Open Access Article
Open Access ArticleDevelopment of light-induced disruptive liposomes (LiDL) as a photoswitchable carrier for intracellular substance delivery†
Taichi
Tsuneishi
a,
Keiichi
Kojima
ab,
Fumika
Kubota
c,
Hideyoshi
Harashima
 c,
Yuma
Yamada
c,
Yuma
Yamada
 c and
Yuki
Sudo
c and
Yuki
Sudo
 *ab
*ab
aGraduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, 700-8530, Japan. E-mail: sudo@okayama-u.ac.jp
bFaculty of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, 700-8530, Japan
cFaculty of Pharmaceutical Sciences, Hokkaido University, Sapporo, 060-0812, Japan
First published on 23rd May 2023
Abstract
Light-driven inward proton pump rhodopsin RmXeR was embedded in pH-sensitive liposomes. Substance release from the proteoliposomes was observed following light illumination both in vitro and in cells, indicating the successful production of light-induced disruptive liposomes (LiDL). Thus, LiDL is a photoswitchable carrier utilized for intracellular substance delivery.
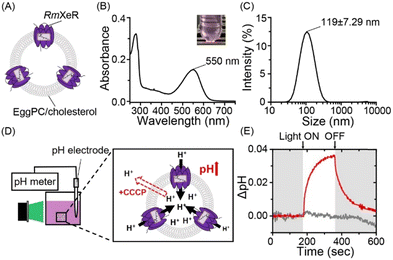
Artificial lipid bilayer vesicle liposomes consist of hydrophobic and hydrophilic molecules such as triglycerides, phospholipids and sterols, and any molecules including chemicals and drugs can be incorporated into those liposomes. The characteristics of liposomes allow them to be used as carriers for intracellular substance delivery.1 It should be noted that liposomes need to have contradictory properties, i.e. high and low stability in extracellular and intracellular compartments, respectively. So far, various techniques have been developed and applied to solve that dilemma. The lipid composition affects the stability of liposomes and its optimization enables the production of stable liposomes at various conditions (e.g., high temperature and low pH), which prevents their degradation in the extracellular compartment.2 In addition, liposomes should have a low stability when they are taken up into cells for efficient intracellular substance delivery. To disrupt liposomes in a condition-dependent manner, several methods have been developed.3 Among them, pH-sensitive liposomes are utilized so far.4,5 For instance, liposomes consisting of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and cholesteryl hemisuccinate (CHEMS) have been established as a method for disrupting the liposome structure in cells due to the phase separation of the DOPE/CHEMS lipid bilayer at an acidic pH (below 5.5) in endosomes, which leads to the release of their contents into the cells (Fig. S1, ESI†).6 In this study, we focused on light as a stimulus having a high spatiotemporal resolution to achieve a more precise release of the contents from pH-sensitive liposomes. To initiate acidification inside liposomes with light, we employed RmXeR (Fig. 1A). RmXeR was identified from a marine bacterium Rubricoccus marinus and phylogenetically categorized into Xenorhodopsin (XeR) functioning as an inward proton pump.7 Microbial rhodopsins, including RmXeR, are membrane-embedded photoreceptor proteins having vitamin-A aldehyde retinal as a chromophore and they show light-dependent molecular functions such as ion transport and signal transduction.8 Among them, RmXeR pumps protons (H+) from the extracellular side to the intracellular side and induces intracellular acidification.7 In fact, a large increase in pH (approximately 0.1 unit) was observed for E. coli cells heterologously expressing RmXeR.7 This means that RmXeR can be an effective tool for facilitating the disruption of pH-sensitive liposomes with light. In this study, we established light-induced disruptive liposomes (LiDL), and used them as a carrier for intracellular substance delivery.
