Open Access Article
Open Access ArticleSynthetic DNA binders for fluorescent sensing of thymine glycol-containing DNA duplexes and inhibition of endonuclease activity†
Yusuke
Sato
 *a,
Yoshihide
Takaku
a,
Toshiaki
Nakano
*a,
Yoshihide
Takaku
a,
Toshiaki
Nakano
 b,
Ken
Akamatsu
*b,
Dai
Inamura
a and
Seiichi
Nishizawa
b,
Ken
Akamatsu
*b,
Dai
Inamura
a and
Seiichi
Nishizawa
 *a
*a
aDepartment of Chemistry, Graduate School of Science, Tohoku University, Sendai 980-8578, Japan. E-mail: yusuke.sato.a7@tohoku.ac.jp; seiichi.nishizawa.c8@tohoku.ac.jp
bDNA damage chemistry research group, Institute for Quantum Life Science, National Institutes for Quantum Science and Technology, Kizugawa, 619-0215, Japan. E-mail: akamatsu.ken@qst.go.jp
First published on 25th April 2023
Abstract
Dimethyllumazine (DML)-thiazole orange (TO) conjugates were developed for fluorescence sensing of thymine glycol (Tg)-containing DNAs based on the selective recognition of the A nucleobase opposite the Tg residue. Additionally, this conjugate has demonstrated an inhibitory activity towards endonuclease III, a DNA repair enzyme, through its competitive binding to Tg-containing DNAs.
Reactive oxygen species (ROS) are known to cause several forms of DNA damage.1 Thymine glycol (5,6-dihydroxy-5,6-dihydrothymine; Tg) is the major oxidized product of thymine under oxidative stress.2 It has been estimated that 400 Tg residues are formed in a normal cell each day, and 10–20% of DNA damage is attributed to the oxidative conversion of thymine to Tg.3 Given its importance in understanding cellular oxidative stress and diagnostic applications,2,4 much attention has been paid to the development of analytical methods for the detection of Tg residues.
While several methods such as HPLC-MS and GC-MS have been developed,5 there are very limited reports on the detection of Tg residues in intact DNA duplexes without degradation.6 On the other hand, synthetic DNA binders that can selectively recognize specific sites in DNA duplexes have significant potential for analyzing non-canonical DNA structures.7 Of particular interest are fluorescence-responsive binders, which can be used to selectively detect abasic sites, mismatched base pairs and damaged nucleobases through the binding-induced fluorescence response;8 however, fluorescent binders for Tg-containing DNA duplexes have yet to be developed due to the challenge of selectively recognizing Tg-DNAs.
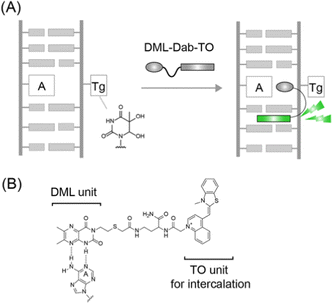
In this work, we report a new strategy for the design of synthetic binders for fluorescence sensing of Tg-DNAs (Fig. 1A). From the NMR analysis of 11-meric DNA duplexes carrying the 5R-Tg residue (A nucleobase opposite the Tg residue) by Bolton's group, the Tg residue was shown to induce the local structural perturbation of DNA duplexes.9 This results in the flipping out of the Tg residue from the DNA helix whereas the opposite A nucleobase attains an intrahelical position.10 We reasoned that the unpaired A nucleobase can be a useful target for molecular recognition by synthetic binders for Tg-DNA sensing. To this aim, dimethyllumazine (DML, Fig. 1B) developed by our group was employed as it can recognize the orphan A opposite the abasic site through pseudo-base pairing in the DNA duplexes.11 We used DML coupled with thiazole orange (TO), one of the well-characterized light-up intercalators,12 with a Dab (L-2,4-diamonobutyric acid) spacer (Fig. 1B), in order to achieve sensitive fluorescence detection of Tg-DNAs.13,14 Such a DML-Dab-TO conjugate was found to serve as a fluorescence-signaling binder with strong affinity and selectivity toward Tg-DNAs. We also showed the inhibitory activity of the conjugate for endonuclease III based on the competitive binding to Tg-DNAs.
