Open Access Article
Open Access ArticleA G-quadruplex structure in microRNA interferes with messenger RNA recognition and controls gene expression†
Kaixin
Lyu
 a and
Chun Kit
Kwok
a and
Chun Kit
Kwok
 *ab
*ab
aDepartment of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon Tong, Hong Kong SAR, China
bShenzhen Research Institute of City University of Hong Kong, Shenzhen, China. E-mail: ckkwok42@cityu.edu.hk; Web: https://twitter.com/kitkwok6
First published on 13th June 2023
Abstract
We identify and characterize an RNA G-quadruplex (rG4) structure motif in the human microRNA 638 (hsa-miR-638). We investigate the formation and role of this rG4 in vitro and in cells, and reveal that it inhibits the miR-638 and MEF2C messenger RNA interaction and controls gene expression at the translational level.
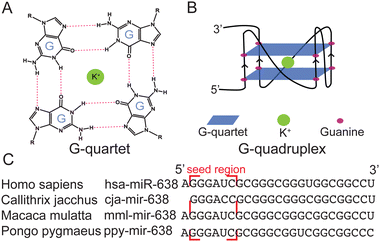
RNA secondary structures have key roles in fundamental biological processes, including RNA synthesis, splicing, processing, and translation.1,2 The guanine (G)-rich sequence in RNA can self-assemble through hydrogen bonds to form G-quartets (Fig. 1A), which can further stack on each other and are linked by connective loops to form RNA G-quadruplexes (rG4s) (Fig. 1B).3 rG4s are stabilized by monovalent cations such as K+ and Na+, but not others like Li+.4 Recently, transcriptome-wide analysis has investigated rG4s in both coding and non-coding (nc) RNAs in the transcriptome.5–7 These rG4s have been proposed to control gene regulation and RNA metabolism and are related to a number of diseases, such as cancers.8,9
miRNAs (or miRs) are a class of small ncRNAs containing about 22 nucleotides and are widely distributed in plants, mammals, and several viruses.10 miRNAs are processed from the cleavage of primary microRNAs (pri-miRNAs) and pre-miRNAs by Drosha and Dicer.11 Each miRNA has a sequence region (seed region) that is reverse complementary to the miRNA recognition element (MRE) on the messenger RNA (mRNA).11,12 It was predicted that more than 60% of mRNAs could be targeted by miRNAs, which control mRNA degradation, protein translation suppression, and other processes.11,12
Compared with coding rG4s, both the identification and characterization of nc rG4s have fallen far behind, and it is only in recent years that research on nc rG4s has gradually increased.1,13,14 Yuan and co-workers showcased rG4 formation in miR-3620-5p,15 miR-5196-5p,16 and miR-1587,17 and reported that the stabilization of miR-3620-5p rG4 impedes the base pairing with its target sequence in vitro.15 Our group has recently performed rG4 computational prediction to identify 166 human miRNAs with high potential to form rG4 and experimentally validated an rG4 formation in miR-765 and the rG4's potential gene regulatory roles in cells.18 Considering the significant biological role of miRNA and the prevalence of rG4 in miRNA, huge gaps remain in the study and understanding of the formation and roles of rG4s in miRNAs in vitro and in cells.
miR-638 is one of the well-studied miRNAs and can regulate diseases including cancers.19 It was reported that the overexpression of miR-638 downregulates the myocyte enhancer factor 2c (MEF2C) expression by targeting the 3′-untranslated region (UTR) of MER2C mRNA.20 Interestingly, this miRNA contained putative rG4 sequences and was also found in the list that we published earlier.18 Based on its biological significance and rG4 forming potential,18,19 we hypothesize that miR-638 is an excellent candidate to help uncover the miRNA G4 structural features and its function in the miRNA-guided RNA silencing pathway. Herein, we first identified the rG4 formation in miR-638 using biophysical and biochemical assays. Next, we characterized that rG4 formation in miR-638 can interfere with the miR-638-MEF2C mRNA interaction in vitro. Last, we showed that rG4 in miR-638 can control miRNA-regulated gene expression in cells.
