 Open Access Article
Open Access ArticleShape control of Au nanostructures using peptides for biotechnological applications
Shuhei
Yoshida
a,
Kin-ya
Tomizaki
b and
Kenji
Usui
 *a
*a
aFaculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, Chuo-ku, Kobe, Hyogo, 6500047, Japan. E-mail: kusui@konan-u.ac.jp; Fax: +81 78 303 1495; Tel: +81 78 303 1418
bDepartment of Materials Chemistry and Innovative Materials and Processing Research Center, Ryukoku University, Seta-Oe, Otsu, Shiga, 5202194, Japan
First published on 19th October 2023
Abstract
Metallic gold (Au) nanostructures have attracted attentions in various fields of materials science and electrical science in terms of catalysts, sensing systems, photonic devices, and drug delivery systems because of their characteristic physical, chemical, and biocompatible properties. Recently, Au nanostructures with near-infrared light absorbing properties have shown potential for applications such as biological imaging and thermotherapy in biotechnological fields. However, fabrication of Au nanostructures with complex shapes often requires the use of highly biotoxic substances such as surfactants and reducing agents. Peptides are promising compounds for controlling the shape of Au nanostructures by mineralization with several advantages for this purpose. In this highlight, we focus on the shapes with respect to the fabrication of Au nanostructures using biocompatible peptides. We classify the peptides that form Au nanostructures into three broad categories: those that bind Au ions, those that reduce Au ions, and those that control the direction of Au crystal growth. Then, we briefly summarize the correlations between peptide sequences and their roles, and propose future strategies for fabricating Au nanostructures using peptides for biotechnological applications.
1. Introduction
Metallic gold (Au) nanostructures have been actively applied in the fields of materials science and electrical science.1,2 In recent years, studies have been conducted on nanosized Au structures for applications in the biotechnology field, such as photothermal therapy3–8 and in vivo sensing and imaging.9,10 These Au nanostructures had surface plasmon resonance (SPR) properties11,12 especially at near infrared (NIR; >800 nm) absorption wavelengths. The desired optical properties can be imparted by precisely controlling the shape of the Au nanostructures.13–15 For example, one of Au nanostructures, Au nanorods, have SPR-derived absorption in the NIR region, which offers high optical permeability in the body16 and is expected to enable photothermal therapy.17,18 Some specific properties, such as chirality, can be influenced by the shapes of Au nanostructures, such as Au nanocube.19 Several review articles discussing shape control and fabrication methods of Au nanoparticles (AuNPs) have been published.20 Although many studies have been conducted to control the shapes of Au nanostructures21 as described above, some problems remain to be solved for the development of biotechnological applications. The problems are as follows: (I) conventional methods have difficulty in strictly controlling the complicated shapes of nanostructures at the nanoscale; (II) conventional methods can use reducing agents, strong acids and bases, surfactants, and other substances that have high environmental impacts and biotoxicity levels; and (III) it is difficult to add functions that nanoparticles cannot exhibit. For example, the nanoparticles must be biocompatible and deliverable to target organs and cells.18Mimicking biomineralization is a powerful method for fabricating Au nanostructures to overcome these problems. Biomineralization is a synthetic reaction by which certain biomolecules, such as proteins and peptides, precipitate inorganic materials with high reproducibility and accuracy. In Au mineralization processes using such biomolecules, the procedure consists of Au ions binding to the molecules, reduction of Au ions, and growth of Au crystals. Peptides are promising compounds for controlling the shape of Au nanostructures by mineralization because they confer several advantages. (1) Sequences for producing Au nanostructures22 can be efficiently obtained by screening systems such as phage display. (2) The affinity for Au ions can be easily adjusted by changing the amino acid sequence.13 (3) Certain peptides can reduce Au ions without reducing agents, which are environmentally hazardous. Thus, the reduction process minimally burdens the environment. (4) Specific peptides used as cell culture substrates and certain peptides with membrane permeability and cell organelle transferability have been reported.17,18,23–25 In addition, in vivo, inorganic structures with various shapes and morphologies have been fabricated by densely arranging inorganic ions using peptides as templates.26 Peptides are found to form various structures, such as fibres,27 tubes,28 vesicles29 and more.30 These structures can be used artificially as templates to control the shapes of Au nanostructures. These biocompatible functions, which inorganic nanostructures generally lack, can be added to shape-controlled Au nanostructures by conjugation of these sequences with those for producing Au nanostructures.