The synthesis of DML-Dab-TO with a Dab (L-2,4-diamonobutyric acid) spacer was carried out essentially as previously described.13 Briefly, the carboxylic group was attached to the N3 position of DML, followed by coupling with the side chain of a Dab unit that was modified with a TO unit on the solid phase (Scheme S2, ESI†). The crude product was purified by reverse-phase HPLC and verified by MALD-TOF-MS (Fig. S1, ESI†). Further details are shown in the ESI.†
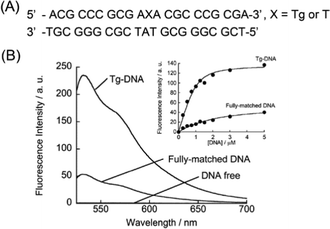
We first examined the fluorescence response of DML-Dab-TO (1.0 μM) for 21-meric model Tg-DNAs at 20 °C in PBS buffer (pH 7.4). The DNA sequences contained a Tg residue flanked by two adenines (Fig. 2A) based on the results obtained by Bolton's groups.10Fig. 2B shows the fluorescence spectra of DML-Dab-TO in the absence and presence of the Tg-DNA that has an A nucleobase opposite the Tg residue. The fluorescence of the TO unit in the probe is negligible in the absence of Tg-DNAs, with a fluorescence quantum yield (Φfl) of less than 0.002. This is attributable to the nonradiative energy loss by free rotation of the benzothiazole and quinoline rings.12 In contrast, we observed strong emission of the TO unit (λem = 533 nm) in the presence of 1.0 μM target Tg-DNA. The emission intensity increased as much as 250-fold. This indicates the significant light-up property of DML-Dab-TO upon binding to Tg-DNA. The Φfl value of DML-Dab-TO bound to Tg-DNA (Φbound) reaches 0.32. Significantly, the light-up response is 4.4-fold larger than that for fully-matched DNA carrying a thymine nucleobase instead of Tg (Fig. 2A). The results reveal that DML-Dab-TO selectively binds to Tg-DNAs. It was found that the nucleobase located opposite the Tg residue has a significant impact on the fluorescence response of DML-Dab-TO (Fig. S2, ESI†). The response for Tg-DNA with the opposite A was superior compared to those with other opposite nucleobases (G, C, or T). This observation can be explained by the A-selectivity of the DML unit due to the formation of the complementary hydrogen bonds.11 Hence, DML-Dab-TO binding for target Tg-DNA is driven by the selective recognition of unpaired A opposite the Tg residue. The observed light-up response (cf.Fig. 2) thus results from the intercalation of the TO unit into the double-stranded region upon the binding of the DML unit to the unpaired A (cf.Fig. 1). The binding affinity of DML-Dab-TO for target Tg-DNA was assessed by fluorescence titration experiments (Inset, Fig. 2B). The obtained titration curve was well fitted by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model,14 yielding the Kd value of 130 ± 21 nM (N = 3). This affinity was one order of magnitude stronger than that for the normal DNA (Kd = 1510 nM; Inset, Fig. 2B). These results showed that DML-Dab-TO had strong binding affinity toward Tg-DNA. It should be noted that the spacer length had a large impact on the binding affinity for Tg-DNAs (Fig. S3, ESI†).
1 binding model,14 yielding the Kd value of 130 ± 21 nM (N = 3). This affinity was one order of magnitude stronger than that for the normal DNA (Kd = 1510 nM; Inset, Fig. 2B). These results showed that DML-Dab-TO had strong binding affinity toward Tg-DNA. It should be noted that the spacer length had a large impact on the binding affinity for Tg-DNAs (Fig. S3, ESI†).
