Open Access Article
Open Access ArticleSilver cluster-assembled materials for label-free DNA detection†
Saikat
Das
a,
Taishu
Sekine
a,
Haruna
Mabuchi
a,
Sakiat
Hossain
a,
Subhabrata
Das
 b,
Shun
Aoki
c,
Shuntaro
Takahashi
b,
Shun
Aoki
c,
Shuntaro
Takahashi
 *b and
Yuichi
Negishi
*b and
Yuichi
Negishi
 *a
*a
aDepartment of Applied Chemistry, Faculty of Science, Tokyo University of Science, Kagurazaka, Shinjuku-ku, Tokyo 162-8601, Japan. E-mail: negishi@rs.tus.ac.jp
bChemical Materials Development Department, TANAKA KIKINZOKU KOGYO K.K., Tsukuba Technical Center, 22 Wadai, Tsukuba, Ibaraki 300-4247, Japan. E-mail: s-tak@ml.tanaka.co.jp
cBio Chemical Development Department, TANAKA KIKINZOKU KOGYO K.K., Hiratsuka Technical Center, 2-73, Shinmachi, Hiratsuka, Kanagawa 254-0076, Japan
First published on 27th February 2023
Abstract
Herein, we report two newly synthesized silver cluster-assembled materials (SCAMs), [Ag14(StBu)10(CF3COO)4(bpa)2]n (bpa = 1,2-bis(4-pyridyl)acetylene) and [Ag12(StBu)6(CF3COO)6(bpeb)3]n (bpeb = 1,4-bis(pyridin-4-ylethynyl)benzene) composed of Ag14 and Ag12 chalcogenolate cluster cores, respectively, bridged by acetylenic bispyridine linkers. The linker structures and electrostatic interaction between positively charged SCAMs and negatively charged DNA confer the SCAMs with the ability to suppress the high background fluorescence of single-stranded (ss) DNA probes with SYBR Green I nucleic acid stain, leading to high signal-to-noise ratio for label-free target DNA detection.
Silver nanoclusters1,2 have flourished as enticing nanomaterials by virtue of their remarkable photoemission with quantum yields as high as 95%,3 high catalytic activities,4 and sensing properties.5 That said, research on silver nanoclusters has been mostly limited to fundamental studies focusing on the synthesis and isolation of new clusters, resolving their geometric structures, and exploring different surface organic ligands and anion templates as stabilizing/directing agents. The primary concern limiting its applicability is its proneness to oxidization, thereby imparting it with a compromised stability. In light of the foregoing, the transformation of molecular clusters into the extended structure regime have endowed silver cluster-assembled materials (SCAMs)6 with the advantages of structural diversity, precise designability, and superior stability. SCAMs are crystalline frameworks consisting of polynuclear silver clusters linked together by organic linkers.
The first report on SCAM appeared from Mak and co-workers who exchanged the acetonitrile (CH3CN) ligands in [(Ag12(StBu)6(CF3COO)6(CH3CN)6]·CH3CN cluster with 4,4′-bipyridine (bpy) ligands to construct the two-dimensional (2D) SCAM [(Ag12(StBu)8(CF3COO)4(bpy)4)]n that exhibited significantly improved stability over one year and 60-fold greater quantum yield.7 Since then, the library of SCAMs has expanded by combining the diverse geometry and composition of silver cluster nodes with a wide array of organic linkers.8–12 The combination of silver clusters and organic linkers has not only endowed the SCAMs with augmented stabilities but also with unprecedented performance such as enhanced photoluminescence quantum yields13–16 and superior photocatalytic bacterial inactivation,17 among others. However, acetylenic bispyridine linkers have not hitherto been reported to construct the SCAMs. In this study, two novel SCAMs, [Ag14(StBu)10(CF3COO)4(bpa)2]n (hereinafter Ag14bpa) and [Ag12(StBu)6(CF3COO)6(bpeb)3]n (hereinafter Ag12bpeb) were successfully obtained utilizing two acetylenic linkers, bpa and bpeb (bpa = 1,2-bis(4-pyridyl)acetylene, bpeb = 1,4-bis(pyridin-4-ylethynyl)benzene). Interestingly, the SCAMs, by merit of π-stacking and electrostatic interaction between negatively charged sugar-phosphate backbone of DNA and positively charged cluster assembly, can contribute to fluorescence background quenching of ssDNA probes with fluorescent dye, entailing significant improvement in signal-to-noise ratio (SNR) for label-free DNA detection.