From these points of view, research on peptide-based mineralization for the fabrication of nanostructures has been active since the 2000s.31 However, no systematic summary of the roles of peptides in the fabrication of Au nanostructures (binding, reduction and crystal growth) has been reported. In this highlight, we focus on the roles of peptides in the fabrication of peptide-based Au nanostructures and the morphology of Au nanostructures reported thus far. We classify the peptides that form Au nanostructures into three broad categories: those that bind Au ions, those that reduce Au ions, and those that control the direction of Au crystal growth. Then, we briefly summarize the correlations between peptide sequences and their roles, and propose future strategies for fabricating Au nanostructures using peptides for biotechnological applications.
2. Au ion-binding peptides
To fabricate Au nanostructures, Au ion binding and accumulating are first needed. We have summarized the representative Au ion-binding peptides reported thus far in Table 1. The Au nanostructures fabricated with Au ion-binding peptides formed spheres, ribbons and superstructures. Review articles discussing superstructures have been published by other authors (see a representative review32).| Name | Sequence | Pepetide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Au ion (mol) Au ion (mol) |
Ref. |
|---|---|---|---|
| Spherical structure | |||
| A3 | H-AYSSGAPPMPPF-OH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
33 |
| Aβ25–35 | H-GSNKGAIIGLM-OH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
35 |
| GBP1 | H-MHGKTQATSGTIQS-OH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
37 |
| Z1 | H-KHKHWHW-OH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
34 |
| R8-AuBP | H-RRRRRRRR-AYSSGAPPMPPF-NH2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
38 |
| Ribbon-like structure | |||
| Aβ25–35 | H-GSNKGAIIGLM-OH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
35 |
| R8-AuBP | H-RRRRRRRR-AYSSGAPPMPPF-NH2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 50 |
38 |
One of the Au ion-binding peptides that forms spherical Au nanostructures is A3.33 A3 was identified from a phage peptide display library. A3 has methionine (Met) in the sequence, as shown by phage display to strongly interact with the zerovalent Au surface. A3 forms hydrophobic interactions and hydrogen bonds on the Au surface. Detailed analysis revealed that Met bound to Au ions. When Au ions were added to A3 peptides in the presence of buffer solution, spherical Au particles formed (Fig. 1a).33 Peptides that bind Au ions, such as A3 peptides, must reduce Au ions by buffer addition conditions,34 reducing agent addition conditions,35 and ultraviolet (UV) irradiation conditions36 to fabricate Au nanostructures because the peptides do not have reducing abilities. In addition to Met, these peptides contain serine (Ser), threonine (Thr), lysine (Lys), glutamine (Gln), and histidine (His), which have been shown to form hydrogen bonds with Au0 and Au ions.33,37 For GBP-1, KTQATS, especially QAT, is important for binding to Au ions.37 For Aβ25–35, amines in the peptide sequence, are used to capture Au ions.35 AuNPs fabricated using A3 exhibited a relatively uniform particle size, and absorbed SPR at approximately 520 nm. However, these AuNPs have no absorption in the NIR region, hindering their biological applications. Consequently, highly complex nanostructures are required.
