 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Precision nanoengineering for functional self-assemblies across length scales†
Nonappa

Facutly of Engineering and Natural Sciences, Tampere University, FI-33720, Tampere, Finland. E-mail: nonappa@tuni.fi
First published on 17th October 2023
Abstract
As nanotechnology continues to push the boundaries across disciplines, there is an increasing need for engineering nanomaterials with atomic-level precision for self-assembly across length scales, i.e., from the nanoscale to the macroscale. Although molecular self-assembly allows atomic precision, extending it beyond certain length scales presents a challenge. Therefore, the attention has turned to size and shape-controlled metal nanoparticles as building blocks for multifunctional colloidal self-assemblies. However, traditionally, metal nanoparticles suffer from polydispersity, uncontrolled aggregation, and inhomogeneous ligand distribution, resulting in heterogeneous end products. In this feature article, I will discuss how virus capsids provide clues for designing subunit-based, precise, efficient, and error-free self-assembly of colloidal molecules. The atomically precise nanoscale proteinic subunits of capsids display rigidity (conformational and structural) and patchy distribution of interacting sites. Recent experimental evidence suggests that atomically precise noble metal nanoclusters display an anisotropic distribution of ligands and patchy ligand bundles. This enables symmetry breaking, consequently offering a facile route for two-dimensional colloidal crystals, bilayers, and elastic monolayer membranes. Furthermore, inter-nanocluster interactions mediated via the ligand functional groups are versatile, offering routes for discrete supracolloidal capsids, composite cages, toroids, and macroscopic hierarchically porous frameworks. Therefore, engineered nanoparticles with atomically precise structures have the potential to overcome the limitations of molecular self-assembly and large colloidal particles. Self-assembly allows the emergence of new optical properties, mechanical strength, photothermal stability, catalytic efficiency, quantum yield, and biological properties. The self-assembled structures allow reproducible optoelectronic properties, mechanical performance, and accurate sensing. More importantly, the intrinsic properties of individual nanoclusters are retained across length scales. The atomically precise nanoparticles offer enormous potential for next-generation functional materials, optoelectronics, precision sensors, and photonic devices.
1. Introduction
Achieving atomic precision across length scales is one of the fundamental challenges in modern nanotechnology. The future of optoelectronics, catalysis, biosensing, nanomedicine, and solutions for environmental challenges relies heavily on the ability to fabricate materials with highly precise structures and reproducible properties.1–4 Such precision could offer programmable control over the shape, strength, conductivity, mechanical properties, and optical responses with high sensitivity.5–7 While the potential benefits are clear, methods that allow atomic precision at the macroscopic scale are limited. Nevertheless, several attempts have been made to fabricate precise structures across length scales in recent years.8 Large-scale three-dimensional (3D) crystals can be considered atomically precise macroscopic objects. However, crystals have limitations due to their poor mechanical properties. Notably, recent reports have demonstrated mechanically strong and flexible crystals of small molecules.9 Furthermore, reversible two-dimensional (2D) polymer sheets have been demonstrated using the photochemical polymerization of monomer crystals.10 The above reports are limited to certain molecules or materials under highly controlled and narrow experimental conditions. Notably, building materials atom by atom is a laborious way to produce structures in scalable quantities.Molecular engineering is a well-established area of research. It allows precise control over the structure, functional groups, geometry, and directional interactions of molecular building blocks for the design and fabrication of materials, and systems with specific properties and functions.11,12 It focuses on manipulating molecular properties, behaviour, and interactions to improve or fabricate new materials and their applications. Therefore, molecular engineering has contributed tremendously to the success of molecular self-assembly, offering a wide range of morphologically distinct functional architectures.13,14 Despite huge success in molecular self-assembly, obtaining large-scale structures with high precision is challenging.15 Furthermore, achieving strictly monodisperse building blocks is far more challenging for progressively larger structures.16 Moreover, non-specific binding at larger scales results in a mixture of different structures rather than the desired product.17 Nevertheless, at higher length scales, block-copolymers, and even molecular polymer brushes, have been reported to achieve structures with precise topography under controlled and narrow experimental conditions.18,19
Beyond molecular and supramolecular scales, nanoscale building blocks have enabled colloidal and supracolloidal self-assembly.20 Specifically, noble metal nanoparticles (MNPs) have emerged as attractive building blocks for self-assembly and hierarchical structure formation.21 The size and shape-dependent optoelectronic properties make noble metal nanoparticles and their self-assembled structures attractive for various applications, including catalysis, sensing, imaging, drug delivery, hydrogen evolution reactions, and photothermal therapy.22 Furthermore, self-assembled superstructures allow nanoconfinement for selective encapsulation and can act as nanoflasks for chemical reactions.23 Despite tremendous progress in plasmonic nanoparticle-based self-assembly, several challenges exist.24 Importantly, the design of MNPs with well-defined shapes, sizes, and surface chemistry with precision still needs to be improved. Due to their high surface-to-volume ratio, MNPs have high surface energy. To minimize the surface energy, they undergo uncontrolled aggregation. Furthermore, MNPs display polydispersity and lack directional interactions.25 Importantly, underlying driving forces and the role of the solvent and other environmental factors that control the nanoparticle interactions are not fully understood. The complexity of the inter-particle interactions and the lack of mechanistic insights are major challenges in achieving controlled self-assembly.26 For example, van der Waals, electrostatic, and steric interactions play a crucial role in nanoparticle self-assembly. However, the exact contribution of these forces and their dependence on nanoparticle size, shape, and surface chemistry remain to be determined.27 The high surface energy of MNPs also destabilizes certain structures, limiting access to self-assembled 1D or 2D layered structures in a template or template-free manner.28 Due to their isotropic nature, the self-assembly of spherical MNPs and other colloidal particles often results in 3D structures. Achieving 1D or 2D structures requires anisotropic interactions between the neighboring nanoparticles.29 The inhomogeneous distribution of surface charge allows electrical dipoles. This approach can be utilized to achieve the dipole-induced self-assembly of nanoparticles into 1D structures provided the inter-nanoparticle interaction strength exceeds the thermal fluctuation energy and Coulombic repulsion.30 External fields such as magnetic and electrical fields, selective ligand exchange, DNA functionalization, and spatial and shear confinement approaches have been utilized to achieve MNP-based 1D and 2D structures.29–31
Computational simulations have been used to study the self-assembly pathway and intermediates of metal nanoparticles.32 Specifically, molecular dynamics (MD), coarse-grained (CG), and Monte Carlo (MC) simulations have been employed. CG and all-atom MD simulations have been used to gain insights into the effects of ligand density on self-assembly and intermolecular forces at the nanoscale.33 Recent studies using all-atom MD simulations have revealed that for gold nanoparticles (AuNPs) coated with dodecanethiol in a toluene-ethanol/water interface, self-assembly is dominated by interactions between the ligands, specifically van der Waals interactions.34 Furthermore, self-assembly depends on the evaporation rate: slow evaporation results in two-dimensional (2D) arrays. Replacing dodecanethiol ligands with mercaptoundecanoic acid results in a roughly ordered, close-packed array with faster aggregation independent of the evaporation rate. This is attributed to the strong polarizing nature of mercaptoundecanoic acid molecules and the strong interactions between them.
Importantly, over the past two decades, size and shape-controlled syntheses have allowed a range of stable noble metal nanoparticles,35 providing routes for 3D crystals,36 2D layered structures,37 and elastic membranes.38 More detailed theoretical insights and experimental approaches for MNP-based 1D, 2D, and 3D assemblies are beyond the scope of this article and have been discussed in several recent reviews.30,31 Moreover, MNP-based structures have been considered inorganic protein mimics.39 Metallic helical structures,40 chiral superlattices,41 and isomeric clusters,42 have also been developed. However, even with narrow size dispersed metal nanoparticles, the batch-to-batch variations, and inhomogeneous ligand density lead to irreproducible mechanical properties, poor device performance, and inaccurate sensing.43,44
The progress in molecular engineering, nanomaterials, fabrication methods, and tools for atomic-level manipulation have contributed to the development of atomically precise manufacturing (APM) approaches.45 The potential applications of APM towards molecular electronics devices, single electron transistors, quantum computing, and nanoresonators have been demonstrated.46 A sophisticated positional control approach produces complex 3D objects but with precision for only a few atoms.47 Furthermore, super-resolution nanolithography and combinatorial patterning have been reported for precision assemblies.48 Despite numerous efforts, large-scale structure formation with atomic-level precision has remained challenging.
In colloidal science, polymer microparticles with controlled size, shape, and surface functionalities have been viewed as “big atoms” as an analogy to atoms.49 Furthermore, it has been shown that discrete clusters of colloidal microparticles can be achieved with a wide range of geometries.50 The resulting finite clusters of colloidal particles are referred to as “colloidal molecules” as they mimic the symmetry of molecular structures.51 The colloidal clusters sustain Brownian motion and allow real-time imaging. Therefore, they act as model systems to investigate the structure, dynamics, and self-assembly properties at the mesoscale. Block-co-polymers, such as triblock co-polymers, offer microparticles with rich surface chemistry, anisotropy, and patchiness.52 Recent experimental studies and computational simulations have shown that patchy colloidal particles offer routes for designing new assemblies with emerging optical properties.53–55
Despite rapid increases in experimental studies and computational simulation in colloidal clusters and plasmonic nanoparticles, identifying the exact number of interacting units at atomic-level precision remains a major challenge. Resolving this issue by using “strictly monodisperse” nanoscale building blocks with engineered properties would represent a significant advance in nanoengineering. This would enable new assembly methods for low-cost, large-area, and flexible devices using solution processing methods to meet the ever-growing demand to integrate next-generation functional devices. In this context, precision nanoengineering, i.e., engineering atomically precise nanoscale building blocks and developing methods for their fabrication into large-scale structures can overcome the challenges faced in manufacturing precision across length scale. Furthermore, the approach will bridge the gap between molecular and colloidal self-assemblies.
Precision nanoengineering has no formalized definition. However, in the context of this feature article, it encompasses the design, synthesis, and characterization of atomically precise nanoscale building blocks for the fabrication of materials, devices, and systems across length scales. The approach will provide routes for self-assembled discrete structures, composites, and large-scale defect-free structures. Due to the atomic-level precision of the building blocks, the position, orientation, and interaction of each unit can be specified. The fabrication may involve one, two, or more than two types of building blocks or components. When two or more building blocks are involved, at least one of the components is atomically precise, while the other is narrowly size dispersed (<5% dispersity). The components can be either synthetic (e.g., atomically precise metal nanoparticles, organic nanoparticles, or organic–inorganic hybrids), biological nanoparticles (e.g., proteins, virus particles, or DNA origamis), or narrowly size dispersed synthetic nanoparticles. Unlike APM, here, the focus is to utilize the atomically precise nanoparticle building blocks for higher-order structure formation. Since the building blocks are atomically precise and at the colloidal level, self-assembly will result in atomically precise higher-order structures. In this context, recent progress in the size and shape-controlled synthesis of monolayer thiol-protected atomically precise noble nanoclusters (NCs) provide new routes to realize precision nanoengineering-enabled self-assemblies across length scales.56,57
In the next section of this feature article, I will discuss the following: (i) the state-of-the-art challenges in the self-assembly of atomically precise NCs; (ii) how to obtain clues from biological colloidal molecules to achieve efficient and error-free self-assembly across length scales, i.e., structures that span multiple size regimes, from the nanoscale to the macroscale; (iii) how to exploit the patchy and anisotropic distribution of ligands on the NC surface for self-assembled 2D colloidal crystals, macroscopic membranes, capsids, and composite cages; (iv) new methods for NC-based reversible frameworks using dynamic covalent chemistry; and (v) in situ synthesis and site-selective doping of photothermally stable NCs on colloidal templates. Specifically, the focus is on atomically precise gold (Au) and silver (Ag) NC-based self-assembled structures. Beyond structure formation, the goal is to design systems with specific properties and functions. Therefore, these self-assembled structures are referred to as functional self-assemblies. Accordingly, the self-assembly-induced amplification or emergence of photophysical properties, catalytic efficiency, and mechanical properties are discussed. The need for building blocks with atomic precision in controlled structure formation, reproducible optomechanical properties, and sensing is highlighted. The importance of self-assembly in the amplification of photoluminescence quantum yield, catalytic efficiency, bioimaging, sensing, and photothermal stability is also presented.