To investigate whether the G-rich sequence is conserved, we obtained the sequences of miR-638 in miRbase and the National Library of Medicine, performed comparative sequence analysis, and found that this putative G-rich sequence is conserved in several primates (Fig. 1C). To inspect the sequence further, we employed G4 prediction by G4RNA scanner and obtained the cGcC,21 G4H,22 and G4NN23 scores among the species (Table S1, ESI†). It was found that all of them have a high possibility of forming G4s according to the G4NN, which is developed specifically for rG4 detection. Interestingly, we found that hsa-miR-638 had G4 scores above the threshold in all three programs, suggesting that it is likely to fold into an rG4 motif.
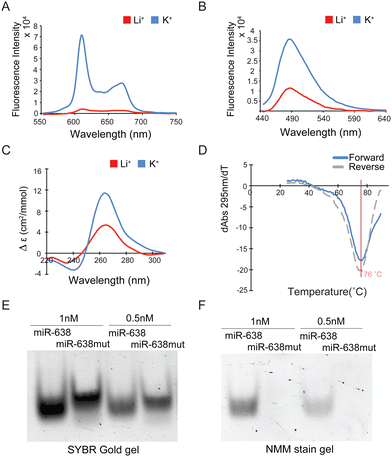
To test if the miR-638 sequence folds into an rG4, various biophysical and biochemical assays were performed. First, we designed the miR-638mut oligonucleotide by mutating all the Gs to As in miR-638, except for those in the seed region (Table S2, ESI†). N-methyl mesoporphyrin IX (NMM) and Thioflavin T (ThT) are well-known G4-specific fluorescent turn-on ligands.24 We employed these two ligands to carry out a ligand-enhanced fluorescence assay. Large fluorescence enhancement was detected for both ligands on miR-638, but not miR-638mut, when comparing K+ over Li+ (Fig. 2A, B and Fig. S1A, B, ESI†). Second, we conducted a circular dichroism (CD) assay. The CD spectrum under K+ displayed a more intense positive peak at around 264 nm and a negative peak at around 240 nm compared with the Li+ condition on miR-638 but not miR-638mut (Fig. 2C and Fig. S1C, ESI†), supporting the formation of a parallel G4 in miR-638. Third, thermal melting monitored by UV absorbance (UV-melting) was conducted, and the melting temperature (Tm) was determined to be 76 °C under 150 mM K+ on miR-638, indicating that the G4 structure is highly thermostable (Fig. 2D). miR-638mut showed no hypochromic shift at 295 nm in UV melting, confirming no G4 formation in this mutated construct (Fig. S1D, ESI†). Last, SYBR Gold nucleic acid stain was used to stain both miR-638 and miR-638mut (Fig. 2E and Fig. S2A, ESI†). We found that the miR-638 bands migrated slightly faster than miR-638mut on the native gel, and this could be explained by the fact that the rG4 structure is folded in miR-638, but not in miR-638mut, thus making miR-638 compacted and it ran faster. To verify this, RNAs were stained by NMM, and only miR-638 bands can be visualized on the gel, which is consistent with our NMM-enhanced fluorescence assay (Fig. 2A), further supporting rG4 formation in miR-638 (Fig. 2F and Fig. S2B, ESI†). Together, these analyses reveal the formation of rG4 in miR-638 under physiologically relevant K+ conditions and temperature.