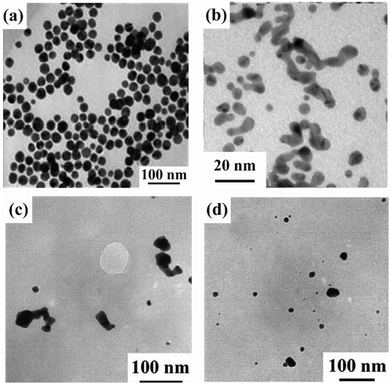 | ||
| Fig. 1 TEM images of the mineralized Au nanostructures obtained using (a) A3, (b) Aβ25–35, and (c) and (d) R8-AuBP. (c) 10 μM R8-AuBP containing 0.5 mM HAuCl4 and (d) 1 μM R8-AuBP containing 0.5 mM HAuCl4. Modified and reproduced from ref. 33, 35 and 38 with permission, copyright 2005, 2018 and 2021, from Wiley-VCH GmbH, American Chemical Society and the Royal Society of Chemistry, respectively. | ||
Peptides that can be fabricated in non-spherical shapes have been reported. One Au nanostructure that can be fabricated using peptides is the ribbon-like structure. A ribbon-like structure has been prepared using Aβ25–35, which is a partial sequence of the aggregating protein amyloid beta (Aβ). In Aβ25–35, ribbon-like structures of Au are prepared using a two-step reaction involving peptide self-assembly and a reduction process.35. The shape of the peptide structures formed by Aβ25–35 varies depending on the self-assembly reaction time and solvent conditions. The Au nanostructures are prepared using these peptide structures as templates. First, Aβ25–35 was self-assembled to produce peptide ribbon-like structures. Then, Au ions were added to the self-assembled peptide structures, and the amines of the peptides were used to place the Au ions on the peptides. The Au nanoribbon structure was then fabricated by adding a reducing agent to reduce the Au ions (Fig. 1b). SPR absorption was observed at 520 nm for Au nanoribbons fabricated with Aβ25–35, although the shape changed. That is, SPR absorption derived from ribbon-like structures cannot be confirmed. Thus reducing agents should be required, which makes biological applications difficult. In addition, in 2021, we reported the use of R8-AuBP (the AuBP sequence was same as that of A333), a complex of the cell-permeable peptide R8 and the Au ion-binding peptide (AuBP), to fabricate ribbon-like structures directly in cells.38 The Au nanoribbons could be fabricated directly into the cell using the intracellular reducing environment. We attempted to control Au mineralization in cells by changing the peptide concentrations. The lower peptide concentration showed only spherical particles (Fig. 1c and d). The shape of the peptide-only structures that formed extracellularly was consistent with the Au nanostructures in cells. Thus, Au nanostructures are formed using the peptide as a template. However, none of the Au nanostructures are sufficiently crystalline to be applied to photothermal therapy. To improve their crystallinity and utilize them in certain biofield applications, such as photothermal therapy in the future, it is necessary to adjust their reduction ability using Au ion reduction peptides.
3. Au ion-reducing peptides
Peptides that can both bind and reduce Au ions have been reported. We describe Au nanostructures fabricated using peptides that can reduce Au ions. We have summarized the representative Au ion-reducing peptides reported thus far in Table 2. The Au nanostructures fabricated with Au ion-reducing peptides formed spheres and ribbons and superstructures.An Au-ion-reducing peptide that forms spherical Au nanostructures is AuBP1.39 The sequence of AuBP1 was isolated by phage display. AuBP1 contains tryptophan (Trp) in the sequence and can reduce Au ions without using a reducing agent.39,40 Furthermore, the Au spherical particles reduced by AuBP1 exhibited catalytic activity and excellent dispersibility13,41,42 (Fig. 2a). In experiments where the number of Trp residues in AuBP1 was increased, the diameters of the AuNPs decreased as the number of Trp residues in the sequences increased.13 Thus, the reducing power of Au ions can be controlled by the number of π-electrons in the aromatic ring of the Trp residue in the peptide sequence. That is, by changing the number and type of aromatic rings in the array, the nucleation rate of Au ions can be varied, and the size of the AuNPs can be controlled. AuNPs prepared using AuBP1 have relatively uniform particle sizes and exhibit SPR absorption at approximately 545 nm. However, these AuNPs do not absorb light in the NIR region, making their biological applications as challenging as those using Au ion-binding peptides.
