Open Access Article
Open Access ArticleSmall molecule modulators of immune pattern recognition receptors†
Taku
Tsukidate‡
 a,
Charles W.
Hespen‡
a,
Charles W.
Hespen‡
 a and
Howard C.
Hang
*ab
a and
Howard C.
Hang
*ab
aLaboratory of Chemical Biology and Microbial Pathogenesis, The Rockefeller University, New York, New York 10065, USA
bDepartment of Immunology and Microbiology and Department of Chemistry, Scripps Research, La Jolla, California 92037, USA
First published on 23rd October 2023
Abstract
Pattern recognition receptors (PRRs) represent a re-emerging class of therapeutic targets for vaccine adjuvants, inflammatory diseases and cancer. In this review article, we summarize exciting developments in discovery and characterization of small molecule PRR modulators, focusing on Toll-like receptors (TLRs), NOD-like receptors (NLRs) and the cGAS-STING pathway. We also highlight PRRs that are currently lacking small molecule modulators and opportunities for chemical biology and therapeutic discovery.
Introduction
The ability of animals and plants to sense microbes provides an important means to discriminate self from non-self and initiate immune responses to establish homeostasis as well as defend against potentially lethal infections. To do so, animals and plants have acquired and evolved pattern recognition receptors (PRRs) that detect and respond to common microbe, pathogen and damage associated molecular patterns (MAMPs, PAMPs and DAMPs, respectively).1,2 These molecular patterns are composed of molecules foreign to a healthy host yet common among different microbes or damaged host cells. These molecules vary drastically in structure and complexity. MAMPs and PAMPs can include components of the bacterial cell wall and membrane, bacterial and viral nucleic acids, particulates, and potassium efflux in the host. DAMPs include molecules associated with the breakdown vital cellular components like organelles and the extracellular matrix. The ability to sense diverse types of molecules allow the host to mount an immune response and initiate clearance of infected or damaged cells.Several classes of PRRs are expressed in humans that signal for MAMPs, PAMPs and DAMPs. These include but are not limited to toll-like receptors (TLRs),3 NOD-like receptors (NLRs),4 and the cGAMP signaling (cGAS-STING) pathway.5 Activation of these receptors leads to downstream production of inflammatory cytokines, antimicrobial factors, and cell death factors. Disfunction of these receptors can lead to immune disorders and diseases resulting in chronic inflammation. For example, improper function of the NLR, nucleotide-binding oligomerization domain-containing protein 2 (NOD2), is associated with Crohn's disease and Blau syndrome,6,7 and disfunction of the NLR-family pyrin domain containing 3 (NLRP3) inflammasome is associated with a myriad of disorders ranging from diabetes to Alzheimer's disease.8
Agonists of PRRs can be powerful immunostimulants and are used as adjuvants. One of the earliest discovered immune activators is Freund's adjuvant, which is composed of heat inactivated mycobacteria in a water/oil emulsion.9 The active molecule of Freund's adjuvant was later identified to be muramyl dipeptide (MDP), which is the ligand of NOD2. MDP and its derivatives that activate NOD2 have been found to be potent immunostimulants that serve important roles in pathogen clearance and increasing the efficacy of cancer immunotherapy.10,11 Numerous immunologic adjuvants exist that target several PRRs. For example, aluminum acts as a NLRP3 agonist, Lipid A derivatives activate TLR4, and CpG oligonucleotides activate TLR9.12
As each PRR has evolved to signal for specific MAMPs and PAMPS, structural models of these receptors deepen the understanding of ligand specificity and activation mechanisms. Reliable structures of PRRs can be used to assist the development of next generation immunotherapies. This review seeks to summarize natural PRR agonists from a structural and chemical perspective and describe synthetic activators and inhibitors of these receptors. Additionally, this review will highlight opportunities for chemical biology and therapeutic discovery and complement existing reviews on this topic.13,14
Toll-like receptors
There are 10 TLR genes in humans and 12 in mice.2,3 TLRs 1–9 are conserved between the two species. TLR10 is expressed in humans but is a pseudogene in mice. TLRs 11–13 are expressed in mice but are pseudo genes in humans. Each TLR recognizes a distinct set of molecular patterns that are atypical of healthy host cells. TLR10 remains to be an orphan receptor. TLRs-1, 2, 4, 5, and 6 are localized on the plasma membrane, while TLRs-3, 7, 8, and 9 are localized on the endosomal membrane (Fig. 1A). TLRs are single-pass membrane proteins consisting of an ectodomain, a transmembrane domain, and a Toll–IL-1 receptor (TIR) domain (Fig. 1B). The ectodomain contains 18–25 copies of leucine rich repeat (LRR) and typically binds ligands. The TIR domain interfaces the cytoplasm and interacts with other TIR-type domains in signaling proteins. Agonistic ligands induce receptor dimerization and bring the two TIR domains together, which allows them to interact with the TIR domains of cytoplasmic adapter molecules to trigger intracellular signaling. There are four such adaptors used by TLRs: MyD88, MAL (also known as TIRAP), TRIF, and TRAM. Most TLRs interact with MyD88 except for TLR3, which interacts with TRIF. All TLRs induce NF-κB–mediated cytokine production, whereas endosomal TLRs also induce IRF-mediated production of type-I interferons. TLRs are intricately involved in various aspects of health and disease and are important therapeutic targets for sepsis, lupus, and vaccine adjuvants to name a few.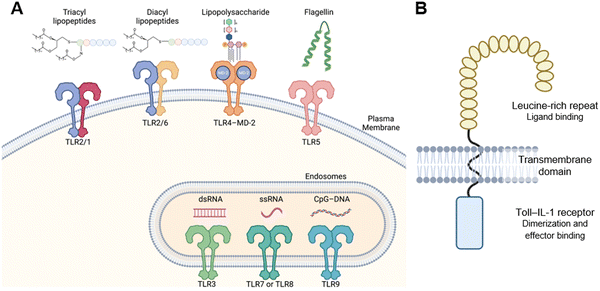 | ||
| Fig. 1 Toll-like receptors. (A) Sub-cellular localization and MAMPs, PAMPs and/or DMAPs of TLRs. (B) Domain structure of TLRs. | ||
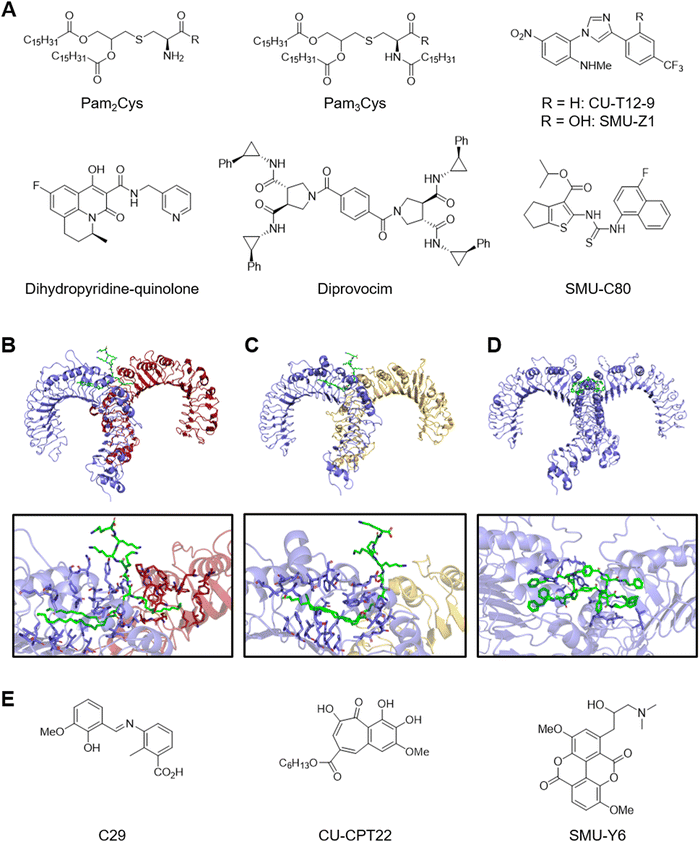
Co-crystal structures of Pam2CSK4 or Pam3CSK4 bound to the TLR2–TLR6 or TLR2–TLR1 heterodimers show exactly how lipopeptides induce dimerization. The TLR2–TLR1 heterodimer assumes an “m” shape with the two C-termini converging at the middle and binds a single Pam3CSK4 at the dimerization interface which serves as a molecular glue (Fig. 2B).22 The two ester-bound lipid chains of Pam3CSK4 insert into an internal pocket of TLR2 from its opening on the convex surface and the amide-bound lipid chain interacts with a channel formed on the convex surface of TLR1. In addition, the glycerol and Cys-Ser backbone of the ligand fits tightly in the dimerization interface. Amino acids from TLR2 and TLR1 also form hydrophobic, hydrogen-bonding, and ionic interactions to further stabilize the heterodimer. Similarly, the TLR2-TLR6 heterodimer binds a single Pam2CSK4 at the dimerization interface (Fig. 2C).23 The lack of the amide-bound lipid chain appears to be compensated by stronger protein–protein interactions between TLR2 and TLR6. These structure analyses advance further understanding and development of the structure–activity relationship of lipopeptide analogues.24
Recent studies have identified small molecule TLR2 agonists that are structurally unrelated to the lipopeptides (Fig. 2A).25 For example, high-throughput screening with reporter gene assays yielded several compounds containing the 1,4-diphenyl-1H-imidazole core such as CU-T12-926,27 and SMU-Z1.28 CU-T12-9 and SMU-Z1 specifically activated TLR2-TLR1 heterodimer and presumably bind the same site as Pam3Cys as suggested by in vitro competitive binding assays. Similar effort also led to the identification of tricyclic dihydropyridine-quinolone compounds.29 Interestingly, the agonist activity was highly dependent on the chirality of the methyl substituent. Diprovocim is a class of potent synthetic TLR2–TLR1 agonists that emerged from the screening of a unique chemical library designed for promoting cell surface receptor dimerization.30 Interestingly, co-crystal structure analysis of diprovocim-1–bound TLR2 homodimer revealed that two molecules of diprovocim-1 bind the ligand-binding pocket formed by the two TLR2 ectodomains, which offers the first insight into how non-lipopeptide ligands interact with TLR2 (Fig. 2D).31 In addition, diprovocim-1 synergized with anti–PD-L1 treatment to inhibit melanoma growth in mice.32 Alternatively, structure-based virtual screening of the ZINC database against the Pam3CSK-bound TLR2–TLR1 heterodimer led to the identification of SMU127 and the potency-optimized compound SMU-C80.33,34 It is noteworthy that SMU-C80 contains the N-aryl-N′-(thiophen-2-yl)thiourea core that had been previously identified through a high-throughput screening campaign.26 These studies demonstrate that TLR2 is a tractable target for small and medium-sized molecules.
Recent studies have also identified several TLR2 antagonists (Fig. 2E). High-throughput screening led to the identification of the first TLR2-selective antagonist CU-CPT22.35 CU-CPT22 competitively binds TLR2–TLR1 dimer in vitro and inhibits Pam3CSK-induced NO and inflammatory cytokine production in RAW 264.7 macrophages. Similar effort led to the identification of taspine and the derivative SMU-Y6.36 Virtual screening approaches yielded several TLR2 antagonists.37–39 For example, docking screening against a pocket located in the TLR2 TIR domain led to the identification of C29.39 Interestingly, ortho-vanillin which is generated from C29 imine hydrolysis turned out to be the active species. Another study reported structure–activity relationship of related N-benzylideneaniline derivatives.40 However, readers should note that most TLR2 antagonists in literature contain alert structures for assay interference and stability.41–43
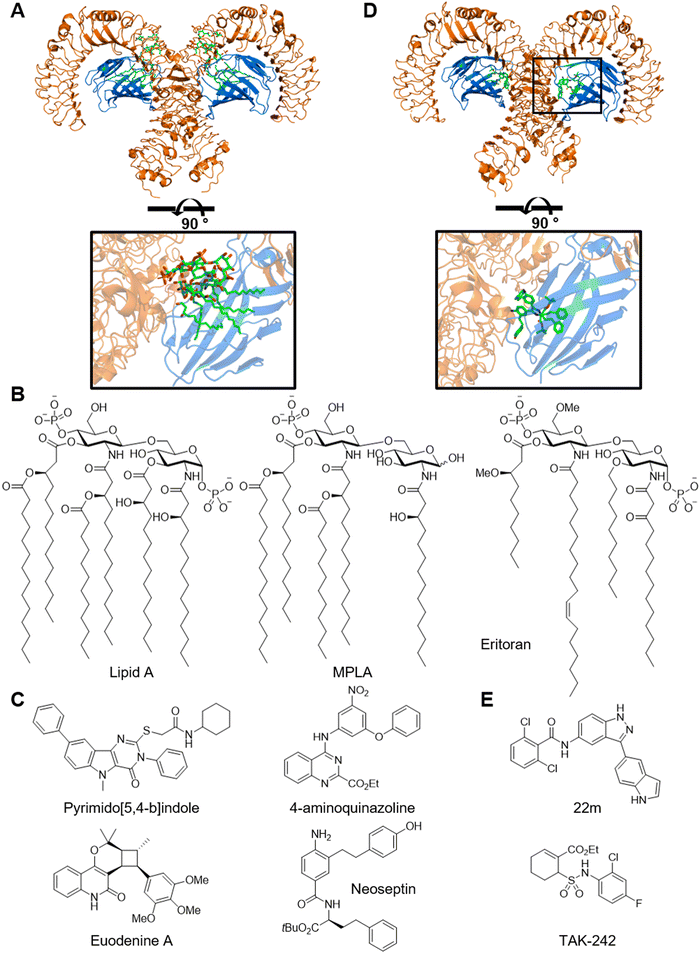
Structure–activity relationship of lipid A analogues has been extensively studied, delineating the impacts of the number and length of lipid chains as well as the identity and substitution pattern of disaccharide scaffold (Fig. 3B).54 For example, monophosphoryl lipid A (MPLA) lacks the 1-phosphono and 3-O-(R)-3-hydroxytetradecanoyl groups. MPLA is a milder TLR4 agonist and is less endotoxic compared to LPS, perhaps due to TRIF-biased downstream signaling.55 MPLA is clinically approved by FDA for the use as a vaccine adjuvant in humans. It is noteworthy that lipid A analogues inspired the development of non-glycolipid amphiphilic TLR4 agonists such as E6020 as a vaccine adjuvant.56 In contrast, E553157 and Eritoran,51 which is also known as E5564 and B1287, are penta- and tetra-lipidated analogues, respectively, and potent TLR4 antagonists. Both compounds entered clinical trials to treat sepsis but were discontinued. In addition, a recent study identified a complex glucorhamnan polysaccharide from a culture medium of Ruminococcus gnavus that activates TLR4 and induces inflammatory cytokines.58 This molecule may contribute to the association between R. gnavus and Crohn's disease.
Recent studies have identified small molecule TLR4 agonists that are structurally unrelated to the lipopeptides (Fig. 3C). For example, high-throughput screening with a reporter gene assay led to the identification of pyrimido[5,4-b]indoles and 4-aminoquinazolines as TLR4 agonists.59,60 Optimization of the pyrimido[5,4-b]indole series achieved ∼20-fold improvement in activity.61 The pyrimido[5,4-b]indoles and 4-aminoquinazolines require MD-2 for activity and are presumed to bind the hydrophobic pocket inside MD-2 based on docking studies. Similarly, high-throughput screening of a nontraditional compound libraries including an α-helix mimetic library led to the discovery of neoseptins.62 Neoseptins exhibit flat structure–activity relationship and low potency in vitro but are effective adjuvants in vivo. Co-crystal structure analysis revealed that the overall conformation of the TLR4–MD-2 complex and the local conformation of the MD-2 ligand-binding pocket were similar between neoseptin-3 and lipid A (Fig. 3D).63 Interestingly, two molecules of neoseptin-3 bound to each 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 TLR4–MD-2 heterodimer with each molecule adopting different conformations and interacting with different areas of TLR4 and MD-2. In addition, the two neoseptins bind to each other via π–π interaction and two hydrogen bonds. Screening a small library of 750 pure natural products led to the identification of Euodenine A.64 Structure–activity relationship development around the cyclobutane ring resulted in a 10-fold increase in potency. Alternatively, a cascade of ligand-based and structure-based virtual screenings yielded 2,3-(9,10-dihydroanthracene-9,10-diyl)succinimides.65 These studies highlight a unique and unexpected binding mechanism of a synthetic ligand with little structural similarity with the natural ligand and should inspire new approaches in library design and computer-assisted drug design.
