it to crystallise in a polar monoclinic structure encompassing a quaternary ritonavir oxalate salt oxalic acid co-crystal acetone solvate with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5
0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 stoichiometry i.e. with one ritonavir cation, one oxalate anion, one half molecule of acetone and one half molecule of oxalic acid, within the asymmetric unit (Fig. 5) and with the latter two being disordered but centred approximately at the same lattice site. The associated crystallographic data are given in Table 2 and the unit cell shown in Fig. 5.
0.5 stoichiometry i.e. with one ritonavir cation, one oxalate anion, one half molecule of acetone and one half molecule of oxalic acid, within the asymmetric unit (Fig. 5) and with the latter two being disordered but centred approximately at the same lattice site. The associated crystallographic data are given in Table 2 and the unit cell shown in Fig. 5.
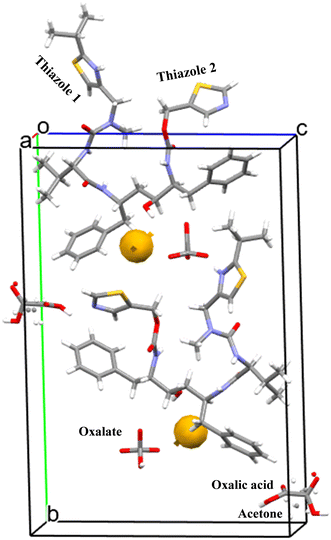 | ||
| Fig. 5 The unit cell of the new oxalate salt form which contains two ritonavir cations, two oxalate anions together with one oxalic acid molecule and one acetone molecule, and also the void space (0.3% as shown in Table 2). | ||
| Material descriptor | Form I | Form II | Form IIIb | Oxalate salt form(this study) |
|---|---|---|---|---|
| Note that void space (%) based on probe radius: 1.0 Å & approximate grid spacing: 0.5 Å. Average packing coefficient for organic crystals: 0.68.a Calculated with ritonavir molecule only.b Based on molecules in the symmetric unit (1 ritonavir, 1 oxalate, 0.5 acetone and 0.5 oxalic acid). | ||||
| Refcode | YIGPIO021 | YIGPIO031 | YIGPIO049 | 2009282 Deposition no. |
| Molecular volume (Å3) | 721.24 | 676.74 | 678.58 | 695.73a (870.51b) |
| Molecular weight (g mol−1) | 720.94 | 720.94 | 720.94 | 720.94a (885.03b) |
| Molecular surface area (Å2) | 656.46 | 645.09 | 634.94 | 617.51a (717.47b) |
| Melting point (°C) | 1231 | 1261 | 114.69 | 125.3 |
| Space group |
P21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
P212121![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
C29 | P21 |
| Z/Z' | 2/11 | 4/11 | 4/19 | 2/1 |
| a (Å) | 13.433(1)1 | 10.0236(3)1 | 23.3307(19)9 | 5.2615(2) |
| b (Å) | 5.293(2)1 | 18.6744(4)1 | 4.9511(5)9 | 25.2098(9) |
| c (Å) | 27.092(4)1 | 20.2692(7)1 | 33.638(3)9 | 16.6111(6) |
| α (°) | 901 | 901 | 909 | 90 |
| β (°) | 103.102(9)1 | 901 | 91.274(6)9 | 91.994(3) |
| γ (°) | 901 | 901 | 909 | 90 |
| Cell volume (Å3) | 1876.0(8)1 | 3831.5(2)1 | 3884.6(6)9 | 2201.99(14) |
| Packing coefficient | 0.77 | 0.71 | 0.7149 | 0.79 |
| Void space (%) | 0.8 | 1.0 | 0.369 | 0.3 |
| Density (g cm−3) | 1.281 | 1.251 | 1.3259 | 1.33 |
As shown in Fig. 5, the growth morphology is actually dominated by stacking of the benzene and thiazole groups in the oxalate salt form along the crystallographic a axis, which is unusual considering the stronger ionic salt interactions were found to be formed in the b, c directions. This might suggest salt formation to be the likely initial driver for the nucleation of this solid form which allows the molecules to assemble for subsequent orientation followed by stacking of the salt dimer/tetramer dominating the growth process.