Firstly, liposomes consisting of Egg yolk phosphatidylcholine (EggPC) and cholesterol at a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio were prepared using the lipid hydration method,9 as a model for pH-insensitive liposomes, where purified RmXeR was incorporated into liposomes using the dilution method (Fig. 1A).10 As seen in Fig. 1B, the sample showed a purple color corresponding to visible absorption around 550 nm. This value is almost identical to that of RmXeR in n-dodecyl-β-D-maltoside (DDM) detergent micelles reported previously (i.e., 550 nm),7 indicating the successful production of the desired proteoliposomes shown in Fig. 1A. The size and distribution of those proteoliposomes were estimated by dynamic light scattering (DLS) measurements as 119 ± 7.29 nm and monodispersity (polydispersity index (PdI) = 0.20 ± 0.019), respectively (Fig. 1C and Table S1, ESI†). We then tested the inward proton pump activity of those proteoliposomes by monitoring the environmental pH in a solution containing 100 mM NaCl. The sample was illuminated with green light (550 ± 10 nm) for 3 min at an intensity of 0.080 mW mm−2, as schematically presented in Fig. 1D. The pH was increased approximately 0.035 units, and the changes in pH slowly returned to around the initial value after stopping the light (red line in Fig. 1E). The increase in pH was not observed for that sample in the presence of the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (gray line in Fig. 1E), indicating the light-driven inward proton transport activity of RmXeR in the proteoliposomes. We had confirmed the linearity between the initial slope amplitude and the light intensity below 0.14 mW mm−2 previously,7 indicating that these experiments are quantitative. Thus, we successfully prepared proteoliposomes, which could induce acidification within them upon light illumination. The results also allowed us to estimate the changes in pH value inside the proteoliposomes from the size of the proteoliposomes (i.e., 120 nm). For estimation, several parameters were used as follows; surface area of the lipid bilayer (outer lipid bilayer; 74 × 10−20 m2 and inner lipid bilayer; 61 × 10−20 m2 for EggPC), thickness (37 × 10−10 m for EggPC),11 surface area of cholesterol in the hydrated dipalmitoyl phosphatidylcholine (DPPC) bilayers containing 25–40% cholesterol (27 × 10−20 m2)12 and the size of the bacteriorhodopsin monomer (corresponding to 40 lipid molecules) (Fig. S2, ESI†).13 Using these parameters, we estimated that the inside of the proteoliposomes was acidified from pH 7.0 to 4.8. It has been previously reported that pH-sensitive liposomes are disrupted below pH 5.5.6 Therefore, we assumed that proteoliposomes containing RmXeR have enough of a change in pH to disrupt the pH-sensitive liposomes.
3 molar ratio were prepared using the lipid hydration method,9 as a model for pH-insensitive liposomes, where purified RmXeR was incorporated into liposomes using the dilution method (Fig. 1A).10 As seen in Fig. 1B, the sample showed a purple color corresponding to visible absorption around 550 nm. This value is almost identical to that of RmXeR in n-dodecyl-β-D-maltoside (DDM) detergent micelles reported previously (i.e., 550 nm),7 indicating the successful production of the desired proteoliposomes shown in Fig. 1A. The size and distribution of those proteoliposomes were estimated by dynamic light scattering (DLS) measurements as 119 ± 7.29 nm and monodispersity (polydispersity index (PdI) = 0.20 ± 0.019), respectively (Fig. 1C and Table S1, ESI†). We then tested the inward proton pump activity of those proteoliposomes by monitoring the environmental pH in a solution containing 100 mM NaCl. The sample was illuminated with green light (550 ± 10 nm) for 3 min at an intensity of 0.080 mW mm−2, as schematically presented in Fig. 1D. The pH was increased approximately 0.035 units, and the changes in pH slowly returned to around the initial value after stopping the light (red line in Fig. 1E). The increase in pH was not observed for that sample in the presence of the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (gray line in Fig. 1E), indicating the light-driven inward proton transport activity of RmXeR in the proteoliposomes. We had confirmed the linearity between the initial slope amplitude and the light intensity below 0.14 mW mm−2 previously,7 indicating that these experiments are quantitative. Thus, we successfully prepared proteoliposomes, which could induce acidification within them upon light illumination. The results also allowed us to estimate the changes in pH value inside the proteoliposomes from the size of the proteoliposomes (i.e., 120 nm). For estimation, several parameters were used as follows; surface area of the lipid bilayer (outer lipid bilayer; 74 × 10−20 m2 and inner lipid bilayer; 61 × 10−20 m2 for EggPC), thickness (37 × 10−10 m for EggPC),11 surface area of cholesterol in the hydrated dipalmitoyl phosphatidylcholine (DPPC) bilayers containing 25–40% cholesterol (27 × 10−20 m2)12 and the size of the bacteriorhodopsin monomer (corresponding to 40 lipid molecules) (Fig. S2, ESI†).13 Using these parameters, we estimated that the inside of the proteoliposomes was acidified from pH 7.0 to 4.8. It has been previously reported that pH-sensitive liposomes are disrupted below pH 5.5.6 Therefore, we assumed that proteoliposomes containing RmXeR have enough of a change in pH to disrupt the pH-sensitive liposomes.