1 TLR4–MD-2 heterodimer with each molecule adopting different conformations and interacting with different areas of TLR4 and MD-2. In addition, the two neoseptins bind to each other via π–π interaction and two hydrogen bonds. Screening a small library of 750 pure natural products led to the identification of Euodenine A.64 Structure–activity relationship development around the cyclobutane ring resulted in a 10-fold increase in potency. Alternatively, a cascade of ligand-based and structure-based virtual screenings yielded 2,3-(9,10-dihydroanthracene-9,10-diyl)succinimides.65 These studies highlight a unique and unexpected binding mechanism of a synthetic ligand with little structural similarity with the natural ligand and should inspire new approaches in library design and computer-assisted drug design.
Small molecule TLR4 antagonist have also been pursued.66 For example, the 3-(indol-5-yl)-indazole weakly 22m binds TLR4 and MD-2 in vitro and inhibits LPS-induced cytokine release (Fig. 3E). The authors demonstrated selectivity against kinases with the KinomeScan assay.67 Another interesting example is TAK-242.68 TAK-242 potently inhibits the production of inflammatory cytokines and nitric oxide induced by LPS in vitro and in vivo and protects mice against LPS-induced lethality.68,69 Mechanistically, TAK-242 is a non-competitive, covalent inhibitor and binds the intracellular domain of TLR4 via Cys747.70 TAK-242 does not affect the dimerization of TLR4 but instead disrupts interactions between TLR4 and the adapter proteins TIRAP and TRAM.71 This case study highlights the potential of modulating pattern recognition receptor functions through covalent mechanisms.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
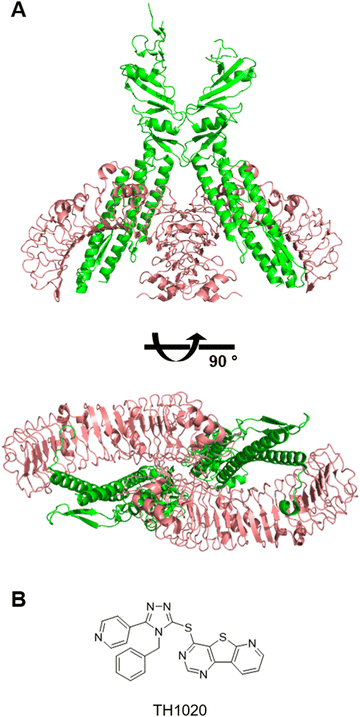
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex with flagellin and recognizes the ligand mostly on the lateral surface (Fig. 4A).74 Entolimod also known as CBLB502 is a pharmacologically-optimized flagellin derivative.75 Numerous studies demonstrated its anti-tumour efficacy in animal models.76 Development of small molecule TLR5 modulators remains challenging. The antagonist TH1020 is the only compound that has been reported in literature (Fig. 4B).77 Its mechanism of action remains unclear.
2 complex with flagellin and recognizes the ligand mostly on the lateral surface (Fig. 4A).74 Entolimod also known as CBLB502 is a pharmacologically-optimized flagellin derivative.75 Numerous studies demonstrated its anti-tumour efficacy in animal models.76 Development of small molecule TLR5 modulators remains challenging. The antagonist TH1020 is the only compound that has been reported in literature (Fig. 4B).77 Its mechanism of action remains unclear.
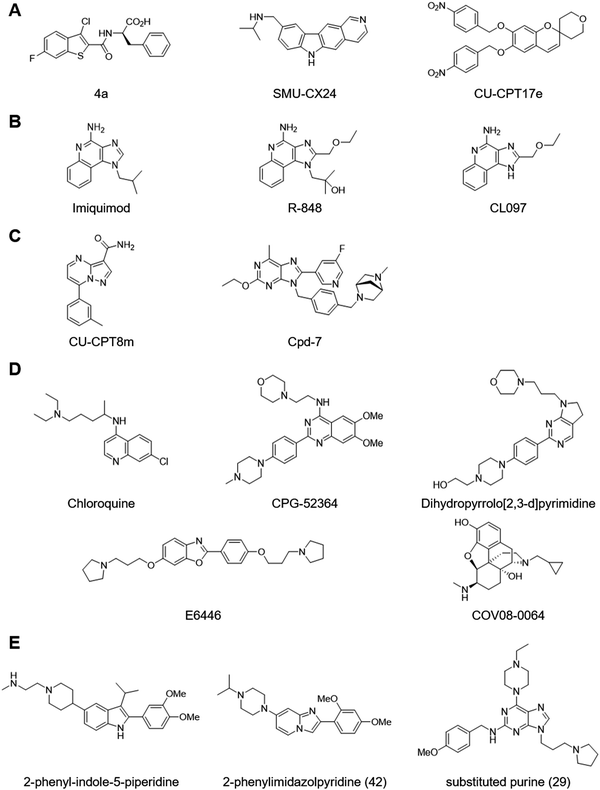
TLR3 remains a challenging target for small molecule modulator development (Fig. 5A). Structure-based virtual screening led to the identification of compound 4a as TLR3-selective antagonist.81 Compound 4a competed with poly I:C for TLR3 binding and inhibited poly I:C–induced NO and inflammatory cytokine production. High-throughput screening with a cell-based assay led to the identification of the potent TLR3-selective antagonist SMU-CX24.82 Notably, SMU-CX24 directly bound TLR3 with high affinity (Kd ∼ 20 nM) and exhibited in vivo efficacy in an atherosclerosis model. Similarly, high-throughput screening with a reporter gene assay yielded a chromene chemical series such as CU-CPT17e that represented the first and only small molecule TLR3 agonists.83 CU-CPT17e also activated TLR7 and TLR8. However, a comprehensive structure–activity relationship study of the series by another group failed to identify any analogues that displayed significant activity and to reproduce the activity for CU-CPT17e that was initially reported.84
Recent structural analyses have revealed mechanisms of TLR7 and TLR8 activation. TLR8 exists as a pre-formed dimer and binds synthetic agonists such as CL097 at a dimerization interface.93 This site also binds uridine derived from ssRNAs, whereas another site located at the concave surface binds short oligonucleotides.94 Agonist ligands induce a conformational change that brings the two C-terminal TIR domains closer and enables downstream signaling. On the other hand, TLR7 exists as a monomer in the absence of ligands and forms a dimer upon binding synthetic agonists such as R-848 or upon binding guanosine and polyuridine.95 Synthetic agonists and guanosine occupy a site at the dimerization surface in TLR7 that corresponds to TLR8's binding site for synthetic agonists and guanosine. Polyuridine binds another site in TLR7 consisting of the concave surface and a dimerization interface, which is distinct from TLR8's binding site for short oligonucleotides. These studies will facilitate not only further understanding of functional roles of TLR7 and TLR8 but also facilitate the development of synthetic ligands.
Development of selective antagonists for TLR7 and TLR8 lagged compared to agonist development (Fig. 5C). A recent study identified the potent and selective TLR8 antagonist CU-CPT8m through a high-throughput screening with a reporter gene assay and structural optimisation.96 Co-crystal structure analysis revealed that CU-CPT8m binds a hydrophobic site located at a dimerization interface and stablises the resting-state conformation. This binding site is nearby, but distinct from, the synthetic agonist–binding site and is only formed by the pre-formed dimer in the resting state. Another recent study identified the potent and selective TLR7 antagonist Cpd-7 through structure-based design.97 Co-crystal structure analysis of the agonist ligand 8-oxadenine derivative suggested that the substitution of the 8-oxo group to fill an additional space at the dimerization interface would convert the agonist into an antagonist. The resulting compounds exhibited antagonistic activity in reporter gene assay and blocked TLR7-dependent IFN-α secretion from PBMC. Surprisingly, crystallographic analysis revealed that the antagonist-bound complex adopted an activated dimeric structure with the antagonist occupying the same site as the original agonist. Interestingly, cryo-EM analysis revealed that the antagonist-bound dimer adopted two major forms: a closed form which was also observed in the crystal structure and an open form in which the two C-termini were separated from each other. These studies highlight implications of conformational dynamics in ligand design.
Recent crystal structure analysis revealed that CpG DNA induces the formation of a symmetric TLR9-CpG DNA complex with 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry.103 In the active complex, the CpG DNA wraps around the N-terminal region of one LRR domain from the lateral surface to the concave surface and extends to the C-terminal region of the other LRR domain, acting as a molecular glue to bridge the two TLR9 molecules. The CpG motif is accommodated in the groove on the lateral surface and recognized via interactions with multiple amino acids and via water-mediated hydrogen bonds. On the other hand, inhibitory DNA binds the concave surface that overlaps with the binding site for CpG DNA and prevents dimerization. Another study identified a second DNA-binding site in TLR9 that binds DNA containing cytosine at the second position from the 5′ end.104 This site corresponds to the nucleoside-binding site in TLR7 and TLR8 and cooperatively regulates receptor dimerization and activation.
2 stoichiometry.103 In the active complex, the CpG DNA wraps around the N-terminal region of one LRR domain from the lateral surface to the concave surface and extends to the C-terminal region of the other LRR domain, acting as a molecular glue to bridge the two TLR9 molecules. The CpG motif is accommodated in the groove on the lateral surface and recognized via interactions with multiple amino acids and via water-mediated hydrogen bonds. On the other hand, inhibitory DNA binds the concave surface that overlaps with the binding site for CpG DNA and prevents dimerization. Another study identified a second DNA-binding site in TLR9 that binds DNA containing cytosine at the second position from the 5′ end.104 This site corresponds to the nucleoside-binding site in TLR7 and TLR8 and cooperatively regulates receptor dimerization and activation.
Recent studies identified several small molecule TLR9-selective antagonists (Fig. 5D). For example, the antimalarial drug chloroquine has inspired the development of multiple TLR9 antagonists. Chloroquine inhibits TLR9 selectively over TLR7 and TLR8.89 Chloroquine directly blocks TLR9–CpG DNA interaction in vitro, perhaps by interacting with the nucleic acid.100,105 Regardless of exact mechanism of action, chloroquine's evolved into more potent and selective CPG-52364 and derivatives with the quinazoline core,106 and further optimization led to dihydropyrrolo[2,3-d]pyrimidines.107 Similarly, CpG DNA is a proposed target for benzoxazoles such as E6446.108,109 Alternatively, benzoxazole derivatives may bind the inhibitory DNA site of TLR9, although experimental evidence is lacking.110 Other TLR9-selective antagonists include COV08-0064, which emerged from a screening of morphinans using a reporter gene assay, and thiophene derivatives.111
Dual-inhibition of TLR7 and TLR9 has potential clinical benefit in autoimmune disorders such as lupus.112 High-throughput screening with TLR7 and TLR9 reporter gene assays yielded a 2-pheny-indole-5-piperidine chemical series (Fig. 5E).113 Optimisation efforts led to the lead compound 7f with potent TLR7 and TLR8 inhibitory activity and modest TLR9 inhibitory activity as well as desirable pharmacokinetic and pharmacodynamic profiles. Co-crystal structure analysis of 7f–TLR8 complex revealed that 7f stabilizes an inactive conformation which aligns well with the apo conformation. The lead compound demonstrated efficacy in rodent disease models for psoriasis and lupus. Interestingly, an analogous 2-phenylimidazopyridine chemical series was developed and optimized into the lead compound 42 with potent TLR7 and TLR9 inhibitory activity and modest TLR8 inhibitory activity.114 Alternatively, a purine-based TLR7 agonist was converted to the dual TLR7 and TLR9 antagonist 29.115 Compounds 29 and 42 did not interact with CpG DNA to a detectable level by isothermal titration calorimetry. It remains unclear whether any of small molecule TLR9 antagonists directly bind TLR9 in vitro or in cells.
NOD-like receptors
There are 22 NLR genes in humans.116 Although several human NLR genes have multiple murine paralogs, some human NLR genes lack murine counterparts (Fig. 6A). NLRs are intracellular receptors and respond to diverse PAMPs and DAMPs (Fig. 6B). Some NLRs activate NF-κB to initiate inflammatory responses just like TLRs, while other NLRs trigger a distinct pathway that induces cell death and the production of pro-inflammatory cytokines. NLR typically consists of three functional domains, namely N-terminal signaling, central [NAIP, CIITA, HETE, TP1 (NACHT)], and leucine-rich repeat (LRR) domains. NLRs are classified into subfamilies according to the type of N-terminal signaling domain: those with acidic activation domain (AAD) are called the NLR AAD containing family (NLRA); those with baculovirus inhibitor of apoptosis repeat (BIR) are called the NLR BIR containing family (NLRB); those with caspase recruitment domain (CARD) are called the NLR CARD containing (NLRC) family; those with pyrin domain (PYD) are called the NLR pyrin domain containing (NLRP) family. For example, an NLRC such as NOD1 and NOD2 recruits RIP2 via its CARD upon activation and triggers proinflammatory cytokine production through NF-κB and MAPK activation.2 In contrast, NLRP members interact with ASC via its PYD to recruit procaspase-1 and form oligomers called inflammasomes.117 Inflammasome formation leads to the autocleavage of procaspase-1 to release the active caspase-1, which subsequently proteolytically processes the inflammatory cytokines IL-1β and IL-18. Caspase-1 also induces the inflammatory cell death called pyroptosis by cleaving gasdermin D.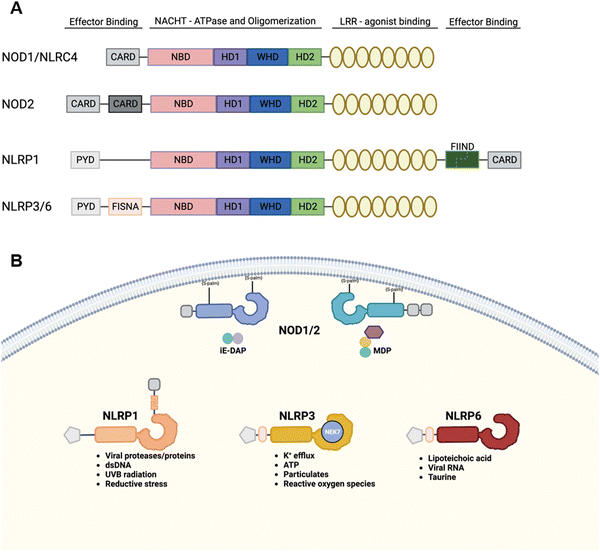 | ||
| Fig. 6 NOD-like receptors. (A) Domain architectures of representative members of the NLR family. (B) Sub-cellular localization and PAMPs/DAMPs of NLRs. | ||
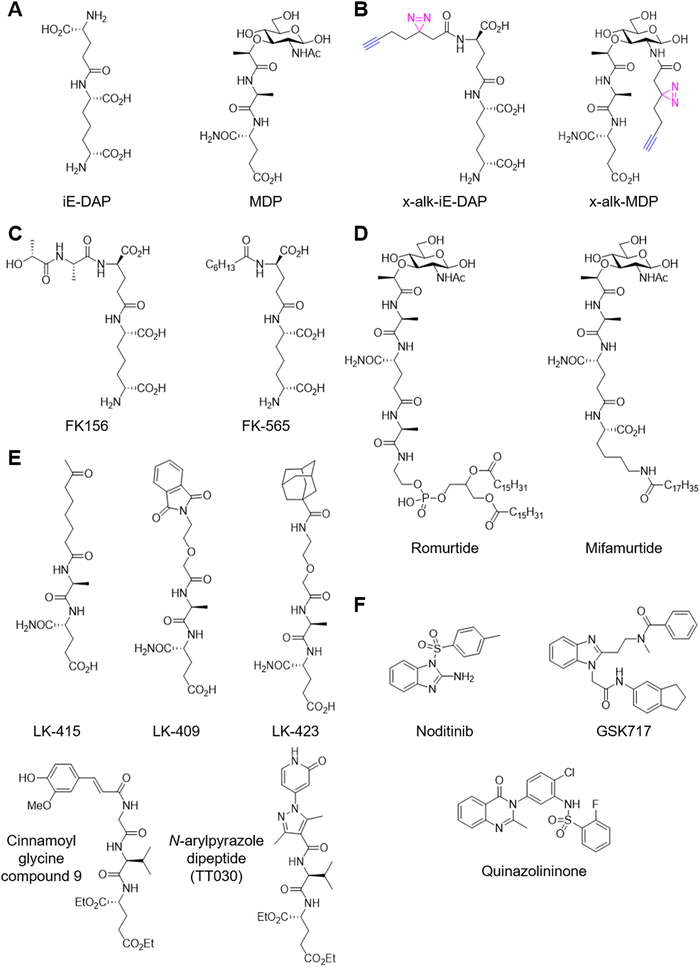
Biochemical evidence supports that NOD1 and NOD2 directly bind iE-DAP and MDP. For example, a biotinylated MDP (of unknown structure) was used to isolate recombinant NOD2 from cell lysate.125 This study suggested that MDP binds to the NOD2 NBD based on a domain truncation analysis, though the specificity of this interaction remains to be evaluated with a diastereomer control and/or competition with unmodified MDP. Another study demonstrated direct binding using surface plasmon resonance employing an MDP-functionalized chip.126 The active MDP-L,D isomer or an inactive MDP-L,L isomer was covalently immobilized to the chip via the 6-amino group and purified full-length NOD2 was flowed over the chip. Surprisingly, NOD2 was found to bind both isomers with similar affinities: KD = 51 nM for the active isomer and KD = 150 nM for the inactive isomer. Subsequent studies from the same group demonstrated that a LRR domain construct binds 6-amino-MDP (KD = 213 nM), 6-amino-GlcNAc (KD = 354 nM), and the dipeptide (KD = 428 nM) using the same setup.127 Binding affinities were weaker for constructs carrying mutations on the concave surface of LRR that had been reported to diminish NOD2 activation, suggesting these residues may be directly involved in MDP binding. In addition, modification via the 1- or 2-positions on MurNAc decreased to affinity to ∼1 μM.128,129 A similar SPR study has also been performed for iE-DAP analogs.130 However, the biotinylated analogues and surface immobilization affect cellular uptake and activity, which precluded target engagement analysis in cells. In this regard, we have developed a series of photo-activatable chemical reporters mimicking iE-DAP and MDP (Fig. 7B).131 We demonstrated diastereo-selective and ligand-competitive crosslinking of NOD1 and NOD2 with these chemical reporters in HEK293T cells and bone marrow-derived macrophages. In addition, chemical proteomics revealed the membrane associated GTPase ARF6 as an unpredicted target of MDP which is recruited to NOD2 upon activation. A recent study demonstrated N-acetylglucosamine kinase (NAGK) is required for the immunostimulatory activity of MDP.132 Mechanistically, NAGK phosphorylates MDP at the C6-hydroxyl group to yiled 6-O-phospho-MDP. While 6-O-phospho-MDP activates NOD2 in NAGK−/− cells, whether it engages NOD2 remains unclear.