The molecular and crystal structural descriptors and some thermodynamic data for forms I and II1,14 and IIIb9 together with the new oxalate salt form structure from this study are provided in Table 2. Previous work9 has shown that form IIIb has a higher molecular volume (Table 2), a higher crystal structure density and a lower void space when compared to the less close-packed form II structure. In this study, the new oxalate salt form was found to have the highest crystal density (1.33 g cm−3) of all these four solid-forms with the lowest void space and highest packing efficiency. The lowest calculated surface area of ritonavir molecule in the oxalate salt form when compared to other three forms, may be attributed to the “stacking” molecular structure shown in Fig. 6 (see also Fig. S4 in the ESI†). Like other conformational polymorphs of complex pharmaceutical molecules, these observations are likely to reflect the result of interplay between the relative strength of the intermolecular and intramolecular energies.36
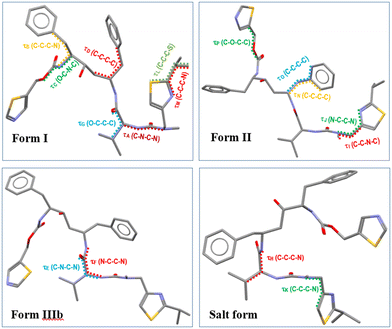 | ||
| Fig. 6 The different molecular conformations of ritonavir in form I, form II, form IIIb and oxalate salt form. | ||
Conformational analysis
The molecular conformation of ritonavir within the new oxalate salt form salt-solvate structure was found to be significantly different from that of forms I, II and IIIb. In the oxalate salt form, the ritonavir molecule is torsioned such to make the two thiazole rings closer to each other (Fig. 6 and see also Fig. S4(ii) in the ESI†) which is much closer to the global minimum conformation “f” calculated by Chakraborty et al.7 Examination of the four torsion angles (τE, τF, τG, τH) involving the rotation of the molecular fragment containing the phenyl–hydroxyl–phenyl–carbamate–thiazole_1 groups, highlights that the new oxalate salt form conformer reveals the molecule to be “folded” into a “U” shape (see Fig. S4(i) in the ESI†). The folded conformation is likely stabilized through intra-molecular dispersive interaction. This is likely due to the polar nature of the crystal structure, resulting in hydrophobic collapse of the ritonavir conformation, where its hydrophobic substituents are shielded through intra-molecular interactions. It was found that the angles of these four torsions in forms I, II and IIIb have differences in the range of 3–20° (see Table S1 in the ESI†), which are quite small. However, when comparing the conformation of the new oxalate salt form with the other three forms (I, II and IIIb), the torsional angle differences of τE, τF, τG and τH become much larger (see Fig. S4(i) and S5 in the ESI†), to a range of 164–165°, 174–178° and 158–162° with regard to form I, form II and form IIIb (see Table S1 in the ESI†), respectively.As shown in the literature,1,9,14 the main molecular conformational differences between the polymorphic forms can be characterised with respect to the ideal conformations around the N-methyl urea and the carbamate functional groups. In this, form I has a cis N-methyl urea conformation and a trans carbamate conformation respectively, whilst form II has a trans N-methyl urea conformation and a cis carbamate conformation. This is caused by the differences between the torsion angles between form I and II: τA and τC being 170.6° and 175.5°,14 respectively (see Fig. S7(i) and Table S1 in the ESI†), leading to the R1 (Fig. 1) of the cis and trans configurations orientated in opposite directions. As shown in Table 3 (see also Fig. S7(ii) in the ESI†), the torsion angle τA of the oxalate salt form (170.6°) is very close to that of form II and form IIIb with only 5° and 6.4° differences respectively, consistent with the N-methyl urea conformation being in a trans configuration for the form IIIb and the new oxalate salt form (see Fig. S7(vi) in the ESI†). The differences of τC between the oxalate salt form (−171.6°) and forms I/IIIb (see Fig. S7(iii and vi) in the ESI†) were found to be only 9.7° and 14.7° respectively, and hence the oxalate salt form has a trans carbamate conformation (Table 3 and see Fig. S7(i) in the ESI†). Therefore the molecular structures of the form IIIb and oxalate salt form have both these groups in the trans configurations comparing to cis and trans conformation in form I, and trans and cis conformation in form II. The detailed comparisons of torsion angles (τA, τB, τC, τD) between forms I, II, IIIb and the new oxalate salt form are shown in the ESI† (see Fig. S7 and Table S1).