To examine the effect of rG4 formation in miR-638 on miRNA–mRNA target interactions, we performed EMSA assays on miR-638 with increasing concentrations of MEF2C mRNA under K+ and Li+ conditions (Fig. 3A and B and Fig. S3A and B, ESI†). The MEF2C mRNA contains the MRE and the flanking sequences (Table S2, ESI†). EMSA results showed that miR-638 exhibited much weaker binding to MEF2C mRNA under 150 mM K+ (too weak to obtain a Kd value) when compared with 150 mM Li+(Kd = 82.19 ± 26.52 nM) (Fig. 3C), highlighting that the rG4 formation in miR-638 inhibits miRNA binding to the mRNA target. As a control, we performed the same experiment on miR-638mut, and the binding was found to be similar under both K+ and Li+ (Fig. S4, ESI†), which verified that the mRNA binding inhibition effect observed in the miR-638 construct is rG4-dependent. We also carried out the EMSA on miR-638 under K+ conditions, adding other 4 miRNAs (miR-150-5p, miR-328-5p, miR-601, and miR-671-5p) as competitors (Fig. S5, ESI†). These four miRNAs do not contain a seed region to target MEF2C, and our result showed that miR-638 interaction with MEF2C was not affected by these miRNA competitors. To support the EMSA results, we also carried out an NMM-enhanced fluorescence assay on miR-638 with varying concentrations of MEF2C mRNA under K+ conditions (Fig. 3D). Increasing MEF2C mRNA displayed NMM away from the miR-638, allowing the formation of an miR 638 – MEF2C mRNA duplex. We found that up to 35% fluorescence intensity decrease was detected upon MEF2C mRNA addition (Fig. 3D). As a control, the same experiment was conducted on miR-638mut and MEF2C mRNA, and no fluorescence change was observed (Fig. 3E), indicating that the fluorescence change is due to the miR-638 G4 formation. As a second control, we performed the assay only on different concentrations of MEF2C (Fig. S6, ESI†) and observed no fluorescence change. In short, these results clearly demonstrate that rG4 formation in miR-638 can control miR-638 miRNA and MEF2C mRNA target interaction in vitro.
To assess whether miR-638 binds to MEF2C and affects its gene expression, we first constructed a short MEF2C plasmid containing the MRE flanking sequence (Table S2, ESI†). It was inserted into the 3′UTR of Renilla luciferase in the psiCHECK2 vector, with the Firefly luciferase signal as the internal control (Fig. 4A, ESI†). The miR-638, miR-638mut, and miR-scramble were co-transfected with the plasmid. miR-638 and miR-638mut share the same seed region, but the remaining Gs were mutated to As in miR-638mut. The results showed that the luciferase signals in miR-638 and miR-638mut were 2.57 ± 0.04 and 2.41 ± 0.03-fold lower than that of miR-Scramble (Fig. S7A, ESI†), suggesting that the seed region from both miR-638 and miR-638mut can bind to the MEF2C MRE and resulted in miRNA-mediated inhibition in gene expression.
To interrogate the impact of miR-638 rG4 formation on miR638-MEF2C targeting in cells, we incubated the cells with NMM while co-transfecting the miRNAs and short MEF2C plasmid. DMSO was added into HEK293T cells as a control. The normalized luciferase activity was measured to be 1.62 ± 0.04-fold higher under NMM conditions than DMSO when cells were transfected with miR-638 (Fig. 4B). No significant difference was observed for miR-638mut or miR-Scramble (Fig. 4B). We reasoned that miR-638 G4 formation is stabilized by NMM, thus reducing the chance for miRNA–mRNA interaction and preventing the gene down-regulation mediated by miRNA, which led to a higher luciferase signal detected. The normalized luciferase signals were reported to be similar for miR-638mut or miR-Scramble under NMM and DMSO treatment (Fig. 4B), illustrating that the NMM effect observed for miR-638 and MEF2C interaction is rG4-specific. To test whether this is acting on the translational or post-transcriptional level, we performed RT-qPCR to detect the Renilla mRNA level. No significant difference was noticed for the Renilla mRNA expression in all groups (Fig. S8, ESI†), underlining that the rG4 in miR-638 affects gene expression on a translational level.
To mimic the native sequence better in cells, we also constructed a full-length 3′UTR MEF2C plasmid, including the 3′UTR of MEF2C (Table S2, ESI†). We performed the similar reporter gene assay described above (Fig. 4A). Both miR-638 and miR-638mut were shown to impede the MEF2C reporter gene expression (Fig. S7B, ESI†). For the NMM effect, a 1.42 ± 0.03-fold higher luciferase signal is detected under NMM compared with DMSO in the miR-638 group, but not miR-638mut or miR-Scramble (Fig. 4C). RT-qPCR was also employed to test the mRNA level, and no significant difference was detected (Fig. S9, ESI†). Collectively, miR-638 rG4 can be stabilized by NMM in both the short and full-length construct, which resulted in the increased luciferase signal, underscoring that rG4 formation in miR-638 can lead to translational regulation in cells.