2. Atomically precise nanoclusters
To utilize any building block for controlled structure formation, well-defined structures, functional groups, directional interactions, and stability are needed. Atomically precise monolayer thiol-protected noble metal NCs with a few to hundreds of atoms have emerged as an important class of nanomaterials.58 Despite more than six decades of history,59,60 their extensive structural characterization, understanding of their optoelectronic properties, and applications of atomically precise nanoclusters have emerged only in the past two decades.61–63 Because of their well-defined structure, various characterization tools are routinely used to determine the molecular structure, composition, and optical and electronic properties of NCs.64 The molecular structure at atomic-level resolution is crucial for understanding the packing of metal atoms, the position and orientation of ligands, and molecular packing in macroscopic crystals. Single-crystal X-ray diffraction (SCXRD) is the most powerful method for complete structure determination. However, X-ray diffraction requires pure and high-quality crystals of NCs. Due to their large size, obtaining single crystals suitable for diffraction is challenging. Despite these challenges, several reports have presented high-resolution single-crystal X-ray structures of NCs. Notably, in 2007, Jadzinsky et al. reported the first crystal structure of monolayer thiol-protected water-soluble Au NC, Au102-pMBA44.65 In 2008, Zhu et al. reported the crystal structure of phenylethane thiol-capped, 25-atom gold cluster, Au25PET18.66 Since then, more than 100 single-crystal X-ray structures of AuNCs, Ag NCs, and alloy NCs have been reported in the literature.67 However, most of the crystal structures reported in the literature are for NCs soluble in organic solvents. The crystallization of water-soluble NCs is still a major challenge. Recently, cryogenic transmission electron microscopy (cryo-TEM) and single-particle reconstruction methods have been employed for atomic-level structure determination of water-soluble m-mercaptobenzoic acid (mMBA)-capped Au68 NC and pMBA-capped Au144 NC.68 However, under cryo-TEM, only the metal core was unambiguously assigned, and identifying ligands is a challenging task. Therefore, other analytical tools with computational modeling were used to predict the number of ligands. In another approach, microcrystal electron diffraction (microED) has been used to determine the structure of Au146-pMBA57 beyond 1.0 Å resolution.69 Electrospray ionization mass spectrometry (ESI-MS), matrix-assisted laser desorption ionization (MALDI-MS), ion mobility coupled to mass spectrometry (IM-MS), and collision-induced dissociation (CID) MS techniques are extensively used to determine the molecular composition, to monitor the reaction kinetics, ligand exchange and ion exchange in NC synthesis.70 One-dimensional (1D) and two-dimensional nuclear magnetic resonance (2D NMR) spectroscopy have been used to study the solution state stability of NCs, ligand composition, charge state, magnetic properties, and their dynamics.71 X-ray absorption spectroscopy (XAS) has been used to determine the local structure and electronic state of a specific element within the NC, in both amorphous and crystalline solids.72a One of the important characterizations is their oxidation state, which can be determined by calculating the binding energy using X-ray photoelectron spectroscopy.72b Unlike plasmonic nanoparticles, due to the quantum confinement effect, NCs display molecule-like optical transitions in the absorption spectra. Based on the number of metal atoms, the UV-Vis absorption spectra of NCs show distinct absorption bands irrespective of the type of ligands.73 Another important property of NCs is their photoluminescence (PL); therefore, photoluminescence spectroscopies, including steady-state, time-resolved, and temperature-dependent PL spectroscopies, have been used to study these properties,73a and to understand the correlations of the ultrafast electron dynamics and structures of NCs, such as femtosecond time-resolved fluorescence up-conversion, and transient absorption.73cBecause of their atomically precise structure and strictly defined number of ligands, NCs are considered “colloidal molecules” with hierarchical structural complexity.74 It is important to clarify that the term “colloidal molecules” used in the literature for colloidal clusters is an analogy to molecular structure, and colloidal clusters do not have the properties of molecules. On the other hand, atomically precise NCs have strictly defined molecular structure, molecular mass, and optical properties despite their colloidal-level size. Therefore, NCs act as a link between organometallic complexes and large plasmonic nanoparticles (Fig. 1a and b). Beyond the atomically precise structure, NCs offer unique optical properties, tunable photoluminescence, photothermal stability, mechanical properties, biocompatibility, and low cytotoxicity (Fig. 1c–e).75 Therefore, NCs offer numerous advanced applications, including catalysis, CO2 reduction, bioimaging, solar concentrators, energy harvesting, photodynamic therapy, high harmonic generation, and dopants for field-effect transistors.76–80 Precision nanoclusters are highly sensitive as compared to plasmonic nanoparticles for the addition or removal of metal atoms. This offers selective doping with amplified luminescence.79 Therefore, a large library of molecularly precise nanoparticle-based building blocks can be prepared with unprecedented properties.
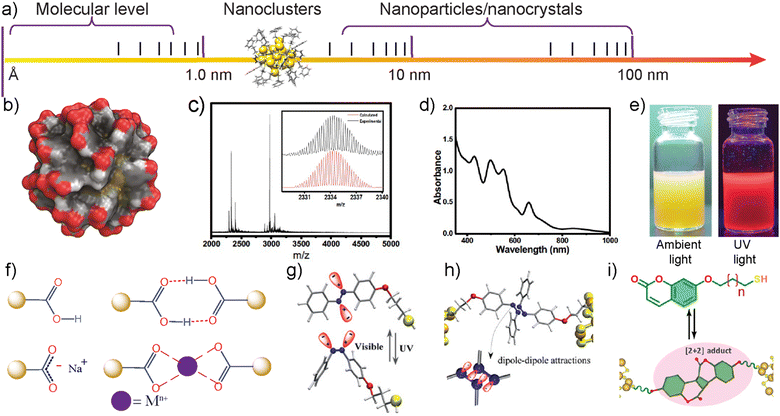 | ||
| Fig. 1 The structure and properties of atomically precise gold and silver NCs. (a) Atomically precise NCs are bridging links between molecules, organometallic complexes, and large plasmonic nanoparticles. (b) Space-filling model showing the structure of Au102-pMBA44 based on the crystal structure reported in ref. 60a. Reproduced with permission from ref. 110 Copyright © 2016 Wiley-VCH. (c) ESI-mass spectra of Na4Ag44-pMBA30. (d) UV-Vis absorption spectra of Na4Ag44-pMBA30 showing molecule-like electronic peaks. Reproduced with permission from ref. 114 Copyright © 2022 Wiley-VCH. (e) Photographs of Au-BSA NCs showing luminescence. Reproduced with permission from ref. 145 Copyright © 2016 American Chemical Society. (f) Carboxylic acid groups on surface ligands allow hydrogen bonding, electrostatic repulsion, and metal chelation-induced assembly. (g) and (h) Dipole–dipole interactions of a thiolated azobenzene ligand using light-triggered isomerization. Reproduced with permission from ref. 141 Copyright © 2020 American Chemical Society. (i) Thiolated coumarin ligands allow dynamic covalent chemistry-assisted assembly via [2+2] photocycloaddition reactions. Reproduced with permission from ref. 143 Copyright © 2023 Wiley-VCH. | ||
Atomically precise NCs have already played a significant role in biological imaging, providing high-resolution information on the location and orientation of subunits in oligomeric enzymes, virus capsids, crystalline sheets of membrane proteins, and thin filaments of muscle.81 NCs are small and stable, making them ideal for labeling biological specimens for electron microscopy imaging. They are also highly versatile and can be used to label a wide variety of targets. For example, undecagold, the first commercial AuNC and the smallest immunogold, was first reported in 1969.82,83 Since then, undecagold and other NCs have been extensively used for immunolabeling and high-resolution imaging of biological structures.84 Excellent accounts of the historical perspectives, synthesis, characterization, optical properties, and applications can be found in several recent reviews.57,58,85 As the field of NCs continues to develop, we can expect to see even more applications of these versatile NCs for new-generation functional materials. However, to achieve such targets, there is a need for focused research on developing approaches for the fabrication of NC-based structures and their integration into devices. Furthermore, NCs alone may not provide all the desired properties. Therefore, it is important to identify various combinations of materials and methods for NC-based composites and hybrid systems. In this context, self-assembly is one of the methods that can be used to achieve this, as it allows readily tunable morphologies.
2.1 State-of-the-art challenges in the self-assembly of atomically precise nanoclusters
Despite the exponentially increasing number of literature reports, the use of atomically precise NCs as a building block for self-assembly has been a recent trend.86 Notably, Schmid et al. made early attempts to assemble phosphine-capped Au NCs on polymer-coated substrates.87 Because of their small size and well-defined surface ligands, NCs have dispersion properties similar to supramolecular complexes.88 Therefore, they are ideal building blocks for self-assemblies. Recent computational studies on experimentally determined gold nanoclusters have shed some light on their unique solubility properties.89 However, their small size also poses significant challenges to achieving stable self-assembled structures in the solution. This is attributed to the fact that the inter-NC interactions are close to the thermal fluctuation energy of the surrounding media.90 Therefore, access to large-scale structures using self-assembly in solution remained a challenge until recently.According to the classical Derjaguin, Landau, Verwey and Overbeek (DLVO) theory, the stability of colloidal particles in a dispersion medium depends on the sum of attractive van der Waals interactions and electrostatic or steric repulsions.91 The DLVO theory was initially formulated for two identical particles. It is based on the assumptions that (i) van der Waals attraction forces and electrostatic double layer forces are independent of each other, (ii) the two forces can be superimposed or added at each interacting distance for two particles, (iii) changes in the ionic strength control the range of the double layer interaction, and (iv) van der Waals forces are independent of ionic strength.92 Therefore, the initial assumptions seriously limit the application of the classical DLVO theory for accurately studying colloidal stability. Several non-DLVO forces, including solvation, solvophobic, and charge correlation forces, were not considered in the original DLVO theory.93 Therefore, several modified or extended DLVO theories have been proposed to study a wide range of colloidal forces.94 Modified DLVO theories have been extended to metal nanoparticles, rod-like particles, and 2D nanomaterials.95 This has contributed tremendously to understanding the self-assembly of polymer-based colloidal particles, metal nanoparticles, and hybrid colloidal assemblies. However, more attempts are needed to develop concepts for self-assemblies for atomically precise NCs.
3. Biological particles provide clues for the assembly of colloidal molecules
Traditional DLVO theory cannot be applied to biological systems such as virus capsid assembly. Short-range interactions and specific ion effects play a major role in biological systems.96 Therefore, understanding how biological colloidal-level molecules such as virus capsids undergo assembly will provide crucial insight into the self-assembly of synthetic colloidal molecules. Because of their unique structure, virus capsids have been an intense topic of research and mathematical puzzle in the mid-20th century.97–100 In 1950 H. R. Crane proposed a subassembly concept to describe the assembly process of the virus capsid.99 This term subassembly was used as an analogy to the automobile factory assembly process at that time. Subsequently, the concept of self-assembly has gained momentum. In 1956 Watson and Crick proposed that plant virus can be considered a molecule because of its high degree of order at atomic level resolution.101 In 1969 an extended essay by Kushner detailed the sub-unit-based self-assembly of virus capsids.102 Each subunit is atomically precise with a well-defined structure. In proteins, the partial double-bond character of peptide bonds imparts conformational rigidity to their primary structure. In their secondary structure, well-defined intramolecular interactions provide structural rigidity to the proteinic subunits.The secondary structure of folded units displays patchy interacting sites, facilitating error-free tertiary structure formation. The patchy interactions are guided by hydrogen bonding, electrostatic repulsion, the hydrophobic effect, and specific interactions between Caspar pairs.103 Each subunit has an interacting distance that is much smaller than a nanometre. Therefore, the self-assembly of nanoparticles below 3 nm can be achieved, provided there are structural and conformational rigidity, patchy interacting sites, and directional supramolecular interactions. Fig. 2a shows the protein subunits and hierarchical assembly of the cowpea chlorotic mottle virus (CCMV) based on the X-ray crystal structure.104 It is important to note that the virus capsid self-assembly is highly complex due to numerous interactions, including nucleic acid (RNA or DNA)–protein interactions and metal chelation that stabilize the protein envelope.104
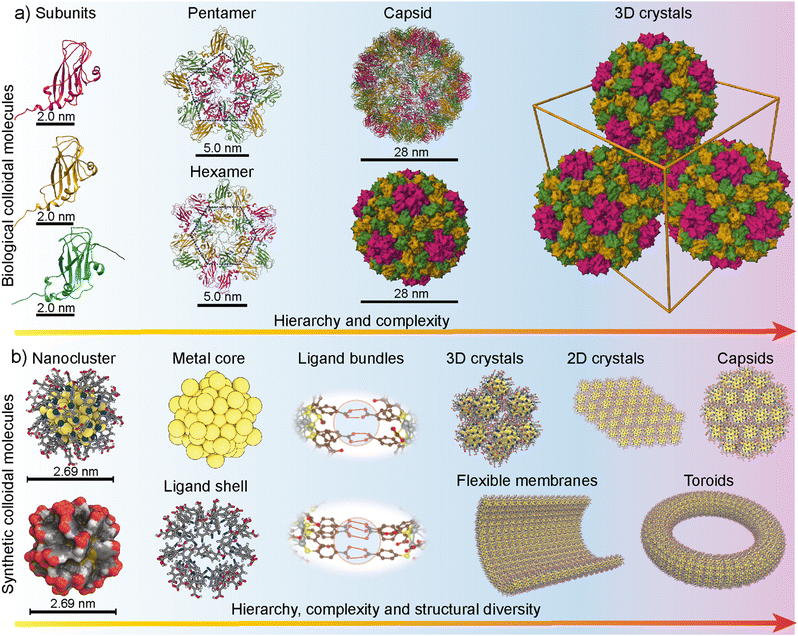 | ||
| Fig. 2 Atomically precise colloidal molecules. (a) Subunit-based self-assembly of the cowpea chlorotic mosaic virus (CCMV) with increasing complexity driven by patchy interactions of subunits based on the crystal structure (PDB ID: ICWP) reported in ref. 104. (b) Metal nanoclusters display rigid core and monolayer ligand shells, patchy ligand bundles and allow hierarchical self-assembly and structural diversity. Reproduced with permission from ref. 86 Copyright © 2018 Wiley-VCH. | ||
In molecular-level building blocks, conformationally rigid surfactants have been shown to display similar self-assembly patterns to that of peptides.105 Importantly, the rigid surfactants and peptides are chemically dissimilar, still producing similar structures. Similarly, identifying the structural features in nanoparticle building blocks that mimic virus capsid proteins and capsids may offer routes for precision structures other than routinely observed 3D crystals. Furthermore, tuning the experimental conditions may result in a diverse set of morphologically distinct structures (Fig. 2b). Therefore, utilizing atomically precise metal nanoparticle-based self-assembly concepts offers precision nanoengineering under a broad range of experimental conditions, including temperature, pH, light, and solvent parameters. Importantly, virus capsid proteins and designer DNA-origamis have been used to hybrid superlattices with plasmonic nanoparticles with unique packing patterns, morphology, and optical properties.106 However, such structures are delicate and are obtained only under very narrow or specific experimental conditions.