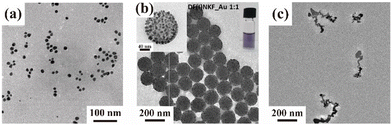 | ||
| Fig. 2 (a)–(c) TEM images of mineralized Au nanostructures obtained using (a) AuBP1, (b) DF(I)NKF, and (c) RU006. Modified and reproduced from ref. 13, 44 and 43 with permission, copyright 2020, 2019 and 2014, from the Royal Society of Chemistry and American Chemical Society, respectively. | ||
Other Au ion-reduced peptides have also been used to create superstructures. C. Pigliacelli et al. used a peptide structure as a template to place AuNPs via iodine substitution of phenylalanine in the cohesive peptide sequence DFNKF (Fig. 2b).43 This superstructure has an absorption at approximately 580 nm and experiences a red-shift greater than that of single spherical AuNPs. In addition, observations of the shape of the structures formed at different peptide concentrations were performed at [DF(I)NKF] = 125, 250, and 500 μM and [HAuCl4] = 500 μM. The results showed that under the lowest peptide concentration condition, single Au NPs of 5–10 nm could be identified and that the AuNPs assembled as the peptide concentration increases. However, although the superstructures fabricated in this report used the peptide structure as a template, a difference was present in the shape of the Au structure and the peptide-only structure.27 Therefore, it is difficult to fabricate Au nanostructures with arbitrary shapes for biotechnology applications. In the future, it will be necessary to elucidate certain mechanisms, such as the accumulation of Au ions on the peptides to control the shape.
Furthermore, peptides that can be fabricated in non-spherical shapes have been reported. Au ion-reducing peptides formed ribbon-like structures, similarily to the Au ion-binding peptides described above. First, we discuss peptides that can reduce Au ions, such as RU006 reported by our group.44 RU006 was designed to provide the driving force for self-assembly into a β-sheet conformation via hydrophobic interactions and π−π stacking. When Au ions were added, the peptides assembled into ribbon-shaped nanostructures by entrapment of Au ions during self-assembly. By using the assembled ribbon-shaped nanostructure as a template, the aromatic amino acids in the sequence reduced the Au ions to form ribbon-like Au structures (Fig. 2c). UV-visible (Vis)-NIR measurements of this ribbon-like structure provided absorption spectra with a broad Vis-near IR band. Given the extensive research on Au nanorods, this result suggests that ribbon-like AuNPs with NIR absorption can be applied to photothermal therapy.
Furthermore, we demonstrated selective Au recovery from an aqueous mixture of HAuCl4 and H2PtCl6 (5.0 × 10−5 M each) by using (Ant6)-RU006 (2.0 × 10−4 M), in which L-2-naphthylalanine in RU006 was replaced with L-2-anthrylalanine. We found that (Ant6)-RU006 selectively reduced and recovered Au with an atomic ratio (Au/Pt) of 7.5.45 With these interesting results in hand, we downsized (Ant6)-RU006 for industrialization from both the N- and C-termini by deleting amino acids individually. The fragment from the 4th to the 8th positions of (Ant6)-RU006 reduced and recovered Au with the same as the original (Ant6)-RU006. It features an anthracene ring as an electron source, two positive charges, moderate hydropathy, and forms hydrogen bonds to reduce Au ions and densify the resulting metallic Au particles, enabling facile separation from a mixture of Au and platinum ions via centrifugation.46 These findings might facilitate the design of low-cost nonpeptidyl molecules for a novel selective Au recovery process.
From the above, the Au nanostructures fabricated with peptides that reduce Au ions exhibit spherical and ribbon-like structures. However, each structure is problematic. The spherical structure is not anisotropic and has no absorption in the NIR region. The ribbon-like structure has an aspect ratio but is less crystalline and does not exhibit SPR-derived absorption. The following studies addressed these issues. CH3–(CH2)14–WWA-OH and CH3–(CH2)14–WWV-OH reported by V. Kumar et al. successfully fabricated Au nanorods by irradiation with a 532 nm laser after spherical AuNP formation.47 The Au nanorods shift in SPR absorption towards longer wavelengths and have a more uniform shape than the ribbon-like structures. Furthermore, after preparing AuNPs using the same peptides, they are irradiated with sunlight; it is confirmed that AuNPs bind to each other.48 Thus, in recent years, examples of the fabrication of anisotropic Au nanostructures by providing light irradiation have been reported.48–50 In the future, it is expected that by controlling the arrangement of AuNPs and by using peptide structures as templates, it is possible to produce anisotropic Au nanostructures using peptides.