Membrane association of NOD1 and NOD2 is critical for their function and is indirectly mediated by post-translational modification.133,134 Indeed, a recent study identified multiple S-palmitoylated cysteine residues and demonstrated that these post-translational modifications are necessary for their membrane localization and ligand-induced signaling.135 The palmitoyltransferase ZDHHC5 was found to be responsible for this critical post-translational modification. In addition, we recently identified the membrane-associated GTPase ARF6 as a component of the MDP–NOD2 complex with the photo-activatable chemical reporter.131 The mutation of an N-terminus myristoylation site of ARF6 from glycine to alanine diminishes MDP–NOD2 binding, presumably due to mis-localization off the plasma membrane. In a follow-up study, we identified the conserved aromatic triad of ARF is necessary for this interaction.136 Interestingly, the lipid-modified MDP analogue L18-MDP induces stronger ARF6–NOD2 association and NF-κB activation than MDP. These studies suggest that membrane-targeting is potentially an effective design strategy for NOD2 ligand development.
NOD1 and NOD2 are attractive targets for therapeutic development.137 For example, our recent studies on the Enterococcus species revealed peptidoglycan remodeling and NOD2 activation as key mechanisms for microbiota-mediated enhancement of immune checkpoint inhibitor therapy.10 Agonist ligand development for these receptors has remained focused on analog design and derivatisation of the peptidoglycan metabolites over the past several decades.137 For example, FK-156 was isolated and synthesized as an immunostimulatory component of Gram-positive Streptomyces olivaceogriseus and S. violaceus strains in 1981 (Fig. 7C).138–142 FK-156 was found to induce proliferation of murine splenocytes, protect against lethal challenge with Escherichia coli, and improve carbon clearance from the blood, an early assay for phagocytic activity in vivo. Through a reductionist approach, a subsequent study identified iE-DAP is the minimal prerequisite structure of FK-156 that elicits activity in 1982, revealing the minimal NOD1 ligand 20 years before the discovery of the receptor.143 Further synthetic studies yielded several lipophilic analogues of iE-DAP including FK-565 which improved overall activity.144,145 Subsequent studies delineated structural elements required for NOD1 activation including the terminal carboxyl and amine groups of DAP and the terminal carboxyl group of glutamic acid.146,147 More recently, rigidification of DAP via the introduction of a double bond retained activity relative to lauroyl iE-DAP while rigidification via cyclisation involving the terminal amine abrogated activity.148,149 Similarly, MDP was found to be the minimal adjuvant molecule from Freund's adjuvant in 1974.150 Over the next decade, its structure–activity relationship was defined and various lipophilic derivatives were synthesized to improve the pharmacological profile.151 These efforts yielded clinical molecules such as romurtide and mifamurtide (Fig. 7D).152 Alternatively, the N-acetylmuramic acid moiety was replaced with various lipophilic surrogates containing adamantyl,153 oxo fatty acid,154 phthalimido,155 or carbocyclic156 groups to yield desmuramyldipeptides (Fig. 7E). More recently, cinnamoylglycine emerged as a promising scaffold to replace MurNAc.157–160 The authors identified a hit compound from a small panel of saccharine- and indole-based desmuramyl dipeptides and empirically morphed it into the optimized scaffold to achieve high nanomolar NOD2 activity in vitro. Further optimization of the series led to lipophilic analogues with the adjuvant activity promoting antibody production in vivo161 and single nanomolar NOD2 activity in vitro.162 In addition, we recently disclosed N-arylpyrazole NOD2 agonists that promote immune checkpoint therapy.163 We identified the N-arylpyrazole dipeptides from a structure-based virtual screening and empirically optimized the hit compounds. Importantly, our N-arylpyrazole NOD2 agonist is enantiomer-specific, effective at promoting immune checkpoint inhibitor therapy and requires NOD2 for activity in vivo.
In contrast, the development of antagonists for NOD1 and NOD2 relies on high-throughput screening (Fig. 7F).164 For example, noditinib-1 emerged from a HTS campaign and is selective for NOD1 over NOD2.165 Similarly, selective inhibitors based on the benzimidazole diamide and several other scaffolds are published.166,167 Protein target of these inhibitors remain unclear.
Decoding danger signals that activate human and/or mouse NLRP1 remains an active research area. An early study reconstituted the human NLRP1 inflammasome in vitro and demonstrated that MDP induced oligomerization and dependent caspase-1 activation.178 Another study also demonstrated that recombinantly expressed human NLRP1 LRR domain binds MDP-functionalized chip surface.129 However, no follow-up studies substantiated MDP-dependent human NLRP1 activation.179 Instead, MDP-driven inflammasome response from THP-1 cells, a monocytic cell line, could stem from NLRP3.180 Rather, recent studies have demonstrated that human NLRP1 senses viral double-stranded RNA (dsRNA),181 viral protein,182 viral protease,183,184 ultraviolet B irradiation,185 and reductive stress,186 whereas mouse NLRP1B senses bacterial and protozoan toxins.187–189 Many of these danger signals such as protease,183,184,188 ultraviolet B irradiation,190 and reductive stress186 lead to the destabilization and accelerated proteasomal degradation of the N-terminal fragment of NLRP1 through post-translational modification of NLRP1 (proteolysis, hyperphosphorylation) or alteration of stabilizing protein–protein interaction. On the other hand, dsRNA directly binds the NACHT-LRR domain and induces ATPase activity.181 dsRNA-induced human NLRP1 activation requires FIIND autoproteolysis and proteasome activity and thus appears to involve the functional degradation mechanism. In contrast, the tegument protein ORF45 from Kaposi sarcoma–associated herpesvirus induces proteasome-independent human NLRP1 activation.182 ORF45 directly binds the disordered domain and drags the N-terminal fragment to the nucleus, which enables the C-terminal fragment to form the inflammasome.
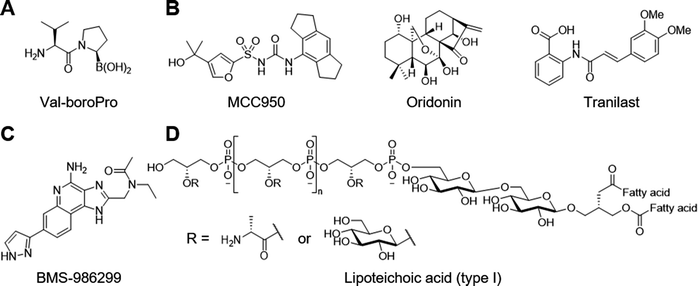
Synthetic inhibitors of DPP8 and DPP9 such as Val-boroPro also known as talabostat and PT-100 activate both human191 and mouse192,193 NLRP1 as well as CARD8175,193 (Fig. 8A). DPP8 and DPP9 are members of the S9B/DPPIV (DPP4) serine protease family, which had been implicated in immune responses.194 These chemical genetics studies employing Val-boroPro revealed their physiological function as a checkpoint that holds innate immune system at bay. Mechanistically, DPP9 bind the NLRP1 FIIND domain and cleave unknown substrates to prevent the inflammasome formation.191 Indeed, cryo-EM analysis shows that DPP9 forms a ternary complex with a full-length NLRP1 and a C-terminal fragment of NLRP1.195,196 The N-terminus of the NLRP1 C-terminal fragment inserts into the DPP9 active site, which is disrupted by Val-boroPro. Similarly, DPP9 forms an inhibitory ternary complex with CARD8; however, the N-terminus of the CARD8 C-terminal fragment does not closely interact with the DPP9 active site.197 Interestingly, Val-boroPro does not dissociate DPP9 from CARD8 in vitro, and it remains unclear whether DPP8 and DPP9 inhibitors need to destabilize the DPP9-CARD8 ternary complex.198 Differences in the regulatory mechanisms of NLRP1 and CARD8 such as this may potentially be exploited to develop selective agonists.199 Alternatively, high-throughput screening with a scintillation proximity assay led to the identification of several ATP-competitive inhibitors.200 These hit compounds were confirmed with fluorescence polarization assay but lacked on-target efficacy in cells.
The active structure of NLRP3 is predicted to be similar to the disc-shaped oligomers observed in active NLRC4.205,206 Activated NLRC4 forms a flat disc with each NLRC4 monomer interacting at the nucleotide binding domain (NBD). The cryo-EM structure of monomeric NLRP3 and NEK7 revealed the binding interfaces in which the C-terminal lobe of NEK7 forms interactions with both the leucine-rich repeat domain (LRR) and helical domain 2 (HD2) of NLRP3.207 Mutations that disrupt binding at either interface diminished inflammasome activation. Although this structure of NLRP3 was in an inactive conformation, the potential interface of active NLRP3 along the NBD surface would not affect binding of NEK7. Inactive oligomeric structures of NLPR3 were also solved by cryo-EM.208,209 Unlike active NLRC4 discs which make inter-NBD interactions, the inactive NLRP3 cages make inter-LRR interactions forming cage-like dodecameric structures that sterically block the N-terminal pyrin domain (PYD) from binding with ASC.
Most recently, the cryo-EM structure of active NLRP3 bound to NEK7 and ASC was solved.210 Active NLRP3 forms a disc structure composed of 10 or 11 NLPR3 monomers similar to the active structures of NLRC4.205,206 NEK7 sits in the concave surface of the LRR, which likely dissolves the inter-LRR interactions observed in the inactive cage structures. Therefore, binding to NEK7 directly breaks up NLRP3 cages freeing the NBD domains to form the flat disc structure of active inflammasomes, which position the PYD to bind ASC and activate caspase 1.
NLRP3 antagonists have broad therapeutic potential in a wide array of autoinflammatory and chronic inflammatory diseases from gout and nonalcoholic steatohepatitis (NASH) to neurovegetative diseases such as the Parkinson's and the Alzheimer's.8 The best characterized NLRP3 antagonist MCC950 emerged from a phenotypic screening of a diarysulfonylurea library against IL-1β secretion from human monocytes (Fig. 8B).211 Initial mechanism of action study proposed GST Omega 1–1 as a functionally relevant target based on affinity labelling and affinity chromatography.212 Further work led to the discovery that these compounds function through NLRP3 inhibition.213 One of these compounds was renamed from CRID3 or CP456,773 to MCC950. MCC950 engages NLRP3 and stabilizes an inactive conformation in cells as demonstrated by various techniques including drug affinity responsive target stabilization, photoaffinity labelling, and bioluminescence resonance energy transfer.214–216 Binding site for MCC950 is located in the NACHT domain. Indeed, crystal structure analysis shows that the binding pocket is formed by the four subdomains of the NACHT domain and that MCC950 acts as an intramolecular glue to lock the protein in an inactive conformation.217 The characterization of the antagonist binding interactions will facilitate the interpretation of structure–activity relationship and structure-based design.218 Alternatively, the natural product oridonin covalently binds to Cys279 in the NLRP3 NACHT domain to block the interaction between NLRP3 and NEK7.219 Mutation of Cys279 to alanine confers resistance to oridonin, which supports this mechanism of action. Similarly, tranilast, which is approved for the treatment of allergy and asthma in Japan, blocks NLRP3 inflammasome formation without affecting the ATPase activity, although exact mechanism of action remains unclear.220 Unlike oridonin and tranilast, most electrophilic NLRP3 antagonists target the ATPase activity.221
In contrast, small molecules that directly bind and activate NLRP3 are currently lacking. Some examples include the TLR7 and TLR8 ligand imiquimod and the related compound CL097, which activate the NLRP3 inflammasome in myeloid cells (Fig. 5B).222 Surprisingly, CL097 activates NLRP3 independently of potassium efflux unlike most NLRP3 agonists.223 Initially proposed mechanism of action is that CL097 inhibits mitochondrial complex I to trigger reactive oxygen species (ROS) production. A recent study confirmed the requirement of the complex I inhibition but found no evidence that mitochondrial ROS are necessary for CL097-induced NLRP3 activation.224 It is plausible that CL097 targets complex I and other unknown molecules to activate the NLRP3 inflammasome. Further mechanism of action study would advance the understanding of NLRP3 regulation. The related imidazoquinoline derivative BMS-986299 entered a phase-I clinical in patients with advanced solid tumour (Fig. 8C).225 This trial was terminated early due to the COVID-19 pandemic but demonstrated modest clinical activity in combination with immune checkpoint inhibitors and carried a manageable toxicity profile.