4. Patchy ligand distribution and symmetry breaking
X-ray crystal structures of monolayer thiol-protected Au and Ag NCs revealed some of the salient features of the NCs (Fig. 2b and 3). Importantly, (i) the metal atoms form the intrinsically rigid core, (ii) ligands are covalently linked to the metal atoms and often present in bundles, (iii) ligands (specifically aromatic ligands) display conformational rigidity due to inter-ligand interactions and orientation, and (iv) ligand bundles result in patchy and anisotropic distribution of interacting sites (Fig. 3b–d). Therefore, NCs have some similarity to the virus capsid subunits in their size, structural features, and patchy interacting sites. These properties can therefore be exploited to fabricate higher-order structures by tuning the experimental conditions and promoting selective interaction between the patchy interacting sites.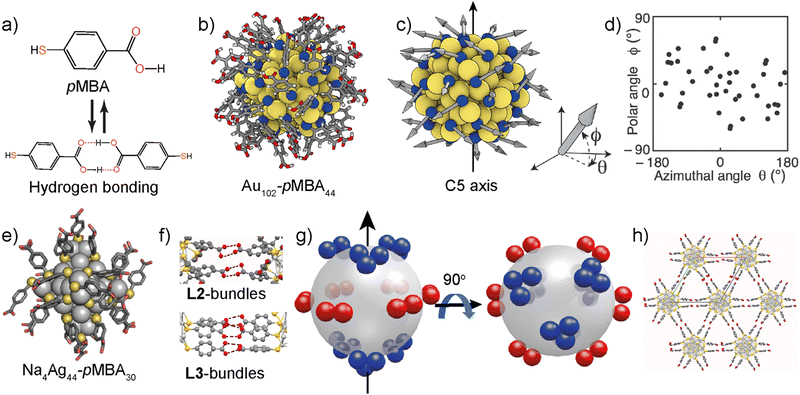 | ||
| Fig. 3 Anisotropic and patchy ligand distribution. (a) Chemical structure of the p-mercaptobenzoic acid (pMBA) ligand and hydrogen bonding dimerization. (b) Crystal structure of Au102-pMBA44; yellow (Au), blue (S), black (C), red (O), and white (H). (c) and (d) The ligands are indicated with arrows to determine their position, orientation, and location using a spherical coordinate system. (d) The ligands are anisotropically and preferentially distributed around the equatorial plane of an imaginary axis. Reproduced with permission from ref. 110 Copyright © 2016 Wiley-VCH. (e) The crystal structure of Na4Ag44-pMBA30. (f) Hydrogen bonding ligand bundles in Na4Ag44-pMBA30. (g) Schematic representation showing the distribution of L2 bundles around the equatorial plane and (h) by preventing interlayer interaction and promoting intralayer hydrogen bonding, 2D structures can be realized. Reproduced with permission from ref. 114 Copyright © 2022 Wiley-VCH. | ||
Jadzinsky et al. reported the single-crystal X-ray structure of p-mercaptobenzoic acid (pMBA)-capped AuNC, Au102-pMBA44.65 The NCs undergo crystallization in acetic acid/methanol solvent mixture.107 More importantly, when the carboxylic acid groups of all ligands are in the protonated form, Au102-pMBA44 is dispersible in methanol but insoluble in water. However, for partial deprotonation, i.e., when ∼50% of the carboxylic acid groups are deprotonated, the NC is water-dispersible but insoluble in methanol. An extensive nuclear magnetic resonance spectroscopy study has revealed that the pMBA ligands in NC are chemically and magnetically non-equivalent.108 Consequently, the deprotonation is not random but results in patchy negative charges due to slight differences in the pKa values of the carboxylic acid groups.109 The patchy and negatively charged surface also imparts amphiphilic properties to the NC.
4.1 2D colloidal crystals and monolayer membranes
Nonappa et al. demonstrated the self-assembly of Au102-pMBA44 NC in aqueous media (Fig. 4a–c).110 A systematic investigation of the solid-state structure using a spherical coordinate system revealed the patchy distribution of the ligands on the nanoparticle surface (Fig. 3b and c). The ligands were anisotropically distributed with a preferential arrangement around the equatorial plane (Fig. 3d). The anisotropic and patchy distribution allows symmetry breaking, offering routes for lower dimensional structures. This was achieved using a dispersion of Au102-pMBA44 in aqueous NaOH, which led to a highly stable dispersion with a zeta potential of −69 mV. When the aqueous solution was sequentially solvent exchanged to methanol via controlled dialysis, 2D crystals were formed (Fig. 4a–c).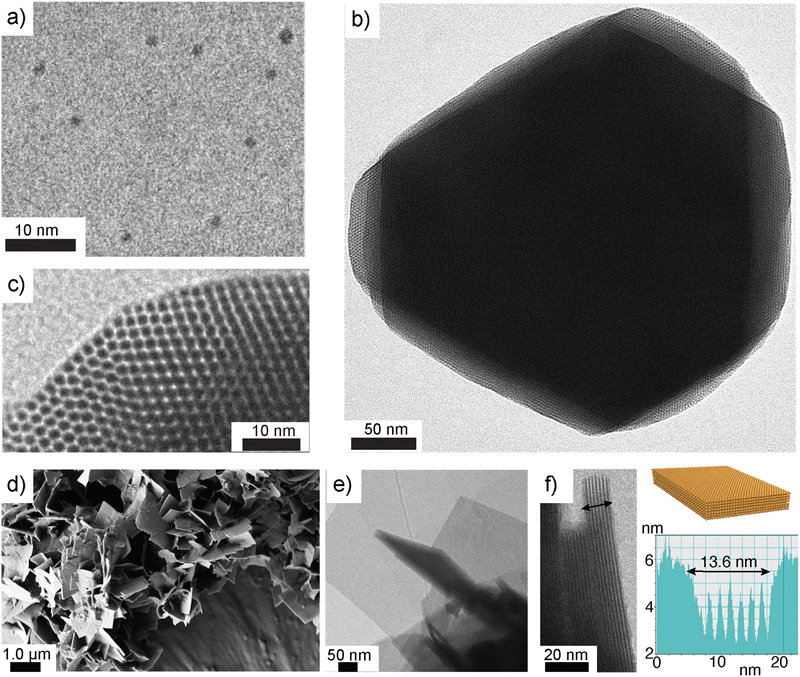 | ||
| Fig. 4 NC-based 2D colloidal crystals. (a) TEM image of Au102-pMBA44 NCs. (b) TEM image of a self-assembled 2D colloidal crystal. (c) TEM image from a selected monolayer showing hexagonally close-packed NCs. Reproduced with permission from ref. 110 Copyright © 2016 Wiley-VCH. (d) SEM image of self-assembled 2D colloidal crystals of Na4Ag44-pMBA30 NC in methanol. (e) and (f) TEM images of self-assembled 2D colloidal crystals suggest that the thickness of each layer is close to the diameter of individual NCs. Reproduced with permission from ref. 114 Copyright © 2022 Wiley-VCH. | ||
The partial deprotonation of the carboxylic acid groups allows a delicate balance between electrostatic repulsion and attractive hydrogen bonding interactions. The electrostatic repulsion prevents uncontrolled aggregation and random structure formation. The carboxylic acid dimerization promotes inter-NC interaction. The 2D colloidal crystals were comprised of a few layers with hexagonal close packing of NCs. Notably, the thickness of each layer corresponds to the diameter of one NC (Fig. 4b and c). This study provided the first evidence that low-dimensional structures can be achieved using solution-state self-assembly of atomically precise gold NCs. However, there are certain limitations to this approach. For example, continued dialysis resulted in further aggregates. Additionally, some heterogeneous populations were observed. Therefore, to achieve large-scale structures, improved methods and concepts are needed.
Desireddy et al. reported the synthesis and characterization of pMBA-capped AgNC, Na4Ag44-pMBA30 (Fig. 3e).111 The ultrastable nature and unique packing patterns in its solids state make this NC an attractive building block for self-assemblies.112 Na4Ag44-pMBA30 forms rhombohedral crystals with the R3C space group. The NCs are hexagonally arranged via inter-NC hydrogen bonding. Unlike Au102-pMBA44 NC, patchy ligand bundles are distinct in Na4Ag44-pMBA30. The crystal structure has layered structures with two types of ligand bundles, viz., bundles of two, L2, and bundles of three, L3 (Fig. 3f). There are six L2 bundles and six L3 bundles. The L2 bundles promote intra-layer hydrogen bonding (Fig. 3f). In contrast, the L3 bundles promote interlayer hydrogen bonding. More importantly, all six L2 bundles are positioned in the equatorial plane. Therefore, each nanocluster forms 24 intralayer H-bonds using L2 bundles (Fig. 3g and h). On the other hand, L3 bundles are distributed in two patches, three at the top and three at the bottom of an imaginary axis. Therefore, they contribute 18 hydrogen bonds each. This leads to a greater number of hydrogen bonds in the equatorial plane. Som et al. utilized Na4Ag44-pMBA30 to prepare crossed-bilayer structures of tellurium nanowires by exploiting the preferential interaction between L3 bundles.113
Recently, Som et al. demonstrated the preparation of strong and elastic large-area monolayer membranes of Na4Ag44-pMBA30 NC.114 Due to the unstable nature of the Na4Ag44-pMBA30 in aqueous media, the self-assembly approach used for Au102-pMBA44 NC was not feasible. Therefore, the dispersion of Na4Ag44-pMBA30 in N,N-dimethyl formamide (DMF) was first tested for self-assembly in alcohols. In all tested lower alcohols (methanol, n-propanol, and n-butanol), a few layered 2D colloidal crystals were formed (Fig. 4d–f). The thickness of each layer corresponds to the size of one nanoparticle. In the next approach, the unique ability of 1-pentanol to form a transient layer when added to an aqueous surface was exploited. For this approach, the NCs were sequentially exchanged by precipitation and redispersion from DMF to 1-pentanol. The dispersion of Na4Ag44-pMBA30 in 1-pentanol was added dropwise onto the surface of water in a container. This resulted in a transient layer of 1-pentanol in which Na4Ag44-pMBA30 NCs were trapped. Due to the predominant intra-layer hydrogen bonding, the NCs encapsulated in the transient layer furnished a stable film (Fig. 5a). This allowed a macroscopic monolayer membrane that could be readily transferred to different substrates without any surface pre-treatment (Fig. 5b). Using UV-vis and Raman spectroscopy, it was shown that the monolayer membranes retained the characteristics peaks of individual Na4Ag44-pMBA30 NCs.
 | ||
| Fig. 5 Free-standing macroscopic monolayer NC membranes. (a) Schematics showing the transient solvent layer-mediated Na4Ag44-pMBA30 NC. (b) Photograph of a large area of self-assembled Na4Ag44-pMBA30 NC membrane on a cover slip. (c) TEM image of the Na4Ag44-pMBA30 membrane stretched over a TEM grid with lacey carbon support film. (d) TEM image suggesting the hexagonal packing of Na4Ag44-pMBA30 NCs (inset shows the FFT). (e) IFFT image from the marked area in image c. (f) Profile showing the inter-nanoparticle distance and periodicity. (g) TEM images of the monolayer membrane stretched over a TEM grid with holey carbon support film. (h) and (i) AFM images of the membrane. (j) A typical force-displacement curve suggesting the elastic nature of the membrane. (k) Overlap of F(δ) curves recorded on three different holes covered with membranes. (l) Histogram showing the Young's modulus (E) values extracted by fitting the F(δ) curves. Reproduced with permission from ref. 114 Copyright © 2016 Wiley-VCH. | ||
However, due to possible electronic coupling between the NCs in membranes, the absorption peaks were slightly broader as compared to that of individual NCs. The TEM imaging of the monolayer film displayed regular hexagonal packing of NCs with a periodicity of 2.01 nm and inter-NC distance of 2.23 nm (Fig. 5c–f). The self-standing nature of the membranes was supported using TEM and atomic force microscopy (AFM) imaging of the membranes placed over a TEM grid with a holey carbon support film (Fig. 5g–i). The AFM-based force spectroscopy revealed that the membranes are strong and elastic with Young's modulus values between 10 and 30 GPa with Gaussian distribution centred at 14.5 ± 0.2 GPa (Fig. 5j–l). Importantly, Young's modulus values of NC membranes displayed a narrow distribution. Notably, the observed Young's modulus values were much higher than plasmonic nanoparticle-based membranes prepared using highly sophisticated DNA nanotechnology.34e Plasmonic gold nanoparticle membranes reported in the literature showed highly scattered Young's modulus values.38b This suggests the importance of atomically precise building blocks with well-defined inter-particle interactions for high mechanical strength and reproducible mechanical properties.
Precision NC-based monolayer membranes offer ultrasmall nanogaps. Such systems can generate very strong and tunable electromagnetic fields for obtaining strong surface-enhanced Raman scattering (SERS) signals from Raman dyes.115 It was shown that Na4Ag44-pMBA30 membranes could detect Raman-active antibiotic molecules such as amphotericin B at a concentration as low as 2.5 μg mL−1. However, when identical experiments were performed using polydisperse Ag nanoparticles, the characteristic peak of amphotericin B was not observed at low concentrations. More importantly, the NC membranes showed highly reproducible SERS signals suggesting the importance of atomic precision in reliable and reproducible sensing.