Secondly, we added the pH-sensitivity into the proteoliposomes. Liposomes consisting of DOPE and CHEMS at a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
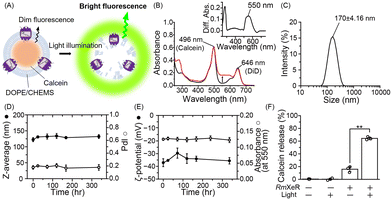
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio were prepared by the lipid hydration method9 as a model for pH-sensitive liposomes, in which the fluorescence probe calcein was incorporated into the liposomes. Calcein is a water soluble fluorescent dye that has a self-quenching property at high concentrations and emits bright fluorescence upon dilution. Purified RmXeR was incorporated into calcein-encapsulated liposomes by the dilution method (Fig. 2A).10 We expected that those proteoliposomes would be disrupted by green light illumination due to the phase separation of the DOPE/CHEMS lipid bilayer at an acidic pH, as schematically shown in Fig. 2A. The absorption spectra of proteoliposomes showed an increase in absorbance around 550 nm compared with liposomes without RmXeR and their difference absorption spectra showed a peak around 550 nm (Fig. 2B), indicating the successful production of the desired proteoliposomes. The size and distribution of the proteoliposomes were estimated as 170 ± 4.16 nm and monodispersity (polydispersity index (PdI) = 0.15 ± 0.019), respectively (Fig. 2C and Table S1, ESI†). Because a suitable size (i.e., 50–200 nm) and monodispersity of liposomes is essential for their efficient incorporation into living cells,14 the constructed proteoliposomes are expected to be suitable for cellular delivery. In addition, for intracellular substance delivery, liposomes should have a high stability against stimuli such as heat. Thus, we examined the stability of those proteoliposomes. As expected, the physicochemical properties (size, PdI, ζ-potential and absorbance of RmXeR at 550 nm) were almost constant for 2 weeks at 4 °C (Fig. 2D and E), indicating the high stability of those proteoliposomes.
3 molar ratio were prepared by the lipid hydration method9 as a model for pH-sensitive liposomes, in which the fluorescence probe calcein was incorporated into the liposomes. Calcein is a water soluble fluorescent dye that has a self-quenching property at high concentrations and emits bright fluorescence upon dilution. Purified RmXeR was incorporated into calcein-encapsulated liposomes by the dilution method (Fig. 2A).10 We expected that those proteoliposomes would be disrupted by green light illumination due to the phase separation of the DOPE/CHEMS lipid bilayer at an acidic pH, as schematically shown in Fig. 2A. The absorption spectra of proteoliposomes showed an increase in absorbance around 550 nm compared with liposomes without RmXeR and their difference absorption spectra showed a peak around 550 nm (Fig. 2B), indicating the successful production of the desired proteoliposomes. The size and distribution of the proteoliposomes were estimated as 170 ± 4.16 nm and monodispersity (polydispersity index (PdI) = 0.15 ± 0.019), respectively (Fig. 2C and Table S1, ESI†). Because a suitable size (i.e., 50–200 nm) and monodispersity of liposomes is essential for their efficient incorporation into living cells,14 the constructed proteoliposomes are expected to be suitable for cellular delivery. In addition, for intracellular substance delivery, liposomes should have a high stability against stimuli such as heat. Thus, we examined the stability of those proteoliposomes. As expected, the physicochemical properties (size, PdI, ζ-potential and absorbance of RmXeR at 550 nm) were almost constant for 2 weeks at 4 °C (Fig. 2D and E), indicating the high stability of those proteoliposomes.