cGAS-STING pathway
The STING pathway responds to cytoplasmic double-stranded DNA that originates from pathogen and self-DNA damage (Fig. 9A).234,235 Cytoplasmic DNA activate the enzyme cyclic GMP–AMP synthase (cGAS) to produce the mixed phosphodiester-linked 2′3′-cyclic-GMP–AMP (cGAMP).236,237 Subsequently, cGAMP binds the ligand binding domain (LBD) of STING and triggers STING activation.238 Bacterial cyclic dinucleotides can also directly activate STING. Upon ligand engagement, STING undergoes conformational change and polymerization239,240 to recruit and activate the downstream effectors TBK1 and IRF3, leading to the production of type-I interferons. The components of this pathway are expressed in a wide variety of cell types and implicated in various physiological processes. STING activation has potential roles in vaccine adjuvants and, more recently, cancer immunotherapy.241,242 On the other hand, aberrant regulation of the STING pathway is associated with autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, and STING-associated vasculopathy with onset in infancy.5 As such, there has been strong interest in the development of both STING agonists and antagonists.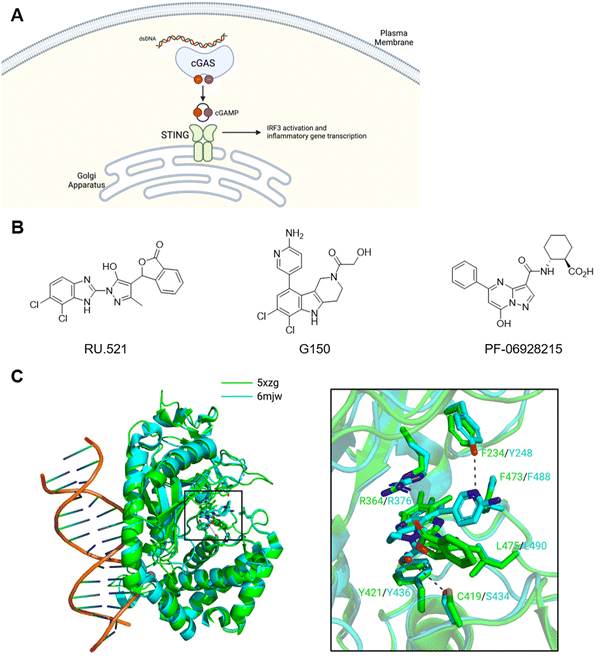 | ||
| Fig. 9 cGAS–STING pathway. (A) Sub-cellular localization and PAMPs/DAMPs of cGAS and STING. (B) Representative cGAS inhibitors. (C) Co-crystal structures of RU.521–cGAS and G150–cGAS complexes. | ||
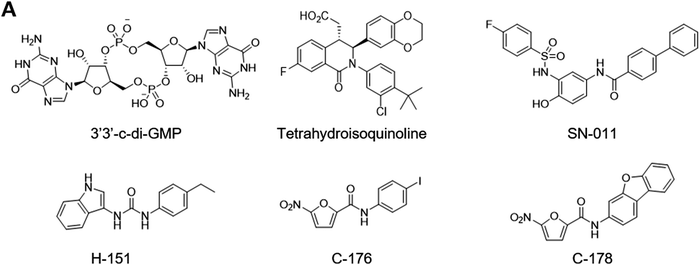
Studies have explored various sites of cGAS as potential target for inhibitory ligand development (Fig. 9B). For example, RU.521 emerged from a biochemical high-throughput screening.243 Crystal structure analysis revealed RU.521 binds the active site and enabled structure-guided optimization. While RU.521 is a potent inhibitor of mouse cGAS both biochemically and in mouse macrophages, it is a poor inhibitor of human cGAS perhaps due to low conservation of active site sequence between the two species. Thus, the same group adapted the biochemical assay for human cGAS and developed G150 (Fig. 9C).244 Interestingly, substitutions on the 4-position of indole differentially impacted the inhibition of human and mouse cGAS. Similarly, PF-06928215 targets the active site but lacks cellular activity.245 The interaction with DNA is an alternative target for inhibition. The DNA binding site contains several key lysine residues that mediate the interaction. Mutation of these residues to glutamine to mimic acetylation or acetyl lysine via amber suppression block cGAS activation and the DNA binding.246 This study also demonstrated that aspirin can acetylate these residues and inhibit cGAS. Several DNA-intercalating compounds such as hydroxychloroquine reportedly inhibit cGAS by blocking the DNA binding.247 The poly-pharmacology of aspirin, hydroxychloroquine, and others248,249 makes it challenging to rationally develop these compounds to achieve cGAS-selective inhibition.
Derivatisation of cGAMP yielded a number of nucleotide-based STING agonists to eliminate some of its liabilities such as poor membrane permeability and susceptibility to enzymatic hydrolysis.250 For example, a bisphosphothioate analogue, 2′3′-cGSASMP, is among the earliest cGMAP analogues (Fig. 10A).251 It is resistant to hydrolysis by the cGAMP hydrolase ENPP1 while retaining a similar affinity for human STING as cGAMP. The related compound cASASMP (also known as ADU-S100 and MIW815) entered dose-escalation phase I clinical trials as a monotherapy252 or a combination therapy with an inhibitory anti-PD-1 mAb253 in patients with advanced/metastatic solid tumour or lymphoma. These treatments were well tolerated but resulted in minimal anti-tumour response. To overcome the poor permeability and instability of nucleotide analogues, several small molecule ligands have been developed (Fig. 10B). For example, the flavonoid compounds such as flavone acetic acid (FAA) and 5,6-dimethylxanthenone-4-acetic acid (DMXAA) possess remarkable anti-tumor activity in mice models.250 DMXAA failed phase III clinical trials due to lack of efficacy. Subsequent studies revealed that these compounds bind mouse STING but not human STING.254 Interestingly, two molecules of DMXAA bind one molecule of STING in the closed conformation.255 On the other hand, amidobenzimidazole (ABZI)-based compounds are the first synthetic small molecule STING agonists that are human active.256 Crystal structure analysis inspired the design of linked compounds with 1000-fold improvement in potency. Similarly, MSA-2257 and SR-717 (Fig. 10C)258 bind STING as non-covalent dimers. These synthetic ligands demonstrated remarkable anti-tumor effects in mouse models and are suitable for systemic administration—a key advantage over cGAMP analogues.
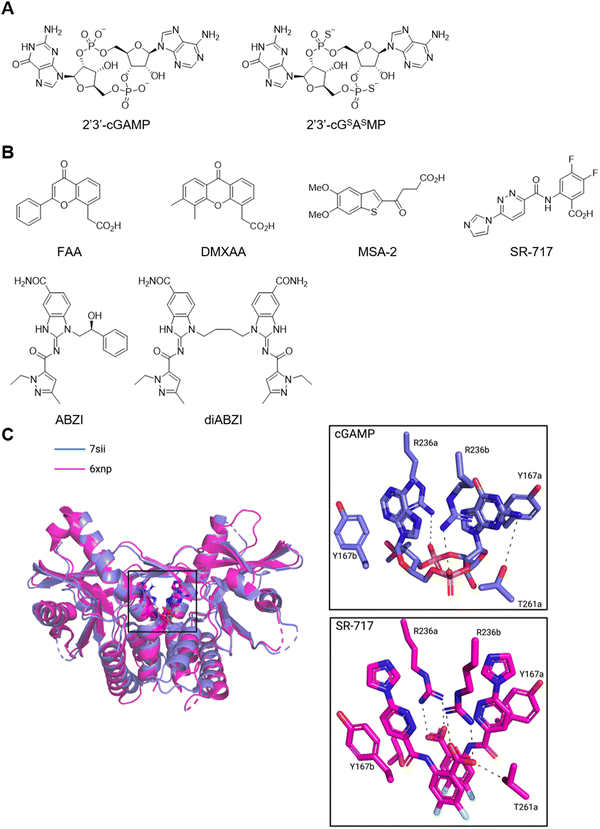 | ||
| Fig. 10 STING ligands. (A) Representative cyclic dinucleotide analogues. (B) Representative synthetic agonist ligands of STING. (C) Co-crystal structures of cGAMP–STING and SR-717–STING complexes. | ||
The development of STING antagonists remains challenging (Fig. 11). This is because antagonists targeting the ligand binding domain would have to stabilize the inactive open conformation and prevent it from binding cGAMP and adopting the active closed conformation. For example, cyclic-di-GMP binds STING in the open conformation and acts as a partial antagonist.239 The open conformer stacks poorly with the closed conformers and interferes with the polymerization and activation. Similarly, a tetrahydroisoquinoline compound binds the open conformation. This compound was designed and crystallographically characterized to capture the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry observed for DMXAA255 and required two-dimensional optimization of protein–ligand and ligand–ligand interactions.259 Additionally, structure-based virtual screening based on the open conformer yielded SN-011.260 The authors confirmed the engagement of LBD with a biotinylated analog. On the other hand, ABZI-based agonists also bind the open conformation but activate STING.256 These observations indicate that the relationship between conformation and activation is more nuanced and merits further research. Alternatively, the S-palmitoylation of transmembrane cysteine residues is necessary for STING activation. The nitrofuran compounds C-176 and C-178 as well as the indolylurea compound H-151 reportedly form covalent adducts with these cysteine residues and inhibit STING activation.261 However, nitrofuran is highly reactive in cellular environment and C-176 and C-178 lack specificity. In addition, the proposed adduct for C-178 appears to be unstable and unlikely. Furthermore, reaction mechanism for H-151 remains unclear. Similarly, nitro fatty acids inhibit STING but lack specificity.262 Nevertheless, these studies proposed an interesting idea of targeting key post-translational modifications with covalent ligands.
1 binding stoichiometry observed for DMXAA255 and required two-dimensional optimization of protein–ligand and ligand–ligand interactions.259 Additionally, structure-based virtual screening based on the open conformer yielded SN-011.260 The authors confirmed the engagement of LBD with a biotinylated analog. On the other hand, ABZI-based agonists also bind the open conformation but activate STING.256 These observations indicate that the relationship between conformation and activation is more nuanced and merits further research. Alternatively, the S-palmitoylation of transmembrane cysteine residues is necessary for STING activation. The nitrofuran compounds C-176 and C-178 as well as the indolylurea compound H-151 reportedly form covalent adducts with these cysteine residues and inhibit STING activation.261 However, nitrofuran is highly reactive in cellular environment and C-176 and C-178 lack specificity. In addition, the proposed adduct for C-178 appears to be unstable and unlikely. Furthermore, reaction mechanism for H-151 remains unclear. Similarly, nitro fatty acids inhibit STING but lack specificity.262 Nevertheless, these studies proposed an interesting idea of targeting key post-translational modifications with covalent ligands.
Conclusions
Early efforts to develop PRR-targeting therapeutics predated the discovery of their PRR targets. Isolation and characterization of immunostimulatory microbial metabolites led to the synthesis of numerous analogues and derivatives including several compounds that continue to be valuable therapeutics today. Genotype analysis, knockout experiments, and co-transfection assay later revealed that PRRs recognize these microbial metabolites. However, several PRRs still lack ligands and their functions remain unclear. Recent studies have begun to dissect mechanisms of host–microbiota interactions and reveal roles of microbiota-derived metabolites.263 Further microbiome analysis may lead to discovery of novel metabolite–receptor pairs involving PRRs. In this regard, chemical proteomics combined with metabolite-based chemical reporters represents a powerful approach.264 Beyond metabolites and their synthetic analogues, PRR ligand development has relied on functional screening of chemical libraries and empirical optimization of screening hits. Non-traditional chemical libraries including dimeric compound collections may lead to interesting discovery.30 In addition, recent studies revealed critical roles of various post-translational modifications in immune regulation265 and ligandability of cysteine residues that could be exploited for immune modulation.266 Covalent ligand may be advantageous to target less druggable sites on PRRs especially for inhibitor campaigns. Recent advance in structural characterization of PRRs has revealed molecular interactions between PRRs and their ligands and illuminated mechanisms of PRR regulation at the atomic level. These studies will continue to inspire structure-based drug design efforts. Finally, target engagement and off-target profile of PRR modulators often remains unestablished. Target engagement and off-target profiling study is not only an important aspect of hit-to-lead effort267 but may also lead to surprising discovery and illuminate new aspects of PRR regulation.131 Chemical proteomics and thermal proteome profiling will facilitate this effort. In summary, discovery and characterization of small molecule ligands for PRRs offers new opportunities for therapeutic development.Conflicts of interest
The authors declare the following competing financial interest(s): T. T. and H. C. H. have filed a patent application for the commercial use of N-arylpyrazole NOD2 agonists for immunotherapy.Acknowledgements
Fig. 1, 6 and 9A were created with BioRender.com. National Institutes of Health grant R01CA245292 to H. C. H.Notes and references
- Y. Kumagai and S. Akira, J. Allergy Clin. Immunol., 2010, 125, 985–992 CrossRef CAS PubMed.
- S. W. Brubaker, K. S. Bonham, I. Zanoni and J. C. Kagan, Annu. Rev. Immunol., 2015, 33, 257–290 CrossRef CAS PubMed.
- L. A. J. O’Neill, D. Golenbock and A. G. Bowie, Nat. Rev. Immunol., 2013, 13, 453–460 CrossRef PubMed.
- J. M. Platnich and D. A. Muruve, Arch. Biochem. Biophys., 2019, 670, 4–14 CrossRef CAS PubMed.
- A. Decout, J. D. Katz, S. Venkatraman and A. Ablasser, Nat. Rev. Immunol., 2021, 21, 548–569 CrossRef CAS PubMed.
- L. Beaugerie, P. Seksik, I. Nion-Larmurier, J. P. Gendre and J. Cosnes, Gastroenterology, 2006, 130, 650–656 CrossRef PubMed.
- P. Sfriso, F. Caso, S. Tognon, P. Galozzi, A. Gava and L. Punzi, Autoimmun. Rev., 2012, 12, 44–51 CrossRef CAS PubMed.
- M. S. J. Mangan, E. J. Olhava, W. R. Roush, H. M. Seidel, G. D. Glick and E. Latz, Nat. Rev. Drug Discovery, 2018, 17, 588–606 CrossRef CAS PubMed.
- J. Freund, J. Casals and E. P. Hosmer, Exp. Biol. Med., 1937, 37, 509–513 CrossRef CAS.
- M. E. Griffin, J. Espinosa, J. L. Becker, J. D. Luo, T. S. Carroll, J. K. Jha, G. R. Fanger and H. C. Hang, Science, 2021, 373, 1040–1046 CrossRef CAS PubMed.
- K. J. Rangan, V. A. Pedicord, Y. C. Wang, B. Kim, Y. Lu, S. Shaham, D. Mucida and H. C. Hang, Science, 2016, 353, 1434–1437 CrossRef CAS PubMed.
- B. Pulendran, P. S. Arunachalam and D. T. O’Hagan, Nat. Rev. Drug Discovery, 2021, 20, 454–475 CrossRef CAS PubMed.
- X. Cao, A. F. Cordova and L. Li, Chem. Rev., 2022, 122, 3414–3458 CrossRef CAS PubMed.
- D. H. O. ’ Donovan, Y. Mao and D. A. Mele, Curr. Med. Chem., 2019, 27, 5654–5674 CrossRef PubMed.
- O. Takeuchi, S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R. L. Modlin and S. Akira, J. Immunol., 2002, 169, 10–14 CrossRef CAS PubMed.
- O. Takeuchi, T. Kawai, P. F. Mühlradt, M. Morr, J. D. Radolf, A. Zychlinsky, K. Takeda and S. Akira, Int. Immunol., 2001, 13, 933–940 CrossRef CAS PubMed.
- P. F. Mühlradt, M. Kieß, H. Meyer, R. Süßmuth and G. Jung, J. Exp. Med., 1997, 185, 1951–1958 CrossRef PubMed.
- W. G. Bessler, M. Cox, A. Lex, B. Suhr, K. H. Wiesmüller and G. Jung, J. Immunol., 1985, 135, 1900–1905 CrossRef CAS.
- V. Braun and K. Rehn, Eur. J. Biochem., 1969, 10, 426–438 CrossRef CAS PubMed.
- U. Zähringer, B. Lindner, S. Inamura, H. Heine and C. Alexander, Immunobiology, 2008, 213, 205–224 CrossRef PubMed.
- M. Bae, C. D. Cassilly, X. Liu, S. M. Park, B. K. Tusi, X. Chen, J. Kwon, P. Filipčík, A. S. Bolze, Z. Liu, H. Vlamakis, D. B. Graham, S. J. Buhrlage, R. J. Xavier and J. Clardy, Nature, 2022, 608, 168–173 CrossRef CAS PubMed.
- M. S. Jin, S. E. Kim, J. Y. Heo, M. E. Lee, H. M. Kim, S. G. Paik, H. Lee and J. O. Lee, Cell, 2007, 130, 1071–1082 CrossRef CAS PubMed.
- J. Y. Kang, X. Nan, M. S. Jin, S. J. Youn, Y. H. Ryu, S. Mah, S. H. Han, H. Lee, S. G. Paik and J. O. Lee, Immunity, 2009, 31, 873–884 CrossRef CAS PubMed.
- B. L. Lu, G. M. Williams and M. A. Brimble, Org. Biomol. Chem., 2020, 18, 5073–5094 RSC.
- A. Kaur, D. Kaushik, S. Piplani, S. K. Mehta, N. Petrovsky and D. B. Salunke, J. Med. Chem., 2021, 64, 233–278 CrossRef CAS PubMed.
- Y. Guan, K. Omueti-Ayoade, S. K. Mutha, P. J. Hergenrother and R. I. Tapping, J. Biol. Chem., 2010, 285, 23755–23762 CrossRef CAS PubMed.
- K. Cheng, M. Gao, J. I. Godfroy, P. N. Brown, N. Kastelowitz and H. Yin, Sci. Adv., 2015, 1, e1400139 CrossRef PubMed.
- X. Cen, G. Zhu, J. Yang, J. Yang, J. Guo, J. Jin, K. S. Nandakumar, W. Yang, H. Yin, S. Liu and K. Cheng, Adv. Sci., 2019, 6, 1802042 CrossRef PubMed.