4.2 Supracolloidal capsids and frameworks
Spherical superstructures using plasmonic nanoparticles are well documented in the literature.23,116 Unlike plasmonic nanoparticles, the presence of directionality in NCs allows high control over their size, shape, and shell thickness. Nonappa et al. demonstrated the self-assembly of Au102-pMBA44 into spherical colloidal capsids with monolayer shells (Fig. 6a–g).110 This was achieved by adding the aqueous dispersion of partially deprotonated Au102-pMBA44 NCs to methanol instead of controlled dialysis. In dialysis, hydrogen bonding dimerization between the protonated carboxylic acid groups provides attractive interactions. The partially deprotonated negatively charged carboxylates facilitate electrostatic repulsion. Therefore, the presence of both attractive and repulsive interactions can overcome the irreversible aggregation, leading to hexagonally packed 2D crystals. On the other hand, methanol is a non-solvent for partially deprotonated Au102-pMBA44 NCs. Therefore, the rapid addition of aqueous dispersions to methanol may result in aggregation and defect-induced curvature formation furnishing spherical superstructures.90,110 Apart from spherical superstructures, ellipsoidal capsids were also observed (Fig. 6e). Importantly, the inter-capsid hydrogen bonding facilitated by an ensemble of nanoclusters led to dimeric, trimeric or higher order structures (Fig. 6f and g). This suggests that the capsids with rich surface functionalities offer hierarchical structure formation with well-defined inter-capsid distance and order. More importantly, atomically precise NCs and narrowly size dispersed nanoparticles behave differently in their structure formation. For example, p-aminobenzoic acid-capped cobalt nanoparticles also formed capsids but with multi-layered shells.117,118 Due to the polydisperse nature of the nanoparticles, there was no regular order or arrangement of nanoparticles in the shell.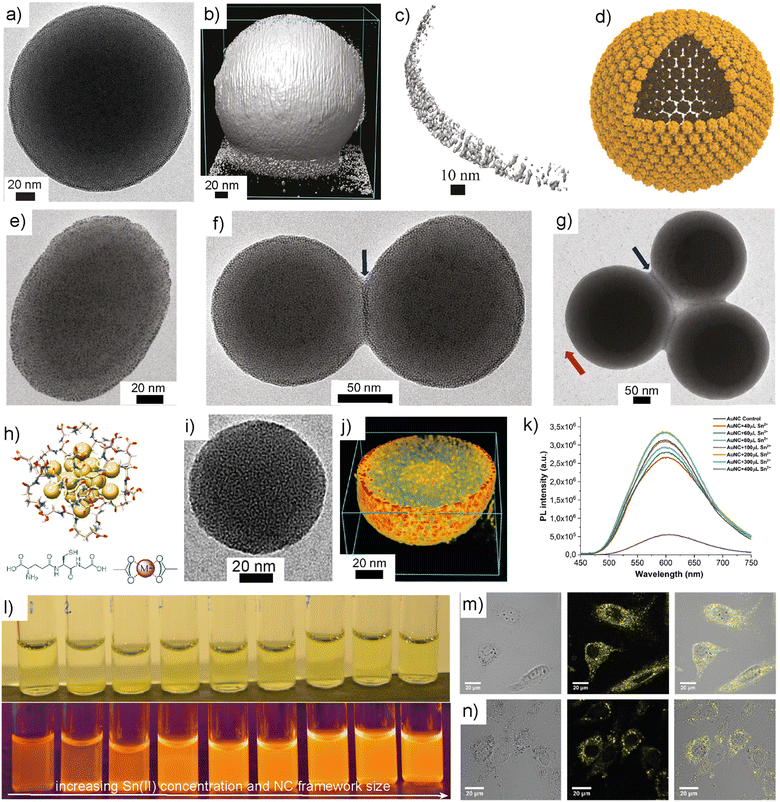 | ||
| Fig. 6 Self-assembled capsids and frameworks. (a) TEM image of a TEM image of self-assembled Au102-pMBA44 capsid. (b)–(d) TEM tomography, a cross-sectional view and schematic representation of a capsid with monolayer shell. (e)–(g) TEM images of ellipsoidal capsid, dimeric and trimeric structures showing inter-capsid bonding. Reproduced with permission from ref. 110 Copyright © 2016 Wiley-VCH. (h) Structure of glutathione capped gold NC. (i) TEM images of metal chelation-induced NC framework. (j) Electron tomographic reconstruction showing a cross-sectional view of NC frameworks. (k) and (l) The PL spectra and photographs of NC frameworks as a function of Sn(II) concentration. (m) and (n) Confocal microscopy images of NIH3T3 cells treated with NC frameworks. Bright-field image (left), confocal fluorescence image (middle), and overlapped image (right). Reproduced with permission from ref. 130 Copyright © 2019 Wiley-VCH. | ||
Bioimaging is one of the important applications of luminescent nanomaterials.119 They offer rapid, low-cost, and sensitive imaging of cells with high spatial resolution. Routinely used fluorescent dyes such as rhodamine 6G show ultrabright fluorescence and high quantum yield. However, they undergo photobleaching, have the tendency to drop out of the cytoplasm, and are cytotoxic.120 Photothermal stability, high quantum yield, bright photoluminescence, and large Stokes shifts are some of the key properties of nanomaterials for bioimaging. Despite the high quantum yield and photothermal stability, semiconductor quantum dots are intrinsically cytotoxic and undergo oxidation, preventing them from extensive use in biological applications.121 Importantly, it has been demonstrated that the size and shape of nanoparticles have a significant effect on cell viability and cytotoxicity.122 In this context, luminescent AuNCs have emerged as attractive nanomaterials for biomedical applications and multimodal imaging. The luminescence properties of Au(I) complexes containing phosphine and thiolate ligands are well documented in the literature.123 The origin of such luminescence is due to strong ligand-to-metal charge transfer (LMCT) and ligand-to-metal-to-metal charge transfer (LMMCT).124 In thiolate complexes, the origin of LMCT is attributed to S–Au(I) interactions, whereas the LMMCT is attributed to the attractive Au(I)⋯Au(I) interactions.125 The attractive Au(I)⋯Au(I) interactions are referred to as aurophilic interactions, typically observed when the distance between adjacent Au atoms is below 3.6 Å. An extensive study by Cha et al. revealed that the Au(I)-alkanethiolate complexes form highly ordered structures, and the luminescence properties depend on the electron-donating ability of thiolate ligands.126 Furthermore, when the Au(I)-alkanethiolate complexes were used as precursors for nanoparticles in the presence of surfactants, the resulting nanoparticles showed no luminescence either in the solid or solution state. This raises the question of whether the luminescence observed in NCs is due to thiolate complexes or the NCs. Unlike plasmonic nanoparticles, in AuNCs, the presence of Au(0) core and Au(I)–S staple units on the surface contribute to the short bonding distances that affects their photoluminescent properties.127 Extensive studies have been performed to understand the size-dependent luminescence properties, the effect of ligands, and aggregation-induced emissions.128 However, the NCs suffer from low efficiency and quantum yield, and the luminescence is restricted to a certain type of NCs with specific ligands and composition. Importantly, the self-assembly of NCs can improve luminescence, stability, and quantum yield.129 For example, metal coordination of functional groups on NC surfaces can rigidify the ligands and restrict their conformational freedom or mobility. This, in turn, amplifies the luminescence and photoluminescent quantum yield of luminescent NCs.
Chandra et al. demonstrated that water-soluble glutathione-capped AuNCs could be assembled into highly luminescent 3D spherical frameworks using metal coordination (Fig. 6h–j).130 Specifically, the 3D frameworks were formed when the NCs interacted with SnCl2. The metal chelation results in the reduction in non-emissive relaxation pathways. The quantum yield was increased from 3.5% for NCs to 25% in NC frameworks. By controlling the metal salt concentration, the size of the framework was tuned from 50 to 200 nm and consequently, the luminescence (Fig. 6k and l). UV-Vis and photoluminescence excitation spectra revealed that the frameworks retained the intrinsic properties of individual NCs. This suggests that the metal chelation-induced assembly is not a random aggregation. The results indicate that the framework formation is predominantly through metallosupramolecular interactions between the surface ligands and metal ions.
When the cell viabilities of NCs and NC frameworks were investigated in NIH3T3 and A549 cell lines, no significant cytotoxicity was observed (Fig. 6m and n). Importantly, in some cases, the viability was higher for frameworks and was pronounced when higher concentrations were used. This was attributed to the fact that NCs result in the generation of reactive oxygen species and the possible aggregation of NCs in the cellular medium before uptake leading to cell death. The bioimaging experiment revealed that the particles were in the cytoplasm and emitted brilliant fluorescent light when excited at 458 nm. Frameworks showed brighter emission than that of the NCs and thus the cells were easier to image. Beyond the amplification of PL, the NC frameworks also displayed improved photocatalytic activity under UV and visible light. The NC frameworks showed a 20-to-25-fold increase in their photocatalytic activity compared to that of NCs when a dye degradation experiment of methylene blue was performed under UV irradiation at 350 nm. Because of their well-defined 3D structures, the NC frameworks demonstrated enhanced adsorption of the dye molecules on their surface and interior, thereby, efficient catalytic activity. Therefore, the self-assembly of NCs not only offers tunable structures but also controllable optical properties, catalysis, biological activity, and imaging.
4.3 Nanoparticle-nanocluster composites
Anisotropic gold nanoparticles such as gold nanorods (AuNRs) display unique surface plasmon resonance peaks and high sensitivity to surrounding chemical environments.131 AuNRs are known for their strong absorption and scattering in the near-infrared (NIR) region. Most human tissues, intrinsic chromophores, haemoglobin, and water have poor absorbance in the NIR region. Therefore, gold nanorods are efficient nanoprobes for in vivo imaging, targeted delivery, and photodynamic therapy.132 Multimodal probes with synergistic properties can be achieved by integrating AuNRs with NCs in a single nanostructure. For example, interacting the AuNRs with organic dye molecules or quantum dots allows unique plasmon-exciton coupling.133 Such interactions can be used to alter optical properties and signal amplification in plasmonic devices.134Because of their molecule-like multiband electronic spectra and surface functionalities, NCs offer a unique opportunity to achieve plasmonic nanoparticle-NC interaction via supramolecular interactions between surface ligands.
Chakraborty et al. demonstrated the hydrogen bonding-directed encapsulation of plasmonic nanorods inside a nanocluster cage.135 The gold nanorods (d ≈ 10 nm, l ≈ 30 nm) capped with pMBA ligands were treated with Na4Ag44-pMBA30 in DMF (Fig. 7). The presence of pMBA on the AuNR surface allows nanoparticle-NC interaction via hydrogen bonding. It was shown that the resulting nanorod-NC composite displayed peaks arising from the nanorod and NCs in their UV-vis spectra. This suggests that the intrinsic properties of both components were retained. However, due to possible electronic interactions between AuNR and NCs in the composites, significant broadening was observed in the NIR region of the spectrum. Using transmission and scanning transmission electron tomographic reconstruction, it was shown that the AuNR-NC co-assembly resulted in an octahedral cage (Fig. 7f and g). Notably, each cage encapsulated a single nanorod, offering a rapid and robust approach to composite supracolloidal cages. The selective encapsulation of nanoparticles was reported using highly sophisticated DNA nanotechnology with predefined size-specific structures with complementary base-pairing. Moreover, DNA nanotechnology has limitations as it requires cages and nanoparticles of precisely defined sizes. However, the self-assembly approach presented using NCs is independent of the nanorod or type of nanoparticles. The structures demonstrate the interaction of precision NCs with plasmonic nanorods with distinct optoelectronic properties. One would expect an amplification of certain properties or the evolution of new behaviour depending on the ligand length, core size, and inter-nanoparticle distance. The interactions can be controlled by modifying the functional groups of the ligands. For example, pMBA-capped AuNCs Au102-pMBA44 and Au250-pMBAn interacted with pMBA-capped AuNRs in aqueous media. Unlike Na4Ag44-pMBA30, the AuNCs produced a monolayer shell around the AuNRs (Fig. 7h–j). This is due to the negatively charged carboxylates on the NC surface, which provided sufficient electrostatic repulsion to stabilize the composite structures. Therefore, by controlling the ligand functional groups and reaction media, the shell thickness and the morphological features of the composites can be tuned.
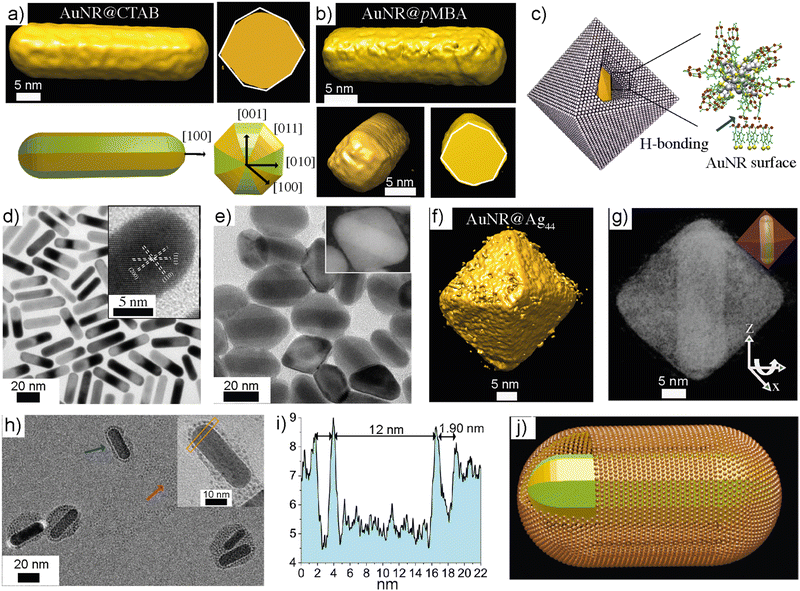 | ||
| Fig. 7 Gold nanorod-NC composite cages. (a) and (b) Electron tomography of gold nanorods before and after ligand exchange. (c) Schematics showing the composite cage structure and nanorod-NC interaction via hydrogen bonding. (d) TEM image of pMBA-capped gold nanorods. (e) TEM image of composite cages (inset shows a DF-STEM image of a single composite cage). (f) and (g) Electron tomogram showing the isosurface and coloured views of a cage with nanorod orientation. (h) Gold nanorods, when interacting with water-soluble Au102-pMBA44 showed monolayer shells around the nanorod. (i) The profile indicating the thickness of the shell. (j) A schematic representation of a composite cage with a monolayer shell. Reproduced with permission from ref. 135 Copyright © 2018 Wiley-VCH. | ||
In a recent study, Chakraborty et al. demonstrated the structural, morphological, and compositional diversity of nanoparticle-NC composites.136 Specifically, gold nanotriangles (AuNTs) interacted with AgNCs with different ligand functionalities such as hexadecyltrimethylammonium chloride (CTAC), dimethyl benzenethiol (DMBT), and 1,2-bis(diphenylphosphino)ethane (DPPE). It was shown that depending on the type of ligand on the NCs, distinct composite structures and compositions were obtained when the NCs were reacted with CTAC-capped AuNTs. For example, the reaction between Ag25(DMBT)18 resulted in the etching of gold atoms from the tips of triangles and Ag-doped AuNTs. On the other hand, when the reaction was performed with Ag25H22(DPPE)8, irregular dendritic shells of Ag were formed around the nanotriangle. Interestingly, faster gold etching was observed when Na4Ag44-pMBA30 was reacted with CTAC-capped AuNTs. Notably, when the AuNT surface was ligand exchanged with pMBA and interacted with Na4Ag44-pMBA30 NCs, a core–shell composite structure was formed (Fig. 8a and b). This suggests that directional hydrogen bonding prevents atom exchange or etching, resulting in a stable composite core–shell structure.
 | ||
| Fig. 8 Effect of ligand chemical structure and interaction on nanoparticle-NC reaction. (a) Shows a schematic representation of a gold nanotriangle (AuNT), ligand exchange, and core–shell structure formation. (b) TEM image showing a core–shell composite of pMBA-coated AuNT and Na4Ag44-pMBA30. Reproduced with permission from ref. 136 Copyright © 2023 Royal Society of Chemistry. (c) Schematic representation of silica-coated gold nanorod. (d) TEM image of the silica-coated gold nanorod. (e) Electron tomography of silica coated gold nanorods. (f) shows absorption spectra of lipoic acid-capped Ag29 NCs, silica-coated AuNR, and AuNR-Silica-NC composite. (g) Photographs of composite AuNR-Ag29 structures (i) without silica coating and (ii) with silica coating under ambient and UV light. (h) Schematic representation of AgNC-coated composite structure. (i) TEM image of AuNR-SiO2-Ag29. (j) Electron tomogram showing the 3D structure of the composite. Reproduced with permission from ref. 140 Copyright © 2022 American Chemical Society. | ||
Plasmonic nanoparticles and nanoclusters are intrinsically sensitive to high-energy electron beams. Furthermore, when directional interactions are not possible, mixing two types of nanoparticles may lead to atom exchange or alloy formation.137,138 Atom transfer and unwanted side reactions can be prevented by growing a protective layer around nanorods with substrates such as silica. The silica coating over nanoparticles provides anchoring for NCs and improves the photothermal stability of nanoparticles. Furthermore, the coating layer between nanorods and NCs also prevents photoluminescence quenching.139 By controlling the thickness of silica layers, the interaction between plasmonic nanoparticles and NCs can be tuned.