Next, we tested whether the proteoliposomes were disrupted by green light illumination by measuring the fluorescence of calcein. As seen in Fig. 2F, the calcein release ratio was estimated as 64.3 ± 1.97% for proteoliposomes exposed to green light, a value that was significantly larger than that without green light illumination (16 ± 3.7%) and of liposomes alone with or without green light illumination (0.4 ± 0.3% and 0.2 ± 1.4%, respectively). These results implied that calcein was released from the proteoliposomes in a light-dependent manner. Thus, we succeeded in producing proteoliposomes that could be disrupted by light, and named them light-induced disruptive liposomes (LiDLs).
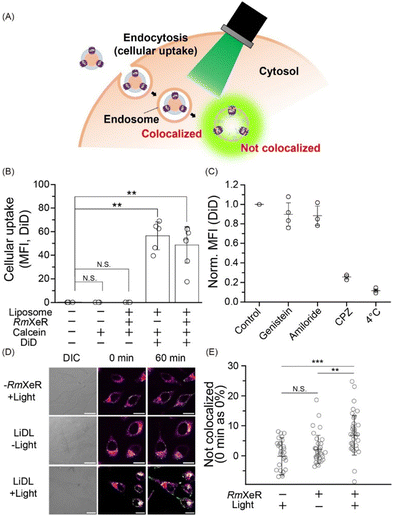
Thirdly, we incorporated LiDL into mammalian HeLa cells to evaluate them as a carrier for intracellular substance delivery (Fig. 3A). To evaluate cellular uptake, LiDL was labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD), which is a lipophilic carbocyanine dye showing red fluorescence that is used to label the lipid bilayer. The cellular uptake was monitored by measuring the change in fluorescence intensity of DiD after 1 h incubation at 37 °C. The results show that liposomes with or without RmXeR showed a significantly larger cellular uptake than the controls (i.e. without liposomes, calcein alone and LiDL without DiD) (Fig. 3B). Similar results were also observed by confocal laser scanning microscopy (CLSM) (Fig. S3, ESI†). These results implied that LiDL effectively entered HeLa cells. It is well-known that cellular uptake is achieved by several endocytosis mechanisms, including clathrin-dependent endocytosis, caveolae-dependent endocytosis, fast endophilin-mediated endocytosis (FEME), macropinocytosis and phagocytosis.15 Thus, we explored the pathway of cellular uptake of LiDL using endocytosis inhibitors, genistein for caveolae-dependent endocytosis,16 amiloride for macropinocytosis17 and chlorpromazine (CPZ) for clathrin-dependent endocytosis.18 Observations at low temperature (4 °C) were also performed as a control for the inhibition of all cellular uptake mechanisms. As seen, the cellular uptake of LiDL was significantly impaired by the addition of CPZ (approx. 25% compared with the control), but not by the addition of genistein or amiloride (89% and 97%, respectively) (Fig. 3C). Similar results were also observed by CLSM (Fig. S4, ESI†). These results suggested that LiDL is mainly taken up into HeLa cells via clathrin-dependent endocytosis.
When the substances were incorporated into the cells, some of them were decomposed in lysosomes, leading to the reduction of their biochemical activities. Therefore, the incorporated substances should escape from immature endosomes (called endosome escape) for efficient intracellular substance delivery.19 Then, to monitor the light-dependent endosome escape, green light (approx. 559 nm) was applied to HeLa cells for 60 min at 4.3 W mm−2 and changes in the localization of calcein and endosomes (lysosomes) were observed by using a fluorescence marker lysotracker. Before the light illumination (i.e., 0 min), all samples (liposomes alone with light [−RmXeR/+light] and LiDL with or without light [LiDL/+light] and [LiDL/−light], respectively) showed white signals corresponding to the colocalization of calcein and endosomes (Fig. 3D). The white signals were also observed after 60 min incubation both for [−RmXeR/+light] and [LiDL/−light]. On the other hand, green fluorescence signals originating from calcein were observed for [LiDL/+light] after 60 min incubation, suggesting intracellular calcein release (i.e., endosome escape).