We studied the influence of the nucleobases flanking the Tg residue on the functions of DML-Dab-TO. Here, we examined three kinds of Tg-DNAs that had an A nucleobase opposite the Tg residue and two flanking nucleobases of either guanine (GTgG), cytosine (CTgC) or thymine (TTgT) (Table S1, ESI†). The Kd and Φbound for the TTgT sequence are determined as 160 ± 30 nM and 0.30, respectively. These values are almost comparable to those for Tg-DNA with two adenine flanking nucleobases (cf.Fig. 2). Meanwhile, we found that DML-Dab-TO had stronger affinity for CTgC and GTgG sequences (Kd/nM: CTgC, 11 ± 3.3; GTgG, 42 ± 9.9), in comparison with ATgA and TTgT ones. In addition, Φbound for the CTgC sequence was larger (Φbound = 0.55) than those for the other three sequences. These findings suggest that DML-Dab-TO can be used for the fluorescence sensing analysis of various Tg-DNA sequences. The variation in the intercalation efficiency of the TO unit is highly likely accountable for the dependence on the flanking nucleobases, given that the binding of the DML monomer is not largely affected by them (Kd = 80–110 μM). We examined the detection sensitivity of DML-Dab-TO based on the binding-induced fluorescence response for Tg-DNAs. DML-Dab-TO exhibited a linear response to the CTgC sequence with a range of concentrations up to 25 nM, from which the limit of detection (LOD) was calculated as 0.20 nM, corresponding to 10 fmol, based on 3 times the standard deviation of the blank (Fig. S4, ESI†). This value is superior to those for the other three sequences (LOD/nM: ATgA, 3.2; GTgG, 2.3; TTgT, 3.7). The LOD value for the CTgC sequence is comparable to that of LC-based methods,15 whereas there are several methods with superior sensitivity such as the GC-MS/MS method.16 These results show the prominent ability of DML-Dab-TO for sensitive detection of Tg-DNAs. It should be noted that DML-Dab-TO shows a fluorescence response for the abasic site-containing DNA duplexes carrying the opposite A nucleobase due to the inherent binding property toward unpaired A in the DNA duplexes.13 In order to accurately detect Tg-DNAs in real samples such as genomic DNAs, it may be necessary to remove any abasic site-containing DNA duplexes through the use of appropriate enzymes.17
Significantly, we found that DML-Dab-TO can differentiate the Tg residue from other thymine lesions, dihydrothymine (DHT) and 5-(hydroxymethyl)uracil (HMU), in the DNA duplexes. Fig. 3A shows the normalized fluorescence response of DML-Dab-TO to DNA duplexes carrying the lesions (5′-ACG CCC GCG A![[Z with combining low line]](https://www.rsc.org/images/entities/char_005a_0332.gif) A CGC CCG CGA-3′/3′-TGC GGG CGC T
A CGC CCG CGA-3′/3′-TGC GGG CGC T![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) T GCG GGC GCT-5′,
T GCG GGC GCT-5′, ![[Z with combining low line]](https://www.rsc.org/images/entities/char_005a_0332.gif) = Tg, HMU, or DHT.