Overall, our computational and comparative sequence analyses have first suggested that the G-rich sequence in miR-638 is conserved and has the potential to form an rG4 motif that could be an RNA structural element in regulating MEF2C gene expression (Fig. 1 and Table S1, ESI†). Next, we used a combination of spectroscopic and gel-based assays to reveal the rG4 formation in miR-638 (Fig. 2) and uncover that this rG4 conformation interferes with the miR-638 from targeting MEF2C mRNA in vitro and in cells, which resulted in altered gene expression at the translational level (Fig. 3 and 4). Previously, rG4s have been reported in pri-miRNAs and pre-miRNAs, and they were found to play significant roles in miRNA maturation and function.25–27 Despite the potential importance of mature miRNA G4 in gene regulation, research on this topic remains limited to identifying and characterizing the miRNA G4 formation in vitro,15–18 with little exploration and investigation into their functional roles in cells. To fill this research and knowledge gap, we have taken the key step to study the rG4 effect on the native miRNA–mRNA binding in vitro and uncover the regulatory role of miR-638 G4 in MEF2C translation in cells for the first time. Given that there are more than 160 of such miRNA G4 candidates in the list we published earlier,18 we think that more rG4s can be experimentally identified and characterized in functionally important miRNAs in the future, as these rG4 motifs may be well suited as potential RNA structural targets for various biological applications. In this work, we have used miR-638 and MEF2C as a proof-of-concept example. It was reported in the literature that the up-regulation and down-regulation of miR-638 were involved in human cancers,19 and miR-638 could modulate the development of MEF2C in endometrial carcinoma (EC).20 NMM is one of the well-studied small molecules shown to bind quadruplexes and stabilizes the G4 activity in cells,28 and in this work we have illustrated that NMM could be used to manipulate rG4-linked gene activity. It is worth noting that many G4 ligands, including NMM, do not have sufficient specificity to distinguish between dG4s versus rG4s, let alone individual G4s of interest in the genome/transcriptome. It will be of great interest to develop and apply new G4-specific tools to modulate the endogenous miR-638 G4 and MERF2C in EC and other cancers in the future.
To sum up, our study reveals that an rG4 structure within miRNA interferes with mRNA recognition, ultimately regulating gene expression. Using miR-638 and MEF2C as an example, we demonstrated that miR-638 contains a thermostable rG4 conformation that can compete with miR-638 – MEF2C mRNA binding, which in turn controls translation. This novel finding highlights the significance of rG4 structures in miRNAs as critical modulators of gene expression, and our multidisciplinary approaches presented here enable the further exploration and study of rG4s in miRNAs and other classes of ncRNAs.
This work was supported by the NSFC Excellent Young Scientists Fund (Hong Kong and Macau) Project [32222089]; Research Grants Council of the Hong Kong SAR, China Projects [CityU 11100222, CityU 11100421, CityU 11101519]; Croucher Foundation Project [9509003]; State Key Laboratory of Marine Pollution Director Discretionary Fund [DDF/0008] and Seed Collaborative Research Fund [SCRF/0037, SCRF/0040]; and City University of Hong Kong projects [9678302] and [9667222] to C. K. K. We thank Dr Tuan Anh Nguyen for the discussion.
Conflicts of interest
There are no conflicts to declare.Notes and references
- K. Lyu, E. Y. Chow, X. Mou, T. F. Chan and C. K. Kwok, Nucleic Acids Res., 2021, 49, 5426–5450 CrossRef CAS PubMed.
- A. Cammas and S. Millevoi, Nucleic Acids Res., 2017, 45, 1584–1595 CAS.
- C. K. Kwok and C. J. Merrick, Trends Biotechnol., 2017, 35, 997–1013 CrossRef CAS PubMed.
- C. K. Kwok, G. Marsico and S. Balasubramanian, Cold Spring Harbor Perspect. Biol., 2018, 10, a032284 CrossRef PubMed.