To quantitatively evaluate the calcein release, we calculated the value of “Not colocalized (%)” by using the Mander coefficient, which is a colocalization indicator for dual-color imaging (see ESI†).20 The values of “not colocalized (%)” were around zero both for liposomes alone with light (−0.153 ± 6.20%) and LiDL without light (1.28 ± 6.34%) (Fig. 3E), while it was significantly increased in LiDL with light (6.80 ± 6.54%), suggesting intracellular calcein release (Fig. 3E). Thus, we concluded that the LiDL developed in this study are effective carriers for intracellular substance delivery.
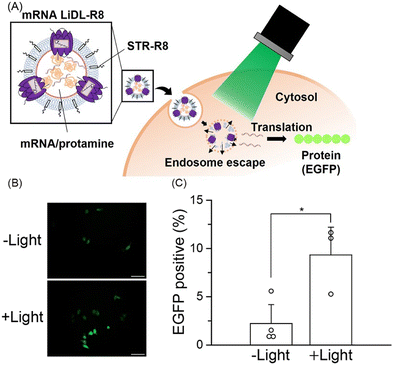
Fourth, we added stearyl octaarginine (STR-R8, 8.5 mol% of lipids), which is a positively charged cell-penetrating peptide,21 into LiDL to improve their intracellular delivery efficiency (Fig. 4A). As a model for the intracellular delivery of substances, a mRNA encoding EGFP was also incorporated into the liposomes by the ethanol dilution method22 after which RmXeR was incorporated into liposomes by the dilution method.10 Proteoliposomes containing STR-R8 were named LiDL-R8 (Fig. 4A). As expected, the surface charge of proteoliposomes was changed from a negative value (−49 mV) for LiDL to a positive value (+29 mV) for LiDL-R8, resulting in a significant increase in cellular uptake (Fig. 4A and Fig. S5, ESI†). Using a RiboGreen assay, we measured the mRNA content and estimated its entrapment efficiency as 24 ± 9.4% (2.3 ± 1.1 μg mL−1 mRNA). It should be noted that, except for the ζ-potential, the physicochemical properties of LiDL-R8 (i.e., size = 218 ± 23.4 nm and PdI = 0.24 ± 0.074) were almost identical to those of LiDL (size = 210 ± 27.1 nm, PdI = 0.24 ± 0.091), suggesting less effect(s) of STR-R8 on the formation of LiDL (Table S2, ESI†). LiDL-R8 was then added into HeLa cells (60–70% confluent) with and without green light illumination (545 ± 10 nm) for 60 min followed by incubation for 23 hr at 37 °C. As shown in Fig. 4B and C, the fluorescence signal of LiDL-R8 was significantly enhanced from 2.2 ± 2.0% to 9.5 ± 2.7% by green light illumination. These results indicated both the release of mRNA encoding EGFP upon light illumination and the translation that produced EGFP protein in the cells (Fig. 4A). Thus, LiDL is useful for intracellular delivery both of chemicals (e.g., hydrophilic and hydrophobic molecules such as calcein and Nile Red, respectively) and biomolecules (e.g., mRNA). To improve the efficiency of the content release, we will characterize the structure of the formed liposomes using various methods such as electron microscopy and optimize the experimental conditions such as buffer concentration in the future.
In conclusion, we developed light-induced disruptive liposomes (LiDLs) that have a high stability in the dark and demonstrated that they are useful, especially for life scientists as an optically controllable carrier for intracellular substance delivery.