Chakraborty et al. developed AuNR-luminescent NC multimodal imaging probes (Fig. 8c–j).140 To fabricate the multimodal probe, a mesoporous silica shell was grown over AuNRs. The silica shell was functionalized using (3-aminopropyl) triethoxysilane (APTES). The lipoic acid (LA)-coated AgNCs (Ag29LA12) were mixed with functionalized silica-coated AuNRs to obtain composite structures (Fig. 8c and h). The resulting composite displayed a nearly two-fold increase in photoluminescence compared to that of NC alone. The approach was not limited to Ag29LA12 NCs, and similar observations were made when bovine serum albumin (BSA)-stabilized AuNCs, Au30BSA was used. In both cases, the PL intensity of the NC was enhanced due to plasmonic coupling, demonstrating the generic nature of the strategy.
4.4 Dynamic covalent chemistry: toroids and supertoroidal frameworks
Most of the reports on nanoparticle-based self-assemblies offer static and equilibrium structures. Importantly, for plasmonic nanoparticles, stimuli-responsive reversible assemblies and dynamic self-assemblies have been studied. However, examples of reversible NC-based assemblies are rare in the literature. Rival et al. developed thiolated azobenzene-capped gold NCs showing light-triggered reversible assemblies.141 In this approach, facile cis–trans isomerization and attractive dipole–dipole interactions between azobenzene molecules in the cis form were exploited to achieve inter-NC interaction. The thiolated azobenzene-functionalized Au25NCs self-assembled into disc-like superstructures (d ∼ 100–1000 nm) when irradiated with UV light at 345 nm. The superstructures disassembled into individual NCs upon irradiation with 435 nm wavelength.In biological systems, amphiphilic water-soluble cytoplasmic β-barrel proteins and transmembrane pore-forming proteins have important functions.142a Their amphiphilic properties have been mimicked using peptide amphiphiles to construct nanorings and microtoroidal structures.142b–d However, toroidal assemblies using metal nanoparticle self-assemblies have not been reported in the literature. Lakshmi et al. reported the dynamic covalent chemistry-driven reversible self-assembly of Au25 NCs into toroids and supertoroidal macroscopic frameworks (Fig. 9a–g).143 The NCs capped with thiolated umbelliferone (7-hydroxycoumarin) ligands allowed a [2+2] cycloaddition reaction when irradiated with UV light at 365 nm (Fig. 9a). This resulted in inter-NC bonding via covalently linked cyclobutene adducts furnishing toroids (Fig. 9b and c). Using extensive electron microscopy imaging and tomographic reconstruction, the growth mechanism of toroid formation was investigated (Fig. 9d). The photocycloaddition initially resulted in spherical aggregates that underwent further rearrangement into toroids. Continued irradiation led to the fusion of the toroids into hexagonally packed macroscopic supertoroidal frameworks (Fig. 9e–g). Due to the dynamic nature of the [2+2] cycloaddition reaction, irradiation of the toroids at 254 nm resulted in their disassembly into individual NCs. The reversible nature of the cycloaddition reaction was exploited for the conjugation of 5-fluorouracil, and controlled release. Overall, this work represents reversible covalent nanocluster-based frameworks across length scales. Furthermore, it offers routes for hierarchically porous macroscopic frameworks.
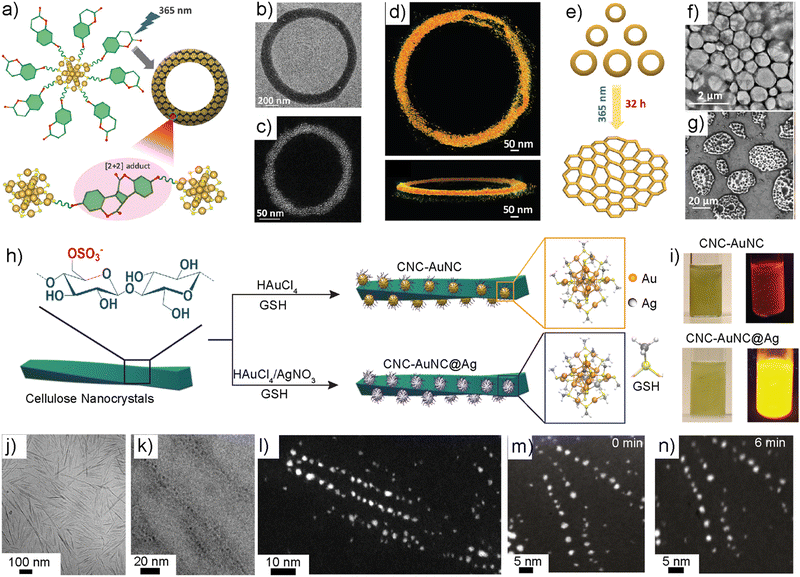 | ||
| Fig. 9 Dynamic covalent chemistry, toroidal frameworks, and templated synthesis. (a) Schematic illustration of thiolated coumarin-capped Au25NC. (b) and (c) TEM and DF-STEM images of a single toroid. (d) Electron tomographic reconstructed structure of a toroid. (e) Schematic illustration of light-triggered super toroidal framework formation. (f) and (g) TEM and SEM images of supertoroidal frameworks. Reproduced with permission from ref. 143 Copyright © 2023 Wiley-VCH. (h) Schematic structure of cellulose nanocrystals and the templated in situ synthesis of NCs. (i) Photographs showing the luminescence of in situ-synthesized NCs. (j) TEM image of cellulose nanocrystals. (k) The Cryo-TEM image of CNC-NC shows the NCs deposited along the CNC surface. (l) DF-STEM image of CNC-NC. (m) and (n) STEM images showing electron beam stability at 0 and 6 minutes of continuous irradiation. Reproduced with permission from ref. 148 Copyright © 2023 Wiley-VCH. | ||
4.5 Colloidal templates and in situ synthesis of luminescent nanoclusters
Noble metal nanoparticles have been studied for hybrid structure formation by mixing a variety of anisotropic colloidal particles. For example, negatively charged cellulose nanocrystals (CNCs) prepared using sulfuric acid hydrolysis have been used for anchoring cationic gold nanoparticles via electrostatic assembly for chiral plasmonics.144,145 CNCs also provide mechanical reinforcement for optomechanically tunable functional gels.146 The rod-like structures with negatively charged sulfate half-ester groups offer a route for the in situ synthesis of NCs. Hynninen et al. demonstrated that glutathione-capped gold NCs can be prepared by interacting the metal precursor (HAuCl4·3H2O) with CNCs.147 The addition of glutathione ligands to the metal ion-deposited templates resulted in highly luminescent NCs. TEM and STEM imaging showed that the NCs were placed along the CNC surface with an inter-NC distance of ∼5 nm. The inter-NC distance corresponds to the relative distance between sulfate groups. The templated NCs were used for mechanically strong luminescent bio-based optical fibers.Selectively replacing the Au atoms with Ag atoms offers a facile route for amplified luminescence in NCs. For example, in Au25NC, the possible doping sites are the central core, outer core and staple motifs. The doping of Ni, Pd and Pt takes place at the central core of AuNC, whereas doping of Ag, Cd, Hg and Mn results in the replacement of Au atoms from the outer core. Wang et al. demonstrated that replacing up to 12 Au atoms in Au25 NCs with Ag atoms resulted in identical optical properties to that of the undoped Au25NC.79a This is due to the exclusive doping of Ag at the outer core. However, when the 13th atom was replaced with Ag, a 200-fold increase in PL was observed due to the replacement of the core including the central Au atom. Substitution at the central core is energetically unfavourable as compared to the outer core. More importantly, NCs with excessive Ag atoms may be cytotoxic due to the leaching of Ag+ ions when used for biological applications. Therefore, there is a need to identify new routes for the selective incorporation of a minimum number of Ag atoms to amplify optical properties.
Chandra et al. reported a CNC-templated approach for the core-selective doping of Au25NCs (Fig. 9h and i).148 This work demonstrated the selective replacement of the central Au atom with a Ag atom without affecting the outer core of Au25NC for the first time. This was achieved by adding Ag and Au precursors to an aqueous solution containing CNCs. The –OSO3– groups on the CNC surface allowed the preferential complexation of Ag(I) ions before accumulating Au(III) complexes, furnishing NCs with the central atom doped with Ag. The resulting core-selective doped NCs displayed an absolute quantum yield of ≈23%. The absolute quantum yield of the templated doped NCs was 6.5 times higher than that of the undoped NCs. The TEM images revealed that the NCs were aligned along the template with an average inter-NC distance of 5.0 nm. Apart from selective doping and amplified optical properties, the NCs displayed resistance from beam damage upon continuous irradiation under STEM imaging conditions (Fig. 9l–n) The MTT assay suggested that the resulting core-doped NCs are non-toxic and non-reactive when studied against Human Dermal Fibroblasts. The role of the –OSO3− group was further studied using DFT calculations. The generic nature of the approach was supported by a similar doping experiment using polystyrene particles. Overall, this study demonstrates, the role of colloidal templates, and the –OSO3 group in selective doping to achieve optically and biologically important NCs.
5. Conclusions
Atomically precise nanoclusters are unique nanoscale building blocks due to their precise structure, composition, optical properties, and stability. Their physical properties can be tuned by adding or removing a single metal atom. While extensive studies on their structure, properties, and characterization are documented in the literature, utilizing them for self-assembled structures with long-range order is still emergent. In this feature article, the author's early attempts to exploit patchy ligand distribution, anisotropic interacting sites, and their potential use for a range of self-assembly are presented along with several examples.It is important to note that plasmonic nanoparticle self-assembly is well established. However, they do not allow the modularity achieved using NCs. Furthermore, large nanoparticles are limited to one assembly type under narrow experimental conditions. However, using one kind of NC, multiple morphologies offering routes for diverse functional structures can be achieved. It is evident that the examples discussed in this manuscript show some of the important properties. For example, understanding the structure and chemical properties of NCs and ligands allows the design of experimental conditions to achieve desired end products. The experimental conditions can be tuned to attain discrete colloidal superstructures with a high degree of control over their size, shape, and optical properties. Since the inter-NC interactions are predominantly guided using the supramolecular interactions through ligands, the intrinsic properties of individual NCs are retained across length scales. At the same time, emergent properties were observed, such as increased quantum yield, improved biological activity, reduced toxicity, and enhanced catalytic properties.
New approaches to utilizing colloidal templates allow the synthesis of NCs that are not possible using conventional methods, paving the way for multifunctional colloidal hybrids. Overall, the self-assembly of NCs is yet to fulfil its potential for further applications such as device fabrication. With the wealth of structural data and increasing self-assembled structures, further efforts are needed to meet the goal of precision nanoengineering. For example, various computational simulation studies have been used to understand nanoclusters' structure and optical properties. However, more effort must be made to use computational simulations to understand or predict the self-assembly of NCs. Computational simulations will play a significant role in predictive structure formation together with rapidly evolving machine learning algorithms and artificial intelligence tools. Combining experimental data and simulations will lead to a breakthrough in precision nanoengineering and provide new avenues for atomically precise manufacturing of macroscopic materials.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Academy of Finland for project funding (No. 352900) and Flagship program on Photonics Research and Innovation (PREIN) are acknowledged. The author thanks all collaborators and co-authors (whose names appear in the cited references) involved in exploring nanocluster self-assembly. Their contributions were crucial in advancing the topics discussed in this feature article.Notes and references
- (a) L. Pokrajac, A. Abbas, W. Chrzanowski, G. M. Dias, B. J. Eggleton, S. Maguire, E. Maine, T. Malloy, J. Nathwani, L. Nazar, A. Sips, J. Sone, A. van den Berg, P. S. Weiss and S. Mitra, ACS Nano, 2021, 15, 18608–18623 CrossRef CAS PubMed; (b) D. J. McClements, ACS Omega, 2020, 5, 29623–29630 CrossRef CAS PubMed; (c) J. Yan, B. K. Teo and N. Zheng, Acc. Chem. Res., 2018, 51(12), 3084–3093 CrossRef CAS PubMed; (d) K. W. Guo, Int. J. Energy Res., 2012, 36, 1–17 CrossRef.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- W. S. Zhi, J. Kibsgaard, F. D. Colin, I. Chorkendorff, K. N. Jens and F. J. Thomas, Science, 2017, 355, eaad4998 CrossRef PubMed.
- (a) R. Jin, Y. Pei and T. Tsukuda, Acc. Chem. Res., 2019, 52, 1 CrossRef CAS PubMed; (b) Q. Yao, Q. Zhang and J. Xie, Ind. Eng. Chem. Res., 2022, 61, 7594–7612 CrossRef CAS.
- D. R. Danielsen, A. Lyksborg-Andersen, K. E. S. Nielsen, B. S. Jessen, T. J. Booth, M.-H. Doan, Y. Zhou, P. Bøggild and L. Gammelgaard, ACS Appl. Mater. Interfaces, 2021, 13, 41886–41894 CrossRef CAS PubMed.
- D. Guo, G. Xie and J. Luo, J. Phy. D: Appl. Phys., 2014, 47, 013001 CrossRef CAS.
- (a) C. M. Aikens, Acc. Chem. Res., 2018, 51, 3065–3073 CrossRef CAS PubMed; (b) M. R. Friedfeld, J. L. Stein, A. Ritchhart and B. M. Cossairt, Acc. Chem. Res., 2018, 51, 2803–2810 CrossRef CAS PubMed; (c) C. Lionello, C. Perego, A. Gardin, R. Klajn and G. M. Pavan, ACS Nano, 2023, 17, 275–287 CrossRef CAS PubMed.
- P. Chakraborty, A. Nag, A. Chakraborty and T. Pradeep, Acc. Chem. Res., 2019, 52, 2–11 CrossRef CAS PubMed.
- (a) Y. Suzuki, G. Cardone, D. Restrepo, P. D. Zavattieri, T. S. Baker and F. A. Tezcan, Nature, 2016, 533, 369–373 CrossRef CAS PubMed; (b) H. Liu, Z. Lu, B. Tang, C. Qu, Z. Zhang and H. Zhang, Angew. Chem., Int. Ed., 2020, 59, 12944–12950 CrossRef CAS PubMed; (c) B. Bhattacharya, D. Roy, S. Dey, A. Puthuvakkal, S. Bhunia, S. Mondal, R. Chowdhury, M. Bhattacharya, M. Mandal, K. Manoj, P. K. Mandal and C. M. Reddy, Angew. Chem., Int. Ed., 2020, 59, 19878–19883 CrossRef CAS PubMed.