- C. K. Kwok, G. Marsico, A. B. Sahakyan, V. S. Chambers and S. Balasubramanian, Nat. Methods, 2016, 13, 841–844 CrossRef CAS PubMed.
- J. Zhao, E. Y. Chow, P. Y. Yeung, Q. C. Zhang, T. F. Chan and C. K. Kwok, BMC Biol., 2022, 20, 257 CrossRef CAS PubMed.
- J. U. Guo and D. P. Bartel, Science, 2016, 353, 6306 CrossRef PubMed.
- N. Kosiol, S. Juranek, P. Brossart, A. Heine and K. Paeschke, Mol. Cancer, 2021, 20, 40 CrossRef CAS PubMed.
- R. Simone, P. Fratta, S. Neidle, G. N. Parkinson and A. M. Isaacs, FEBS Lett., 2015, 589, 1653–1668 CrossRef CAS PubMed.
- L. F. R. Gebert and I. J. MacRae, Nat. Rev. Mol. Cell Biol., 2019, 20, 21–37 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- M. Ha and V. N. Kim, Nat. Rev. Mol. Cell Biol., 2014, 15, 509–524 CrossRef CAS PubMed.
- S. Ghafouri-Fard, A. Abak, A. Baniahmad, B. M. Hussen, M. Taheri, E. Jamali and M. E. Dinger, Cancer Cell Int., 2022, 22, 171 CrossRef CAS PubMed.
- M. Tassinari, S. N. Richter and P. Gandellini, Nucleic Acids Res., 2021, 49, 3617–3633 CrossRef CAS PubMed.
- W. Tan, J. Zhou, J. Gu, M. Xu, X. Xu and G. Yuan, Talanta, 2016, 154, 560–566 CrossRef CAS PubMed.
- W. Tan, L. Zhang, J. Zhou, H. Chen and G. Yuan, Spectrosc. Lett., 2017, 50, 489–493 CrossRef CAS.
- W. Tan, L. Yi, Z. Zhu, L. Zhang, J. Zhou and G. Yuan, Talanta, 2018, 179, 337–343 CrossRef CAS PubMed.
- K. L. Chan, B. Peng, M. I. Umar, C. Y. Chan, A. B. Sahakyan, M. T. N. Le and C. K. Kwok, Chem. Commun., 2018, 54, 10878–10881 RSC.
- Z. X. Chong, S. K. Yeap and W. Y. Ho, Pathol., Res. Pract., 2021, 220, 153351 CrossRef CAS PubMed.
- J. Ni, S. Liang, B. Shan, W. Tian, H. Wang and Y. Ren, Int. J. Mol. Med., 2020, 45, 1753–1770 CAS.
- J. D. Beaudoin, R. Jodoin and J. P. Perreault, Nucleic Acids Res., 2014, 42, 1209–1223 CrossRef CAS PubMed.
- A. Bedrat, L. Lacroix and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1746–1759 CrossRef PubMed.
- J. M. Garant, J. P. Perreault and M. S. Scott, Bioinformatics, 2017, 33, 3532–3537 CrossRef CAS PubMed.
- K. Lyu, S. B. Chen, C. Y. Chan, J. H. Tan and C. K. Kwok, Chem. Sci., 2019, 10, 11095–11102 RSC.
- S. G. Rouleau, J. M. Garant, F. Bolduc, M. Bisaillon and J. P. Perreault, RNA Biol., 2018, 15, 198–206 CrossRef PubMed.
- G. Mirihana Arachchilage, A. C. Dassanayake and S. Basu, Chem. Biol., 2015, 22, 262–272 CrossRef CAS PubMed.
- C. K. Kwok, A. B. Sahakyan and S. Balasubramanian, Angew. Chem., Int. Ed., 2016, 55, 8958–8961 CrossRef CAS PubMed.
- A. Yett, L. Y. Lin, D. Beseiso, J. Miao and L. A. Yatsunyk, J. Porphyrins Phthalocyanines, 2019, 23, 1195–1215 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc01549a |
| This journal is © The Royal Society of Chemistry 2023 |