Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemical control of phase separation in DNA solutions†
Samuel
Hauf
 and
Yohei
Yokobayashi
and
Yohei
Yokobayashi
 *
*
Nucleic Acid Chemistry and Engineering Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, Japan 904-0495, Japan. E-mail: yohei.yokobayashi@oist.jp
First published on 6th March 2023
Abstract
We designed a series of DNA sequences comprising a trinucleotide repeat segment and a small molecule-binding aptamer. Optimization of the DNA sequences and reaction conditions enabled chemical control of phase separation of DNA condensates. Our results demonstrate a new strategy to regulate biomolecular phase transition.
Phase separation is the process by which a solution of previously miscible chemical species becomes unmixed.1 Liquid–liquid phase separation (LLPS) has recently gained recognition as an important process in life, involved in regulatory processes,2,3 stress response,4 or certain diseases,5 among others. Besides its importance in natural phenomena, LLPS is also finding applications in synthetic biology.6,7 The process is usually driven by biopolymers such as proteins and/or nucleic acids forming dense intermolecular networks.
Nucleic acids, especially RNA, are often involved in phase separation processes observed in cells. For example, RNA involved in repeat expansion disorders can form ribonucleoprotein bodies in the cellular nucleus.5 In the nucleolus, DNA, RNA, and proteins phase separate from the surrounding environment to regulate transcription.8 Stress granules are phase separated aggregates that increase fitness under stress conditions.4 How exactly phase separation occurs and how it is influenced by the sequence of the biopolymers are among the subjects of ongoing research. The process is assumed to be tightly controlled by factors such as the concentration and sequence of the phase separating species, posttranslational protein modifications (e.g., SUMOylation),4,9 temperature, and cation species.10,11
For RNA, it has been shown that phase separation depends on sequence, length, and strength of RNA–RNA interaction.5 It has been hypothesized that the phase separation process is driven by intermolecular base pairing resulting in a complex network of RNA molecules.5 Because nucleic acid interactions based on Watson–Crick base pairing are relatively straightforward, it provides an opportunity to design nucleic acid sequences with programmable LLPS properties. Control of LLPS was previously shown using temperature,12,13 enzymatic pH-modulation,14 light,15 small molecules,16,17 and size-specific diffusion of ATP through pores in lipid bilayers.18
We aimed to investigate if sequence-driven phase separation of nucleic acids can be controlled using aptamers. Aptamers are nucleic acid sequences that selectively bind target molecules (ligands) such as small molecules and proteins. Here, we show for the first time that single-stranded oligonucleotide sequences that combine the ability to form phase separated aggregates with the capability to sense a small molecule recognized by an aptamer can be designed. Our system can provide insights into chemical regulation of phase separation, and it can lead to applications in biosensing and drug delivery.
Jain and Vale showed that RNAs consisting of CAG or GGGGCC repeats can phase separate when reaching a minimum length.5 Their results suggested that a minimum valency and strength of interaction is necessary for nucleic acids phase separation without other factors (e.g., proteins). It was hypothesized that CAG repeats drive aggregation by forming intermolecular base pairs and the GGGGCC repeats by forming intermolecular G-quadruplexes.5 G-quadruplex-forming RNAs have also been shown to drive phase separation in the presence of an arginine-rich peptide.19
We started by evaluating if it is possible to combine a DNA aptamer with repeat sequences to control phase separation behavior by the aptamer ligand. For this purpose, we chose the well-studied ATP aptamer20 and combined it with different numbers of CAG repeats. The ATP aptamer sequence can form G-quadruplexes20 which can contribute to phase separation.5 However, its short length may not provide sufficient intermolecular interactions to induce phase separation. We presumed that adding CAG-repeats to the 5′ end of the aptamer would increase the valency of interstrand interactions to trigger phase separation. Upon binding of ATP,21 however, the intermolecular network in the condensed phase is compromised and cannot maintain separate phases (Fig. 1).