- (a) P. Kissel, D. J. Murray, W. J. Wulftange, V. J. Catalano and B. T. King, Nat. Chem., 2014, 6, 774–778 CrossRef CAS PubMed; (b) M. J. Kory, M. Wörle, T. Weber, P. Payamyar, S. W. van de Poll, J. Dshemuchadse, N. Trapp and A. D. Schlüter, Nat. Chem., 2014, 6, 779–784 CrossRef CAS PubMed.
- A. von Hippel, Science, 1956, 123, 315–317 CrossRef CAS PubMed.
- (a) K. E. Drexler, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 5275–5278 CrossRef CAS PubMed; (b) G. Zaldivar, Y. A. P. Sirkin, G. Debais, M. Fiora, L. L. Missoni, E. G. Solveyra and M. Tagliazucchi, ACS Omega, 2022, 7(43), 38109–38121 CrossRef CAS PubMed.
- (a) X. Xing, Z. Man, J. Bian, Y. Yin, W. Zhang and Z. Lu, Nat. Commun., 2020, 11, 6002 CrossRef CAS PubMed; (b) S. Jesse, A. Y. Borisevich, J. D. Fowlkes, A. R. Lupini, P. D. Rack, R. R. Unocic, B. G. Sumpter, S. V. Kalinin, A. Belianinov and O. S. Ovchinnikova, ACS Nano, 2016, 10, 5600–5618 CrossRef CAS PubMed.
- G. M. Whitesides and M. Boncheva, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4769–4774 CrossRef CAS PubMed.
- (a) K. E. B. Doncom, L. D. Blackman, D. B. Wright, M. I. Gibson and R. K. O’Reilly, Chem. Soc. Rev., 2017, 46, 4119–4134 RSC; (b) V. N. Manoharan, Science, 2015, 349, e1253751 CrossRef PubMed.
- S. Hiraoka, Bull. Chem. Soc. Jpn., 2018, 91, 957–978 CrossRef CAS.
- D. Moatsu and R. K. O’Reilly, Chem, 2019, 5, 487–490 Search PubMed.
- (a) O. Ikkala and G. ten Brinke, Chem. Commun., 2004, 2131–2137 RSC; (b) L. Li, K. Raghupathi, C. Song, P. Prasad and S. Thayumanavan, Chem. Commun., 2014, 50, 13417–13432 RSC.
- (a) T. Pelras, C. S. Mahon, Nonappa, O. Ikkala, A. H. Gröschel and M. Müllner, J. Am. Chem. Soc., 2018, 140, 12736–12740 CrossRef CAS PubMed; (b) T. Pelras, Nonappa, C. S. Mahon and M. Müllner, Macromol. Rapid Commun., 2021, 42, 2000401 CrossRef CAS PubMed.
- (a) Y. Xia, T. D. Nguyen, M. Yang, B. Lee, A. Santos, P. Podsiadlo, Z. Tang, S. C. Glotzer and N. A. Kotov, Nat. Nanotechnol., 2011, 6, 580–587 CrossRef CAS PubMed; (b) N. A. Kotov, Europhys. Lett., 2017, 119, 6 CrossRef (c).
- A. Rao, S. Roy, V. Jain and P. Pillai, ACS Appl. Mater. Interfaces, 2022, 15, 25248–25274 CrossRef PubMed.
- (a) M. M. Nigra, J.-M. Ha and A. Katz, Catal. Sci. Technol., 2013, 3, 2976–2983 RSC; (b) Y. Zhao, Y. Huang, H. Zhu, Q. Zhu and Y. Xia, J. Am. Chem. Soc., 2016, 138, 16645–16654 CrossRef CAS PubMed; (c) N. Sarfraz and I. Khan, Chem. – Asian J., 2021, 16, 720–742 CrossRef CAS PubMed; (d) Z. Xu, Y. Li, G. Gao, F. Xie, R. Ju, S. Yu, K. Liu, J. Li, W. Wang, W. Li, T. Li and C.-W. Qiu, APL Photonics, 2023, 8, 020801 CrossRef CAS; (e) K. Bian, H. Schunk, D. Ye, A. Hwang, T. S. Luk, R. Li, Z. Wang and H. Fan, Nat. Commun., 2018, 9, 2365 CrossRef PubMed; (f) T. D. Tran, A. M. T. T. Nguyen, H. V. Le, D. N. Nguyen, Q. D. Truong and P. D. Tran, Chem. Commun., 2018, 54, 3363–3366 RSC; (g) N. S. Thakur, G. Patel, V. Kushwah, S. Jain and U. C. Banerjee, ACS Appl. Bio Mater., 2019, 2, 349–361 CrossRef CAS PubMed; (h) S. Park, W. J. Lee, S. Park, D. Choi, S. Kim and N. Park, Sci. Rep., 2019, 9, 20180 CrossRef CAS PubMed; (i) S. Yang, D. Yao, Y. Wang, W. Yang, B. Zhang and D. Wang, Chem. Commun., 2018, 54, 9841–9844 RSC.
- (a) Y. Wang, O. Zeiri, M. Raula, B. Le Ouay, F. Stellacci and I. A. Weinstock, Nat. Nanotechnol., 2016, 12, 170–176 CrossRef PubMed; (b) H. Zhao, S. Sen, T. Udayabhaskararao, M. Sawczyk, K. Kučanda, D. Manna, P. K. Kundu, J.-W. Lee, P. Král and R. Klajn, Nat. Nanotechnol., 2016, 11, 82–88 CrossRef CAS PubMed; (c) C. Pigliacelli, D. Maiolo, Nonappa, J. S. Haataja, H. Amenitsch, C. Michelet, P. S. Moreno, I. Tirotta, P. Metrangolo and F. B. Bombelli, Angew. Chem., Int. Ed., 2017, 56, 16186–16190 CrossRef CAS PubMed.
- M. S. Lee, D. W. Yee, M. Ye and R. J. Macfarlane, J. Am. Chem. Soc., 2022, 144, 3330–3346 CrossRef CAS PubMed.
- W. Zhang, Nanoparticle Aggregation: Principles and Modeling, in Nanomaterial. Advances in Experimental Medicine and Biology, ed. D. Capco and Y. Chen, Springer, Dordrecht, 2014, 811, pp. 19–43 Search PubMed.
- D. Luo, C. Yan and T. Wang, Small, 2015, 11, 5984–6008 CrossRef CAS PubMed.
- S. Shrestha, B. Wang and P. Dutta, Adv. Colloid Interface Sci., 2020, 279, 102162 CrossRef CAS PubMed.
- X. Zhang, L. Lv, L. Ji, G. Guo, L. Liu, D. Han, B. Wang, Y. Tu, J. Hu, D. Yang and A. Dong, J. Am. Chem. Soc., 2016, 138, 3290–3293 CrossRef CAS PubMed.
- Z. Tang and N. A. Kotov, Adv. Mater., 2005, 17, 951–962 CrossRef CAS.
- L. Chen, B. Su and L. Jang, Chem. Soc. Rev., 2019, 48, 8–21 RSC.
- X. Xue, K. Liu and E. P. Furlani, J. Phys. Chem. C, 2017, 121, 9489–9496 CrossRef CAS.
- S. Cheng and G. S. Grest, ACS Macro Lett., 2016, 5, 694–698 CrossRef CAS PubMed.
- (a) P. Katiyar and J. K. Singh, J. Chem. Phys., 2019, 150, 044708 CrossRef PubMed; (b) S. Cheng and G. S. Grest, J. Chem. Phys., 2013, 138, 064701 CrossRef PubMed; (c) J. Ku, D. M. Aruguete, A. P. Alivisatos and P. L. Geissler, J. Am. Chem. Soc., 2011, 133, 838–848 CrossRef CAS PubMed; (d) E. Rabani, D. R. Reichman, P. L. Geissler and L. E. Brus, Nature, 2003, 426, 271–274 CrossRef CAS PubMed; (e) P. Guo and Y. Gao, Phys. Rev. Lett., 2020, 124, 066101 CrossRef CAS PubMed; (f) Y. Zhou, T.-Y. Tang, B. H.-J. Lee and G. Arya, ACS Nano, 2022, 16, 7457–7470 CrossRef CAS PubMed.
- H. Jia, Y.-F. Zhang, C. Zhang, M. Ouyang and S. Du, J. Phys. Chem. B, 2023, 127, 2258–2266 CrossRef CAS PubMed.
- (a) Y. Sun and Y. Xia, Science, 2002, 298, 2176–2179 CrossRef CAS PubMed; (b) K. An and G. A. Somorjai, ChemCatChem, 2012, 4, 1512–1524 CrossRef CAS; (c) M. L. Personick and C. A. Mirkin, J. Am. Chem. Soc., 2013, 135, 18238–18247 CrossRef CAS PubMed; (d) J. Piella, N. G. Bastús and V. Puntes, Chem. Mater., 2016, 28, 1066–1075 CrossRef CAS.
- (a) A. M. Kalsin, M. Fialkowski, M. Paszewski, S. K. Smoukov, K. J. M. Bishop and B. A. Grzybowski, Science, 2006, 312, 420–424 CrossRef CAS PubMed; (b) R. Klajn, K. J. M. Bishop and B. A. Grzybowski, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 10305–10309 CrossRef CAS PubMed; (c) D. Nykypanchuk, M. M. Maye, D. van der Lelie and O. Gang, Nature, 2008, 451, 549–552 CrossRef CAS PubMed; (d) S. Y. Park, A. K. R. Lytton-Jean, B. Lee, S. Weigand, G. C. Schatz and C. A. Mirkin, Nature, 2008, 451, 553–556 CrossRef CAS PubMed.
- (a) R. A. Alvarez-Puebla, A. Agarwal, P. Manna, B. P. Khanal, P. Aldeanueva-Potel, E. Carbó-Argibay, N. Pazos-Pérez, L. Vigderman, E. R. Zubarev, N. A. Kotov and L. M. Liz-Marzán, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8157–8161 CrossRef CAS PubMed; (b) P. P. Pillai, B. Kowalczyk and B. A. Grzybowski, Nanoscale, 2016, 8, 157–161 RSC; (c) P. J. Santos, P. A. Gabrys, L. Z. Zornberg, M. S. Lee and R. J. Macfarlane, Nature, 2021, 591, 586–591 CrossRef CAS PubMed; (d) J. Wang, T. S. Peled and R. Klajn, J. Am. Chem. Soc., 2023, 145(7), 4098–4108 CrossRef CAS PubMed.
- (a) T. P. Bigioni, X.-M. Lin, T. T. Nguyen, E. I. Corwin, T. A. Witten and H. M. Jaeger, Nat. Mater., 2006, 5, 265–270 CrossRef CAS PubMed; (b) K. E. Mueggenburg, X.-M. Lin, R. H. Goldsmith and H. M. Jaeger, Nat. Mater., 2007, 6, 656–660 CrossRef CAS PubMed; (c) W. Cheng, M. J. Campolongo, J. J. Cha, S. J. Tan, C. C. Umbach, D. A. Muller and D. Luo, Nat. Mater., 2009, 8, 519–525 CrossRef CAS PubMed; (d) A. Dong, J. Chen, P. M. Vora, J. M. Kikkawa and C. B. Murray, Nature, 2010, 466, 474–477 CrossRef CAS PubMed; (e) A. Desireddy, C. P. Joshi, M. Sestak, S. Little, S. Kumar, N. J. Podraza, S. Masillac, R. W. Collins and T. P. Bigioni, Thin Solid Films, 2011, 519, 6077–6084 CrossRef CAS; (f) W. Cheng, M. J. Campolongo, J. J. Cha, S. J. Tan, C. C. Umbach, D. A. Muller and D. Luo, Nat. Mater., 2009, 8, 519 CrossRef CAS PubMed.
- N. A. Kotov, Science, 2010, 330, 188–189 CrossRef CAS PubMed.
- J.-Y. Kim and N. A. Kotov, Science, 2019, 365, 1378–1379 CrossRef CAS PubMed.
- N. A. Kotov, L. M. Liz-Marzán and Q. Wang, Mater. Adv., 2022, 3, 3677–3679 RSC.
- A. Nag, P. Chakraborty, G. Paramasivam, M. Bodiuzzaman, G. Natarajan and T. Pradeep, J. Am. Chem. Soc., 2018, 140, 13590–13593 CrossRef CAS PubMed.
- (a) F. Alibart, S. Pleutin, D. Guérin, C. Novembre, S. Lenfant, K. Lmimouni, C. Gamrat and D. Vuillaume, Adv. Funct. Mater., 2010, 22, 330 CrossRef; (b) F. Alibert, S. Pleutin, O. Bichler, C. Gamrat, T. Serrano-Gotarrendona, B. Linares-Barranco and D. Vuillaume, Adv. Funct. Mater, 2012, 8, 609 CrossRef.
- W. J. Cho, Y. Kim and J. K. Kim, ACS Nano, 2012, 6, 249 CrossRef CAS PubMed.
- (a) J. N. Randall, J. R. Von Ehr, J. Ballard, J. Owen, R. Saini, E. Fuchs, H. Xu and S. Chen, Atomically Precise Manufacturing: The Opportunity, Challenges, and Impact, in Atomic Scale Interconnection Machines. Advances in Atom and Single Molecule Machines, ed. C. Joachim, Springer, Berlin, Heidelberg, 2012, pp. 89–106 Search PubMed; (b) J. N. Randall, J. H. G. Owen, E. Fuchs, J. Lake, J. R. Von Her, J. Ballard and E. Henriksen, Micro Nano Eng., 2018, 1, 1–14 CrossRef; (c) F. Fang, N. Zhang, D. Guo, K. Ehmann, B. Cheung, K. Liu and K. Yamamura, Int. J. Extrem. Manuf., 2019, 1, 012001 CrossRef CAS.
- (a) S. Umbrello and S. D. Baum, Futures, 2018, 100, 63–73 CrossRef; (b) S. Knappe, V. Shah, P. Schwindt, L. Hollberg, J. Kitching, L.-A. Liew and J. Moreland, Appl. Phys. Lett., 2004, 85, 1460–1462 CrossRef CAS.
- D. M. Eigler and E. K. Schweizer, Nature, 1990, 344, 524–526 CrossRef CAS.