| Torsion | (a) Form I | (b) Form II | (c) Form IIIb | (d) Salt form | Δ(a–b) | Δ(a–d) | Δ(b–d) | Δ(c–d) |
|---|---|---|---|---|---|---|---|---|
| Note that the definitions of 4 torsion angles for form I, form II, form IIIb and the new oxalate salt form can be found in Fig. 6. | ||||||||
| τ A | −23.8 | 165.6 | 177.0 | 170.6 | 170.6 | 165.6 | 5.0 | 6.4 |
| τ B | 71.0 | −72.8 | 46.1 | 48.5 | 143.8 | 22.5 | 121.3 | 2.4 |
| τ C | 178.7 | −5.8 | 173.7 | −171.6 | 175.5 | 9.7 | 165.8 | 14.7 |
| τ D | 55.9 | 168.3 | 173.3 | −175.4 | 112.4 | 128.7 | 16.3 | 11.3 |
| τ E | −67.7 | −56.8 | −73.9 | 127.5 | 10.9 | 164.8 | 175.7 | 158.6 |
There are also additional torsion angles which were found to exhibit large differences between these four forms and these are plotted in Fig. S5 and listed in Table S1 (ESI†). By comparing the torsions (τI, τJ, τK) in the molecular structure of the oxalate salt form with forms I, II and IIIb, it was found that only τI in the oxalate salt form is close to form I with a small (21°) angular difference, but all three torsions (τI, τJ, τK) in both form IIIb and oxalate salt form also have small (<33°) angular differences. For the torsions (τL, τM), the isopropyl group in the oxalate salt form rotates about (105–111°), (170–178°) and (102–112°) compared to these in forms I, II and IIIb, whilst the torsions (τN, τO) have about a (160–180°) angle of rotation. Interestingly, as shown in Table 3 (see also Fig. S7(iii) in the ESI†), the ureido and thiazole 1 functional groups are roughly overlaid between form IIIb and new oxalate salt form. However, the rest of the molecular structures of both forms rotate about 160° to each other as evidenced by the torsion angle τE in Table 3. Further detail can be found in ESI.†
Hydrogen bonding network analysis
Ritonavir has a relatively large molecular mass compared to many API's, exhibiting four hydrogen bond donors; one hydroxyl O–H and three amidic N–H groups together with twelve acceptors.14 In the oxalate salt form structure, the hydrogen bond donors and acceptors of ritonavir are in competition with those provided by oxalic acid during co-crystallisation. Hence, its resulting crystal structure encompasses donor–acceptor interactions between API/API, API/co-former and co-former/co-former. Details on the hydrogen bonds and their structure for the ritonavir polymorphs for form I, form II, form IIIb and the new oxalate salt form are presented in Table 4. Examination of the data reveal that there are four hydrogen bonds formed between adjoining ritonavir molecules in the crystal structures of forms I, II and IIIb with forms I and IIIb having their hydrogen bonding donors and acceptors from the exactly same functional groups but with different bond lengths. However, there are only three hydrogen bonds formed within the new oxalate salt form structure with the intermolecular hydrogen bonds involving the hydroxyl group present in forms I, II and IIIb being absent in the new oxalate salt form