4. Au crystal growth-controlling peptides
We describe cysteine (Cys)-containing short-chain peptides that were not used as templates but were used as reagents to control the direction of Au crystal growth. Begining with a study reported in 2018, attempts have been made to fabricate Au nanostructures with complex geometries on their surfaces (Table 3).51–53 In the fabrication of these Au nanostructures, peptides that could control the direction of Au crystal growth were used. At first, a 5-residue peptide was found in 2018, but it could not completely control the shapes.51 Subsequently, even shorter peptides were used to achieve complete shape control. Herein we discuss these 3- and 2-residue peptides in detail.52,53 Scholars demonstrated the fabrication of helicoidal structures, reported in 2018 and 2020. Helicoidal structures were cubic Au nanostructures with regular helical planes on their surface. Helicoidal structures were shown to possess chirality, and were expected to have applications in biotechnology, such as sensing and imaging, using the differences in optical rotation.54 Although scholars could manufacture Au nanostructures with a few complex shapes, many of these peptides required reducing agents.4.1. Glutathione for fabrication of helicoidal structures
In 2018, H.-E. Lee et al. reported that the addition of glutathione (GSH) consisting of three amino acid residues, Au ions, and reducing agents to a presynthesized cubic Au nanostructure resulted in the formation of Au helicoidal structures52 (Fig. 3a and b). When GSH was added, Au crystal growth proceeded in a fan-like fashion, forming 432 helicoid I (Fig. 3a). This phenomenon was consistent with the fact that Cys affected the Au crystal structure, as reported by A. Kühnle et al. in 2005.55 The interaction of the thiol groups of Cys with the Au surface controlled the direction in which the Au ions were supplied, forming a helicoidal structure with a unique surface structure. Scholars performed an experiment in which they added the L and D forms of a single residue of the amino acid Cys instead of GSH to produce a helicoidal structure. Therefore, the Au helicoidal structure was altered by the chirality of Cys to form 432 helicoid II (Fig. 3b). These results suggested that the surface morphologies of Au nanostructures could be controlled by using optical isomeric amino acids to fabricate peptide-based Au nanostructures. Furthermore, since nanostructures with surface structures twisted by T. Greber et al. could identify enantiomers of organic compounds interacting with the nanostructure surface, the Au helicoidal structures were expected to be applied to enantiomer identification. That is, the precise control of the surface structure would be necessary to give Au nanostructures substance specificity.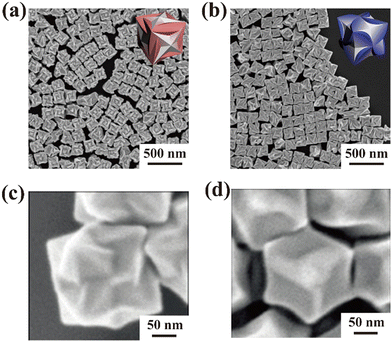 | ||
| Fig. 3 SEM image and illustration of 432 helicoids. (a) 432 helicoid I fabricated by L-GSH. (b) 432 helicoid II fabricated by D-GSH. (c) 432 helicoid I fabricated by γ-Glu-Cys. (d) 432 helicoid V fabricated by Cys-Gly. Modified and reproduced from ref. 52 and 53 with permission, copyright 2018 and 2020, from Nature Publishing Group and Wiley-VCH GmbH, respectively. | ||
4.2. Dipeptides for fabrication of helicoidal structures
In 2020, dipeptides forming helicoidal structures with a shape different from that reported by H.-e. Lee et al. were identified (Fig. 3c and d).53 In the report by H. Kim et al., two different dipeptides were used to create a helicoidal structure. The dipeptides were each GSH minus glycine and formed amide bonds at the α- and γ-positions. Therefore, two types of helicoidal structures were formed, as shown in Fig. 3c and d. Fig. 3c shows 432 helicoid I prepared with γ-Glu-Cys, and Fig. 3d shows 432 helicoid V prepared with Glu-Cys. Due to the difference in the position of the amide bond, the strain position in which Cys could interact changed, forming helicoidal structures with different shapes. There have been an increasing number of reports on the use of peptides to control the direction of Au crystal growth. If the specificity of molecules that could interact with each structure surface shape could be improved, these controls could be achieved more effectively and the Au nanostructures could have potential applications in sensing. While it was proposed that these short peptides interacted simultaneously with Au ions and their nucleus to form cubic Au nanostructures, the details of their interactions have not yet been clarified, and analyses at the atomic and ionic levels are needed in the future. In addition, these peptides could not reduce Au ions; therefore, the addition of a reducing agent would be needed. Consequently, the addition of reducing ability to the peptides is an issue to be addressed in the future.Conclusions
We discussed peptides capable of producing AuNPs of various shapes, and we discussed the role of each peptide. We classified the peptides into three broad categories: those that bound to Au ions, those that reduced Au ions, and those that controlled the direction of Au crystal growth. However, the formed AuNPs had low aspect ratios to prepare spherical particles, low crystallinity in ribbon-like structures, and forming helicoids required reducing agents. These characteristics could pose problems for biological applications in terms of SPR properties especially at NIR (>800 nm) absorption wavelengths. Therefore, we hypothesized that the combination of these peptides could solve these problems. For example, anisotropic Au nanostructures with high crystallinity could be fabricated by combining certain peptide sequences that could form spherical structures to enhance the crystallinity and that form ribbon-like structures.In the future, better shape-controlled Au nanostructures could be fabricated by combining such peptide sequences playing the three roles. Thus, with the advancement of research on the fabrication of Au nanostructures by peptide-based mineralization and with the improved understanding of the atomic and ionic levels, the shape control of Au nanostructures and their applications in the biotechnology field, including photothermal therapy and sensing, would be developed.
Author contributions
All authors contributed to the discussion of the contents and the editing of the manuscript prior to submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Prof. T. Nishikata for valuable discussions and generous support.References
- V. Biju, Chem. Soc. Rev., 2014, 43, 744–764 RSC.
- Z. Fan, X. Huang, C. Tan and H. Zhang, Chem. Sci., 2015, 6, 95–111 RSC.
- Y. Liu, P. Bhattarai, Z. Dai and X. Chen, Chem. Soc. Rev., 2019, 48, 2053–2108 RSC.
- H. Chen, L. Shao, Q. Li and J. Wang, Chem. Soc. Rev., 2013, 42, 2679–2724 RSC.
- J. Qiu, M. Xie, T. Wu, D. Qin and Y. Xia, Chem. Sci., 2020, 11, 12955–12973 RSC.
- J. Zong, S. L. Cobb and N. R. Cameron, Biomater. Sci., 2017, 5, 872–886 RSC.
- J. Zhou, Y. Jiang, S. Hou, P. K. Upputuri, D. Wu, J. Li, P. Wang, X. Zhen, M. Pramanik, K. Pu and H. Duan, ACS Nano, 2018, 12, 2643–2651 CrossRef CAS PubMed.
- M. Hu, J. Chen, Z.-Y. Li, L. Au, G. V. Hartland, X. Li, M. Marquez and Y. Xia, Chem. Soc. Rev., 2006, 35, 1084–1094 RSC.
- C. Lu, L. Han, J. Wang, J. Wan, G. Song and J. Rao, Chem. Soc. Rev., 2021, 50, 8102–8146 RSC.
- J. Qiu, Y. Liu and Y. Xia, Adv. Healthcare Mater., 2021, 10, e2002031 CrossRef PubMed.
- X. Yang, M. Yang, B. Pang, M. Vara and Y. Xia, Chem. Rev., 2015, 115, 10410–10488 CrossRef CAS PubMed.