- (a) X. Xing, Z. Man, J. Bian, Y. Yin, W. Zhang and Z. Lu, Nat. Commun., 2020, 11, 6002 CrossRef CAS PubMed; (b) S. Jesse, A. Y. Borisevich, J. D. Fowlkes, A. R. Lupini, P. D. Rack, R. R. Unocic, B. G. Sumpter, S. V. Kalinin, A. Belianinov and O. S. Ovchinnikova, ACS Nano, 2016, 10, 5600–5618 CrossRef CAS PubMed.
- (a) K. P. Velikov, C. G. Christova, R. P. A. Dullens and A. van Blaaderen, Science, 2002, 296, 106–109 CrossRef CAS PubMed; (b) W. Poon, Science, 2004, 304, 830–831 CrossRef CAS PubMed.
- (a) V. N. Manoharan, M. T. Elsesser and D. J. Pine, Science, 2003, 301, 483–487 CrossRef CAS PubMed; (b) Y. Wang, Y. Wang, D. R. Breed, V. N. Manoharan, L. Feng, A. D. Hollingsworth, M. Weck and D. J. Pine, Nature, 2012, 491, 51–55 CrossRef CAS PubMed; (c) A. van Blaaderen, Science, 2003, 301, 470–471 CrossRef CAS PubMed.
- D. Morphew and D. Chakrabarti, Curr. Opin. Colloid Interface Sci., 2017, 30, 70–80 CrossRef CAS.
- (a) P. L. Biancaniello, A. J. Kim and J. C. Crocker, Phys. Rev. Lett., 2005, 94, 058302 CrossRef PubMed; (b) D. J. Kraft, J. Groenewold and W. K. Kegel, Soft Matter, 2009, 5, 3823–3826 RSC.
- D. Morphew, J. Shaw, C. Avins and D. Chakrabarti, ACS Nano, 2018, 12, 2355–2364 CrossRef CAS PubMed.
- (a) J. Opdam, R. Tuinier, T. Hueckel, T. J. Snoeren and S. Sacanna, Soft Matter, 2020, 16, 7438–7446 RSC; (b) X. Zhou, D. Yao, W. Hua, N. Huang, X. Chen, L. Li, M. He, Y. Zhang, Y. Guo, S. Xiao, F. Bian and H. Liang, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 5617–5623 CrossRef CAS PubMed; (c) B. Luo, A. Kim, J. W. Smith, Z. Ou, Z. Wu, J. Kim and Q. Chen, Nat. Commun., 2019, 10, 1815 CrossRef PubMed.
- (a) A. B. Rao, J. Shaw, A. S. Neophytou, D. Morphew, F. Sciortino, R. L. Johnston and D. Chakrabarti, ACS Nano, 2020, 14, 5348–5359 CrossRef CAS PubMed; (b) A. Neophytou, V. N. Manoharan and D. Chakrabarti, ACS Nano, 2021, 15, 2668–2678 CrossRef CAS PubMed.
- (a) in Protected Metal Clusters from Fundamentals to Applications in Frontiers of Nanoscience ed. T. Tsukuda and H. Häkkinen, Elsevier Inc., 2015, 9, pp. 1–358 Search PubMed; (b) in Atomically Precise Metal Nanoclusters, ed. T. Pradeep, Elsvier Inc., 2022, pp. 1–643 Search PubMed; (c) R. Jin and D. Jiang, Atomically Precise Nanochemistry, Wiley-VCH, 2023, pp. 1–528 CrossRef.
- (a) R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS PubMed; (b) Y. Du, H. Sheng, D. Astruc and M. Zhu, Chem. Rev., 2020, 120, 526–622 CrossRef CAS PubMed; (c) X. Kang, Y. Li, M. Zhu and R. Jin, Chem. Soc. Rev., 2020, 49, 6443–6514 RSC; (d) J. Yang and R. Jin, ACS Mater. Lett., 2019, 1, 482–489 CrossRef CAS; (e) I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- H. Häkkinen, Nat. Chem., 2012, 4, 443–455 CrossRef PubMed.
- (a) L. Malatesta, L. Naldini, G. Simonetta and F. Cariati, Chem. Commun., 1965, 212–213 RSC; (b) L. Naldini, F. Cariati, G. Simonetta and L. Malatesta, Chem. Commun., 1966, 647–648 RSC; (c) M. McPartlin and R. Mason, Chem. Commun., 1969, 334 RSC; (d) P. A. Bartlett and B. B. S. J. Sanger, J. Am. Chem. Soc., 1978, 5085 CrossRef CAS.
- D. Safer, L. Bolinger and J. S. Leigh Jr., J. Inorg. Biochem., 1986, 26, 77–91 CrossRef CAS PubMed.
- (a) G. Schmid, R. Pfeil, R. Boese, F. Bandermann, S. Meyer, G. H. M. Calis and J. W. A. van der Velden, Chem. Ber., 1981, 114, 3634–3642 CrossRef CAS; (b) D. M. P. Mingos, Struct. Bond, 2014, 161, 117–154 CrossRef.
- M. Brust, M. Walker, D. Bethell, D. J. Schiffrin and R. Whyman, J. Chem. Soc., Chem. Commun., 1994, 801–802 RSC.
- Y. Negishi, K. Nobusada and T. Tsukuda, J. Am. Chem. Soc., 2005, 127, 5261–5270 CrossRef CAS PubMed.
- Z. Wu and R. Jin, Atomically Precise Metal Nanoclusters, Springer International Publishing, Cham, 2021, pp. 31–78 Search PubMed.
- P. D. Jadzinsky, G. Calero, C. J. Ackerson, D. A. Bushnell and R. D. Kornberg, Science, 2007, 318, 430–433 CrossRef CAS PubMed.
- M. Zhu, C. M. Aikens, F. J. Hollander, G. C. Schatz and R. Jin, J. Am. Chem. Soc., 2008, 130, 5883–5885 CrossRef CAS PubMed.
- C. Pigliacelli, A. Acocella, I. Díez, L. Moretti, V. Dichiarante, N. Demitri, H. Jiang, M. Maiuri, R. H. A. Ras, F. B. Bombelli, G. Cerullo, F. Zerbetto, P. Metrangolo and G. Terraneo, Nat. Commun., 2022, 13, 2607 CrossRef CAS PubMed.
- (a) M. Azubel, J. Koivisto, S. Malola, D. Bushnell, G. L. Hura, A. L. Koh, H. Tsunoyama, T. Tsukuda, M. Pettersson, H. Häkkinen and R. D. Kornberg, Science, 2014, 345, 909–911 CrossRef CAS PubMed; (b) M. Azubel, A. L. Koh, K. Koyasu, T. Tsukuda and R. D. Kornberg, ACS Nano, 2017, 11, 11866–11871 CrossRef CAS PubMed.
- S. Vergara, D. A. Lukes, M. W. Martynowycz, U. Santiago, G. Plascencia-Villa, S. C. Weiss, M. J. de la Cruz, D. M. Black, M. M. Alvarez, X. López-Lozano, C. O. Barnes, G. Lin, H.-C. Weissker, R. L. Whetten, T. Gonen, M. J. Yacaman and G. Calero, J. Phys. Chem. Lett., 2017, 8, 5523–5530 CrossRef CAS PubMed.
- P. Chakraborthy and T. Pradeep, NPG Asia Mater., 2019, 11, 48 CrossRef.
- K. Salorinne, S. Malola, O. A. Wong, C. D. Rithner, X. Chen, C. J. Ackersonand and H. Häkkinen, Nat. Commun., 2016, 7, 10401 CrossRef CAS PubMed.
- (a) D. M. Chevrier, R. Yang, A. Chatt and P. Zhang, Nanotechnol. Rev., 2015, 4, 193–206 CrossRef CAS; (b) E. Bolli, A. Mezzi, L. Burratti, P. Prosposito, S. Casciardi and S. Kaciulis, Surf. Interface Anal., 2020, 52, 1017–1022 CrossRef CAS.
- (a) H. Kang and M. Zhu, Chem. Soc. Rev., 2019, 48, 2422–2457 RSC; (b) H. Qian, M. Y. Sfeir and R. Jin, J. Phys. Chem. C, 2010, 114, 19935–19940 CrossRef CAS; (c) S. H. Yau, O. Varnavski and T. G. Ill, Acc. Chem. Res, 2013, 46, 1506–1516 CrossRef CAS PubMed.
- C. Zeng, Y. Chen, K. Kirschbaun, K. J. Lambright and R. Jin, Science, 2016, 354, 1580–1584 CrossRef CAS PubMed.
- (a) K. S. Sugi, P. Bandyopadhyay, M. Bodiuzzaman, A. Nag, M. Hridya, W. A. Dar, P. Ghosh and T. Pradeep, Chem. Mater., 2020, 32, 7973–7984 CrossRef CAS; (b) K. S. Sugi, A. Maier and M. Scheele, Chem. Commun., 2022, 58, 6998–7107 RSC.
- X. Du and R. Jin, ACS Nano, 2019, 13, 7383–7738 CrossRef CAS PubMed.
- S. Zhao, R. Jin, Y. Song, H. Zhang, S. D. House, J. C. Yang and R. Jin, Small, 2017, 13, 1701519 CrossRef PubMed.
- (a) S. Li, A. V. Nagarajan, D. R. Alfonso, M. Sun, D. R. Kauffman, G. Mpourmpakis and R. Jin, Angew. Chem., Int. Ed., 2021, 60, 6351–6356 CrossRef CAS PubMed; (b) X. Cai, G. Li, W. Hu and Y. Zhu, ACS Catal, 2022, 12, 10638–10653 CrossRef CAS.
- (a) S. Wang, X. Meng, A. Das, T. Li, Y. Song, T. Cao, X. Zhu, M. Zhu and R. Jin, Angew. Chem., Int. Ed., 2014, 53, 2376 CrossRef CAS PubMed; (b) L. Chen, A. Black, W. J. Parak, C. Klinke and I. Chakraborty, Aggregate, 2022, 3, e132 CrossRef CAS; (c) S. Chandra, A. Sciortino, S. Das, F. L. Ahmed, A. Jana, J. Roy, D. Li, V. Liljeström, H. Jiang, L.-S. Johansson, X. Chen, Nonappa, M. Cannas, T. Pradeep, B. Peng, R. H. A. Ras, Z. Sun, O. Ikkala and F. Messina, Adv. Opt. Mater., 2023, 11, 2202649 CrossRef CAS.
- (a) V. Jeseentharani, N. Pugazhenthiran, A. Mathew, I. Chakraborty, A. Baksi, J. Ghosh, M. Jash, G. S. Anjusree, T. G. Deepak, A. S. Nair and T. Pradeep, ChemistrySelect, 2017, 2, 1454–1463 CrossRef CAS; (b) K. G. Stamplecoskie and A. Swint, J. Mater. Chem. A, 2016, 4, 2075–2081 RSC.
- (a) M. McPartlin and R. Mason, Chem. Commun., 1969, 334 RSC; (b) P. A. Bartlett, B. Bauer and S. J. Singer, J. Am. Chem. Soc., 1978, 100, 5085–5089 CrossRef CAS.
- (a) D. Safer, J. Hainfeld, J. Wall and J. Reardon, Science, 1982, 218, 290 CrossRef CAS PubMed; (b) J. E. Reardon and P. A. Frey, Biochem., 1984, 23, 3849–3856 CrossRef CAS PubMed; (c) D. Safer, L. Bolinger and J. S. Leigh Jr., J. Inorg. Biochem., 1986, 26, 77–91 CrossRef CAS PubMed.
- (a) J. F. Hainfeld, Nature, 1988, 333, 281–282 CrossRef CAS PubMed; (b) T. Takizawa and J. M. Robinson, J. Histochem. Cytochem., 1994, 42, 1615–1623 CrossRef CAS PubMed.
- G. Zuber, E. Weiss and M. Chiper, Nanotechnol., 2019, 30, 352001 CrossRef CAS PubMed.
- J. V. Rival, P. Mymoona, K. M. Lakshmi, Nonappa, T. Pradeep and E. S. Shibu, Small, 2021, 17, 2005718 CrossRef CAS PubMed.
- Nonappa and O. Ikkala, Adv. Funct. Mater., 2018, 28, 1704328 CrossRef.
- (a) G. Schmid, R. Pugin, T. Sawitowski, U. Simon and B. Marler, Chem. Commun., 1999, 1303–1304 RSC; (b) G. Schmid, M. Bäumle and N. Beyer, Angew. Chem., Int. Ed., 2000, 39, 181 CrossRef CAS.
- T. Laaksonen, P. Ahonen, C. Johans and K. Kontturi, ChemPhysChem, 2006, 7, 2143–2149 CrossRef CAS PubMed.
- M. J. Cowan, T. Higaki, R. Jin and G. Mpourmpakis, J. Phys. Chem. C, 2019, 123, 20006–20012 CrossRef CAS.
- (a) J. Kao, P. Bai, J. M. Lucas, A. P. Alivisatos and T. Xu, J. Am. Chem. Soc., 2013, 135, 1680–1683 CrossRef CAS PubMed; (b) F. Schulz, S. Tober and H. Lange, Langmuir, 2017, 33, 14437–14444 CrossRef CAS PubMed.
- (a) B. Derjaguin, Acta Phys. Chim. U.S.S.R., 1939, 10, 333 Search PubMed; (b) B. Derjaguin and L. D. Landau, Acta Phys. Chim. U.S.S.R., 1941, 14, 633 Search PubMed; (c) E. J. W. Verwey and J. T. G. Overbeek, Theory of Stability of Lyophobic Colloids, Elsevier, Amsterdam, The Netherlands, 1948 Search PubMed.
- J. H. Adair, E. Suvaci and J. Sindel, Surface and Colloid Chemistry, ed. K. H. J. Buschow, R. W. Cahn, M. C. Flemings, B. Ilschner, E. J. Kramer, S. Mahajan and P. Veyssière, in Encyclopedia of Materials: Science and Technology, Elsevier, 2001, pp. 1–10 Search PubMed.
- (a) Y. Liang, N. Hilal, P. Langston and V. Starov, Adv. Colloid Interface Sci., 2007, 134–135, 151–166 CrossRef CAS PubMed; (b) Y. S. Lee, Self-Assembly and Nanotechnology: A Force Balance Approach, John Wiley & Sons, Inc., Hoboken, USA, 2008 CrossRef.
- M. Boström, V. Deniz, G. V. Franks and B. W. Ninham, Adv. Colloid Interface Sci., 2006, 123–126, 5–15 CrossRef PubMed.
- T. M. Mohona, A. Gupta, A. Masud, S.-C. Chien, L.-C. Lin, P. C. Nalam and N. Aich, Environ. Sci. Technol., 2019, 53, 4161–4172 CrossRef CAS PubMed.