= Tg, HMU, or DHT. ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) = the nucleobase opposite the lesion). The fluorescence responses for DHT and HMU are smaller by 2.9-fold and 3.2-fold, respectively, in comparison with Tg. We found that this resulted from much weaker affinity for these DNAs (Kd/nM: DHT, 860 nM; HMU, 1410 nM; Fig. S5, ESI†) in comparison with that for Tg-DNA. These results suggest that even small variation in the DNA lesions would influence the binding of DML-Dab-TO. To explore the structural aspects of these DNAs, the thermal stability was assessed by melting temperature (Tm) analysis (Fig. S6, ESI†). The Tm value for Tg-DNA was determined as 74.5 ± 1.4 °C, which is lower by more than 4.5 °C relative to DHT- (Tm = 79.0 ± 1.8 °C) and HMU-carrying DNA (Tm = 81.5 ± 1.8 °C). Thus, Tg-DNA is more unstable relative to the other two DNAs. We next examined the replacement of adenine opposite the lesion with 2-aminopurine (2AP) in order to probe the local structures around these lesions. 2AP is a fluorescent adenine analogue whose emission is quenched when stacked with the flanking nucleobases in the DNA duplexes.18 We measured the fluorescence spectra of the duplexes carrying 2AP opposite the lesion (5′-ACG CCC GCG A
= the nucleobase opposite the lesion). The fluorescence responses for DHT and HMU are smaller by 2.9-fold and 3.2-fold, respectively, in comparison with Tg. We found that this resulted from much weaker affinity for these DNAs (Kd/nM: DHT, 860 nM; HMU, 1410 nM; Fig. S5, ESI†) in comparison with that for Tg-DNA. These results suggest that even small variation in the DNA lesions would influence the binding of DML-Dab-TO. To explore the structural aspects of these DNAs, the thermal stability was assessed by melting temperature (Tm) analysis (Fig. S6, ESI†). The Tm value for Tg-DNA was determined as 74.5 ± 1.4 °C, which is lower by more than 4.5 °C relative to DHT- (Tm = 79.0 ± 1.8 °C) and HMU-carrying DNA (Tm = 81.5 ± 1.8 °C). Thus, Tg-DNA is more unstable relative to the other two DNAs. We next examined the replacement of adenine opposite the lesion with 2-aminopurine (2AP) in order to probe the local structures around these lesions. 2AP is a fluorescent adenine analogue whose emission is quenched when stacked with the flanking nucleobases in the DNA duplexes.18 We measured the fluorescence spectra of the duplexes carrying 2AP opposite the lesion (5′-ACG CCC GCG A![[Z with combining low line]](https://www.rsc.org/images/entities/char_005a_0332.gif) A CGC CCG CGA-3′/3′-TGC GGG CGC T(2AP)T GCG GGC GCT-5′,
A CGC CCG CGA-3′/3′-TGC GGG CGC T(2AP)T GCG GGC GCT-5′, ![[Z with combining low line]](https://www.rsc.org/images/entities/char_005a_0332.gif) = Tg, HMU, or DHT). As shown in Fig. 3B, the emission of 2AP-DNA with Tg is comparable to that of the fully-matched DNA carrying T opposite 2AP. This suggests that the A nucleobase opposite Tg is well stacked with neighbouring nucleobases, which is consistent with the previous NMR results.10 In contrast, the HMU-containing duplex exhibited much lower emission relative to the fully-matched DNA. HMU and its opposite A are thus highly likely to be well stacked in the DNA duplexes. This would be responsible for the observed weak binding (cf. Fig. S5, ESI†) given the limited accessibility of DML-Dab-TO for recognition of the A nucleobase opposite HMU. Instead, we observed larger emission for DHT-carrying DNA. While the structures of DHT-carrying DNAs were not fully clarified, the obtained results show that the structure around DHT is largely disordered. In particular, the target A nucleobase opposite DHT would not be stacked with the adjacent nucleobases, which is unfavorable for DML-Dab-TO binding. These results indicate that the local structures around the lesions have a large impact on the binding of DML-Dab-TO, as can be seen in DNA repair enzymes.19 While further structural analysis such as NMR and X-ray analysis is needed to clarify this, DML-Dab-TO holds unique potential applications, including the analysis of thymine-related damage given its selective response toward Tg over HMU and DHT.
= Tg, HMU, or DHT). As shown in Fig. 3B, the emission of 2AP-DNA with Tg is comparable to that of the fully-matched DNA carrying T opposite 2AP. This suggests that the A nucleobase opposite Tg is well stacked with neighbouring nucleobases, which is consistent with the previous NMR results.10 In contrast, the HMU-containing duplex exhibited much lower emission relative to the fully-matched DNA. HMU and its opposite A are thus highly likely to be well stacked in the DNA duplexes. This would be responsible for the observed weak binding (cf. Fig. S5, ESI†) given the limited accessibility of DML-Dab-TO for recognition of the A nucleobase opposite HMU. Instead, we observed larger emission for DHT-carrying DNA. While the structures of DHT-carrying DNAs were not fully clarified, the obtained results show that the structure around DHT is largely disordered. In particular, the target A nucleobase opposite DHT would not be stacked with the adjacent nucleobases, which is unfavorable for DML-Dab-TO binding. These results indicate that the local structures around the lesions have a large impact on the binding of DML-Dab-TO, as can be seen in DNA repair enzymes.19 While further structural analysis such as NMR and X-ray analysis is needed to clarify this, DML-Dab-TO holds unique potential applications, including the analysis of thymine-related damage given its selective response toward Tg over HMU and DHT.