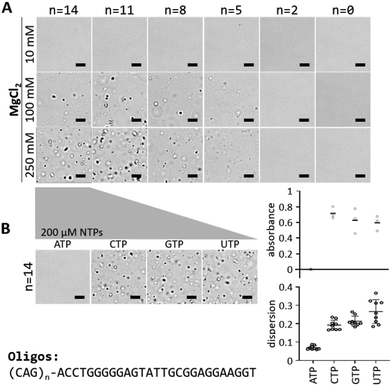
Under the conditions determined by Jain and Vale in their RNA experiments (10 mM MgCl2), we did not observe phase separation with the ATP aptamer fused to up to 14 CAG repeats (Fig. 2A). However, increasing MgCl2 concentration to 100 mM or 250 mM yielded aggregates for constructs with at least 5 CAG repeats (Fig. 2A). At 100 mM MgCl2, the aggregates were small but became clearly visible at 250 mM. This is consistent with the recent observation that higher concentrations of divalent cations enhance phase separation of ssDNA.11,22 The ATP aptamer alone could not form aggregates (Fig. 2A). It thus appears that both the ATP aptamer and the CAG repeats contribute to the observed phase separation. In agreement with previous results,5,22 a minimum length or minimum number of intermolecular interactions appears to be necessary.
The presence of 200 μM ATP in the solution of (CAG)14-ApATP (14 CAG repeats fused to ATP aptamer) with 250 mM MgCl2 prevented the formation of visible aggregates, while presence of other nucleotides (CTP, GTP, UTP) did not affect phase separation (Fig. 2B). This suggests that the structural rearrangement of the ATP aptamer upon ATP binding20 prevents phase separation. The degree of phase separation was quantified by monitoring the absorbance at 500 nm (turbidity) of the solution, and by calculating the dispersion of pixel intensities from the microscopic images (Fig. 2B). Phase separation (turbidity) strongly depends on the Mg2+ as well as the DNA concentrations used (Fig. S1, ESI†). The aggregates were stained by SYBR Green I, indicating that they contain DNA (Fig. S2, ESI†). Preformed aggregates were rapidly dissolved upon addition of ATP, while addition of other NTPs had no effect (Movies S1–S5, ESI†). In agreement with previous reports on DNA-based systems,13,23 fluorescence recovery after photobleaching (FRAP) experiments showed that the aggregates behave like gels at room temperature (23 °C) and more like liquid at 43 °C (Fig. S3 and S4, ESI†). At 38 °C, fusion of droplets could be observed in the presence of 5% ethylene glycol indicating a liquid-like state, while aggregate formation was still prevented by addition of 1 mM ATP (Fig. S5, ESI†). Taken together, these results suggest that it is possible to chemically control the phase separation behavior of short DNA by incorporating an aptamer into the sequence.
Thus far, DNA phase separation was enhanced by a high Mg2+ concentration. Molecular crowding is also known to influence LLPS.24,25 Therefore, effects of the addition of uncharged crowding agents PEG200 and DMSO were investigated. Indeed, it was possible to enhance phase separation at 100 mM MgCl2 by adding ≥2.5% PEG200 or ≥5% DMSO (Fig. S6A, ESI†). PEG200 was very efficient, but the aggregates formed no longer responded to ATP. Aggregates formed in the presence of DMSO, however, remained sensitive to ATP (Fig. S6B, ESI†). PEG200 is known to enhance phase separation and stabilize G-quadruplex structures.26 Therefore, it is possible that the aggregates are too stable to respond to ATP in the presence of PEG200. Alternatively, PEG200 may result in DNA precipitation.27 Next, we tested the effect of Tris–HCl (pH 7.0) and NaCl concentration on aggregate formation in the presence of 10% DMSO. High concentration of Na+ is known to inhibit phase separation of nucleic acids.5,15 In our case, though, Na+ concentration had a negligible effect, while Tris–HCl at concentrations >100 mM inhibited phase separation (Fig. S6C, ESI†). The identity of the alkali cation used can influence the stability of the aggregates by affecting G-quadruplex stability.28 We therefore tested if the aggregates behave differently in the presence of Li+ or K+ compared to Na+. We found that K+ stabilizes the aggregates so that they no longer respond to ATP, while Li+ and Na+ destabilize the aggregates at the highest concentration tested (200 mM, Fig. S7, ESI†).
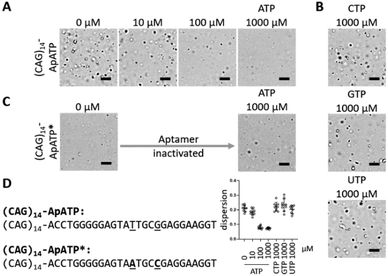
Next, the concentration dependence on ATP was investigated. To minimize pH variation due to the different ATP and high Mg2+ concentrations, the experiments were performed at a higher Tris–HCl concentration (100 mM each of Tris–HCl (pH 7.0), NaCl, MgCl2, 10% DMSO). A marked decrease in aggregate formation was observed between 100–1000 μM ATP, while CTP, GTP, and UTP had no effect at the highest concentration tested (1000 μM) (Fig. 3A, B, and D). Additionally, inactivation of the aptamer by mutation of the conserved residues29 abolished the reaction to ATP, but it also negatively impacted the rate of aggregation probably due to negative effects on intermolecular interactions (Fig. 3C). These results demonstrate that the phase separation process can be fine-tuned by the addition of the aptamer ligand.
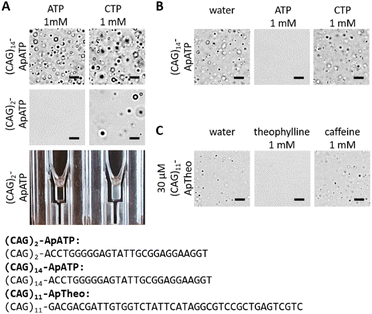
When working with DNA (instead of RNA), Jain and Vale used spermine to drive DNA LLPS.5 Spermine is a tetravalent cation at neutral pH and has been shown to drive aggregation of G-quadruplex-forming sequences.30 It was also shown that the properties of the resulting aggregates depend on the sequences used.5,30 In our case, LLPS of (CAG)14-ApATP could be triggered by 5 mM spermine, but the resulting aggregates did not react to ATP (Fig. 4A, top). The ATP aptamer alone did not phase separate in the presence of spermine (data not shown), but the construct (CAG)2-ApATP did form aggregates that respond to ATP (Fig. 4A, middle). The change in turbidity is clearly visible to the naked eye offering a convenient read-out of the chemical regulation of phase separation (Fig. 4A, bottom) which may be useful for analytical applications.
Shorter DNA sequences can undergo phase separation in the presence of spermine compared to Mg2+. This suggests that the tetravalent cation spermine is a powerful driver of DNA LLPS. Therefore, a window in which the DNA aggregates respond to the aptamer ligand appears at shorter lengths.
It was recently shown that Ca2+ can also trigger the formation of phase separated aggregates that are more stable than aggregates formed with Mg2+.22 Indeed, our construct also formed aggregates with 100 mM CaCl2 (Fig. 4B). Interestingly, these aggregates still react to ATP. Nucleic acid aggregates produced by phase separation with calcium thus seem to be more stable than with magnesium, but they can still react to sequence-encoded triggers.
Finally, we tested whether the approach is extendable to other aptamers. For this purpose, a theophylline DNA aptamer was chosen.31 A construct with 11 CAG repeats fused to the aptamer ((CAG)11-ApTheo) phase separate when heated to 46 °C even in the presence of 1 mM caffeine, but not in the presence of 1 mM theophylline (Fig. 4C). In this case, preformed aggregates were not dissolved by the addition of the ligand. When heated to 95 °C, the difference between the condition with or without theophylline was not as pronounced (data not shown). This suggests that the ligand might affect the second transition temperature at which the phase separation occurs.22 The findings with the theophylline aptamer highlight the highly sequence- and condition-dependent behavior of nucleic acids and suggest that the chemical response may be improved through further optimization, although more work is needed to explain the underlying mechanism that causes sequence-dependent phase separation of DNA.
In summary, we have shown that it is possible to regulate the phase separation process of DNA by a small molecule by combining a trinucleotide repeat sequence and a small molecule aptamer. The oligo DNAs designed here form chemically controllable microdroplets under appropriate conditions. The chemically responsive phase separation behavior observed depends on a number of factors. First, the DNA sequences must be able to form sufficient intermolecular base pairing or tertiary interactions (e.g. G-quadruplex) to drive phase separation. Second, aptamer-ligand interaction must be able to substantially compromise the intermolecular network to dissolve or prevent phase separation. This objective can be achieved through DNA sequence design and/or optimizing reaction conditions. For example, longer DNA can favor phase separation22 but may inhibit sensitivity to aptamer-ligand binding. Similarly, a chemical environment that enhances DNA phase separation may negatively affect ligand response, but it can also allow shorter DNA sequences to form aggregates that respond to the ligand. While our systems are based on DNA, the results may also apply to RNA. It is also worth noting that Deng and Walther recently designed ATP-fuelled DNA coacervates that use ATP as a fuel for DNA ligation.32
Although previously not reported, it is possible that small molecule metabolites regulate LLPS in living cells through interaction with nucleic acids. Chemical regulation of microdroplets containing nucleic acids may find applications in synthetic biology and medicine, for example, for diagnostic (aptamer-based sensors) or therapeutic (drug delivery) tools. Alternatively, nucleic acid-based aggregates may be useful for the study of protocells or artificial cell models.7,12 Further refinements of the nucleic acid sequence design and analysis methods toward these goals are the subjects of future research.
We would like to thank the OIST Imaging Section, in particular Paolo Barzaghi, for support with FRAP experiments and video acquisition. This work was supported by funds from Okinawa Institute of Science and Technology Graduate University and Deutsche Forschungsgemeinschaft (DFG) - Projektnummer 452628014, Geschätszeichen: HA9374 to S. H.
Conflicts of interest
There are no conflicts to declare.References
- J. B. Clarke, J. W. Hastie, L. H. E. Kihlborg, R. Metselaar and M. M. Thackeray, Pure Appl. Chem., 1994, 66, 577–594 CrossRef.
- S. Chong, C. Dugast-Darzacq, Z. Liu, P. Dong, G. M. Dailey, C. Cattoglio, A. Heckert, S. Banala, L. Lavis, X. Darzacq and R. Tjian, Science, 2018, 361, eaar2555 CrossRef PubMed.
- D. Hnisz, K. Shrinivas, R. A. Young, A. K. Chakraborty and P. A. Sharp, Cell, 2017, 169, 13–23 CrossRef CAS PubMed.
- J. A. Riback, C. D. Katanski, J. L. Kear-Scott, E. V. Pilipenko, A. E. Rojek, T. R. Sosnick and D. A. Drummond, Cell, 2017, 168, 1028–1040 CrossRef CAS PubMed.
- A. Jain and R. D. Vale, Nature, 2017, 546, 243–247 CrossRef CAS PubMed.
- C. D. Crowe and C. D. Keating, Interface Focus, 2018, 8, 20180032 CrossRef PubMed.
- C. Xu, N. Martin, M. Li and S. Mann, Nature, 2022, 609, 1029–1037 CrossRef CAS PubMed.
- S. Ide, R. Imai, H. Ochi and K. Maeshima, Sci. Adv., 2020, 6, eabb5953 CrossRef CAS PubMed.
- S. F. Banani, A. M. Rice, W. B. Peeples, Y. Lin, S. Jain, R. Parker and M. K. Rosen, Cell, 2016, 166, 651–663 CrossRef CAS.
- J. H. Jung, A. D. Barbosa, S. Hutin, J. R. Kumita, M. Gao, D. Derwort, C. S. Silva, X. Lai, E. Pierre, F. Geng, S. B. Kim, S. Baek, C. Zubieta, K. E. Jaeger and P. A. Wigge, Nature, 2020, 585, 256–260 CrossRef CAS PubMed.
- P. Pullara, I. Alshareedah and P. R. Banerjee, Soft Matter, 2022, 18, 1342–1349 RSC.
- A. Samanta, V. Sabatino, T. R. Ward and A. Walther, Nat. Nanotechnol., 2020, 15, 914–921 CrossRef CAS.
- Y. Sato, T. Sakamoto and M. Takinoue, Sci. Adv., 2020, 6, eaba3471 CrossRef CAS PubMed.
- H. Karoui, M. J. Seck and N. Martin, Chem. Sci., 2021, 12, 2794–2802 RSC.
- N. Martin, L. Tian, D. Spencer, A. Coutable-Pennarun, J. L. R. Anderson and S. Mann, Angew. Chem., Int. Ed., 2019, 58, 14594–14598 CrossRef CAS PubMed.
- W. M. Babinchak, B. K. Dumm, S. Venus, S. Boyko, A. A. Putnam, E. Jankowsky and W. K. Surewicz, Nat. Commun., 2020, 11, 5574 CrossRef CAS PubMed.
- M. Ramesh, C. Balachandra, P. Baruah and T. Govindaraju, J. Pept. Sci., 2022, e3465 Search PubMed.
- S. Deshpande, F. Brandenburg, A. Lau, M. G. F. Last, W. K. Spoelstra, L. Reese, S. Wunnava, M. Dogterom and C. Dekker, Nat. Commun., 2019, 10, 1800 CrossRef PubMed.
- M. Tsuruta, T. Torii, K. Kohata, K. Kawauchi, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Chem. Commun., 2022, 58, 12931–12934 RSC.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef CAS PubMed.
- C. H. Lin and D. J. Patel, Chem. Biol., 1997, 4, 817–832 CrossRef CAS PubMed.
- W. Liu, A. Samanta, J. Deng, C. O. Akintayo and A. Walther, Angew. Chem., Int. Ed., 2022, 61, e202208951 CAS.
- R. Merindol, S. Loescher, A. Samanta and A. Walther, Nat. Nanotechnol., 2018, 13, 730–738 CrossRef CAS PubMed.
- J. Hochmair, C. Exner, M. Franck, A. Dominguez-Baquero, L. Diez, H. Brognaro, M. L. Kraushar, T. Mielke, H. Radbruch, S. Kaniyappan, S. Falke, E. Mandelkow, C. Betzel and S. Wegmann, EMBO J., 2022, 41, e108882 CrossRef CAS PubMed.
- S. Park, R. Barnes, Y. Lin, B.-J. Jeon, S. Najafi, K. T. Delaney, G. H. Fredrickson, J.-E. Shea, D. S. Hwang and S. Han, Commun. Chem., 2020, 3, 83 CrossRef CAS.
- R. Hansel, F. Lohr, S. Foldynova-Trantirkova, E. Bamberg, L. Trantirek and V. Dotsch, Nucleic Acids Res., 2011, 39, 5768–5775 CrossRef.
- A. Schmitz and D. Riesner, Anal. Biochem., 2006, 354, 311–313 CrossRef CAS.
- E. Largy, J.-L. Mergny and V. Gabelica, in The Alkali Metal Ions: Their Role for Life, ed. A. Sigel, H. Sigel and R. K. O. Sigel, Springer International Publishing, Cham, 2016, pp. 203–258 DOI:10.1007/978-3-319-21756-7_7.
- R. Nutiu and Y. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- A. M. Williams, R. R. Poudyal and P. C. Bevilacqua, Biochemistry, 2021, 60, 2715–2726 CrossRef CAS PubMed.
- P. J. Huang and J. Liu, ACS Chem. Biol., 2022, 17, 2121–2129 CrossRef CAS PubMed.
- J. Deng and A. Walther, Chem, 2020, 6, 3329–3343 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods, and supplementary figures. See DOI: https://doi.org/10.1039/d2cc06901f |
| This journal is © The Royal Society of Chemistry 2023 |