- M. Boström, D. R. M. Williams and B. W. Ninham, Phys. Rev. Lett., 2001, 87, 168103 CrossRef PubMed.
- A. Klug and D. L. D. Caspar, Adv. Virus Res., 1960, 13, 1–63 Search PubMed.
- M. G. Rossmann, Quarter. Rev. Biophys., 2013, 46, 133–180 CrossRef CAS PubMed.
- H. R. Crane, Sci. Monthly, 1950, 70, 376–389 Search PubMed.
- (a) D. L. D. Caspar, Adv. Prot. Chem., 1963, 18, 37–121 CrossRef CAS PubMed; (b) D. L. D. Caspar and A. Klug, Cold Spring Harb. Symp. Quant. Biol., 1962, 27, 1–24 CrossRef CAS PubMed.
- F. H. C. Crick and J. D. Watson, Nature, 1956, 177, 473–475 CrossRef CAS PubMed.
- D. J. Kushner, Bacteriol. Rev., 1969, 33, 302–345 CrossRef CAS PubMed.
- (a) D. L. D. Caspar, Adv. Protein Chem., 1964, 18, 37 CrossRef PubMed; (b) W. H. Roos, R. Bruinsma and G. J. L. Wuite, Nat. Phys., 2010, 6, 733–743 Search PubMed.
- J. A. Speir, S. Munshi, G. Wang, T. S. Baker and J. E. Johnson, Structure, 1995, 3, 63–78 CrossRef CAS PubMed.
- (a) L. Travaglini, A. D’Annibale, M. C. di Gregorio, K. Schillén, U. Olsson, S. Sennato, N. V. Pavel and L. Galantini, J. Phys. Chem. B, 2013, 117, 9248–9257 CrossRef CAS PubMed; (b) Z. Özdemir, D. Šaman, K. Bertula, M. Lahtinen, L. Bednárová, M. Pazderková, L. Rárová, Nonappa and Z. Wimmer, Langmuir, 2021, 37(8), 2693–2706 CrossRef PubMed; (c) Z. Özdemir, D. Saman, L. Bednarova, M. Pazderkova, L. Janovska, Nonappa and Z. Wimmer, ACS Appl. Nano Mater., 2022, 5, 3799–3810 CrossRef; (d) Z. Özdemir, Nonappa and Z. Wimmer, ACS Appl. Nano Mater., 2022, 5, 16264–16277 CrossRef.
- V. Linko, H. Zhang, Nonappa, M. A. Kostiainen and O. Ikkala, Acc. Chem. Res., 2022, 55, 1785–1795 CrossRef CAS PubMed.
- Y. Levi-Kalisman, P. D. Jadzinsky, N. Kalisman, H. Tsunoyama, T. Tsukuda, D. A. Bushnell and R. D. Kornberg, J. Am. Chem. Soc., 2011, 133, 2976–2982 CrossRef CAS PubMed.
- K. Salorinne, S. Malola, O. A. Wong, C. D. Rithner, X. Chen, C. J. Ackerson and H. Häkkinen, Nat. Commun., 2016, 7, 10401 CrossRef CAS PubMed.
- J. Koivisto, X. Chen, S. Donnini, T. Lahtinen, H. Häkkinen, G. Groenhof and M. Pettersson, J. Phys. Chem. C, 2016, 120, 10041–10050 CrossRef CAS.
- Nonappa, T. Lahtinen, J. S. Haataja, T.-R. Tero, H. Häkkinen and O. Ikkala, Angew. Chem., Int. Ed., 2016, 55, 16035–16038 CrossRef CAS PubMed.
- A. Desireddy, B. E. Conn, J. Guo, B. Yoon, R. N. Barnett, B. M. Monahan, K. Kirschbaum, W. P. Griffith, R. L. Whetten, U. Landman and T. P. Bigioni, Nature, 2013, 501, 399–402 CrossRef CAS PubMed.
- B. Yoon, W. D. Luedtke, R. N. Barnett, J. Gao, A. Desireddy, B. E. Conn, T. Bigioni and U. Landman, Nat. Mater., 2014, 13, 807–811 CrossRef CAS PubMed.
- A. Som, I. Chakraborty, T. A. Maark, S. Bhat and T. Pradeep, Adv. Mater., 2016, 28, 2827–2833 CrossRef CAS PubMed.
- A. Som, A. Griffo, I. Chakraborty, H. Hähl, B. Mondal, A. Chakraborty, K. Jacobs, P. Laaksonen, O. Ikkala, T. Pradeep and Nonappa, Small, 2022, 18, 2201707 CrossRef CAS PubMed.
- J.-M. Nam, J.-W. Oh, H. Lee and Y. D. Suh, Acc. Chem. Res., 2016, 49, 2746–2755 CrossRef CAS PubMed.
- (a) A. K. Boal, F. Ilhan, J. E. DeRouchey, T. Thurn-Albrecht, T. P. Russell and V. M. Rotello, Nature, 2000, 404, 746–748 CrossRef CAS PubMed; (b) D. Liu, F. Zhou, C. Li, T. Zhang, H. Zhang, W. Cai and Y. Li, Angew. Chem., Int. Ed., 2015, 54, 9596–9600 CrossRef CAS PubMed; (c) C. Pigliacelli, K. B. Sanjeeva, Nonappa, A. Pizzi, A. Gori, F. B. Bombelli and P. Metrangolo, ACS Nano, 2019, 13, 2158–2166 CAS; (d) V. Dichiarante, C. Pigliacelli, P. Metrangolo and F. B. Bombelli, Chem. Sci., 2021, 12, 1632–1646 RSC.
- Nonappa, J. S. Haataja, J. V. I. Timonen, S. Malola, P. Engelhardt, N. Houbenov, M. Lahtinen, H. Häkkinen and O. Ikkala, Angew. Chem., Int. Ed., 2017, 56, 6473–6477 CrossRef CAS PubMed.
- Nonappa and P. Engelhardt, Imaging Microsc., 2019, 21, 22–24 Search PubMed.
- Nonappa, Beilstein J. Nanotechnol., 2020, 11, 533–546 CrossRef CAS PubMed.
- (a) C. Eggeling, J. Widengren, R. Rigler and C. A. M. Seidel, Anal. Chem., 1998, 13(70), 2651–2659 CrossRef PubMed; (b) B. A. Kairdolf, A. M. Smith, T. H. Stokes, M. D. Wang, A. N. Young and S. Nie, Ann. Rev. Chem., 2013, 6, 143–162 CrossRef CAS PubMed; (c) M. Montalti, A. Cantellia and G. Battistellia, Chem. Soc. Rev., 2015, 44, 4853–4921 RSC; (d) Y. Wang and A. Hu, J. Mater. Chem. C, 2014, 2, 6921–6939 RSC.
- (a) Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941 CrossRef CAS PubMed; (b) A. Woźniak, A. Malankowska, G. Nowaczyk, B. F. Grześkowiak, K. Tuśnio, R. Słomski, A. Zaleska-Medynska and S. Jurga, J. Mater. Sci.: Mater. Med., 2017, 28, 92 CrossRef PubMed.
- (a) N. Goswami, F. Lin, Y. Liu, D. T. Leong and J. Xie, Chem. Mater., 2016, 28(11), 4009–4016 CrossRef CAS; (b) N. Goswami, Q. Yao, Z. Luo, J. Li, T. Chen and J. Xie, J. Phys. Chem. Lett., 2016, 7(6), 962–975 CrossRef CAS PubMed; (c) D. Bera and N. Goswami, J. Phys. Chem. Lett., 2021, 12, 9033–9046 CrossRef CAS PubMed.
- J. M. Forward, D. Bohmann, J. P. Fackler, Jr. and R. J. Staples, Inorg. Chem., 1995, 34, 6330–6336 CrossRef CAS.
- Y. Huang, L. Fuksman and J. Zheng, Dalton Trans., 2018, 47, 6267–6273 RSC.
- H. Schmidbaur, Gold Bull., 1990, 23, 11–21 CrossRef CAS.
- S.-H. Cha, J.-U. Kim, K.-H. Kim and J.-C. Lee, Chem. Mater., 2007, 19, 6297–6303 CrossRef CAS.
- G. A. Palermo, M. Tarannum and S. Egusa, J. Phys. Chem. C, 2020, 124, 11248–11255 CrossRef CAS PubMed.
- Y. Huang, L. Fuksman and J. Zheng, Dalton Trans., 2018, 47, 6267–6273 RSC.
- D. Bera, M. Baruah, A. K. Dehury, A. Samanta, Y. S. Chaudhary and N. Goswami, J. Phys. Chem. Lett., 2022, 13, 9411–9421 CrossRef CAS PubMed.
- S. Chandra, Nonappa, G. Beaune, A. Som, S. Zhou, J. Lahtinen, H. Jiang, J. V. I. Timonen, O. Ikkala and R. H. A. Ras, Adv. Opt. Mater., 2019, 7, 1900620 CrossRef CAS.
- H. Chen, L. Shao, Q. Lia and J. Wang, Chem. Soc. Rev., 2013, 42, 2679–2724 RSC.
- A. M. Alkilany, L. B. Thompson, S. P. Boulos, P. N. Sisco and C. J. Murphy, Adv. Drug. Delivery Rev., 2012, 64, 190–199 CrossRef CAS PubMed.
- H. Yu, Y. Peng, Y. Yang and Z.-Y. Li, npj Comput. Mater., 2019, 5, 45 CrossRef.
- H.-X. Xia, X.-Q. Yang, J.-T. Song, J. Chen, M.-Z. Zhang, D.-M. Yan, L. Zhang, M.-Y. Qin, L.-Y. Bai, Y.-D. Zhao and Z.-Y. Ma, J. Mater. Chem. B, 2014, 2, 1945–1953 RSC.
- A. Chakraborty, A. C. Fernandez, A. Som, B. Mondal, G. Natarajan, G. Paramasivam, T. Lahtinen, H. Häkkinen, Nonappa and T. Pradeep, Angew. Chem., Int. Ed., 2018, 57, 6522–6526 CrossRef CAS PubMed.
- A. Chakraborty, M. M. Stanley, B. Mondal, Nonappa, M. Bodiuzzaman, P. Chakraborty, M. P. Kannan and T. Pradeep, Nanoscale, 2023, 15, 2690–2699 RSC.
- P. Bose, P. Chakraborty, J. S. Mohanty, Nonappa, A. R. Chowdhuri, E. Khatun, T. Ahuja, A. Mahendranath and T. Pradeep, Nanoscale, 2020, 12, 22116–22128 RSC.
- J. Roy, B. Mondal, G. Vishwakarma, Nonappa, N. V. Sridharan, P. Krishnamurthi and T. Pradeep, Nanoscale, 2023, 15, 8225–8234 RSC.
- P. Reineck, D. Gómez, S. H. Ng, M. Karg, T. Bell, P. Mulvaney and U. Bach, ACS Nano, 2013, 7(8), 6636–6648 CrossRef CAS PubMed.
- A. Chakraborty, H. Dave, B. Mondal, Nonappa, E. Khatun and T. Pradeep, J. Phys. Chem. B, 2022, 126, 1842–1851 CrossRef CAS PubMed.
- J. V. Rival, Nonappa and E. S. Shibu, ACS Appl. Mater. Interfaces, 2020, 12, 14569–14577 CrossRef CAS PubMed.
- (a) W. C. Wimley, Curr. Opin. Struct. Biol., 2003, 13, 404–411 CrossRef CAS PubMed; (b) I. S. Park, Y. R. Yoon, M. Jung, K. Kim, S. Park, S. Shin, Y. B. Lim and M. Lee, Chem. – Asian J., 2011, 6, 452–458 CrossRef CAS PubMed; (c) N. A. Yewdall, T. M. Allison, F. G. Pearce, C. V. Robinson and J. A. Gerrard, Chem. Sci., 2018, 9, 6099–6106 RSC; (d) C. Du, Z. Li, X. Zhu, G. Ouyang and M. Liu, Nat. Nanotechnol., 2022, 17, 1294–1302 CrossRef CAS PubMed.
- K. M. Lakshmi, J. V. Rival, S. R. Nambiar, P. Sreeraj, C. Jeyabharathi, Nonappa and E. S. Shibu, Small, 2023, 19, 2207119 CrossRef CAS PubMed.
- (a) J. Majoinen, J. Hassinen, J. S. Haataja, H. T. Rekola, E. Kontturi, M. A. Kostiainen, R. H. A. Ras, P. Törmä and O. Ikkala, Adv. Mater., 2016, 28, 5262–5267 CrossRef CAS PubMed; (b) A. Chakraborty, Nonappa, B. Mondal, K. Chaudhari, H. Rekola, V. Hynninen, M. A. Kostiainen, R. H. A. Ras and T. Pradeep, J. Phys. Chem. C, 2021, 125, 3256–3267 CrossRef CAS.
- N. Mohammed, A. Baidya, V. Murugesan, A. A. Kumar, M. A. Ganayee, J. S. Mohanty, K. C. Tam and T. Pradeep, ACS Sustain. Chem. Eng., 2016, 4, 6167–6176 CrossRef CAS.
- (a) V. Hynninen, S. Hietala, J. R. McKee, L. Murtomäki, O. J. Rojas, O. Ikkala and Nonappa, Biomacromolecules, 2018, 19, 2795–2804 CrossRef CAS PubMed; (b) V. Hynninen, P. Mohammadi, W. Wagermaier, S. Hietala, M. B. Linder, O. Ikkala and Nonappa, Eur. Polym. J., 2019, 112, 334–345 CrossRef CAS; (c) V. Hynninen, J. Patrakka and Nonappa, Materials, 2021, 14, 5137 CrossRef CAS PubMed.
- V. Hynninen, S. Chandra, S. Das, M. Amini, Y. Dai, S. Lepikko, P. Mohammadi, S. Hietala, R. H. A. Ras, Z. Sun, O. Ikkala and Nonappa, Small, 2021, 17, 2005205 CrossRef CAS PubMed.
- S. Chandra, A. Sciortino, S. Shandilya, L. Fang, X. Chen, Nonappa, H. Jiang, L.-S. Johansson, M. Cannas, J. Ruokolainen, R. H. A. Ras, F. Messina, B. Peng and O. Ikkala, Adv. Opt. Mater., 2023, 11, 2201901 CrossRef CAS.
Footnote |
| † Dedicated to Professor Olli Ikkala on the occasion of his 70th Birthday. |
| This journal is © The Royal Society of Chemistry 2023 |