structure. However, these are replaced by O–H (hydroxyl)⋯O (oxalate) interactions at a similar inter-atomic distance. The thiazole 2 acceptor N forms a hydrogen bond with one of the hydroxyl groups in the oxalic acid. N–H (ureido) switched its acceptor from O (ureido) in form I/IIIb or O (carbamate) in form II to O (amide). Hence both N–H (amide) and N–H (ureido) in the new form act as hydrogen bond donors to the O (amide) acceptor. In contrast the corresponding O (ureido) hydrogen bonds to one of the oxalic acid hydroxyl groups. A new donor: N–H+ (thiazole 1) forms the ionic salt interaction with the other O− ion in the oxalate molecule.| Compound | Hydrogen bond | d (H⋯A) (Å) | d (D⋯A) (Å) | θ (°) (D–H⋯A) | ||
|---|---|---|---|---|---|---|
| Donor (D–H) | ⋯ | Acceptor (A) | ||||
| Form I1 | N–H (amide) | ⋯ | O (amide) | 1.960 | 3.029(6) | 160.3 |
| N–H (ureido) | ⋯ | O (ureido) | 2.222 | 3.275(8) | 157.5 | |
| N–H (carbamate) | ⋯ | O (carbamate) | 2.131 | 3.261(6) | 148.1 | |
| O–H (hydroxyl) | ⋯ | N (thiazole 2) | 2.141 | 2.989(6) | 131.0 | |
| Form II1 | N–H (amide) | ⋯ | O (hydroxyl) | 2.245 | 2.999(6) | 146.2 |
| N–H (ureido) | ⋯ | O (carbamate) | 2.122 | 2.933(6) | 157.0 | |
| N–H (carbamate) | ⋯ | O (amide) | 2.219 | 2.883(6) | 134.0 | |
| O–H (hydroxyl) | ⋯ | O (ureido) | 1.900 | 2.688(5) | 160.7 | |
| Form IIIb9 | N–H (amide) | ⋯ | O (amide) | 1.946 | 2.773(12) | 160.9 |
| N–H (ureido) | ⋯ | O (ureido) | 2.120 | 2.963(2) | 166.1 | |
| N–H (carbamate) | ⋯ | O (carbamate) | 1.972 | 2.800(14) | 161.4 | |
| O–H (hydroxyl) | ⋯ | N (thiazole 2) | 2.252 | 3.049(16) | 164.5 | |
| Oxalate salt form | N–H (amide) | ⋯ | O (amide) | 2.390 | 3.390(5) | 166.5 |
| N–H (ureido) | ⋯ | O (amide) | 2.116 | 2.900(5) | 151.2 | |
| N–H (carbamate) | ⋯ | O (carbamate) | 2.344 | 3.187(5) | 167.1 | |
| O–H (hydroxyl) | ⋯ | O (oxalate) | 2.029 | 2.849(5) | 179.9 | |
| N–H+ (thiazole 1) | ⋯ | O− (oxalate) | 1.810 | 2.664(5) | 171.6 | |
| O–H (oxalate) | ⋯ | O− (oxalate) | 1.635 | 2.443(5) | 167.9 | |
| O–H (oxalic acid) | ⋯ | O (ureido) | 1.703 | 2.506(9) | 166.3 | |
| O–H (oxalic acid) | ⋯ | N (thiazole 2) | 1.983 | 2.604(10) | 132.0 | |
The N3–H3A (amide)⋯O4 (amide), N4–H4 (ureido)⋯O4 (amide) and N2–H2 (carbamate)⋯O2 (carbamate) hydrogen bonds of ritonavir and the O9–H9A (oxalate)⋯O6− (oxalate) hydrogen bond (with the shortest bond length of 1.635 Å), as shown in Fig. 7 and summarised in Table 4, form the layered 2D hydrogen-bonding network structure with stacking in the a axis direction. It would be expected that the strong stacking structure involving 4 hydrogen bonds would lead to fast crystal growth in the a axis direction. The chain hydrogen bonding structures with ritonavir and the oxalate in the b axis direction, and ritonavir and the oxalic acid in the c axis direction would produce similar growth rates but slower than the a axis direction. This would be consistent with the crystallisation of block-like crystals; similar to those shown in Fig. 2.
Overall, three functional groups (amide, ureido and carbamate) of ritonavir form three hydrogen bonds with another ritonavir molecule with the oxalate ion also forming one hydrogen bond with another molecule of itself in the same a axis direction, leading overall to a 2D hydrogen bonding network. The oxalate also acts as an acceptor forming two hydrogen bonds with the hydroxyl and thiazole 1 groups of two separate ritonavir molecules in the b axis direction, whilst the oxalic acid donates hydrogens to both the ureido and thiazole 2 groups from two separate ritonavir molecules to form two hydrogen bonds in the c axis direction. Hence, a chain hydrogen bonding network is formed in both the b and c axis directions.
Crystal chemistry and local coordination within the quaternary structure
The corresponding calculated pKa values of the two thiazole groups in ritonavir (thiazole 1 = 4.05 and thiazole 2 = 1.10) as shown in Fig. 1, show that these weakly basic nitrogen atoms are the only groups in the molecule which are likely to form salt interactions with oxalic acid due to the ΔpKa values being 2.78 and −0.17 respectively (further details in Table S3 (ESI†)). A recent study into >6000 acid–base pairs in the Cambridge Structural Database, provided convincing evidence that the ΔpKa > 2–3 rule, holds for the large majority of molecules and that there were three distinct regions; zone 1 ΔpKa < −1 where non-ionised hydrogen bonded pairs dominate the crystal structures, zone 2 ΔpKa = −1 to 4 where a combination of non-ionised hydrogen bonded and salt complexes can form and zone 3 ΔpKa > 4 where the vast majority of crystal structures are salts.20 Ritonavir when paired with oxalic acid falls into zone 2 of these categories and perhaps explains to some extent why ritonavir forms both a salt and co-crystal complex with oxalic acid.
Concluding remarks
In this paper, a new solid form of ritonavir (salt co-crystal solvate) was isolated through co-crystallisation with oxalic acid in supersaturated acetone solutions. Single crystal X-ray diffraction analysis reveals the molecular conformation of the new oxalate salt form structure to be associated with the folding of the ritonavir molecule, hence forming a stacking or “U” shape molecular structure similar to that predicted as the global minimum conformation.7 This folding of the molecular conformation not only stabilizes the structure through intra-molecular dispersive interactions, but also exposes its polar atoms to be able to form strong, directional hydrogen bonding and ionic interactions. Both the N-methyl urea conformation and the carbamate conformation in the new oxalate salt form structure were found to be in a trans and trans conformation in contrast to cis and trans, and trans and cis for forms I and II, respectively.14 This might, in turn, suggest that another new form might be feasible having cis and cis conformations, albeit there has been no direct evidence of this to date. However recent discovery of the structure of form IIIb,9 also in a trans and trans conformation, may provide opportunities for further analysis in this regard. The new oxalate salt form co-crystal was also found to have the highest crystal density of all four polymorphs (I, II, IIIb and oxalate salt form) with known structures. There was no observed degradation of the sample upon heating with no obvious loss of structural solvent in the DSC traces. However, in future studies the possibility of removing the structural solvent from the solid-state during, for example, long term storage or at elevated temperatures, would reveal whether these types of materials have utility as pharmaceutical precursors. The crystal habit of the new oxalate salt form structure displays as a blocky-needle where the longer needle axis was found to be the short a crystallographic axis and corresponds to close π–π stacking of the benzene and thiazole rings along this lattice direction.List of symbols and abbreviations