- Y. Imura, R. Akiyama, S. Furukawa, R. Kan, C. Morita-Imura, T. Komatsu and T. Kawai, Chem. – Asian J., 2019, 14, 547–552 CrossRef CAS PubMed.
- M. Ozaki, S. Yoshida, M. Oura, T. Tsuruoka and K. Usui, RSC Adv., 2020, 10, 40461–40466 RSC.
- S. E. Skrabalak, J. Chen, Y. Sun, X. Lu, L. Au, C. M. Cobley and Y. Xia, Acc. Chem. Res., 2008, 41, 1587–1595 CrossRef CAS PubMed.
- T. H. Chow, Y. Lai, X. Cui, W. Lu, X. Zhuo and J. Wang, Small, 2019, 15, e1902608 CrossRef PubMed.
- B. Nikoobakht and M. A. EL-Sayed, Chem. Mater., 2003, 15, 1957–1962 CrossRef CAS.
- H. J. Moon, M. Ku, H. Lee, N. Yoon, J. Yang and K. W. Bong, Sci. Rep., 2018, 8, 13683 CrossRef PubMed.
- J. Wan, J.-H. Wang, T. Liu, Z. Xie, X.-F. Yu and W. Li, Sci. Rep., 2015, 5, 11398 CrossRef CAS PubMed.
- F. Lu, Y. Tian, M. Liu, D. Su, H. Zhang, A. O. Govorov and O. Gang, Nano Lett., 2013, 13, 3145–3151 CrossRef CAS PubMed.
- X. Hu, Y. Zhang, T. Ding, J. Liu and H. Zhao, Front. Bioeng. Biotechnol., 2020, 8, 990 CrossRef PubMed.
- C. M. Cobley, J. Chen, E. C. Cho, L. V. Wang and Y. Xia, Chem. Soc. Rev., 2011, 40, 44–56 RSC.
- M. Grzelczak, J. Perez-Juste, P. Mulvaney and L. M. Liz-Marzan, Chem. Soc. Rev., 2008, 37, 1783–1791 RSC.
- S. Futaki, T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda and Y. Sugiura, J. Biol. Chem., 2001, 276, 5836–5840 CrossRef CAS PubMed.
- S. Futaki and I. Nakase, Acc. Chem. Res., 2017, 50, 2449–2456 CrossRef CAS PubMed.
- N. Pfanner, Curr. Biol., 2000, 10, R412–R415 CrossRef CAS PubMed.
- L. C. Palmer, C. J. Newcomb, S. R. Kaltz, E. D. Spoerke and S. I. Stupp, Chem. Rev., 2008, 108, 4754–4783 CrossRef CAS PubMed.
- A. Bertolani, L. Pirrie, L. Stefan, N. Houbenov, J. S. Haataja, L. Catalano, G. Terraneo, G. Giancane, L. Valli, R. Milani, O. Ikkala, G. Resnati and P. Metrangolo, Nat. Commun., 2015, 6, 7574 CrossRef PubMed.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CrossRef CAS PubMed.
- X. Yan, Q. He, K. Wang, L. Duan, Y. Cui and J. Li, Angew. Chem., Int. Ed., 2007, 46, 2431–2434 CrossRef CAS PubMed.
- X. Yan, P. Zhu and J. Li, Chem. Soc. Rev., 2010, 39, 1877–1890 RSC.
- W. J. Crookes-Goodson, J. M. Slocik and R. R. Naik, Chem. Soc. Rev., 2008, 37, 2403–2412 RSC.
- S. Mokashi-Punekar, Y. Zhou, S. C. Brooks and N. L. Rosi, Adv. Mater., 2020, 32, e1905975 CrossRef PubMed.
- J. M. Slocik, M. O. Stone and R. R. Naik, Small, 2005, 1, 1048–1052 CrossRef CAS PubMed.
- B. R. Peelle, E. M. Krauland, K. D. Wittrup and A. M. Belcher, Langmuir, 2005, 21, 6929–6933 CrossRef CAS PubMed.
- Y. Feng, H. Wang, J. Zhang, Y. Song, M. Meng, J. Mi, H. Yin and L. Liu, Biomacromolecules, 2018, 19, 2432–2442 CrossRef CAS PubMed.
- R. Djalali, J. Samason and H. Mastui, J. Am. Chem. Soc., 2004, 126, 7935–7939 CrossRef CAS PubMed.
- J. L. Kulp III, M. Sarikaya and J. S. Evans, J. Mater. Chem., 2004, 14, 2325–2332 RSC.
- M. Ozaki, S. Yoshida, T. Tsuruoka and K. Usui, Chem. Commun., 2021, 57, 725–728 RSC.
- C. J. Munro, Z. E. Hughes, T. R. Walsh and M. R. Knecht, J. Phys. Chem. C, 2016, 120, 18917–18924 CrossRef CAS.
- S. Si and T. K. Mandal, Chem. – Eur. J., 2007, 13, 3160–3168 CrossRef CAS PubMed.
- C. J. Munro and M. R. Knecht, Langmuir, 2017, 33, 13757–13765 CrossRef CAS PubMed.
- N. M. Bedford, Z. E. Hughes, Z. Tang, Y. Li, B. D. Briggs, Y. Ren, M. T. Swihart, V. G. Petkov, R. R. Naik, M. R. Knecht and T. R. Walsh, J. Am. Chem. Soc., 2016, 138, 540–548 CrossRef CAS PubMed.
- C. Pigliacelli, K. B. Sanjeeva Nonappa, A. Pizzi, A. Gori, F. B. Bombelli and P. Metrangolo, ACS Nano, 2019, 13, 2158–2166 CAS.
- K.-Y. Tomizaki, S. Wakizaka, Y. Yamaguchi, A. Kobayashi and T. Imai, Langmuir, 2014, 30, 846–856 CrossRef CAS PubMed.
- K.-Y. Tomizaki, T. Okamoto, T. Tonoda, T. Imai and M. Asano, Int. J. Mol. Sci., 2020, 21, 5060 CrossRef CAS PubMed.
- K.-Y. Tomizaki, T. Tonoda, S. Teramura, H. Okazaki, T. Imai and M. Asano, Processes, 2021, 9, 2010 CrossRef CAS.
- V. Kumar, N. K. Mishra, S. Gupta and K. B. Joshi, ChemistrySelect, 2017, 2, 211–218 CrossRef CAS.
- K. Kesharwani, A. Kautu, S. Sharma, R. Singh, V. Kumar, S. K. Tripathi, P. Shukla and K. B. Joshi, Chem. Commun., 2022, 58, 13815–13818 RSC.
- N. K. Mishra, V. Kumar and K. B. Joshi, RSC Adv., 2015, 5, 64387–64394 RSC.
- N. K. Mishra, K. B. Joshi and S. Verma, J. Colloid Interface Sci., 2015, 455, 145–153 CrossRef CAS PubMed.
- H.-E. Lee, J. Lee, M. Ju, H.-Y. Ahn, Y. Y. Lee, H.-S. Jang and K. T. Nam, Mol. Syst. Des. Eng., 2018, 3, 581–590 RSC.
- H.-E. Lee, H.-Y. Ahn, J. Mun, Y. Y. Lee, M. Kim, N. H. Cho, K. Chang, W. S. Kim, J. Rho and K. T. Nam, Nature, 2018, 556, 360–365 CrossRef CAS PubMed.
- H. Kim, S. W. Im, N. H. Cho, D. H. Seo, R. M. Kim, Y.-C. Lim, H.-E. Lee, H.-Y. Ahn and K. T. Nam, Angew. Chem., Int. Ed., 2020, 59, 12976–12983 CrossRef CAS PubMed.
- T. Greber, Z. Sljivancanin, R. Schillinger, J. Wider and B. Hammer, Phys. Rev. Lett., 2006, 96, 056103 CrossRef CAS PubMed.
- A. Kuhnle, T. R. Linderoth and F. Besenbacher, J. Am. Chem. Soc., 2006, 128, 1076–1077 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |



