Open Access Article
Open Access ArticleFluorescent indicator displacement assay for the discovery of UGGAA repeat-targeted small molecules†
Tomonori
Shibata
 *a,
Yasumasa
Matsumoto
b,
Akiko
Iihara
b,
Kazunori
Yamada
b,
Hiroshi
Ochiai
b,
Ryo
Saito
b,
Shinichi
Kusaka
b and
Toshiyuki
Kume
b
*a,
Yasumasa
Matsumoto
b,
Akiko
Iihara
b,
Kazunori
Yamada
b,
Hiroshi
Ochiai
b,
Ryo
Saito
b,
Shinichi
Kusaka
b and
Toshiyuki
Kume
b
aDepartment of Regulatory Bioorganic Chemistry, SANKEN (The Institute of Scientific and Industrial Research), Osaka University, Ibaraki, Osaka 567-0047, Japan. E-mail: shibata@sanken.osaka-u.ac.jp
bSohyaku. Innovative Research Division, Mitsubishi Tanabe Pharma Corporation, 1-1-1, Marunouchi, Chiyoda-ku, Tokyo 100-8205, Japan
First published on 31st March 2023
Abstract
We report that a selective fluorescent indicator NBD-NCD for UGGAA repeats resulted in fluorescence quenching upon binding to RNA and recovered the fluorescence by displacing NBD-NCD with UGGAA repeat-targeted small molecules. The fluorescent indicator displacement assay using NBD-NCD can detect the interaction of small molecules with UGGAA repeats.
RNA is an important factor in diverse cellular functions involving biological processes. As understanding of the RNA functions in cells and their relevance to many human diseases has progressed,1–3 RNA has attracted the interest of researchers as a druggable target. Oligonucleotide therapeutics using antisense oligonucleotides and small interfering RNAs is a therapeutic modality for RNA-targeted therapies.4–6 The success of RNA-targeted drugs based on oligonucleotide therapeutics has demonstrated that RNA is a promising therapeutic target. Apart from oligonucleotide therapeutics, the discovery of small molecules that bind to RNAs and regulate their functions is highly desirable in drug discovery of RNA-targeted small molecules.7–9 Researchers have focused on the exploratory study of RNA-targeted small molecules utilizing various screening methods such as the fluorescent indicator displacement (FID) assay,10–15 small molecule microarray-based screening,16–18 affinity-selection mass spectroscopy,19,20 surface plasmon resonance (SPR)-based screening,21in silico screening,22 fragment-based screening,23–25 and DNA-encoded library technology.26,27
The FID assay is a simple and high-throughput method that can detect the interactions between small molecules and RNAs without fluorescent labelling of both target RNAs and compounds used in screening. In general, fluorescent indicators that change their fluorescence properties upon binding to RNA are used for FID assays. Previously, it has been reported that a fluorescent indicator, 2,7-disubstituted 9H-xanthen-9-one derivative (X2S), quenches upon RNA binding and emits fluorescence by the displacement of X2S with RNA-binding small molecules.11 TO-PRO1 and thiazole orange have also been used for FID assays.12,28 These dyes result in the increase of the fluorescence upon binding to RNA but quench upon the displacement of the dyes with RNA-binding small molecules. In addition to fluorescent indicators based on dyes, Tat peptide labelled with a Förster Resonance Energy Transfer (FRET) pair has also been used in displacement assays for screening of the molecules binding to HIV TAR RNAs.10 In this assay, the FRET efficiency of the Tat peptide probe increases upon RNA binding, whereas the displacement of the Tat peptide probe from RNA upon the binding of molecules decreases the FRET efficiency. An FID assay has been used to screen small molecules targeting various structured RNAs including bacterial rRNA A-site, HIV-1-TAR, Rev response element, enterovirus RNA structure, and disease-causing repeat RNAs.10–15,29,30 Since fluorescent indicators used in RNA-targeted FID assays bind to secondary structures including hairpins, bulges, and internal loops with low sequence selectivity, screening of RNA-binding small molecules based on the displacement of non-selective fluorescent indicators may lead to a decrease of the accuracy in FID assays. Therefore, the development of fluorescent indicators that selectively bind to target RNAs could provide a rigorous screening system with high accuracy. Several groups have reported fluorescent indicators that selectively bind to the RNA motif of interest.28,31
Disease-causing repeat RNAs known as toxic RNAs32 can be a potential therapeutic target for the treatment of repeat expansion disorders. Since targeting the disease-causing repeat RNAs with small molecules has been identified as a promising approach to inhibit its toxic functions, several groups have reported bioactive small molecules binding to the disease-causing repeat RNAs.33–35 Small molecules targeting disease-causing repeat RNAs have been identified by various screening methods.36–38 It has been reported that high-throughput screening by FID assay using TO-PRO-1 has successfully discovered a bioactive small molecule to a disease-causing repeat RNA.30 On the other hand, fluorescent indicators with binding selectivity to disease-causing repeat RNAs have not been well-studied. Here, we report a selective fluorescent indicator that can be used in FID assays for UGGAA repeats causing spinocerebellar ataxia type 31 (SCA31).
Previously, we have reported that a small molecule, naphthyridine carbamate dimer (NCD; Fig. 1a), which binds to SCA31-causing UGGAA repeats, alleviated RNA toxicity in the Drosophila model of SCA31.38 To develop the selective fluorescent indicator for UGGAA repeats, we designed and synthesized nitrobenzoxadiazole-labelled NCD (NBD-NCD; Fig. 1a and Scheme S1, ESI†) based on the fact that fluorescent dyes are efficiently quenched by guanosine residues.39 The nucleobase-specific quenching of fluorescent dyes is a well-known phenomenon, which is used in the probe design for the specific detection of DNA sequences.40 Therefore, we expected that the fluorescence of NBD-NCD would be quenched upon binding to UGGAA repeats containing guanosine residues and recovered by the dissociation of NBD-NCD from the complex accompanied by the binding of UGGAA repeat-targeted small molecules (Fig. 1b). Firstly, we investigated the binding of NBD-NCD to UGGAA repeats by thermal melting temperature (Tm) and circular dichroism (CD) measurements (Fig. 2). The UV melting curve of r(UGGAA)9 had an unclear melting transition, whereas the UV melting profile in the presence of NBD-NCD showed a clear melting curve with an inflection point around 52 °C (Fig. 2a). The CD spectrum of r(UGGAA)9 upon the addition of NBD-NCD showed induced CD bands at 320–370 nm and 420–520 nm attributed to NCD and NBD, respectively (Fig. 2b). Importantly, the induced CD bands at 320–370 nm were almost identical to those observed in the CD spectrum of r(UGGAA)9 with NCD,37 suggesting that the binding mode of NBD-NCD is likely similar to that of NCD. The binding stoichiometry of NBD-NCD-bound r(UGGAA)5 complexes was examined by cold-spray ionization mass spectrometry (CSI-MS). The complexes with the binding stoichiometry ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 were observed under the condition of 10 μM r(UGGAA)5 and 50–100 μM NBD-NCD (Fig. 2c, Fig. S1, and Table S1, ESI†).
5 were observed under the condition of 10 μM r(UGGAA)5 and 50–100 μM NBD-NCD (Fig. 2c, Fig. S1, and Table S1, ESI†).
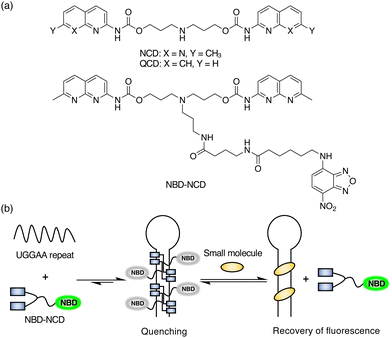 | ||
| Fig. 1 (a) Chemical structures of NCD, QCD, and NBD-NCD. (b) Schematic illustration of the FID assay using NBD-NCD and UGGAA repeats. | ||
To investigate the fluorescence quenching of NBD-NCD upon binding to UGGAA repeats, we performed titration experiments of r(UGGAA)n (n = 5, 9, 20) to NBD-NCD. The fluorescence intensity (FI) of NBD-NCD was decreased upon the addition of r(UGGAA)n in an RNA concentration- and repeat size-dependent manner, which reached saturation at lower RNA concentrations with increasing the repeat size (Fig. 3a). Since saturation of fluorescence quenching was observed under the conditions of 200 nM r(UGGAA)9 and 1 μM NBD-NCD, we selected this condition for our assays. Under the same conditions, the titration experiments of r(UGGAA)9 to already-reported fluorescent indicators such as X2S11 and TO-PRO-112 required >5-fold RNA concentration (> 1 μM) for reaching saturation (Fig. S2, ESI†). This suggested that the binding affinity of NBD-NCD for UGGAA repeats was higher than those of X2S and TO-PRO-1. We also confirmed that the fluorescence polarization (FP) of NBD-NCD was increased in an RNA concentration-dependent manner (Fig. 3b). In addition, titration experiments of r(UAGAA)9 and r(UAAAA)9 to NBD-NCD showed almost no change in FI and FP (Fig. S3, ESI†). These results indicated the selective binding of NBD-NCD to UGGAA repeats. We next examined whether displacement of NBD-NCD with UGGAA repeat-binding small molecules leads to an increase in the fluorescence of NBD-NCD. The FID assay with the addition of NCD as a positive control demonstrated that the fluorescence of NBD-NCD increased in an NCD concentration-dependent manner and eventually led to a 4-fold increase in FI (Fig. 3c). As a negative control, we also tested quinoline carbamate dimer (QCD; Fig. 1a), which does not bind to UGGAA repeats,38 and confirmed that the addition of QCD did not increase the fluorescence of NBD-NCD. The addition of NCD decreased the FP of NBD-NCD in a concentration-dependent manner but not QCD (Fig. 3d), implying the dissociation of NBD-NCD from UGGAA repeats upon the binding of NCD.
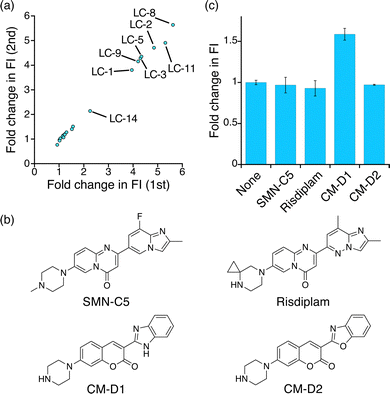
The result of the FID assay using NCD prompted us to perform the screening of small molecule libraries. In this assay, we used an in-house chemical library containing 20 compounds (LC-1–LC-20), which was previously used in SPR assay-based screening for UGGAA repeat-binding small molecules (Fig. S4, ESI†),38 and performed two independent experiments (Fig. 4a). The fold changes in FI upon the addition of LC-1–LC-20 obtained from the two independent experiments are in good agreement with each other. Among the 20 compounds that we tested, fold changes in FI above 3.0 were observed in 7 compounds discovered by previous SPR assay-based screening. The addition of these compounds resulted in a decrease of FP (Fig. S5, ESI†), indicating that the binding of the hit compounds dissociated NBD-NCD from UGGAA repeats. In addition, we found that LC-14 exhibited an approximately 2.2-fold change in FI. The increase in FI and the decrease in FP were LC-14 concentration-dependent (Fig. S6, ESI†), suggesting that NBD-NCD is dissociated from r(UGGAA)9 upon LC-14 binding. We also confirmed that the Tm of r(UGGAA)9 was increased upon the addition of LC-14 (Fig. S7, ESI†). SPR assay for the r(UGGAA)9-immobilized surface indicated that several non-hit compounds showed a significant increase in response unit (Fig. S8, ESI†). This suggested that several compounds can interact with UGGAA repeats but not displace NBD-NCD upon ligand-binding.
Finally, we investigated whether our FID assay could detect an unidentified interaction of a small molecule with UGGAA repeats. We selected SMN-C5, risdiplam, and two coumarin derivatives (CM-D1 and CM-D2) as tested compounds (Fig. 4b). Since it has been reported that SMN-C5 and their derivatives selectively bind to purine-rich RNA motifs,41,42 these small molecules could have potential for binding to purine-rich UGGAA repeats. We performed the FID assay for SMN-C5, risdiplam, CM-D1, and CM-D2 (Fig. 4c and Fig. S9, ESI†). SMN-C5, risdiplam, and CM-D2 showed negligible change in FI, whereas approximately 1.6-fold change was observed in the presence of CM-D1. The addition of CM-D1 to r(UGGAA)9 increased FI and decreased FP in a concentration-dependent manner (Fig. S10, ESI†). The binding of SMN-C5, risdiplam, CM-D1, and CM-D2 to r(UGGAA)9 was investigated by Tm measurement and SPR assay, suggesting that CM-D1 likely bound to r(UGGAA)9 with the highest affinity among the 4 compounds (Fig. S11 and S12, ESI†). Together, these results demonstrated that our FID assay is capable of detecting unidentified interactions between small molecules with UGGAA repeats.
In conclusion, we designed and synthesized a selective fluorescent indicator NBD-NCD for UGGAA repeats by fluorescently labelling NCD with NBD. The fluorescence quenching of NBD-NCD selectively occurred upon binding to UGGAA repeats, whereas its fluorescence was recovered by the displacement of NBD-NCD with UGGAA repeat-binding small molecules. In the FID assay using NBD-NCD and UGGAA repeats, we succeeded in identifying not only UGGAA repeat-binding small molecules from the in-house chemical library but also unknown UGGAA repeat binder CM-D1. The FID assay described here is useful for high-throughput screening to detect the interactions between small molecules and UGGAA repeats.
This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific research (B) (20H02880) to T. S., JST, CREST Grant Number JPMJCR20E6. Project MEET, Osaka University Graduate School of Medicine, and Mitsubishi Tanabe Pharma Corporation.
Conflicts of interest
There are no conflicts to declare.Notes and references
- T. A. Cooper, L. Wan and G. Dreyfuss, Cell, 2009, 136, 777–793 CrossRef CAS PubMed.
- E. Lekka and J. Hall, FEBS Lett., 2018, 592, 2884–2900 CrossRef CAS PubMed.
- A. Neueder, J. Mol. Biol., 2019, 431, 1780–1791 CrossRef CAS PubMed.
- A. Khvorova and J. K. Watts, Nat. Biotechnol., 2017, 35, 238–248 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- M. Winkle, S. M. El-Daly, M. Fabbri and G. A. Calin, Nat. Rev. Drug Discovery, 2021, 20, 629–651 CrossRef CAS PubMed.
- K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discovery, 2018, 17, 547–558 CrossRef CAS PubMed.
- J. P. Falese, A. Donlic and A. E. Hargrove, Chem. Soc. Rev., 2021, 50, 2224–2243 RSC.
- J. L. Childs-Disney, X. Yang, Q. M. R. Gibaut, Y. Tong, R. T. Batey and M. D. Disney, Nat. Rev. Drug Discovery, 2022, 21, 736–762 CrossRef CAS PubMed.
- C. Matsumoto, K. Hamasaki, H. Mihara and A. Ueno, Bioorg. Med. Chem. Lett., 2000, 10, 1857–1861 CrossRef CAS PubMed.
- J. Zhang, S. Umemoto and K. Nakatani, J. Am. Chem. Soc., 2010, 132, 3660–3661 CrossRef CAS PubMed.
- P. N. Asare-Okai and C. S. Chow, Anal. Biochem., 2011, 408, 269–276 CrossRef CAS PubMed.
- R. del Villar-Guerra, R. D. Gray, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2018, 46, e41 CrossRef CAS PubMed.
- N. N. Patwardhan, Z. Cai, C. N. Newson and A. E. Hargrove, Org. Biomol. Chem., 2019, 17, 1778–1786 RSC.
- J. Li, Y. Y. Fan, M. Wang, H. L. Duan, J. Zhang, F. Q. Dang, L. Zhang and Z. Q. Zhang, Anal. Chem., 2020, 92, 13532–13538 CrossRef CAS PubMed.
- L. P. Labuda, A. Pushechnikov and M. D. Disney, ACS Chem. Biol., 2009, 4, 299–307 CrossRef CAS PubMed.
- J. Sztuba-Solinska, S. R. Shenoy, P. Gareiss, L. R. H. Krumpe, S. F. J. Le Grice, B. R. O’Keefe and J. S. Schneekloth, J. Am. Chem. Soc., 2014, 136, 8402–8410 CrossRef CAS PubMed.
- C. M. Connelly, F. A. Abulwerdi and J. S. Schneekloth Jr, Methods Mol. Biol., 2017, 1518, 157–175 CrossRef CAS PubMed.
- N. F. Rizvi, J. A. Howe, A. Nahvi, D. J. Klein, T. O. Fischmann, H. Y. Kim, M. A. McCoy, S. S. Walker, A. Hruza, M. P. Richards, C. Chamberlin, P. Saradjian, M. T. Butko, G. Mercado, J. Burchard, C. Strickland, P. J. Dandliker, G. F. Smith and E. B. Nickbarg, ACS Chem. Biol., 2018, 13, 820–831 CrossRef CAS PubMed.
- R. Aguilar, K. B. Spencer, B. Kesner, N. F. Rizvi, M. D. Badmalia, T. Mrozowich, J. D. Mortison, C. Rivera, G. F. Smith, J. Burchard, P. J. Dandliker, T. R. Patel, E. B. Nickbarg and J. T. Lee, Nature, 2022, 604, 160–166 CrossRef CAS PubMed.
- T. Fukuzumi, A. Murata, H. Aikawa, Y. Harada and K. Nakatani, Chem. – Eur. J., 2015, 21, 16859–16867 CrossRef CAS PubMed.
- L. R. Ganser, J. Lee, A. Rangadurai, D. K. Merriman, M. L. Kelly, A. D. Kansal, B. Sathyamoorthy and H. M. Al-Hashimi, Nat. Struct. Mol. Biol., 2018, 25, 425–434 CrossRef CAS PubMed.
- B. Tam, D. Sherf, S. Cohen, S. A. Eisdorfer, M. Perez, A. Soffer, D. Vilenchik, S. R. Akabayov, G. Wagner and B. Akabayov, Chem. Sci., 2019, 10, 8764–8767 RSC.
- B. M. Suresh, W. Li, P. Zhang, K. W. Wang, I. Yildirim, C. G. Parker and M. D. Disney, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 33197–33203 CrossRef CAS PubMed.
- K. P. Lundquist, V. Panchal, C. H. Gotfredsen, R. Brenk and M. H. Clausen, ChemMedChem, 2021, 16, 2588–2603 CrossRef CAS PubMed.
- R. I. Benhamou, B. M. Suresh, Y. Tong, W. G. Cochrane, V. Cavett, S. Vezina-Dawod, D. Abegg, J. L. Childs-Disney, A. Adibekian, B. M. Paegel and M. D. Disney, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, 1–8 CrossRef PubMed.
- Q. M. R. Gibaut, Y. Akahori, J. A. Bush, A. Taghavi, T. Tanaka, H. Aikawa, L. S. Ryan, B. M. Paegel and M. D. Disney, J. Am. Chem. Soc., 2022, 144, 21972–21979 CrossRef CAS PubMed.
- Y. Sato, S. Yajima, A. Taguchi, K. Baba, M. Nakagomi, Y. Aiba and S. Nishizawa, Chem. Commun., 2019, 55, 3183–3186 RSC.
- J. Davila-Calderon, N. N. Patwardhan, L. Y. Chiu, A. Sugarman, Z. Cai, S. R. Penutmutchu, M. L. Li, G. Brewer, A. E. Hargrove and B. S. Tolbert, Nat. Commun., 2020, 11, 4775 CrossRef CAS PubMed.
- A. Ursu, K. W. Wang, J. A. Bush, S. Choudhary, J. L. Chen, J. T. Baisden, Y. J. Zhang, T. F. Gendron, L. Petrucelli, I. Yildirim and M. D. Disney, ACS Chem. Biol., 2020, 15, 3112–3123 CrossRef CAS PubMed.
- B. Das, A. Murata and K. Nakatani, Nucleic Acids Res., 2021, 49, 8462–8470 CrossRef CAS PubMed.
- B. Swinnen, W. Robberecht and L. Van Den Bosch, EMBO J., 2020, 39, e101112 CrossRef CAS PubMed.
- J. Y. Lee, Y. Bai, U. V. Chembazhi, S. Peng, K. Yum, L. M. Luu, L. D. Hagler, J. F. Serrano, H. Y. Edwin Chan, A. Kalsotra and S. C. Zimmerman, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 8709–8714 CrossRef CAS PubMed.
- S. G. Rzuczek, L. A. Colgan, Y. Nakai, M. D. Cameron, D. Furling, R. Yasuda and M. D. Disney, Nat. Chem. Biol., 2017, 13, 188–193 CrossRef CAS PubMed.
- J. A. Bush, S. M. Meyer, R. Fuerst, Y. Tong, Y. Li, R. I. Benhamou, H. Aikawa, P. R. A. Zanon, Q. M. R. Gibaut, A. J. Angelbello, T. F. Gendron, Y. Zhang, L. Petrucelli, T. H. Jensen, J. L. Childs-Disney and M. D. Disney, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2210532119 CrossRef CAS PubMed.
- W. Y. Yang, R. Gao, M. Southern, P. S. Sarkar and M. D. Disney, Nat. Commun., 2016, 7, 11647 CrossRef CAS PubMed.
- K. M. Green, U. J. Sheth, B. N. Flores, S. E. Wright, A. B. Sutter, M. G. Kearse, S. J. Barmada, M. I. Ivanova and P. K. Todd, J. Biol. Chem., 2019, 294, 18624–18638 CrossRef CAS PubMed.
- T. Shibata, K. Nagano, M. Ueyama, K. Ninomiya, T. Hirose, Y. Nagai, K. Ishikawa, G. Kawai and K. Nakatani, Nat. Commun., 2021, 12, 236 CrossRef CAS PubMed.
- C. A. M. Seidel, A. Schulz and M. H. M. Sauer, J. Phys. Chem., 1996, 100, 5541–5553 CrossRef CAS.
- T. Heinlein, J. P. Knemeyer, O. Piestert and M. Sauer, J. Phys. Chem. B, 2003, 107, 7957–7964 CrossRef CAS.
- J. Wang, P. G. Schultz and K. A. Johnson, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4604–E4612 CAS.
- M. Sivaramakrishnan, K. D. McCarthy, S. Campagne, S. Huber, S. Meier, A. Augustin, T. Heckel, H. Meistermann, M. N. Hug, P. Birrer, A. Moursy, S. Khawaja, R. Schmucki, N. Berntenis, N. Giroud, S. Golling, M. Tzouros, B. Banfai, G. Duran-Pacheco, J. Lamerz, Y. Hsiu Liu, T. Luebbers, H. Ratni, M. Ebeling, A. Cléry, S. Paushkin, A. R. Krainer, F. H. T. Allain and F. Metzger, Nat. Commun., 2017, 8, 1476 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc00837a |
| This journal is © The Royal Society of Chemistry 2023 |