| a, b, c, α, β, γ | Crystal unit cell parameters |
| λ | Radiation wavelength |
| τ | Interatomic torsion angle of molecular fragment |
| Z/Z′ | Number of molecules in the unit cell/asymmetric unit |
| AIDS | Acquired immunodeficiency syndrome |
| API | Active pharmaceutical ingredient |
| BCS | Biopharmaceutics classification system |
| DFT | Density functional theory |
| DSC | Differential scanning calorimetry |
| H-bond | Hydrogen bond |
| OX A | Oxalate |
| OX B | Oxalic acid |
| pKa | Acid dissociation constant at logarithmic scale |
| PXRD | Powder X-ray diffraction |
| Å | Angstrom 10−10 m |
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
One of us (C. W.) acknowledges funding support from the China Scholarship Council. This work was also supported by the Advanced Manufacturing Supply Chain Initiative ‘Advanced Digital Design of Pharmaceutical Therapeutics’ (ADDoPT) project (Grant No. 14060). This research also builds upon previous work on morphological modelling which was supported by EPSRC grant EP/I028293/1. We also gratefully acknowledge EPSRC for the long-term support of crystallisation research at Leeds notably recently through the Critical Mass Project grant ‘Molecules, Clusters and Crystals’ (Grant references EP/IO14446/1 and EP/IO13563/1) which was in collaboration with the University of Manchester. Two of us (R. S. H. and A. S.) are employees of AbbVie and may own AbbVie stock. AbbVie contributed to the design of study; interpretation of data, and in reviewing, and approving of the final publication.References
- J. Bauer, S. Spanton, R. Henry, J. Quick, W. Dziki, W. Porter and J. Morris, Pharm. Res., 2001, 18, 859–866 CrossRef CAS PubMed.
- S. Chemburkar, R. J. Bauer, K. Deming, H. Spiwek, K. Patel, J. Morris and J. Quick, J. Am. Chem. Soc., 2000, 4, 413–417 CAS.
- J. Bernstein, presented in part at the Engineering of Crystalline Materials Properties: State of the Art in Modeling, Design and Applications, NATO Science for Peace and Security Series B-Physics and Biophysics, Dordrecht, 2008 Search PubMed.
- W. Du, Q. Yin, H. Hao, Y. Bao, X. Zhang, J. Huang and J. Gong, Ind. Eng. Chem. Res., 2014, 53, 5652–5659 CrossRef CAS.
- A. Maher, D. M. Croker, Å. C. Rasmuson and B. K. Hodnett, Cryst. Growth Des., 2012, 12, 6151–6157 CrossRef CAS.
- A. Maher, D. M. Croker, C. C. Seaton, Å. C. Rasmuson and B. K. Hodnett, Cryst. Growth Des., 2014, 14, 3967–3974 CrossRef CAS.
- D. Chakraborty, N. Sengupta and D. J. Wales, J. Phys. Chem. B, 2016, 120, 4331–4340 CrossRef CAS PubMed.
- S. L. Morissette, S. Soukasene, D. Levinson, M. J. Cima and O. Almarsson, Proc. Natl. Acad. Sci., 2003, 100, 2180–2184 CrossRef CAS PubMed.
- X. Yao, R. F. Henry and G. G. Z. Zhang, J. Pharm. Sci., 2023, 112, 237–242 CrossRef CAS PubMed.
- S. D. Parent, P. A. Smith, D. K. Purcell,, D. T. Smith, S. J. Bogdanowich-Knipp, A. S. Bhavsar, L. R. Chan, J. M. Croom, H. C. Bauser, A. McCalip, S. R. Byrn and A. Radocea, Cryst. Growth Des., 2023, 23(1), 320–325 CrossRef CAS.
- K. Kawakami, J. Pharm. Sci., 2015, 104, 276–279 CrossRef CAS PubMed.
- K. Kawakami, T. Harada, K. Miura, Y. Yoshihashi, E. Yonemochi, K. Terada and H. Moriyama, Mol. Pharmaceutics, 2014, 11, 1835–1843 CrossRef CAS PubMed.
- K. R. Chaudhari, J. K. Savjani, K. T. Savjani and H. Shah, Drug Dev. Ind. Pharm., 2021, 47, 1633–1642 CrossRef CAS PubMed.
- C. Wang, I. Rosbottom, T. D. Turner, S. Laing, A. G. P. Maloney, A. Y. Sheikh, R. Docherty, Q. Yin and K. J. Roberts, Pharm. Res., 2021, 38, 971–990 CrossRef CAS PubMed.
- N. K. Duggirala, M. L. Perry, Ö. Almarsson and M. J. Zaworotko, Chem. Commun., 2016, 52, 640–655 RSC.
- L. S. Reddy, S. J. Bethune, J. W. Kampf and N. Rodriguez-Hornedo, Cryst. Growth Des., 2009, 9, 378–385 CrossRef CAS.
- N. Venu, B. R. Sreekanth, T. Ram and S. Devarakonda, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2008, 64, o290–o292 CrossRef CAS PubMed.
- S. R. Byrn, R. Pfeiffer and J. Stowell, Solid-State Chemistry of Drugs, SSCI Inc., West Lafayette, 2nd edn, 1999 Search PubMed.
- B. R. Bhogala, S. Basavoju and A. Nangia, CrystEngComm, 2005, 7, 551–562 RSC.
- A. J. Cruz-Cabeza, CrystEngComm, 2012, 14, 6362–6365 RSC.
- A. Denninger, U. Westedt, J. Rosenberg and K. G. Wagner, Pharmaceutics, 2020, 12, 237–260 CrossRef CAS.
- R. I. Gelb, Anal. Chem., 1971, 43, 1110–1113 CrossRef CAS.
- P. T. Galek, F. H. Allen, L. Fábián and N. Feeder, CrystEngComm, 2009, 11, 2634–2639 RSC.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 CrossRef PubMed.
- A. D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang and R. A. Friesner, Int. J. Quantum Chem., 2013, 113, 2110–2142 CrossRef CAS.
- A. D. Bochevarov, M. A. Watson, J. R. Greenwood and D. M. Philipp, J. Chem. Theory Comput., 2016, 12, 6001–6019 CrossRef CAS PubMed.
- Schrödinger Release 2022-3: MacroModel, Schrödinger, LLC, New York, NY, 2021 Search PubMed.
- C. Lu, C. Wu, D. Ghoreishi, W. Chen, L. Wang, W. Damm, G. A. Ross, M. K. Dahlgren, E. Russell, C. D. Von Bargen, R. Abel, R. A. Friesner and E. D. Harder, J. Chem. Theory Comput., 2021, 17, 4291–4300 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS.
- J. S. Binkley, J. A. Pople and W. J. Hehre, J. Am. Chem. Soc., 1980, 102, 939–947 CrossRef CAS.
- R. A. Kendall, T. H. Dunning and R. J. Harrison, J. Chem. Phys., 1992, 96, 6796–6806 CrossRef CAS.
- C. M. Cortis and R. A. Friesner, J. Comput. Chem., 1997, 18, 1570–1590 CrossRef CAS.
- C. Wang, PhD Thesis, Tianjing University, 2020 Search PubMed.
- S. L. Morissette, O. Almarsson and S. Soukasene, Solvates and polymorphs of ritonavir and methods of making and using the same, US Pat., 7205413, 2007 Search PubMed.
- A. Y. Sheikh, A. Mattei, R. Miglani Bhardwaj, R. S. Hong, N. S. Abraham, G. Schneider-Rauber, K. M. Engstrom, M. Diwan, R. F. Henry, Y. Gao, V. Juarez, E. Jordan, D. A. DeGoey and C. W. Hutchins, J. Am. Chem. Soc., 2021, 143, 17479–17491 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2009282 contains the supplementary crystallographic data for this paper. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2ce01612e |
| ‡ School of Chemistry, University of Leeds, Woodhouse Lane, Leeds, LS2 9JT, UK. |
| This journal is © The Royal Society of Chemistry 2023 |