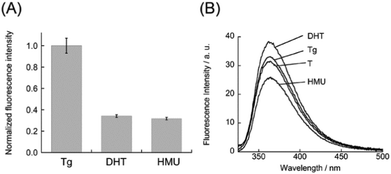 | ||
| Fig. 3 (A) Normalized fluorescence response of DML-Dab-TO (1.0 μM) for the lesion-containing DNA duplexes (1.0 μM). Excitation, 512 nm. Analysis, 533 nm. Temperature, 20 °C. (B) Fluorescence spectra of DNA duplexes carrying 2AP with its opposite lesion (Tg, DHT, or HMU) or T nucleobase (2.0 μM). Excitation, 310 nm. Temperature, 20 °C. Other solution conditions were the same as those given in Fig. 2. | ||
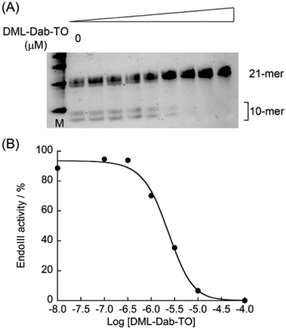
As demonstrated above, DML-Dab-TO has strong binding affinity for Tg-DNAs. We then investigated whether DML-Dab-TO could compete with the binding of Escherichia coli endonuclease III (EndoIII), a base excision repair enzyme that can recognize the Tg residue in the DNA duplex, to Tg-DNAs.20 We conducted polyacrylamide gel electrophoresis (PAGE) experiments to examine the cleavage of 21-meric Tg-DNAs by Endo III in the absence and presence of DML-Dab-TO. When Endo III cleaves upon recognizing the Tg residue, two kinds of 10-meric DNA fragments can be generated. Fig. 4A shows the PAGE results for CTgC sequences that DML-Dab-TO could strongly bind (Kd = 11 nM, cf. Table S1, ESI†). We observed a decrease in the band intensity of the cleavage products as the concentration of DML-Dab-TO increased. The half maximal inhibitory concentration (IC50) value was determined as 1.9 ± 0.35 μM (N = 3, Fig. 4B),21a which indicates the efficient inhibition of Endo III activity by DML-Dab-TO. When ATgA/TAT sequences (Kd = 130 nM, Fig. 2) were utilized, it was found that the inhibition activity of DML-Dab-TO significantly declined (Fig. S7, ESI;† IC50 > 20 μM). This indicates that the strong binding of DML-Dab-TO is crucial for the competitive binding toward Tg-DNAs to achieve efficient inhibition of Endo III. As for endonuclease inhibition, several small molecules have been successfully developed as effective inhibitors that can directly interact with target enzymes.19 In contrast, DML-Dab-TO is distinctive in that it inhibits enzyme activity by binding to Tg-DNAs, not enzymes, through the mechanism of competitive binding referred to as the DNA masking effect.22 While the activity of DML-Dab-TO depends on the DNA sequences, the IC50 value for CTgC sequences is indeed comparable to those of enzyme binding-based inhibition (0.78 μM for NTH1;21a 4.0 μM for NEIL121b). This represents, to the best of our knowledge, the first report of inhibition of DNA repair enzymes by Tg-DNA binding molecules. This class of molecules therefore holds therapeutic potential for modulating the enzyme activity.23
In conclusion, we have developed a new class of synthetic DNA binders with selective fluorescence sensing of Tg-DNAs. DML-Dab-TO exhibited a significant light-up response upon strong binding to the unpaired A nucleobase opposite the Tg residue in the DNA duplexes. In addition, we demonstrated the inhibitory activity of DML-Dab-TO for Endo III based on the competitive binding to Tg-DNAs. Given very limited reports on the molecular tools targeting Tg-DNAs, we envision that the obtained results provide valuable insights into the design of Tg-DNA-targeting binders with a view toward various applications in the DNA repair field. For example, according to our previous study,24 we conducted preliminary experiments to examine the potential of DML-Dab-TO for direct observation of Tg lesions in target DNAs using atomic force microscopy (AFM) (ESI†). We found that the Tg lesion could be visualized in combination with biotin-streptavidin interactions (Fig. S8, ESI†). Hence, this would provide information on the Tg distribution for the understanding of the complexity of multiple Tg lesions.24 We are undertaking further studies in this direction.