Conflicts of interest
There are no conflicts to declare.Notes and references
- K. Maruyama, Adv. Drug Delivery Rev., 2011, 63, 161–169 CrossRef CAS PubMed.
- M. Taira, N. Chiaramoni, K. Pecuch and S. Alonso-Romanowski, Drug Delivery, 2004, 11, 123–128 CrossRef CAS PubMed.
- A. Scomparin, H. F. Florindo, G. Tiram, E. L. Ferguson and R. Satchi-Fainaro, Adv. Drug Delivery Rev., 2017, 118, 52–64 CrossRef CAS PubMed.
- Y. Sato, Chem. Pharm. Bull., 2021, 69, 1141–1159 CrossRef CAS PubMed.
- H. Ding, P. Tan, S. Fu, X. Tian, H. Zhang, X. Ma, Z. Gu and K. Luo, J. Controlled Release, 2022, 348, 206–238 CrossRef CAS PubMed.
- Y. Fan, C. Chen, Y. Huang, F. Zhang and G. Lin, Colloids Surf., B, 2017, 151, 19–25 CrossRef CAS PubMed.
- S. Inoue, S. Yoshizawa, Y. Nakajima, K. Kojima, T. Tsukamoto, T. Kikukawa and Y. Sudo, Phys. Chem. Chem. Phys., 2018, 20, 3172–3183 RSC.
- O. P. Ernst, D. T. Lodowski, M. Elstner, P. Hegemann, L. S. Brown and H. Kandori, Chem. Rev., 2014, 114, 126–163 CrossRef CAS PubMed.
- Y. Yamada, H. Akita, H. Kamiya, K. Kogure, T. Yamamoto, Y. Shinohara, K. Yamashita, H. Kobayashi, H. Kikuchi and H. Harashima, Biochim. Biophys. Acta, Biomembr., 1778, 2008, 423–432 Search PubMed.
- J.-L. Rigaud and D. Lévy, Methods Enzymol., 2003, 372, 65–86 CAS.
- C. Huang and J. Mason, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 308–310 CrossRef CAS PubMed.
- C. Hofsäβ, E. Lindahl and O. Edholm, Biophys. J., 2003, 84, 2192–2206 CrossRef PubMed.
- M. Orwick-Rydmark, J. E. Lovett, A. Graziadei, L. Lindholm, M. R. Hicks and A. Watts, Nano Lett., 2012, 12, 4687–4692 CrossRef CAS PubMed.
- D. Guimarães, A. Cavaco-Paulo and E. Nogueira, Int. J. Pharm., 2021, 601, 120571 CrossRef PubMed.
- J. J. Rennick, A. P. Johnston and R. G. Parton, Nat. Nanotechnol., 2021, 16, 266–276 CrossRef CAS PubMed.
- T. Akiyama, J. Ishida, S. Nakagawa, H. Ogawara, S.-I. Watanabe, N. Itoh, M. Shibuya and Y. Fukami, J. Biol. Chem., 1987, 262, 5592–5595 CrossRef CAS PubMed.
- T. R. Kleyman and E. J. Cragoe Jr, J. Membr. Biol., 1988, 105, 1–21 CrossRef CAS PubMed.
- Z. Wang, A. B. Asenjo and D. D. Oprian, Biochemistry, 1993, 32, 2125–2130 CrossRef CAS PubMed.
- J. Rink, E. Ghigo, Y. Kalaidzidis and M. Zerial, Cell, 2005, 122, 735–749 CrossRef CAS PubMed.
- E. Manders, F. Verbeek and J. Aten, J. Microsc., 1993, 169, 375–382 CrossRef CAS PubMed.
- S. Futaki, W. Ohashi, T. Suzuki, M. Niwa, S. Tanaka, K. Ueda, H. Harashima and Y. Sugiura, Bioconjugate Chem., 2001, 12, 1005–1011 CrossRef CAS PubMed.
- Y. Yamada, K. Somiya, A. Miyauchi, H. Osaka and H. Harashima, Sci. Rep., 2020, 10, 1–13 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc02056h |
| This journal is © The Royal Society of Chemistry 2023 |