- Z. Hu, J. Banothu, M. Beesu, C. J. Gustafson, M. J. H. Brush, K. L. Trautman, A. C. D. Salyer, B. Pathakumari and S. A. David, ACS Med. Chem. Lett., 2019, 10, 132–136 CrossRef CAS PubMed.
- M. Morin, Y. Wang, B. T. Jones, Y. Mifune, L. Su, H. Shi, E. M. Y. Moresco, H. Zhang, B. Beutler and D. L. Boger, J. Am. Chem. Soc., 2018, 140, 14440–14454 CrossRef CAS PubMed.
- L. Su, Y. Wang, J. Wang, Y. Mifune, M. D. Morin, B. T. Jones, E. M. Y. Moresco, D. L. Boger, B. Beutler and H. Zhang, J. Med. Chem., 2019, 62, 2938–2949 CrossRef CAS PubMed.
- Y. Wang, L. Su, M. D. Morin, B. T. Jones, Y. Mifune, H. Shi, K. Wen Wang, X. Zhan, A. Liu, J. Wang, X. Li, M. Tang, S. Ludwig, S. Hildebrand, K. Zhou, D. J. Siegwart, E. M. Y. Moresco, H. Zhang, D. L. Boger and B. Beutler, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E8698–E8706 CAS.
- Z. Chen, X. Cen, J. Yang, X. Tang, K. Cui and K. Cheng, Chem. Commun., 2018, 54, 11411–11414 RSC.
- Z. Chen, L. Zhang, J. Yang, L. Zheng, F. Hu, S. Duan, K. S. Nandakumar, S. Liu, H. Yin and K. Cheng, J. Med. Chem., 2021, 64, 7371–7389 CrossRef CAS PubMed.
- K. Cheng, X. Wang, S. Zhang and H. Yin, Angew. Chem., Int. Ed., 2012, 51, 12246–12249 CrossRef CAS PubMed.
- J. Yang, Y. Pan, X. Zeng, S. Liu, Z. Chen and K. Cheng, Acta Pharm. Sin. B, 2023, 13, 3782–3801 CrossRef CAS PubMed.
- M. S. Murgueitio, P. Henneke, H. Glossmann, S. Santos-Sierra and G. Wolber, ChemMedChem, 2014, 9, 813–822 CrossRef CAS PubMed.
- Z. Zhong, L. J. Liu, Z. Q. Dong, L. Lu, M. Wang, C. H. Leung, D. L. Ma and Y. Wang, Chem. Commun., 2015, 51, 11178–11181 RSC.
- P. Mistry, M. H. W. Laird, R. S. Schwarz, S. Greene, T. Dyson, G. A. Snyder, T. S. Xiao, J. Chauhan, S. Fletcher, V. Y. Toshchakov, A. D. MacKerell and S. N. Vogel, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5455–5460 CrossRef CAS PubMed.
- S. Cai, G. Zhu, X. Cen, J. Bi, J. Zhang, X. Tang, K. Chen and K. Cheng, Bioorganic Med. Chem., 2018, 26, 2041–2050 CrossRef CAS PubMed.
- M. Bermudez, M. Grabowski, M. S. Murgueitio, M. Tiemann, P. Varga, T. Rudolf, G. Wolber, G. Weindl and J. Rademann, ChemMedChem, 2020, 15, 1364–1371 CrossRef CAS PubMed.
- J. Baell and M. A. Walters, Nature, 2014, 513, 481–483 CrossRef CAS PubMed.
- C. Aldrich, C. Bertozzi, G. I. Georg, L. Kiessling, C. Lindsley, D. Liotta, K. M. Merz, A. Schepartz and S. Wang, ACS Cent. Sci., 2017, 3, 143–147 CrossRef CAS PubMed.
- R. Medzhitov, P. Preston-Hurlburt and C. A. Janeway, Nature, 1997, 388, 394–397 CrossRef CAS PubMed.
- S. T. Qureshi, L. Larivière, G. Leveque, S. Clermont, K. J. Moore, P. Gros and D. Malo, J. Exp. Med., 1999, 189, 615–625 CrossRef CAS PubMed.
- A. Poltorak, X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton and B. Beutler, Science, 1998, 282, 2085–2088 CrossRef CAS PubMed.
- K. Hoshino, O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda and S. Akira, J. Immunol., 1999, 162, 3749–3752 CrossRef CAS.
- R. Shimazu, S. Akashi, H. Ogata, Y. Nagai, K. Fukudome, K. Miyake and M. Kimoto, J. Exp. Med., 1999, 189, 1777–1782 CrossRef CAS PubMed.
- A. B. Schromm, E. Lien, P. Henneke, J. C. Chow, A. Yoshimura, H. Heine, E. Latz, B. G. Monks, D. A. Schwartz, K. Miyake and D. T. Golenbock, J. Exp. Med., 2001, 194, 79–88 CrossRef CAS PubMed.
- B. S. Park, D. H. Song, H. M. Kim, B. S. Choi, H. Lee and J. O. Lee, Nature, 2009, 458, 1191–1195 CrossRef CAS PubMed.
- R. R. Ingalls, B. G. Monks, R. Savedra, W. J. Christ, R. L. Delude, A. E. Medvedev, T. Espevik and D. T. Golenbock, J. Immunol., 1998, 161, 5413–5420 CrossRef CAS.
- H. M. Kim, B. S. Park, J. I. Kim, S. E. Kim, J. Lee, S. C. Oh, P. Enkhbayar, N. Matsushima, H. Lee, O. J. Yoo and J. O. Lee, Cell, 2007, 130, 906–917 CrossRef CAS PubMed.
- U. Ohto, K. Fukase, K. Miyake and T. Shimizu, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7421–7426 CrossRef CAS PubMed.
- J. Gao and Z. Guo, Med. Res. Rev., 2018, 38, 556–601 CrossRef CAS PubMed.
- C. R. Casella and T. C. Mitchell, Cell. Mol. Life Sci., 2008, 65, 3231–3240 CrossRef CAS PubMed.
- S. T. Ishizaka and L. D. Hawkins, Expert Rev. Vaccines, 2007, 6, 773–784 CrossRef CAS PubMed.
- W. J. Christ, O. Asano, A. L. C. Robidoux, M. Perez, Y. Wang, G. R. Dubuc, W. E. Gavin, L. D. Hawkins, P. D. McGuinness, M. A. Mullarkey, M. D. Lewis, Y. Kishi, T. Kawata, J. R. Bristol, J. R. Rose, D. P. Rossignol, S. Kobayashi, L. Hishinuma, A. Kimura, N. Asakawa, K. Katayama and I. Yamatsu, Science, 1995, 268, 80–83 CrossRef CAS PubMed.
- M. T. Henke, D. J. Kenny, C. D. Cassilly, H. Vlamakis, R. J. Xavier and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12672–12677 CrossRef CAS PubMed.
- A. Nour, T. Hayashi, M. Chan, S. Yao, R. I. Tawatao, B. Crain, I. F. Tsigelny, V. L. Kouznetsova, A. Ahmadiiveli, K. Messer, M. Pu, M. Corr, D. A. Carson and H. B. Cottam, Bioorganic Med. Chem. Lett., 2014, 24, 4931–4938 CrossRef CAS PubMed.
- M. Chan, T. Hayashi, R. D. Mathewson, A. Nour, Y. Hayashi, S. Yao, R. I. Tawatao, B. Crain, I. F. Tsigelny, V. L. Kouznetsova, K. Messer, M. Pu, M. Corr, D. A. Carson and H. B. Cottam, J. Med. Chem., 2013, 56, 4206–4223 CrossRef CAS PubMed.
- M. Chan, Y. Kakitsubata, T. Hayashi, A. Ahmadi, S. Yao, N. M. Shukla, S. Y. Oyama, A. Baba, B. Nguyen, M. Corr, Y. Suda, D. A. Carson, H. B. Cottam and M. Wakao, J. Med. Chem., 2017, 60, 9142–9161 CrossRef CAS PubMed.
- M. D. Morin, Y. Wang, B. T. Jones, L. Su, M. M. R. P. Surakattula, M. Berger, H. Huang, E. K. Beutler, H. Zhang, B. Beutler and D. L. Boger, J. Med. Chem., 2016, 59, 4812–4830 CrossRef CAS PubMed.
- Y. Wang, L. Su, M. D. Morin, B. T. Jones, L. R. Whitby, M. M. R. P. Surakattula, H. Huang, H. Shi, J. H. Choi, K. W. Wang, E. M. Y. Moresco, M. Berger, X. Zhan, H. Zhang, D. L. Boger and B. Beutler, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E884–E893 CAS.
- J. E. Neve, H. P. Wijesekera, S. Duffy, I. D. Jenkins, J. A. Ripper, S. J. Teague, M. Campitelli, A. Garavelas, G. Nikolakopoulos, P. V. Le, P. De, A. Leone, N. B. Pham, P. Shelton, N. Fraser, A. R. Carroll, V. M. Avery, C. McCrae, N. Williams and R. J. Quinn, J. Med. Chem., 2014, 57, 1252–1275 CrossRef CAS PubMed.
- J. Honegr, D. Malinak, R. Dolezal, O. Soukup, M. Benkova, L. Hroch, O. Benek, J. Janockova, K. Kuca and R. Prymula, Eur. J. Med. Chem., 2018, 146, 38–46 CrossRef CAS PubMed.
- Y. Zhang, X. Liang, X. Bao, W. Xiao and G. Chen, Eur. J. Med. Chem., 2022, 235, 114291 CrossRef CAS PubMed.
- Z. Liu, L. Chen, P. Yu, Y. Zhang, B. Fang, C. Wu, W. Luo, X. Chen, C. Li and G. Liang, J. Med. Chem., 2019, 62, 5453–5469 CrossRef CAS PubMed.
- M. Ii, N. Matsunaga, K. Hazeki, K. Nakamura, K. Takashima, T. Seya, O. Hazeki, T. Kitazaki and Y. Iizawa, Mol. Pharmacol., 2006, 69, 1288–1295 CrossRef CAS PubMed.
- T. Sha, M. Sunamoto, T. Kitazaki, J. Sato, M. Ii and Y. Iizawa, Eur. J. Pharmacol., 2007, 571, 231–239 CrossRef CAS PubMed.
- K. Takashima, N. Matsunaga, M. Yoshimatsu, K. Hazeki, T. Kaisho, M. Uekata, O. Hazeki, S. Akira, Y. Iizawa and M. Ii, Br. J. Pharmacol., 2009, 157, 1250–1262 CrossRef CAS PubMed.
- N. Matsunaga, N. Tsuchimori, T. Matsumoto and M. Ii, Mol. Pharmacol., 2011, 79, 34–41 CrossRef CAS PubMed.
- A. T. Gewirtz, T. A. Navas, S. Lyons, P. J. Godowski and J. L. Madara, J. Immunol., 2001, 167, 1882–1885 CrossRef CAS PubMed.
- F. Hayashi, K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill and A. Aderem, Nature, 2001, 410, 1099–1103 CrossRef CAS PubMed.
- S. Il Yoon, O. Kurnasov, V. Natarajan, M. Hong, A. V. Gudkov, A. L. Osterman and I. A. Wilson, Science, 2012, 335, 859–864 CrossRef PubMed.
- L. G. Burdelya, V. I. Krivokrysenko, T. C. Tallant, E. Strom, A. S. Gleiberman, D. Gupta, O. V. Kurnasov, F. L. Fort, A. L. Osterman, J. A. Didonato, E. Feinstein and A. V. Gudkov, Science, 2008, 320, 226–230 CrossRef CAS PubMed.
- C. M. Brackett, B. Kojouharov, J. Veith, K. F. Greene, L. G. Burdelya, S. O. Gollnick, S. I. Abrams and A. V. Gudkov, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E874–E883 CrossRef CAS PubMed.
- L. Yan, J. Liang, C. Yao, P. Wu, X. Zeng, K. Cheng and H. Yin, ChemMedChem, 2016, 11, 822–826 CrossRef CAS PubMed.
- L. Alexopoulou, A. Holt, R. Medzhitov and R. Flavell, Nature, 2001, 413, 732–738 CrossRef CAS PubMed.
- K. A. Fitzgerald, D. C. Rowe, B. J. Barnes, D. R. Caffrey, A. Visintin, E. Latz, B. Monks, P. M. Pitha and D. T. Golenbock, J. Exp. Med., 2003, 198, 1043–1055 CrossRef CAS PubMed.
- L. Liu, I. Botos, Y. Wang, J. N. Leonard, J. Shiloach, D. M. Segal and D. R. Davies, Science, 2008, 320, 379–381 CrossRef CAS PubMed.
- K. Cheng, X. Wang and H. Yin, J. Am. Chem. Soc., 2011, 133, 3764–3767 CrossRef CAS PubMed.
- X. Cen, B. Wang, Y. Liang, Y. Chen, Y. Xiao, S. Du, K. S. Nandakumar, H. Yin, S. Liu and K. Cheng, Acta Pharm. Sin. B, 2022, 12, 3667–3681 CrossRef CAS PubMed.
- L. Zhang, V. Dewan and H. Yin, J. Med. Chem., 2017, 60, 5029–5044 CrossRef CAS PubMed.
- A. Sarkar, A. C. G. Kankanamalage, Q. Zhang, H. Cheng, P. Sivaprakasam, J. Naglich, C. Xie, S. Gangwar and D. L. Boger, Med. Chem. Res., 2021, 30, 1377–1385 CrossRef CAS PubMed.
- S. S. Diebold, T. Kaisho, H. Hemmi, S. Akira and C. Reise Sousa, Science, 2004, 303, 1529–1531 CrossRef CAS PubMed.
- F. Heil, H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner and S. Bauer, Science, 2004, 303, 1526–1529 CrossRef CAS PubMed.
- H. Hemmi, T. Kaisho, O. Takeuchi, S. Sato, H. Sanjo, K. Hoshino, T. Horiuchi, H. Tomizawa, K. Takeda and S. Akira, Nat. Immunol., 2002, 3, 196–200 CrossRef CAS PubMed.
- M. Jurk, F. Heil, J. Vollmer, C. Schetter, A. M. Krieg, H. Wagner, G. Lipford and S. Bauer, Nat. Immunol., 2002, 3, 499 CrossRef CAS PubMed.
- J. Lee, T. H. Chuang, V. Redecke, L. She, P. M. Pitha, D. A. Carson, E. Raz and H. B. Cottam, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 6646–6651 CrossRef CAS PubMed.
- F. Heil, P. Ahmad-Nejad, H. Hemmi, H. Hochrein, F. Ampenberger, T. Gellert, H. Dietrich, G. Lipford, K. Takeda, S. Akira, H. Wagner and S. Bauer, Eur. J. Immunol., 2003, 33, 2987–2997 CrossRef CAS PubMed.
- A. Talukdar, D. Ganguly, S. Roy, N. Das and D. Sarkar, J. Med. Chem., 2021, 64, 8010–8041 CrossRef CAS PubMed.
- D. Kaushik, A. Kaur, N. Petrovsky and D. B. Salunke, RSC Med. Chem., 2021, 12, 1065–1120 RSC.
- H. Tanji, U. Ohto, T. Shibata, K. Miyake and T. Shimizu, Science, 2013, 339, 1426–1429 CrossRef CAS PubMed.
- H. Tanji, U. Ohto, T. Shibata, M. Taoka, Y. Yamauchi, T. Isobe, K. Miyake and T. Shimizu, Nat. Struct. Mol. Biol., 2015, 22, 109–116 CrossRef CAS PubMed.
- Z. Zhang, U. Ohto, T. Shibata, E. Krayukhina, M. Taoka, Y. Yamauchi, H. Tanji, T. Isobe, S. Uchiyama, K. Miyake and T. Shimizu, Immunity, 2016, 45, 737–748 CrossRef CAS PubMed.
- S. Zhang, Z. Hu, H. Tanji, S. Jiang, N. Das, J. Li, K. Sakaniwa, J. Jin, Y. Bian, U. Ohto, T. Shimizu and H. Yin, Nat. Chem. Biol., 2018, 14, 58–64 CrossRef CAS PubMed.
- S. Tojo, Z. Zhang, H. Matsui, M. Tahara, M. Ikeguchi, M. Kochi, M. Kamada, H. Shigematsu, A. Tsutsumi, N. Adachi, T. Shibata, M. Yamamoto, M. Kikkawa, T. Senda, Y. Isobe, U. Ohto and T. Shimizu, Nat. Commun., 2020, 11, 4–6 CrossRef PubMed.