This work was supported by JSPS KAKENHI Grant Numbers JP20H02761, JP22H02099.
Conflicts of interest
There are no conflicts to declare.Notes and references
- U. S. Srinnivas, B. W. Q. Tan, B. A. Vellayappan and A. D. Jeyasekharan, Redox Biol., 2019, 25, 9716927 CrossRef.
- N. G. Dolinnaya, E. A. Kubereva, E. A. Romanova, R. M. Trikin and T. S. Oretskaya, Biochimie, 2013, 95, 134–147 CrossRef CAS PubMed.
- S. D. Wang, L. A. Eriksson and R. B. Zhang, J. Chem. Inf. Model., 2022, 62, 386–398 CrossRef CAS PubMed.
- I. Mahmud, F. G. Pintop, V. Y. Rubio, B. Lee, C. P. Pavlovih, R. J. Perera and T. J. Garrett, Anal. Chem., 2021, 93, 7774–7780 CrossRef CAS PubMed.
- (a) Y. Wang, Chem. Res. Toxicol., 2002, 15, 671–676 Search PubMed; (b) J. Cadet and H. Poulsen, Free Radical Biol. Med., 2010, 48, 1457–1459 CrossRef CAS PubMed; (c) D. B. Naritsin and S. P. Markey, Anal. Biochem., 1996, 241, 35–41 CrossRef CAS PubMed.
- D. Luvino, D. Gasparutto, S. Reynaud, M. Smietana and J.-J. Vasseur, Tetrahedron Lett., 2008, 49, 6075–6078 CrossRef CAS.
- (a) A. Granzhan, N. Kotera and M.-P. Teulade-Fichou, Chem. Soc. Rev., 2014, 43, 3630–3665 RSC; (b) P. D. Dayanidhi and V. G. Vaidyanathan, Dalton Trans., 2021, 50, 5691–5712 RSC.
- (a) Y. Sato, S. Nishizawa, K. Yoshimoto, T. Seino, T. Ichihashi, K. Morita and N. Teramae, Nucleic Acids Res., 2009, 37, 1411–1422 CrossRef CAS PubMed; (b) M. H. Lim, H. Song, E. D. Olmon, E. E. Dervan and J. K. Barton, Inorg. Chem., 2009, 48, 5392–5397 CrossRef CAS PubMed; (c) R. K. Verma, F. Takei and K. Nakatani, Org. Lett., 2016, 18, 3170–3173 CrossRef PubMed; (d) J. Peng, Y. Shao, L. Liu, L. Zhang and H. Liu, Dalton Trans., 2014, 43, 1534–1541 RSC; (e) Y. Taniguchi, R. Kawaguchi and S. Sasaki, J. Am. Chem. Soc., 2011, 133, 7272–7275 CrossRef CAS PubMed.
- J. Y. Kao, I. Goljier, T. A. Phan and P. H. Bolton, J. Biol. Chem., 1993, 268, 17787–17793 CrossRef CAS.
- H. C. Kunh and P. H. Bolton, J. Biol. Chem., 1997, 272, 9227–9236 CrossRef PubMed.
- Z. Ye, B. Rajendar, D. Qing, S. Nishizawa and N. Teramae, Chem. Commun., 2008, 6588–6590 RSC.
- (a) B. A. Armitage, Top. Curr. Chem., 2005, 253, 55–76 CrossRef CAS; (b) J. Nygren, N. Svanvik and M. Kubista, Biopolymers, 1998, 46, 39–51 CrossRef CAS PubMed.
- Y. Sato, H. Saito, D. Aoki, N. Teramae and S. Nishizawa, Chem. Commun., 2016, 52, 14446–14449 RSC.
- Y. Sato, M. Kudo, Y. Toriyabe, S. Kuchitsu, C.-X. Wang, S. Nishizawa and N. Teramae, Chem. Commun., 2014, 50, 515–517 RSC.
- (a) M. Dizdaroglu, P. Jaruga and H. Rodriguez, Nucleic Acids Res., 2001, 29, e12 CrossRef CAS PubMed; (b) P. Jaruga, M. Birincioglu, H. Rodriguez and M. Dizdaroglu, Biochemistry, 2002, 41, 3703–3711 CrossRef CAS PubMed.
- O. Aybastier and C. Demir, J. Pharm. Biomed. Anal., 2021, 200, 114068 CrossRef CAS PubMed.
- F. Tang, J. Yuan, B.-F. Yuan and Y. Wang, J. Am. Chem. Soc., 2022, 144, 454–462 CrossRef CAS PubMed.
- (a) J. T. Stivers, Nucleic Acids Res., 1998, 26, 3837–3844 CrossRef CAS; (b) K. Onizuka, K. Ishida, E. Mano and F. Nagatsugi, Org. Lett., 2019, 21, 2833–2837 CrossRef CAS PubMed; (c) M. Li, Y. Sato, S. Nishizawa, T. Seino, K. Nakamura and N. Teramae, J. Am. Chem. Soc., 2009, 131, 2448–2449 CrossRef CAS PubMed; (d) L. M. Wilhelmsson, Q. Rev. Biophys., 2010, 43, 159–183 CrossRef CAS PubMed; (e) R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579–2619 CrossRef CAS PubMed.
- (a) K. Asagoshi, H. Odawara, H. Nakano, T. Miyano, H. Terato, Y. Ohyama, S. Seki and H. Ide, Biochemistry, 2000, 39, 11389–11398 CrossRef CAS PubMed; (b) H. Ide, Prog. Nucelic Acid Res. Mol. Biol., 2001, 68, 207–221 CAS.
- N. Matsumoto, T. Toga, R. Hayashi, K. Sugasawa, K. Katayanagi, H. Ide, I. Kuraoka and S. Iwai, Nucleic Acids Res., 2010, 38, e101 CrossRef PubMed.
- (a) W. W. Jun, D. L. Wilson, A. M. Kietrys, E. R. Lotsof, S. G. Conlon, S. S. David and E. T. Kool, Angew. Chem., Int. Ed., 2020, 59, 7450–7455 CrossRef PubMed; (b) A. C. Jacobs, M. J. Calkins, A. Jadhav, D. Dorjsuren, D. Maloney, A. Simeonov, P. Jaruga, M. Dizdaroglu, A. K. McCullough and R. S. Lloyd, PLoS One, 2013, 8, e81667 CrossRef PubMed.
- N. Kotera, F. Poyer, A. Granzhan and M.-P. Teulade-Fichou, Chem. Commun., 2015, 51, 15948–15951 RSC.
- D. L. Wilson and E. T. Kool, ACS Chem. Biol., 2018, 13, 1721–1733 CrossRef CAS PubMed.
- (a) X. Xu, T. Nakano, M. Tsuda, R. Kanamoto, R. Hirayama, A. Uzawa and H. Ide, Nucleic Acids Res., 2020, 48, e18 CrossRef PubMed; (b) T. Nakano, K. Akamatsu, M. Tsuda, A. Tsujimoto, R. Hiramoto, T. Tamada, H. Ide and N. Shikazono, Proc. Natl. Acad. Sc. U. S. A., 2022, 119, e2119132119 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, fluorescence response, melting temperature, inhibitory activity, AFM imaging. See DOI: https://doi.org/10.1039/d3cc01501g |
| This journal is © The Royal Society of Chemistry 2023 |