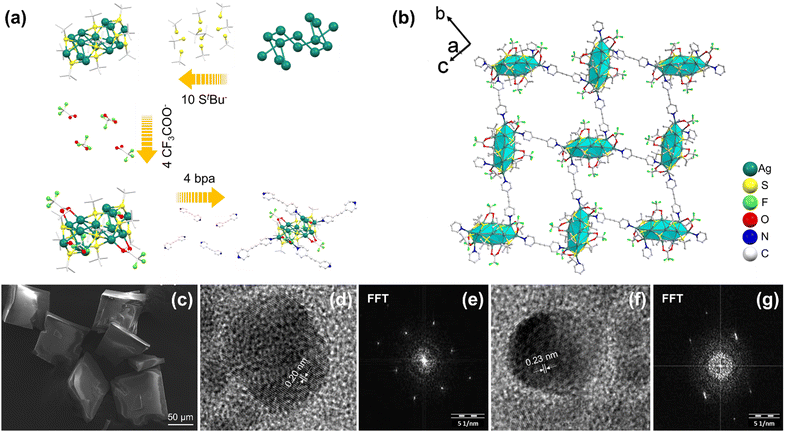
Ag14bpa and Ag12bpeb were prepared in high yields through the one-pot reaction of metal salts (AgStBu, CF3COOAg) and the organic linkers (bpa or bpeb) in a mixture of solvents (acetonitrile/chloroform for Ag14bpa or dimethylacetamide/toluene for Ag12bpeb). First, the reaction took place between AgStBu with CF3COOAg in the aforementioned solvent mixtures. Next, the solutions containing the organic linkers bpa or bpeb were added dropwise. Following solvent removal by slow evaporation, high-quality yellow and colorless single crystals were obtained for Ag14bpa and Ag12bpeb, respectively (ESI† includes full details). The structure of Ag14bpa SCAM was determined by single-crystal X-ray diffraction (SCXRD) studies that exhibited monoclinic crystal system, space-group type C2/c (Table S1, ESI†). Similar to the situation in Ag14apy,15 the core of the Ag14 cluster in Ag14bpa adopts the shape of a slightly distorted ortho-bielongated square pyramid (Fig. S1, ESI†) in which the Ag5 atoms are located at the apical sites (Fig. S2, ESI†) and the Ag14 core is held by AgI⋯AgI argentophilic interactions (2.92–3.22 Å, Table S2, ESI†), longer than the Ag–Ag bond length (2.89 Å) of bulk silver,18 but shorter than the sum of two standard AgI van der Waals radii (3.44 Å).19 The Ag14 cluster node features eight square faces and eight triangular faces and is stabilized by ten StBu− and four CF3COO− ligands (Fig. 1a). The four μ3-S (S2, S5) bind to Ag atoms (Ag2, Ag3, Ag4, Ag7) on the square faces with average Ag–S bond length of 2.50 Å (Fig. S2 and Table S3, ESI†) and Ag–S–Ag bond angle in the range of 71.24–128.77° (Fig. S3, ESI†). The two μ4-S (S4) bind to Ag atoms (Ag1, Ag4, Ag6, Ag7) on the square faces with average Ag–S bond length of 2.65 Å (Fig. S2 and Table S3, ESI†) and Ag–S–Ag bond angle in the range of 64.78–111.47° (Fig. S4, ESI†). The four μ3-S (S3, S1) bind to Ag atoms (Ag1, Ag2, Ag3, Ag5, Ag6) on the triangular faces with average Ag–S bond length of 2.46 Å (Fig. S2 and Table S4, ESI†) and Ag–S–Ag bond angle in the range of 75.08–124.08° (Fig. S5, ESI†). As can be seen from Fig. S2 and Table S5 (ESI†), the two μ2-CF3COO− (O1, O2) bind to Ag atoms (Ag2, Ag7) on the square faces with average Ag–O bond length of 2.36 Å. The two μ2-CF3COO− (O3, O4) bind to Ag atoms (Ag5, Ag6) on the triangular faces with average Ag–O bond length of 2.40 Å. The Ag14 clusters are linked by four bpa linkers with average Ag–N bond length of 2.31 Å (Table S6, ESI†), resulting in a 2D rectangular net with ABA stacking pattern.
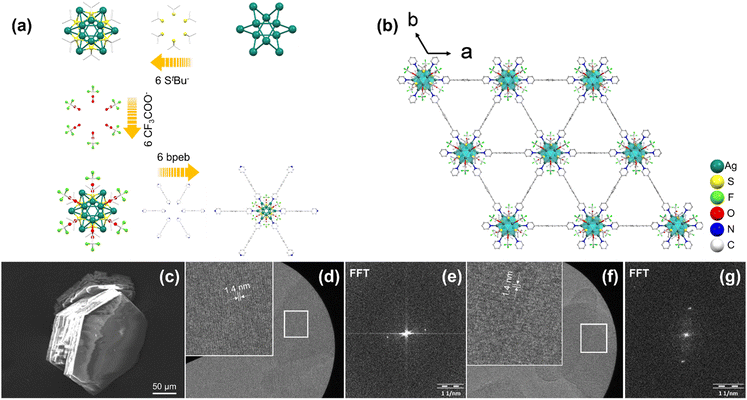
Ag12bpeb SCAM crystallizes in the trigonal crystal system with the space group of R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) m (Table S7, ESI†). The structure of Ag12bpeb is built from the secondary building units (SBUs) Ag12(StBu)6(CF3COO)6, which are connected to each other through bpeb linkers (Fig. 2a). The cuboctahedron-shaped Ag12 core features two equilateral and six isosceles triangles (Fig. S6 and S7, ESI†). In the two equilateral triangles, the average Ag1–Ag1 bond length is 2.96 Å and Ag1–Ag1–Ag1 bond angle is 60° (Fig. S8, ESI†). In the six isosceles triangles, the average Ag1–Ag2 bond length is 3.15 Å, Ag2–Ag1–Ag2 bond angle 74.88°, and Ag1–Ag2–Ag2 bond angle 57.48° (Fig. S9, ESI†). The six μ4-S (S1) bind to Ag atoms (Ag1, Ag2) with average Ag–S bond length of 2.50 Å and Ag–S–Ag bond angle in the range of 73.48-133.52° (Fig. S10, ESI†). The six μ1-CF3COO− (O2) bind to Ag1 atoms with average Ag–O bond length of 2.41 Å. The Ag12 clusters are linked by six bpeb linkers with average Ag–N bond length of 2.29 Å to afford a high-symmetry extended 2D hxl net that displays the ABCA stacking pattern.
m (Table S7, ESI†). The structure of Ag12bpeb is built from the secondary building units (SBUs) Ag12(StBu)6(CF3COO)6, which are connected to each other through bpeb linkers (Fig. 2a). The cuboctahedron-shaped Ag12 core features two equilateral and six isosceles triangles (Fig. S6 and S7, ESI†). In the two equilateral triangles, the average Ag1–Ag1 bond length is 2.96 Å and Ag1–Ag1–Ag1 bond angle is 60° (Fig. S8, ESI†). In the six isosceles triangles, the average Ag1–Ag2 bond length is 3.15 Å, Ag2–Ag1–Ag2 bond angle 74.88°, and Ag1–Ag2–Ag2 bond angle 57.48° (Fig. S9, ESI†). The six μ4-S (S1) bind to Ag atoms (Ag1, Ag2) with average Ag–S bond length of 2.50 Å and Ag–S–Ag bond angle in the range of 73.48-133.52° (Fig. S10, ESI†). The six μ1-CF3COO− (O2) bind to Ag1 atoms with average Ag–O bond length of 2.41 Å. The Ag12 clusters are linked by six bpeb linkers with average Ag–N bond length of 2.29 Å to afford a high-symmetry extended 2D hxl net that displays the ABCA stacking pattern.
The powder X-ray diffraction (PXRD) patterns of Ag14bpa and Ag12bpeb are in good agreement with the simulated patterns, indicative of the phase purity of the SCAMs (Fig. S11 and S12, ESI†). Scanning electron microscopy (SEM) and optical microscopy characterization of the SCAMs revealed single crystals with well-defined morphology (Fig. 1c, 2c and Fig. S13, ESI†). The lattice fringes of the SCAMs could be clearly observed from the high-resolution transmission electron microscopy (HRTEM) images (Fig. 1d, f and 2d, f). Thermogravimetric analysis (TGA) showed that the Ag14bpa and Ag12bpeb remained stable up to ∼200 °C and 150 °C, respectively (Fig. S14 and S15, ESI†). To assess the chemical stability of Ag14bpa and Ag12bpeb, we immersed the SCAMs in different organic solvents and water. As evident from the PXRD patterns (Fig. S16 and S17, ESI†), the structural integrity of the SCAMs was well-maintained after the treatment.
The unique structural features of the SCAMs intrigued us to explore their potential as label-free DNA sensors. As illustrated in Fig. S20 (ESI†), the DNA detection assay was prepared by hybridizing the probe DNA (40 nM) with the target DNA (40 nM) in 10 mM phosphate buffer (pH 7.4) at 37 °C for 30 min. Subsequently, 0.245 μM SYBR Green I dye was added and incubated at room temperature. Finally, Ag-SCAM (2.0 mg mL−1) was added to achieve the desired concentration. The final mixture was incubated at room temperature for another 30 min, and then centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 minutes. The supernatant obtained was directly used for fluorescence measurement. The fluorescence measurements were captured with an excitation wavelength of 490 nm, and the emission wavelength was recorded at 521 nm.
000 × g for 5 minutes. The supernatant obtained was directly used for fluorescence measurement. The fluorescence measurements were captured with an excitation wavelength of 490 nm, and the emission wavelength was recorded at 521 nm.
Consistent with the literature,20 the principle of recognition of target DNA goes as (Fig. 3a): the ssDNA probe with fluorescent dye is adsorbed onto the surface of SCAMs, the latter acting as an effective quencher for the fluorescence signal of the former. When the ssDNA probe encounters a target DNA strand, probe hybridization separates the double-stranded DNA from the SCAMs making it inaccessible for quenching, thereby producing signal amplification and high SNR.
As can be seen from Fig. 3b, the SNR of the DNA sensor system is poor when unaccompanied by the SCAMs, the high background fluorescence of ssDNA probes with SYBR Green I nucleic acid stain being the central concern. With the addition of the SCAMs, the background fluorescence shows a steady decline, which in turn enhances the SNR. For Ag14bpa, the SNR reveals a value of ca. 42.5 at a Ag14bpa concentration of 5 μg mL−1, a 2.5-fold increase in SNR compared to that before the addition of Ag14bpa. In case of Ag12bpeb, the SNR attained a value of ca. 32.5 at a Ag12bpeb concentration of 10 μg mL−1, also a 2.5-fold enhancement in SNR than that prior to the addition of Ag12bpeb. As the concentration of the SCAMs was increased more, the SNR starts declining owing to the adsorption of further double-stranded DNA by the SCAMs.
We attribute the competency of the SCAMs to suppress the high background fluorescence of ssDNA probes with fluorescent dye to the π-stacking and electrostatic interaction. Based on linker structures, the SCAMs reveal a π-stacked conformation (Fig. 3c). We also tested the zeta potentials of Ag14bpa and Ag12bpeb with and without ssDNA probe and SYBR Green I that elucidated the negatively charged backbone of DNA and positively charged cluster assembly (Fig. 3d).
To conclude, Ag14 and Ag12 chalcogenolate cluster cores were successfully reticulated with acetylenic bispyridine linkers into extended frameworks. We also developed a simple and label-free assay for HIV-1 DNA detection utilizing a DNA probe to hybridize with target DNA sequences and the silver cluster-based frameworks as quencher to diminish the high background fluorescence of ssDNA probes with SYBR Green I nucleic acid stain. This work could chart the way towards harnessing cluster-assembled materials in DNA-based diagnostics and molecular biology.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. Yang and R. Jin, ACS Mater. Lett., 2019, 1, 482–489 CrossRef CAS.
- Y.-P. Xie, Y.-L. Shen, G.-X. Duan, J. Han, L.-P. Zhang and X. Lu, Mater. Chem. Front., 2020, 4, 2205–2222 RSC.
- Z. Han, X.-Y. Dong, P. Luo, S. Li, Z.-Y. Wang, S.-Q. Zang and T. C. W. Mak, Sci. Adv., 2020, 6, eaay0107 CrossRef CAS PubMed.
- K. Shimizu, M. Nishimura and A. Satsuma, ChemCatChem, 2009, 1, 497–503 CrossRef CAS.
- X. Shen, X. Yang, C. Su, J. Yang, L. Zhang, B. Liu, S. Gao, F. Gai, Z. Shao and G. Gao, J. Mater. Chem. C, 2018, 6, 2088–2094 RSC.
- A. Ebina, S. Hossain, H. Horihata, S. Ozaki, S. Kato, T. Kawawaki and Y. Negishi, Nanomaterials, 2020, 10, 1105 CrossRef CAS PubMed.
- R.-W. Huang, Y.-S. Wei, X.-Y. Dong, X.-H. Wu, C.-X. Du, S.-Q. Zang and T. C. W. Mak, Nat. Chem., 2017, 9, 689–697 CrossRef CAS PubMed.
- Y.-H. Li, R.-W. Huang, P. Luo, M. Cao, H. Xu, S.-Q. Zang and T. C. W. Mak, Sci. China: Chem., 2019, 62, 331–335 CrossRef CAS.
- X.-H. Ma, J.-Y. Wang, J.-J. Guo, Z.-Y. Wang and S.-Q. Zang, Chin. J. Chem., 2019, 37, 1120–1124 CrossRef CAS.
- C.-H. Gong, Z.-B. Sun, M. Cao, X.-M. Luo, J. Wu, Q.-Y. Wang, S.-Q. Zang and T. C. W. Mak, Chem. Commun., 2022, 58, 9806–9809 RSC.
- Z. Wei, X.-H. Wu, P. Luo, J.-Y. Wang, K. Li and S.-Q. Zang, Chem. – Eur. J., 2019, 25, 2750–2756 CrossRef CAS PubMed.
- J.-J. Zhu, P. Hu, K.-K. Zhou, B. Li and T. Zhang, Dalton Trans., 2017, 46, 6663–6669 RSC.
- Y.-M. Wang, J.-W. Zhang, Q.-Y. Wang, H.-Y. Li, X.-Y. Dong, S. Wang and S.-Q. Zang, Chem. Commun., 2019, 55, 14677–14680 RSC.
- M. I. Rogovoy, A. S. Berezin, D. G. Samsonenko and A. V. Artem‘ev, Inorg. Chem., 2021, 60, 6680–6687 CrossRef CAS PubMed.
- A. K. Das, S. Biswas, A. Thomas, S. Paul, A. S. Nair, B. Pathak, M. S. Singh and S. Mandal, Mater. Chem. Front., 2021, 5, 8380–8386 RSC.
- X.-H. Wu, P. Luo, Z. Wei, Y.-Y. Li, R.-W. Huang, X.-Y. Dong, K. Li, S.-Q. Zang and B. Z. Tang, Adv. Sci., 2019, 6, 1801304 CrossRef PubMed.
- M. Cao, S. Wang, J.-H. Hu, B.-H. Lu, Q.-Y. Wang and S.-Q. Zang, Adv. Sci., 2022, 9, 2103721 CrossRef CAS PubMed.
- C.-G. Shi, J.-H. Jia, Y. Jia, G. Li and M.-L. Tong, CCS Chem., 2022 DOI:10.31635/ccschem.022.202201960.
- H. Schmidbaur and A. Schier, Angew. Chem., Int. Ed., 2014, 53, 2–41 CrossRef.
- J. M. Fang, F. Leng, X. J. Zhao, X. L. Hu and Y. F. Li, Analyst, 2014, 139, 801–806 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2231764 and 2231770. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2cc06933d |
| This journal is © The Royal Society of Chemistry 2023 |