- H. Hemmi, O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda and S. Akira, Nature, 2000, 408, 740–745 CrossRef CAS PubMed.
- J. Lund, A. Sato, S. Akira, R. Medzhitov and A. Iwasaki, J. Exp. Med., 2003, 198, 513–520 CrossRef CAS PubMed.
- M. Rutz, J. Metzger, T. Gellert, P. Luppa, G. B. Lipford, H. Wagner and S. Bauer, Eur. J. Immunol., 2004, 34, 2541–2550 CrossRef CAS PubMed.
- T. Kawai, S. Sato, K. J. Ishii, C. Coban, H. Hemmi, M. Yamamoto, K. Terai, M. Matsuda, J. I. Inoue, S. Uematsu, O. Takeuchi and S. Akira, Nat. Immunol., 2004, 5, 1061–1068 CrossRef CAS PubMed.
- J. Scheiermann and D. M. Klinman, Vaccine, 2014, 32, 6377–6389 CrossRef CAS PubMed.
- U. Ohto, T. Shibata, H. Tanji, H. Ishida, E. Krayukhina, S. Uchiyama, K. Miyake and T. Shimizu, Nature, 2015, 520, 702–705 CrossRef CAS PubMed.
- U. Ohto, H. Ishida, T. Shibata, R. Sato, K. Miyake and T. Shimizu, Immunity, 2018, 48, 649–658 CrossRef CAS PubMed.
- A. Kužnik, M. Benčina, U. Švajger, M. Jeras, B. Rozman and R. Jerala, J. Immunol., 2011, 186, 4794–4804 CrossRef PubMed.
- B. Paul, O. Rahaman, S. Roy, S. Pal, S. Satish, A. Mukherjee, A. R. Ghosh, D. Raychaudhuri, R. Bhattacharya, S. Goon, D. Ganguly and A. Talukdar, Eur. J. Med. Chem., 2018, 159, 187–205 CrossRef CAS PubMed.
- M. Watanabe, M. Kasai, H. Tomizawa, M. Aoki, K. Eiho, Y. Isobe and S. Asano, ACS Med. Chem. Lett., 2014, 5, 1235–1239 CrossRef CAS PubMed.
- B. S. Franklin, S. T. Ishizaka, M. Lamphier, F. Gusovsky, H. Hansen, J. Rose, W. Zheng, M. A. Ataíde, R. B. De Oliveira, D. T. Golenbock and R. T. Gazzinelli, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3689–3694 CrossRef CAS PubMed.
- M. Lamphier, W. Zheng, E. Latz, M. Spyvee, H. Hansen, J. Rose, M. Genest, H. Yang, C. Shaffer, Y. Zhao, Y. Shen, C. Liu, D. Liu, T. R. Mempel, C. Rowbottom, J. Chow, N. C. Twine, M. Yu, F. Gusovsky and S. T. Ishizaka, Mol. Pharmacol., 2014, 85, 429–440 CrossRef PubMed.
- S. Roy, A. Mukherjee, B. Paul, O. Rahaman, S. Roy, G. Maithri, B. Ramya, S. Pal, D. Ganguly and A. Talukdar, Eur. J. Med. Chem., 2017, 134, 334–347 CrossRef CAS PubMed.
- R. Hoque, A. Farooq, A. Malik, B. N. Trawick, D. W. Berberich, J. P. McClurg, K. P. Galen and W. Mehal, J. Immunol., 2013, 190, 4297–4304 CrossRef CAS PubMed.
- S. R. Christensen, J. Shupe, K. Nickerson, M. Kashgarian, R. A. A. Flavell and M. J. Shlomchik, Immunity, 2006, 25, 417–428 CrossRef CAS PubMed.
- C. P. Mussari, D. S. Dodd, R. K. Sreekantha, L. Pasunoori, H. Wan, S. L. Posy, D. Critton, S. Ruepp, M. Subramanian, A. Watson, P. Davies, G. L. Schieven, L. M. Salter-Cid, R. Srivastava, D. M. Tagore, S. Dudhgaonkar, M. A. Poss, P. H. Carter and A. J. Dyckman, ACS Med. Chem. Lett., 2020, 11, 1751–1758 CrossRef CAS PubMed.
- N. Das, P. Bandopadhyay, S. Roy, B. P. Sinha, U. G. Dastidar, O. Rahaman, S. Pal, D. Ganguly and A. Talukdar, J. Med. Chem., 2022, 65, 11607–11632 CrossRef CAS PubMed.
- B. Kundu, D. Raychaudhuri, A. Mukherjee, B. P. Sinha, D. Sarkar, P. Bandopadhyay, S. Pal, N. Das, D. Dey, K. Ramarao, K. Nagireddy, D. Ganguly and A. Talukdar, J. Med. Chem., 2021, 64, 9279–9301 CrossRef CAS PubMed.
- J. P. Y. Ting, R. C. Lovering, E. S. Alnemri, J. Bertin, J. M. Boss, B. K. Davis, R. A. Flavell, S. E. Girardin, A. Godzik, J. A. Harton, H. M. Hoffman, J. P. Hugot, N. Inohara, A. MacKenzie, L. J. Maltais, G. Nunez, Y. Ogura, L. A. Otten, D. Philpott, J. C. Reed, W. Reith, S. Schreiber, V. Steimle and P. A. Ward, Immunity, 2008, 28, 285–287 CrossRef CAS PubMed.
- P. Broz and V. M. Dixit, Nat. Rev. Immunol., 2016, 16, 407–420 CrossRef CAS PubMed.
- R. Caruso, N. Warner, N. Inohara and G. Núñez, Immunity, 2014, 41, 898–908 CrossRef CAS PubMed.
- S. K. Kuss-Duerkop and A. M. Keestra-Gounder, Infect. Immun., 2020, 88, 1–19 CrossRef PubMed.
- S. E. Girardin, I. G. Boneca, L. A. M. Carneiro, A. Antignac, M. Jéhanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zähringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti and D. J. Philpott, Science, 2003, 300, 1584–1587 CrossRef CAS PubMed.
- M. Chamaillard, M. Hashimoto, Y. Horie, J. Masumoto, S. Qiu, L. Saab, Y. Ogura, A. Kawasaki, K. Fukase, S. Kusumoto, M. A. Valvano, S. J. Foster, T. W. Mak, G. Nuñez and N. Inohara, Nat. Immunol., 2003, 4, 702–707 CrossRef CAS PubMed.
- N. Inohara, Y. Ogura, A. Fontalba, O. Gutierrez, F. Pons, J. Crespo, K. Fukase, S. Inamura, S. Kusumoto, M. Hashimoto, S. J. Foster, A. P. Moran, J. L. Fernandez-Luna and G. Nuñez, J. Biol. Chem., 2003, 278, 5509–5512 CrossRef CAS PubMed.
- S. E. Girardin, I. G. Boneca, J. Viala, M. Chamaillard, A. Labigne, G. Thomas, D. J. Philpott and P. J. Sansonetti, J. Biol. Chem., 2003, 278, 8869–8872 CrossRef CAS PubMed.
- Y. G. Kim, J. H. Park, T. Reimer, D. P. Baker, T. Kawai, H. Kumar, S. Akira, C. Wobus and G. Núñez, Cell Host Microbe, 2011, 9, 496–507 CrossRef CAS PubMed.
- J. Mo, J. P. Boyle, C. B. Howard, T. P. Monie, B. K. Davis and J. A. Duncan, J. Biol. Chem., 2012, 287, 23057–23067 CrossRef CAS PubMed.
- C. L. Grimes, L. D. Z. Ariyananda, J. E. Melnyk and E. K. O’Shea, J. Am. Chem. Soc., 2012, 134, 13535–13537 CrossRef CAS PubMed.
- M. L. Lauro, E. A. D’Ambrosio, B. J. Bahnson and C. L. Grimes, ACS Infect. Dis., 2017, 3, 264–270 CrossRef CAS PubMed.
- K. M. Lazor, J. Zhou, K. E. DeMeester, E. A. D’Ambrosio and C. L. Grimes, ChemBioChem, 2019, 20, 1369–1375 CrossRef CAS PubMed.
- E. A. D’Ambrosio, K. L. Bersch, M. L. Lauro, C. L. Grimes and C. L. Grimes, J. Am. Chem. Soc., 2020, 142, 10926–10930 CrossRef PubMed.
- H. Laroui, Y. Yan, Y. Narui, S. A. Ingersoll, S. Ayyadurai, M. A. Charania, F. Zhou, B. Wang, K. Salaita, S. V. Sitaraman and D. Merlin, J. Biol. Chem., 2011, 286, 31003–31013 CrossRef CAS PubMed.
- Y. C. Wang, N. P. Westcott, M. E. Griffin and H. C. Hang, ACS Chem. Biol., 2019, 14, 405–414 CrossRef CAS PubMed.
- C. A. Stafford, A.-M. Gassauer, C. C. de Oliveira Mann, M. C. Tanzer, E. Fessler, B. Wefers, D. Nagl, G. Kuut, K. Sulek, C. Vasilopoulou, S. J. Schwojer, A. Wiest, M. K. Pfautsch, W. Wurst, M. Yabal, T. Fröhlich, M. Mann, N. Gisch, L. T. Jae and V. Hornung, Nature, 2022, 609, 590–596 CrossRef CAS PubMed.
- N. Barnich, J. E. Aguirre, H. C. Reinecker, R. Xavier and D. K. Podolsky, J. Cell Biol., 2005, 170, 21–26 CrossRef CAS PubMed.
- A. K. Schaefer, J. E. Melnyk, M. M. Baksh, K. M. Lazor, M. G. Finn and C. L. Grimes, ACS Chem. Biol., 2017, 12, 2216–2224 CrossRef CAS PubMed.
- Y. Lu, Y. Zheng, É. Coyaud, C. Zhang, A. Selvabaskaran, Y. Yu, Z. Xu, X. Weng, J. S. Chen, Y. Meng, N. Warner, X. Cheng, Y. Liu, B. Yao, H. Hu, Z. Xia, A. M. Muise, A. Klip, J. H. Brumell, S. E. Girardin, S. Ying, G. D. Fairn, B. Raught, Q. Sun and D. Neculai, Science, 2019, 366, 460–467 CrossRef CAS PubMed.
- C. W. Hespen, X. Zhao and H. C. Hang, Chem. Commun., 2022, 58, 6598–6601 RSC.
- M. E. Griffin, C. W. Hespen, Y. C. Wang and H. C. Hang, Clin. Transl. Immunol., 2019, 8, 1–18 Search PubMed.
- K. Hemmi, H. Takeno, S. Okada, O. Nakaguchi, Y. Kitaura and M. Hashimoto, J. Am. Chem. Soc., 1981, 103, 7026–7028 CrossRef CAS.
- T. Gotoh, K. Nakahara, T. Nishiura, M. Hashimoto, T. Kino, Y. Kuroda, M. Okuhara, M. Kohsaka, H. Aoki and H. Imanaka, J. Antibiot., 1982, 35, 1286–1292 CrossRef CAS PubMed.
- T. Gotoh, K. Nakahara, M. Iwami, H. Aoki and H. Imanaka, J. Antibiot., 1982, 35, 1280–1285 CrossRef CAS PubMed.
- Y. Kawai, K. Nakahara, T. Gotoh, I. Uchida, H. Tanaka and H. Imanaka, J. Antibiot., 1982, 35, 1293–1299 CrossRef CAS PubMed.
- K. Hemmi, M. Aratani, H. Takeno, S. Okada, Y. Miyazaki, O. Nakaguchi, Y. Kitaura and M. Hashimoto, J. Antibiot., 1982, 35, 1300–1311 CrossRef CAS PubMed.
- Y. Kitaura, O. Nakaguchi, H. Takeno, S. Okada, S. Yonishi, K. Hemmi, J. Mori, H. Senoh, Y. Mine and M. Hashimoto, J. Med. Chem., 1982, 25, 335–337 CrossRef CAS PubMed.
- Y. Kitaura, H. Takeno, M. Aratani, S. Okada, S. Yonishi, K. Hemmi, O. Nakaguchi and M. Hashimoto, Experientia, 1982, 38, 1101–1103 CrossRef CAS PubMed.
- Y. Fujimoto, A. R. Pradipta, N. Inohara and K. Fukase, Nat. Prod. Rep., 2012, 29, 568–579 RSC.
- M. A. Wolfert, A. Roychowdhury and G. J. Boons, Infect. Immun., 2007, 75, 706–713 CrossRef CAS PubMed.
- G. Agnihotri, R. Ukani, S. S. Malladi, H. J. Warshakoon, R. Balakrishna, X. Wang and S. A. David, J. Med. Chem., 2011, 54, 1490–1510 CrossRef CAS PubMed.
- Ž. Jakopin, M. Gobec, J. Kodela, T. Hazdovac, I. Mlinarič-Raščan and M. Sollner Dolenc, Eur. J. Med. Chem., 2013, 69, 232–243 CrossRef PubMed.
- S. Guzelj and Ž. Jakopin, Molecules, 2020, 25, 5228 CrossRef CAS PubMed.
- F. Ellouz, A. Adam, R. Ciorbaru and E. Lederer, Biochem. Biophys. Res. Commun., 1974, 59, 1317–1325 CrossRef CAS PubMed.
- A. Adam and E. Lederer, Med. Res. Rev., 1984, 4, 111–152 CrossRef CAS PubMed.
- S. V. Guryanova and R. M. Khaitov, Front. Immunol., 2021, 12, 607178 CrossRef CAS PubMed.
- B. Vranešić, J. Tomašić, S. Smerdel, D. Kantoci and F. Benedetti, Helv. Chim. Acta, 1993, 76, 1752–1758 CrossRef.
- M. Sollner, V. Kotnik Pečar, A. Štalc, S. Simčič, L. Povšič, B. Herzog-Wraber, L. Klampfer, A. Ihan and P. Grosman, Agents Actions, 1993, 38, 273–280 CrossRef CAS PubMed.
- U. Urleb, A. Krbavčič, M. Sollner, D. Kikelj and S. Pečar, Arch. Pharm., 1995, 328, 113–117 CrossRef CAS PubMed.
- D. Kikelj, S. Pečar, V. Kotnik, A. Štalc, B. Wraber-Herzog, S. Simčič, A. Ihan, L. Klamfer, L. Povšič, R. Grahek, E. Suhadolc, M. Hočevar, H. Hönig and R. Rogi-Kohlenprath, J. Med. Chem., 1998, 41, 530–539 CrossRef CAS PubMed.
- Ž. Jakopin, E. Corsini, M. Gobec, I. Mlinarič-Raščan and M. S. Dolenc, Eur. J. Med. Chem., 2011, 46, 3762–3777 CrossRef PubMed.
- Ž. Jakopin, M. Gobec, I. Mlinarič-Raščan and M. Sollner Dolenc, J. Med. Chem., 2012, 55, 6478–6488 CrossRef PubMed.
- M. Gobec, I. Mlinarič-Raščan, M. S. Dolenc and Ž. Jakopin, Eur. J. Med. Chem., 2016, 116, 1–12 CrossRef CAS PubMed.
- M. Gobec, T. Tomašič, A. Štimac, R. Frkanec, J. Trontelj, M. Anderluh, I. Mlinarič-Raščan and Ž. Jakopin, J. Med. Chem., 2018, 61, 2707–2724 CrossRef CAS PubMed.
- S. Guzelj, S. Nabergoj, M. Gobec, S. Pajk, V. Klančič, B. Slütter, R. Frkanec, A. Štimac, P. Šket, J. Plavec, I. Mlinarič-Raščan and Ž. Jakopin, J. Med. Chem., 2021, 64, 7809–7838 CrossRef CAS PubMed.
- S. Guzelj, Š. Bizjak and Ž. Jakopin, ACS Med. Chem. Lett., 2022, 13, 1270–1277 CrossRef CAS PubMed.
- M. E. Griffin, T. Tsukidate and H. C. Hang, ACS Chem. Biol., 2023, 18, 1368–1377 CrossRef CAS PubMed.
- Ž. Jakopin, J. Med. Chem., 2014, 57, 6897–6918 CrossRef PubMed.
- P. M. Khan, R. G. Correa, D. B. Divlianska, S. Peddibhotla, E. H. Sessions, G. Magnuson, B. Brown, E. Suyama, H. Yuan, A. Mangravita-Novo, M. Vicchiarelli, Y. Su, S. Vasile, L. H. Smith, P. W. Diaz, J. C. Reed and G. P. Roth, ACS Med. Chem. Lett., 2011, 2, 780–785 CrossRef CAS PubMed.
- D. J. Rickard, C. A. Sehon, V. Kasparcova, L. A. Kallal, X. Zeng, M. N. Montoute, T. Chordia, D. D. Poore, H. Li, Z. Wu, P. M. Eidam, P. A. Haile, J. Yu, J. G. Emery, R. W. Marquis, P. J. Gough and J. Bertin, PLoS One, 2013, 8, e69619 CrossRef CAS PubMed.
- D. J. Rickard, C. A. Sehon, V. Kasparcova, L. A. Kallal, P. A. Haile, X. Zeng, M. N. Montoute, D. D. Poore, H. Li, Z. Wu, P. M. Eidam, J. G. Emery, R. W. Marquis, P. J. Gough and J. Bertin, PLoS One, 2014, 9, e96737 CrossRef PubMed.
- F. Martinon, K. Burns and J. Tschopp, Mol. Cell, 2002, 10, 417–426 CrossRef CAS PubMed.
- J. Lilue, A. G. Doran, I. T. Fiddes, M. Abrudan, J. Armstrong, R. Bennett, W. Chow, J. Collins, S. Collins, A. Czechanski, P. Danecek, M. Diekhans, D. D. Dolle, M. Dunn, R. Durbin, D. Earl, A. Ferguson-Smith, P. Flicek, J. Flint, A. Frankish, B. Fu, M. Gerstein, J. Gilbert, L. Goodstadt, J. Harrow, K. Howe, X. Ibarra-Soria, M. Kolmogorov, C. J. Lelliott, D. W. Logan, J. Loveland, C. E. Mathews, R. Mott, P. Muir, S. Nachtweide, F. C. P. Navarro, D. T. Odom, N. Park, S. Pelan, S. K. Pham, M. Quail, L. Reinholdt, L. Romoth, L. Shirley, C. Sisu, M. Sjoberg-Herrera, M. Stanke, C. Steward, M. Thomas, G. Threadgold, D. Thybert, J. Torrance, K. Wong, J. Wood, B. Yalcin, F. Yang, D. J. Adams, B. Paten and T. M. Keane, Nat. Genet., 2018, 50, 1574–1583 CrossRef CAS PubMed.
- A. D’Osualdo, C. X. Weichenberger, R. N. Wagner, A. Godzik, J. Wooley and J. C. Reed, PLoS One, 2011, 6, e27396 CrossRef PubMed.
- A. Sandstrom, P. S. Mitchell, L. Goers, E. W. Mu, C. F. Lesser and R. E. Vance, Science, 2019, 364, eaau1330 CrossRef CAS PubMed.
- A. J. Chui, M. C. Okondo, S. D. Rao, K. Gai, A. R. Griswold, D. C. Johnson, D. P. Ball, C. Y. Taabazuing, E. L. Orth, B. A. Vittimberga and D. A. Bachovchin, Science, 2019, 364, 82–85 CrossRef CAS PubMed.
- H. Xu, J. Shi, H. Gao, Y. Liu, Z. Yang, F. Shao and N. Dong, EMBO J., 2019, 38, 1–11 Search PubMed.
- F. L. Zhong, O. Mamaï, L. Sborgi, L. Boussofara, R. Hopkins, K. Robinson, I. Szeverényi, T. Takeichi, R. Balaji, A. Lau, H. Tye, K. Roy, C. Bonnard, P. J. Ahl, L. A. Jones, P. J. Baker, L. Lacina, A. Otsuka, P. R. Fournie, F. Malecaze, E. B. Lane, M. Akiyama, K. Kabashima, J. E. Connolly, S. L. Masters, V. J. Soler, S. S. Omar, J. A. McGrath, R. Nedelcu, M. Gribaa, M. Denguezli, A. Saad, S. Hiller and B. Reversade, Cell, 2016, 167, 187–202 CrossRef CAS PubMed.
- D. C. Johnson, C. Y. Taabazuing, M. C. Okondo, A. J. Chui, S. D. Rao, F. C. Brown, C. Reed, E. Peguero, E. de Stanchina, A. Kentsis and D. A. Bachovchin, Nat. Med., 2018, 24, 1151–1156 CrossRef CAS PubMed.
- A. Linder, S. Bauernfried, Y. Cheng, M. Albanese, C. Jung, O. T. Keppler and V. Hornung, EMBO J., 2020, 39, 1–16 CrossRef PubMed.
- D. C. Johnson, M. C. Okondo, E. L. Orth, S. D. Rao, H.-C. Huang, D. P. Ball and D. A. Bachovchin, Cell Death Dis., 2020, 11, 628 CrossRef CAS PubMed.
- B. Faustin, L. Lartigue, J. M. Bruey, F. Luciano, E. Sergienko, B. Bailly-Maitre, N. Volkmann, D. Hanein, I. Rouiller and J. C. Reed, Mol. Cell, 2007, 25, 713–724 CrossRef CAS PubMed.
- S. Bauernfried and V. Hornung, J. Exp. Med., 2021, 219, 1–8 Search PubMed.
- F. Martinon, L. Agostini, E. Meylan and J. Tschopp, Curr. Biol., 2004, 14, 1929–1934 CrossRef CAS PubMed.
- S. Bauernfried, M. J. Scherr, A. Pichlmair, K. E. Duderstadt and V. Hornung, Science, 2021, 371, eabd0811 CrossRef CAS PubMed.
- X. Yang, J. Zhou, C. Liu, Y. Qu, W. Wang, M. Z. X. Xiao, F. Zhu, Z. Liu and Q. Liang, Nat. Immunol., 2022, 23, 916–926 CrossRef CAS PubMed.
- B. V. Tsu, C. Beierschmitt, A. P. Ryan, R. Agarwal, P. S. Mitchell and M. D. Daugherty, eLife, 2021, 10, 1–76 CrossRef PubMed.
- R. Planès, M. Pinilla, K. Santoni, A. Hessel, C. Passemar, K. Lay, P. Paillette, A. L. C. Valadão, K. S. Robinson, P. Bastard, N. Lam, R. Fadrique, I. Rossi, D. Pericat, S. Bagayoko, S. A. Leon-Icaza, Y. Rombouts, E. Perouzel, M. Tiraby, Q. Zhang, P. Cicuta, E. Jouanguy, O. Neyrolles, C. E. Bryant, A. R. Floto, C. Goujon, F. Z. Lei, G. Martin-Blondel, S. Silva, J. L. Casanova, C. Cougoule, B. Reversade, J. Marcoux, E. Ravet and E. Meunier, Mol. Cell, 2022, 82, 2385–2400 CrossRef PubMed.
- G. Fenini, S. Grossi, E. Contassot, T. Biedermann, E. Reichmann, L. E. French and H. D. Beer, J. Invest. Dermatol., 2018, 138, 2644–2652 CrossRef CAS PubMed.
- D. P. Ball, L. P. Tsamouri, A. E. Wang, H.-C. Huang, C. D. Warren, Q. Wang, I. H. Edmondson, A. R. Griswold, S. D. Rao, D. C. Johnson and D. A. Bachovchin, Sci. Immunol., 2022, 7, 1–16 Search PubMed.
- S. E. Ewald, J. Chavarria-Smith and J. C. Boothroyd, Infect. Immun., 2014, 82, 460–468 CrossRef PubMed.
- J. Chavarría-Smith and R. E. Vance, PLoS Pathog., 2013, 9, e1003452 CrossRef PubMed.
- K. M. Cirelli, G. Gorfu, M. A. Hassan, M. Printz, D. Crown, S. H. Leppla, M. E. Grigg, J. P. J. Saeij and M. Moayeri, PLoS Pathog., 2014, 10, e1003927 CrossRef PubMed.
- K. S. Robinson, G. A. Toh, P. Rozario, R. Chua, S. Bauernfried, Z. Sun, M. J. Firdaus, S. Bayat, R. Nadkarni, Z. S. Poh, K. C. Tham, C. R. Harapas, C. K. Lim, W. Chu, C. W. S. Tay, K. Y. Tan, T. Zhao, C. Bonnard, R. Sobota, J. E. Connolly, J. Common, S. L. Masters, K. W. Chen, L. Ho, B. Wu, V. Hornung and F. L. Zhong, Science, 2022, 377, 328–335 CrossRef CAS PubMed.
- F. L. Zhong, K. Robinson, D. E. T. Teo, K. Y. Tan, C. Lim, C. R. Harapas, C. H. Yu, W. H. Xie, R. M. Sobota, V. B. Au, R. Hopkins, A. D’Osualdo, J. C. Reed, J. E. Connolly, S. L. Masters and B. Reversade, J. Biol. Chem., 2018, 293, 18864–18878 CrossRef CAS PubMed.
- M. C. Okondo, S. D. Rao, C. Y. Taabazuing, A. J. Chui, S. E. Poplawski, D. C. Johnson and D. A. Bachovchin, Cell Chem. Biol., 2018, 25, 262–267 CrossRef CAS PubMed.
- M. C. Okondo, D. C. Johnson, R. Sridharan, E. Bin Go, A. J. Chui, M. S. Wang, S. E. Poplawski, W. Wu, Y. Liu, J. H. Lai, D. G. Sanford, M. O. Arciprete, T. R. Golub, W. W. Bachovchin and D. A. Bachovchin, Nat. Chem. Biol., 2017, 13, 46–53 CrossRef CAS PubMed.
- H. Zhang, Y. Chen, F. M. Keane and M. D. Gorrell, Mol. Cancer Res., 2013, 11, 1487–1496 CrossRef CAS PubMed.
- M. Huang, X. Zhang, G. A. Toh, Q. Gong, J. Wang, Z. Han, B. Wu, F. Zhong and J. Chai, Nature, 2021, 592, 773–777 CrossRef CAS PubMed.
- L. R. Hollingsworth, H. Sharif, A. R. Griswold, P. Fontana, J. Mintseris, K. B. Dagbay, J. A. Paulo, S. P. Gygi, D. A. Bachovchin and H. Wu, Nature, 2021, 592, 778–783 CrossRef CAS PubMed.
- H. Sharif, L. R. Hollingsworth, A. R. Griswold, J. C. Hsiao, Q. Wang, D. A. Bachovchin and H. Wu, Immunity, 2021, 54, 1392–1404 CrossRef CAS PubMed.
- A. R. Griswold, D. P. Ball, A. Bhattacharjee, A. J. Chui, S. D. Rao, C. Y. Taabazuing and D. A. Bachovchin, ACS Chem. Biol., 2019, 14, 2424–2429 CrossRef CAS PubMed.
- S. D. Rao, Q. Chen, Q. Wang, E. L. Orth-He, M. Saoi, A. R. Griswold, A. Bhattacharjee, D. P. Ball, H. C. Huang, A. J. Chui, D. J. Covelli, S. You, J. R. Cross and D. A. Bachovchin, Nat. Chem. Biol., 2022, 18, 565–574 CrossRef CAS PubMed.
- P. A. Harris, C. Duraiswami, D. T. Fisher, J. Fornwald, S. J. Hoffman, G. Hofmann, M. Jiang, R. Lehr, P. M. McCormick, L. Nickels, B. Schwartz, Z. Wu, G. Zhang, R. W. Marquis, J. Bertin and P. J. Gough, Bioorganic Med. Chem. Lett., 2015, 25, 2739–2743 CrossRef CAS PubMed.
- K. V. Swanson, M. Deng and J. P. Y. Ting, Nat. Rev. Immunol., 2019, 19, 477–489 CrossRef CAS PubMed.
- J. L. Schmid-Burgk, D. Chauhan, T. Schmidt, T. S. Ebert, J. Reinhardt, E. Endl and V. Hornung, J. Biol. Chem., 2016, 291, 103–109 CrossRef CAS PubMed.
- Y. He, M. Y. Zeng, D. Yang, B. Motro and G. Núñez, Nature, 2016, 530, 354–357 CrossRef CAS PubMed.
- H. Shi, Y. Wang, X. Li, X. Zhan, M. Tang, M. Fina, L. Su, D. Pratt, C. Hui Bu, S. Hildebrand, S. Lyon, L. Scott, J. Quan, Q. Sun, J. Russell, S. Arnett, P. Jurek, D. Chen, V. V. Kravchenko, J. C. Mathison, E. M. Y. Moresco, N. L. Monson, R. J. Ulevitch and B. Beutler, Nat. Immunol., 2016, 17, 250–258 CrossRef CAS PubMed.
- L. Zhang, S. Chen, J. Ruan, J. Wu, A. B. Tong, Q. Yin, Y. Li, L. David, A. Lu, W. L. Wang, C. Marks, Q. Ouyang, X. Zhang, Y. Mao and H. Wu, Science, 2015, 350, 404–409 CrossRef CAS PubMed.
- J. L. Tenthorey, E. M. Kofoed, M. D. Daugherty, H. S. Malik and R. E. Vance, Mol. Cell, 2014, 54, 17–29 CrossRef CAS PubMed.
- H. Sharif, L. Wang, W. L. Wang, V. G. Magupalli, L. Andreeva, Q. Qiao, A. V. Hauenstein, Z. Wu, G. Núñez, Y. Mao and H. Wu, Nature, 2019, 570, 338–343 CrossRef CAS PubMed.
- L. Andreeva, L. David, S. Rawson, C. Shen, T. Pasricha, P. Pelegrin and H. Wu, Cell, 2021, 184, 6299–6312 CrossRef CAS PubMed.
- U. Ohto, Y. Kamitsukasa, H. Ishida, Z. Zhang, K. Murakami, C. Hirama, S. Maekawa and T. Shimizu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2121353119 CrossRef CAS PubMed.
- L. Xiao, V. G. Magupalli and H. Wu, Nature, 2023, 613, 595–600 CrossRef CAS PubMed.
- D. G. Perregaux, P. Mcniff, R. Laliberte, N. Hawryluk, H. Peurano, E. Stam, J. Eggler, R. Griffiths, M. A. Dombroski and C. A. Gabel, J. Pharmacol. Exp. Ther., 2001, 299, 187–197 CAS.
- R. E. Laliberte, D. G. Perregaux, L. R. Hoth, P. J. Rosner, C. K. Jordan, K. M. Peese, J. F. Eggler, M. A. Dombroski, K. F. Geoghegan and C. A. Gabel, J. Biol. Chem., 2003, 278, 16567–16578 CrossRef CAS PubMed.
- R. C. Coll, A. A. B. Robertson, J. J. Chae, S. C. Higgins, R. Muñoz-Planillo, M. C. Inserra, I. Vetter, L. S. Dungan, B. G. Monks, A. Stutz, D. E. Croker, M. S. Butler, M. Haneklaus, C. E. Sutton, G. Núñez, E. Latz, D. L. Kastner, K. H. G. Mills, S. L. Masters, K. Schroder, M. A. Cooper and L. A. J. O’Neill, Nat. Med., 2015, 21, 248–257 CrossRef CAS PubMed.
- A. Tapia-Abellán, D. Angosto-Bazarra, H. Martínez-Banaclocha, C. de Torre-Minguela, J. P. Cerón-Carrasco, H. Pérez-Sánchez, J. I. Arostegui and P. Pelegrin, Nat. Chem. Biol., 2019, 15, 560–564 CrossRef PubMed.
- R. C. Coll, J. R. Hill, C. J. Day, A. Zamoshnikova, D. Boucher, N. L. Massey, J. L. Chitty, J. A. Fraser, M. P. Jennings, A. A. B. Robertson and K. Schroder, Nat. Chem. Biol., 2019, 15, 556–559 CrossRef CAS PubMed.
- L. Vande Walle, I. B. Stowe, P. Šácha, B. L. Lee, D. Demon, A. Fossoul, F. Van Hauwermeiren, P. H. V. Saavedra, P. Šimon, V. Šubrt, L. Kostka, C. E. Stivala, V. C. Pham, S. T. Staben, S. Yamazoe, J. Konvalinka, N. Kayagaki and M. Lamkanfi, PLoS Biol., 2019, 17, 1–24 CrossRef PubMed.
- C. Dekker, H. Mattes, M. Wright, A. Boettcher, A. Hinniger, N. Hughes, S. Kapps-Fouthier, J. Eder, P. Erbel, N. Stiefl, A. Mackay and C. J. Farady, J. Mol. Biol., 2021, 433, 167309 CrossRef CAS PubMed.
- C. McBride, L. Trzoss, D. Povero, M. Lazic, G. Ambrus-Aikelin, A. Santini, R. Pranadinata, G. Bain, R. Stansfield, J. A. Stafford, J. Veal, R. Takahashi, J. Ly, S. Chen, L. Liu, M. Nespi, R. Blake, A. Katewa, T. Kleinheinz, S. Sujatha-Bhaskar, N. Ramamoorthi, J. Sims, B. McKenzie, M. Chen, M. Ultsch, M. Johnson, J. Murray, C. Ciferri, S. T. Staben, M. J. Townsend and C. E. Stivala, J. Med. Chem., 2022, 65, 14721–14739 CrossRef CAS PubMed.
- H. He, H. Jiang, Y. Chen, J. Ye, A. Wang, C. Wang, Q. Liu, G. Liang, X. Deng, W. Jiang and R. Zhou, Nat. Commun., 2018, 9, 1–12 CrossRef PubMed.
- Y. Huang, H. Jiang, Y. Chen, X. Wang, Y. Yang, J. Tao, X. Deng, G. Liang, H. Zhang, W. Jiang and R. Zhou, EMBO Mol. Med., 2018, 10, 1–15 CrossRef PubMed.
- M. Bertinaria, S. Gastaldi, E. Marini and M. Giorgis, Arch. Biochem. Biophys., 2019, 670, 116–139 CrossRef CAS PubMed.
- T. D. Kanneganti, N. Özören, M. Body-Malapel, A. Amer, J. H. Park, L. Franchi, J. Whitfield, W. Barchet, M. Colonna, P. Vandenabeele, J. Bertin, A. Coyle, E. P. Grant, S. Akira and G. Núñez, Nature, 2006, 440, 233–236 CrossRef CAS PubMed.
- C. J. Groß, R. Mishra, K. S. Schneider, G. Médard, J. Wettmarshausen, D. C. Dittlein, H. Shi, O. Gorka, P. A. Koenig, S. Fromm, G. Magnani, T. Ćiković, L. Hartjes, J. Smollich, A. A. B. Robertson, M. A. Cooper, M. Schmidt-Supprian, M. Schuster, K. Schroder, P. Broz, C. Traidl-Hoffmann, B. Beutler, B. Kuster, J. Ruland, S. Schneider, F. Perocchi and O. Groß, Immunity, 2016, 45, 761–773 CrossRef PubMed.
- L. K. Billingham, J. S. Stoolman, K. Vasan, A. E. Rodriguez, T. A. Poor, M. Szibor, H. T. Jacobs, C. R. Reczek, A. Rashidi, P. Zhang, J. Miska and N. S. Chandel, Nat. Immunol., 2022, 23, 692–704 CrossRef CAS PubMed.
- B. E. Nelson, R. Sheth, K. Kelly, H. Park, S. P. Patel, D. McKinley, S. O’Brien, J. L. Parra Palau and S. A. Piha-Paul, J. Clin. Oncol., 2023, 41, e14584–e14584 CrossRef.
- D. Angosto-Bazarra, C. Molina-López and P. Pelegrín, Commun. Biol., 2022, 5, 1–8 CrossRef PubMed.
- M. Levy, C. A. Thaiss, D. Zeevi, L. Dohnalová, G. Zilberman-Schapira, J. A. Mahdi, E. David, A. Savidor, T. Korem, Y. Herzig, M. Pevsner-Fischer, H. Shapiro, A. Christ, A. Harmelin, Z. Halpern, E. Latz, R. A. Flavell, I. Amit, E. Segal and E. Elinav, Cell, 2015, 163, 1428–1443 CrossRef CAS PubMed.
- P. Lemire, S. J. Robertson, H. Maughan, I. Tattoli, C. J. Streutker, J. M. Platnich, D. A. Muruve, D. J. Philpott and S. E. Girardin, Cell Rep., 2017, 21, 3653–3661 CrossRef CAS PubMed.
- M. Mamantopoulos, F. Ronchi, F. Van Hauwermeiren, S. Vieira-Silva, B. Yilmaz, L. Martens, Y. Saeys, S. K. Drexler, A. S. Yazdi, J. Raes, M. Lamkanfi, K. D. McCoy and A. Wullaert, Immunity, 2017, 47, 339–348 CrossRef CAS PubMed.
- H. Hara, S. S. Seregin, D. Yang, K. Fukase, M. Chamaillard, E. S. Alnemri, N. Inohara, G. Y. Chen and G. Núñez, Cell, 2018, 175, 1651–1664 CrossRef CAS PubMed.
- P. Wang, S. Zhu, L. Yang, S. Cui, W. Pan, R. Jackson, Y. Zheng, A. Rongvaux, Q. Sun, G. Yang, S. Gao, R. Lin, F. You, R. Flavell and E. Fikrig, Science, 2015, 350, 826–830 CrossRef CAS PubMed.
- C. Shen, R. Li, R. Negro, J. Cheng, S. M. Vora, T. M. Fu, A. Wang, K. He, L. Andreeva, P. Gao, Z. Tian, R. A. Flavell, S. Zhu and H. Wu, Cell, 2021, 184, 5759–5774 CrossRef CAS PubMed.
- H. R. Kilgore and R. A. Young, Nat. Chem. Biol., 2022, 18, 1298–1306 CrossRef CAS PubMed.
- H. Ishikawa, Z. Ma and G. N. Barber, Nature, 2009, 461, 788–792 CrossRef CAS PubMed.
- H. Ishikawa and G. N. Barber, Nature, 2008, 455, 674–678 CrossRef CAS PubMed.
- A. Ablasser, M. Goldeck, T. Cavlar, T. Deimling, G. Witte, I. Röhl, K. P. Hopfner, J. Ludwig and V. Hornung, Nature, 2013, 498, 380–384 CrossRef CAS PubMed.
- L. Sun, J. Wu, F. Du, X. Chen and Z. J. Chen, Science, 2013, 339, 786–791 CrossRef CAS PubMed.
- X. Zhang, H. Shi, J. Wu, X. Zhang, L. Sun, C. Chen and Z. J. Chen, Mol. Cell, 2013, 51, 226–235 CrossRef CAS PubMed.
- S. L. Ergun, D. Fernandez, T. M. Weiss and L. Li, Cell, 2019, 178, 290–301 CrossRef CAS PubMed.
- G. Shang, C. Zhang, Z. J. Chen, X. Chen Bai and X. Zhang, Nature, 2019, 567, 389–393 CrossRef CAS PubMed.
- C. Vanpouille-Box, J. A. Hoffmann and L. Galluzzi, Nat. Rev. Drug Discovery, 2019, 18, 845–867 CrossRef CAS PubMed.
- K. M. Garland, T. L. Sheehy and J. T. Wilson, Chem. Rev., 2022, 122, 5977–6039 CrossRef CAS PubMed.
- J. Vincent, C. Adura, P. Gao, A. Luz, L. Lama, Y. Asano, R. Okamoto, T. Imaeda, J. Aida, K. Rothamel, T. Gogakos, J. Steinberg, S. Reasoner, K. Aso, T. Tuschl, D. J. Patel, J. F. Glickman and M. Ascano, Nat. Commun., 2017, 8, 1–12 CrossRef PubMed.
- L. Lama, C. Adura, W. Xie, D. Tomita, T. Kamei, V. Kuryavyi, T. Gogakos, J. I. Steinberg, M. Miller, L. Ramos-Espiritu, Y. Asano, S. Hashizume, J. Aida, T. Imaeda, R. Okamoto, A. J. Jennings, M. Michino, T. Kuroita, A. Stamford, P. Gao, P. Meinke, J. F. Glickman, D. J. Patel and T. Tuschl, Nat. Commun., 2019, 10, 1–14 CrossRef CAS PubMed.
- J. Hall, A. Brault, F. Vincent, S. Weng, H. Wang, D. Dumlao, A. Aulabaugh, D. Aivazian, D. Castro, M. Chen, J. Culp, K. Dower, J. Gardner, S. Hawrylik, D. Golenbock, D. Hepworth, M. Horn, L. Jones, P. Jones, E. Latz, J. Li, L. L. Lin, W. Lin, D. Lin, F. Lovering, N. Niljanskul, R. Nistler, B. Pierce, O. Plotnikova, D. Schmitt, S. Shanker, J. Smith, W. Snyder, T. Subashi, J. Trujillo, E. Tyminski, G. Wang, J. Wong, B. Lefker, L. Dakin and K. Leach, PLoS One, 2017, 12, 1–16 Search PubMed.
- J. Dai, Y. J. Huang, X. He, M. Zhao, X. Wang, Z. S. Liu, W. Xue, H. Cai, X. Y. Zhan, S. Y. Huang, K. He, H. Wang, N. Wang, Z. Sang, T. Li, Q. Y. Han, J. Mao, X. Diao, N. Song, Y. Chen, W. H. Li, J. H. Man, A. L. Li, T. Zhou, Z. G. Liu, X. M. Zhang and T. Li, Cell, 2019, 176, 1447–1460 CrossRef CAS PubMed.
- J. An, J. J. Woodward, T. Sasaki, M. Minie and K. B. Elkon, J. Immunol., 2015, 194, 4089–4093 CrossRef CAS PubMed.
- M. Wang, M. A. Sooreshjani, C. Mikek, C. Opoku-Temeng and H. O. Sintim, Future Med. Chem., 2018, 10, 1301–1317 CrossRef CAS PubMed.
- R. Padilla-Salinas, L. Sun, R. Anderson, X. Yang, S. Zhang, Z. J. Chen and H. Yin, J. Org. Chem., 2020, 85, 1579–1600 CrossRef CAS PubMed.
- H. Zhang, Q. You and X. Xu, J. Med. Chem., 2020, 63, 3785–3816 CrossRef CAS PubMed.
- L. Li, Q. Yin, P. Kuss, Z. Maliga, J. L. Millán, H. Wu and T. J. Mitchison, Nat. Chem. Biol., 2014, 10, 1043–1048 CrossRef CAS PubMed.
- F. Meric-Bernstam, R. F. Sweis, F. S. Hodi, W. A. Messersmith, R. H. I. Andtbacka, M. Ingham, N. Lewis, X. Chen, M. Pelletier, X. Chen, J. Wu, S. M. McWhirter, T. Müller, N. Nair and J. J. Luke, Clin. Cancer Res., 2022, 28, 677–688 CrossRef CAS PubMed.
- F. Meric-Bernstam, R. F. Sweis, S. Kasper, O. Hamid, S. Bhatia, R. Dummer, A. Stradella, G. V. Long, A. Spreafico, T. Shimizu, N. Steeghs, J. J. Luke, S. M. McWhirter, T. Müller, N. Nair, N. Lewis, X. Chen, A. Bean, L. Kattenhorn, M. Pelletier and S. Sandhu, Clin. Cancer Res., 2023, 29, 110–121 CrossRef CAS PubMed.
- S. Kim, L. Li, Z. Maliga, Q. Yin, H. Wu and T. J. Mitchison, ACS Chem. Biol., 2013, 8, 1396–1401 CrossRef CAS PubMed.
- P. Gao, M. Ascano, T. Zillinger, W. Wang, P. Dai, A. A. Serganov, B. L. Gaffney, S. Shuman, R. A. Jones, L. Deng, G. Hartmann, W. Barchet, T. Tuschl and D. J. Patel, Cell, 2013, 154, 748–762 CrossRef CAS PubMed.
- J. M. Ramanjulu, G. S. Pesiridis, J. Yang, N. Concha, R. Singhaus, S. Y. Zhang, J. L. Tran, P. Moore, S. Lehmann, H. C. Eberl, M. Muelbaier, J. L. Schneck, J. Clemens, M. Adam, J. Mehlmann, J. Romano, A. Morales, J. Kang, L. Leister, T. L. Graybill, A. K. Charnley, G. Ye, N. Nevins, K. Behnia, A. I. Wolf, V. Kasparcova, K. Nurse, L. Wang, Y. Li, M. Klein, C. B. Hopson, J. Guss, M. Bantscheff, G. Bergamini, M. A. Reilly, Y. Lian, K. J. Duffy, J. Adams, K. P. Foley, P. J. Gough, R. W. Marquis, J. Smothers, A. Hoos and J. Bertin, Nature, 2018, 564, 439–443 CrossRef CAS PubMed.
- B.-S. Pan, S. A. Perera, J. A. Piesvaux, J. P. Presland, G. K. Schroeder, J. N. Cumming, B. W. Trotter, M. D. Altman, A. V. Buevich, B. Cash, S. Cemerski, W. Chang, Y. Chen, P. J. Dandliker, G. Feng, A. Haidle, T. Henderson, J. Jewell, I. Kariv, I. Knemeyer, J. Kopinja, B. M. Lacey, J. Laskey, C. A. Lesburg, R. Liang, B. J. Long, M. Lu, Y. Ma, E. C. Minnihan, G. O’Donnell, R. Otte, L. Price, L. Rakhilina, B. Sauvagnat, S. Sharma, S. Tyagarajan, H. Woo, D. F. Wyss, S. Xu, D. J. Bennett and G. H. Addona, Science, 2020, 369, 993–999 CrossRef PubMed.
- E. N. Chin, C. Yu, V. F. Vartabedian, Y. Jia, M. Kumar, A. M. Gamo, W. Vernier, S. H. Ali, M. Kissai, D. C. Lazar, N. Nguyen, L. E. Pereira, B. Benish, A. K. Woods, S. B. Joseph, A. Chu, K. A. Johnson, P. N. Sander, F. Martínez-Peña, E. N. Hampton, T. S. Young, D. W. Wolan, A. K. Chatterjee, P. G. Schultz, H. M. Petrassi, J. R. Teijaro and L. L. Lairson, Science, 2020, 369, 993–999 CrossRef CAS PubMed.
- T. Siu, M. D. Altman, G. A. Baltus, M. Childers, J. M. Ellis, H. Gunaydin, H. Hatch, T. Ho, J. Jewell, B. M. Lacey, C. A. Lesburg, B. S. Pan, B. Sauvagnat, G. K. Schroeder and S. Xu, ACS Med. Chem. Lett., 2019, 10, 92–97 CrossRef CAS PubMed.
- Z. Hong, J. Mei, C. Li, G. Bai, M. Maimaiti, H. Hu, W. Yu, L. Sun, L. Zhang, D. Cheng, Y. Liao, S. Li, Y. You, H. Sun, J. Huang, X. Liu, J. Lieberman and C. Wang, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2105465118 CrossRef CAS PubMed.
- S. M. Haag, M. F. Gulen, L. Reymond, A. Gibelin, L. Abrami, A. Decout, M. Heymann, F. G. Van Der Goot, G. Turcatti, R. Behrendt and A. Ablasser, Nature, 2018, 559, 269–273 CrossRef CAS PubMed.
- A. L. Hansen, G. J. Buchan, M. Rühl, K. Mukai, S. R. Salvatore, E. Ogawa, S. D. Andersen, M. B. Iversen, A. L. Thielke, C. Gunderstofte, M. Motwani, C. T. Møller, A. S. Jakobsen, K. A. Fitzgerald, J. Roos, R. Lin, T. J. Maier, R. Goldbach-Mansky, C. A. Miner, W. Qian, J. J. Miner, R. E. Rigby, J. Rehwinkel, M. R. Jakobsen, H. Arai, T. Taguchi, F. J. Schopfer, D. Olagnier and C. K. Holm, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E7768–E7775 CAS.
- K. A. Krautkramer, J. Fan and F. Bäckhed, Nat. Rev. Microbiol., 2021, 19, 77–94 CrossRef CAS PubMed.
- T. Tsukidate, Q. Li and H. C. Hang, Curr. Opin. Chem. Biol., 2020, 54, 19–27 CrossRef CAS PubMed.
- C. Diskin, T. A. J. Ryan and L. A. J. O’Neill, Immunity, 2021, 54, 19–31 CrossRef CAS PubMed.
- E. V. Vinogradova, X. Zhang, D. Remillard, D. C. Lazar, R. M. Suciu, Y. Wang, G. Bianco, Y. Yamashita, V. M. Crowley, M. A. Schafroth, M. Yokoyama, D. B. Konrad, K. M. Lum, G. M. Simon, E. K. Kemper, M. R. Lazear, S. Yin, M. M. Blewett, M. M. Dix, N. Nguyen, M. N. Shokhirev, E. N. Chin, L. L. Lairson, B. Melillo, S. L. Schreiber, S. Forli, J. R. Teijaro and B. F. Cravatt, Cell, 2020, 182, 1009–1026 CrossRef CAS PubMed.
- J. Stefaniak and K. V. M. Huber, ACS Med. Chem. Lett., 2020, 11, 403–406 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cb00096f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |