Open Access Article
Open Access ArticlePhotoreactive bioorthogonal lipid probes and their applications in mammalian biology
Karthik
Shanbhag
,
Kavita
Sharma
and
Siddhesh S.
Kamat
 *
*
Department of Biology, Indian Institute of Science Education and Research (IISER), Dr Homi Bhabha Road, Pashan, Pune, 411008, Maharashtra, India. E-mail: siddhesh@iiserpune.ac.in
First published on 12th December 2022
Abstract
Lipids are an important class of biological molecules that possess many critical physiological functions, which enable the optimal survival of all organisms, including humans. While the role of lipids in the formation of biological cellular membranes and as a source of energy is fairly well understood, the cellular signalling pathways that lipids modulate in mammals are, in comparison, poorly characterized mechanistically and/or largely unknown. In an effort to dissect these mammalian cellular pathways regulated by signalling lipids and map hitherto unknown protein–lipid interactions, the last two decades have seen tremendous progress in the development of multifunctional lipid probes that, in conjunction with well-established bioorthogonal chemistries and chemoproteomics platforms, has almost exponentially expanded our knowledge in this field. In this review, we focus on the various photoreactive bioorthogonal lipid probes described in the literature, and briefly summarize the different photo-crosslinking groups and bioorthogonal chemistries used by them. Furthermore, we report specific case examples of such photoreactive bioorthogonal lipid probes, and discuss the new biological pathways and insights that have emerged from their use through chemoproteomics in mammalian cells. Finally, we highlight the challenges associated with the use of lipid probes in biological systems, and highlight their importance in the discovery and mechanistic understanding of lipid signalling pathways in the years to come.
Introduction
Lipids are an essential building block for all forms of life, and this important class of biomolecules has critical physiological functions that enable the survival of cells and, in turn, complex multicellular organisms like humans.1,2 Historically, amongst the innumerable indispensable functions of lipids, perhaps the most studied one is their ability to form membranous mono- and bi-layers that define cellular boundaries and the internal compartments of the cell (e.g., organelles), thus providing a hydrophobic physical barrier against detrimental environmental factors that are largely aqueous in nature, and by doing so, the survival of cells across various physiological conditions and challenges is greatly promoted.1,3 Popularly and extensively studied from a metabolic standpoint is the ability of lipids to function as an important source of energy in cells and tissues via the catabolic β-oxidation pathway, when carbohydrates (especially glucose) become limiting.4 In addition to these popularly studied functions, in recent years, several lipid classes, especially signalling lipids (e.g., sphingosine 1-phosphate,5,6 endocannabinoids,7,8 prostaglandins,9,10 and lysophospholipids11–13), have been physiologically shown to have hormone-like effects and modulate diverse cellular signalling and immunological pathways. Furthermore, by lipidating (e.g., via protein palmitoylation) important proteins (e.g., receptors and ion channels), cellular fatty acids are able to regulate their localization in cells, and, by doing so, spatiotemporally regulate their functions.14,15Not surprisingly, given their central role in important physiological processes in humans, dysregulation in their cellular metabolism leads to numerous pathophysiological conditions such as dyslipidaemia, diabetes, metabolic syndrome, atherosclerosis and cardiovascular diseases, autoimmune diseases, cancers and neurodegenerative disorders, to list a few.16–19 It is of note that, although deregulated lipid metabolism has been implicated in an array of human diseases, only a handful of small molecule drugs have been developed and/or used in patients and/or in different stages of clinical trials that target the known metabolic (biosynthetic or degradative) enzymes and/or cognate receptors for these aforementioned lipids. This disconnect stems largely from our current (poor) mechanistic understanding of the cellular pathways that lipids globally regulate, and the distinct set of protein ligands that they interact with, during diverse physiological processes.
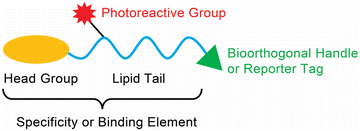
Given their hydrophobic and fairly inert chemical nature, synthetic methodologies towards making lipids in general has been challenging over the years. However, with the advent of newer bioorthogonal chemical reactions, coupled with tremendous advances in mass spectrometry and chemoproteomics technologies, the past two decades has seen a real emergence in the development of lipid probes that have enabled extensive profiling and mapping of the signalling pathways that are regulated by different lipid classes in various mammalian cells and tissues.20–22 Generically, most lipid probes described in literature have three major components: (i) specificity or binding element(s) for a particular lipid class; (ii) a photoreactive chemical crosslinking group; and (iii) a bioorthogonal handle or reporter tag (e.g., a fluorophore or biotin) (Fig. 1). As part of this review, we aim to summarize the various photoreactive chemical crosslinking groups used in lipid probes, along with the bioorthogonal chemistries used in profiling these probes in lysates and mammalian cells. In addition to this, we discuss specific classes of lipid probes and describe, briefly, the studies carried out using them to tease out novel biological pathways in mammalian systems.
Photoreactive chemical crosslinkers
The role of a photoreactive crosslinking group in a bioorthogonal lipid probe is to make protein–lipid interactions that are usually non-covalent (ionic or hydrophobic) in nature, permanent via the formation of stable covalent bonds. This covalently bonded protein–lipid probe complex enables researchers to ensure that such cellular protein–lipid interactions can be studied and characterized in more detail. With the advent of photochemical reactions, to this end, most lipid probes described in the literature have been appended with photoreactive chemical crosslinking groups (also referred to as photo-crosslinking groups) that are chemically stable and efficiently form a reactive intermediate via photolysis upon exposure to light of a defined wavelength range. Recently, Backus and co-workers23 elegantly described in significant detail the photoreactive groups used across diverse chemical probes, their respective chemistries upon photolysis, and deliberated on how the selection of these chemical moieties is important, so as to not alter the properties of the ligand itself. Hence, here, we briefly summarize the three photo-crosslinking groups, namely benzophenone, arylazide, and alkyldiazirine, that have been prominently used in lipid probes, and list their advantages and drawbacks (Table 1).| Photoreactive group | Reactive species | Advantages | Drawbacks |
|---|---|---|---|
| Benzophenone24,25 |
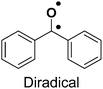
|
1. Biocompatible | 1. Bulky size |
| 2. Diverse photo-reactivity | 2. Non-specific protein labeling due to the short life of the radical | ||
| 3. High photo-crosslinking efficiency | |||
| Arylazide26,27 |
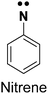
|
1. Chemically stable | 1. Low UV activation, not amenable to live cell studies |
| 2. Low photo-crosslinking efficiency due to the longer half-life of the radical | |||
| Alkyldiazirene28–32 |
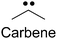
|
1. Small size | 1. Non-specific protein labeling due to the short life of the radical |
| 2. Biocompatible | |||
| 3. High photo-crosslinking efficiency |
Bioorthogonal reactions
Whilst designing a ligand-affinity based probe (e.g., lipid probe), it is important that the eventual probe must closely mimic the physical, chemical and structural properties of the parent ligand. Therefore, studies from lipid (and other ligand) probes have found that the addition of bulky reporter tags (e.g., fluorophores, biotin) hinders the binding of the probe to the “correct” (desired) protein, and often has many off-targets during downstream pulldown experiments. The rapid progress in the evolution of bioorthogonal (chemical) reactions has largely helped to circumvent this limitation, and has enabled the development of high-quality lipid probes with smaller appendages that offer better selectivity, biocompatibility and amenability to various type of studies (in vitro, ex vivo and in vivo) in mammalian cells and tissues. Over the past decade, bioorthogonal reactions have been the subject of numerous very well written reviews33–35 that discuss these reactions and their biological applications in significant detail, and hence, here, we briefly summarize only those bioorthogonal reactions that have been used with lipid probes (Table 2). Given their wide applications across diverse biological systems, for pioneering the development of various bioorthogonal reactions, Carolyn Bertozzi, K. Barry Sharpless and Morten Meldal were jointly awarded the 2022 Nobel Prize in Chemistry (https://www.nobelprize.org/prizes/chemistry/2022/summary/).| Reaction | Advantages | Drawbacks |
|---|---|---|
| Copper-catalyzed azide–alkyne cycloaddition (CuAAC)36–40 (click chemistry) | 1. Fast reaction kinetics | 1. Not suitable for live cell studies due to toxicity from copper [Cu(I)] |
| 2. Efficient under mild reaction (physiological) conditions | ||
| 3. Amenable to various protein profiling platforms | ||
| Copper-free azide–cycloalkyne cycloaddition41–45 | 1. Suitable for live cell studies | 1. Slow reactivity compared with CuAAC |
| 2. Efficient under mild reaction (physiological) conditions | ||
| Staudinger–Bertozzi ligation46–51 | 1. Suitable for live cell studies | 1. Slower rates of ligation |
| 2. Efficient under mild reaction (physiological) conditions | 2. Phosphine is prone to degradation by air oxidation and metabolic enzymes | |
| Inverse electron demand Diels–Alder (IEDDA) cycloaddition52,53 | 1. Fast reaction kinetics | 1. Strained alkyne ring is susceptible to isomerization |
| 2. Efficient under mild reaction (physiological) conditions | 2. Dienophile (tetrazine) is prone to degradation | |
| 3. Amenable to live cell studies |
Photoreactive bioorthogonal lipid probes
Historically, protein–lipid interactions were identified and characterized using conventional biochemical techniques like affinity purification, isothermal calorimetry, thermal shift assays or by computational approaches (e.g., molecular docking). However, while these techniques have proved useful over the years in specific cases, they have had a poor proteome wide coverage, and most of the studies using these conventional methods were restricted to recombinantly purified proteins. Over the past two decades, coupled with the advent in multiplexing mass spectrometry based chemoproteomics, ligand-photo-affinity based protein profiling techniques have provided an edge over the aforementioned conventional approaches in mapping protein–ligand interactions, in that these multifunctional lipid probes enable the screening and identification of hitherto unknown protein ligands of lipids at a proteome wide scale under various physiological conditions in mammalian cells and tissues.While lipid probes studied in the past have employed multiple strategies,54–57 such as appending a photoreactive group to a radioactive58,59 or fluorophore conjugated lipid tail,60–63 in this section, we aim to specifically discuss lipid probes that possess both a photoreactive group and a bioorthogonal handle, and how they have been profiled in mammalian systems to discover and characterize protein–lipid interactions. Commonly in such chemoproteomics experiments involving a lipid probe that has a photoreactive group and a bioorthogonal handle, as the first step, after incubation with the probe, the cellular lysates or live cells are irradiated with UV light of an appropriate wavelength and for a suitable exposure time, to ensure efficient crosslinking of the lipid probe with the protein(s) of interest. Subsequently, a bioorthogonal reaction is performed using a complementary reporter tag, which is either a fluorophore (for gel-based imaging or microscopy based live cell imaging applications) or an affinity group (e.g., biotin, for the enrichment of target proteins using avidin chromatography coupled to LC–MS based proteomics applications downstream) (Fig. 2).
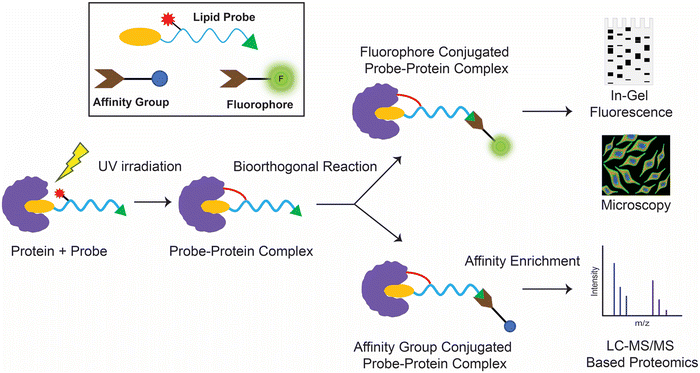 | ||
| Fig. 2 Schematic workflow for the typical enrichment and detection of protein(s) of interest using photo-crosslinking bioorthogonal lipid probes. | ||
Bifunctional lipid probes
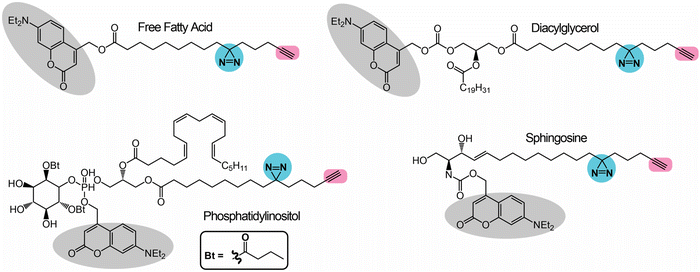
The first class of lipids for which photoreactive bioorthogonal (bifunctional) probes were developed were phosphatidylcholines (PCs).25,27 Towards synthesizing these PC probes, both of the lipid tails of this molecule were appended with a terminal azide, and the choline moiety of the head group was conjugated with benzophenone or arylazide (Fig. 3A). Both the benzophenone-PC and phenylazide-PC probes were profiled in yeast mitochondrial membrane proteomes using gel and LC–MS based chemoproteomics approaches, and, interestingly, it was found that the two photoreactive groups resulted in the enrichment of different sets of proteins, suggesting that the photo-crosslinking moiety did impart a degree of specificity to the proteins enriched by the probe. Although this study was performed in yeast proteomes, it has since laid the foundation for several subsequent studies involving the development of bifunctional probes and their applications in mammalian systems. Following up on these studies, Best and co-workers also developed bifunctional phosphatidylinositol (3,4,5)-triphosphate (PI(3,4,5)P3) probes containing benzophenone as the photoreactive crosslinking group, and an alkyne bioorthogonal handle (Fig. 3B), profiled these in different mammalian cells and found that >250 proteins bound to the PI(3,4,5)P3 lipid head group, several of which were novel PI(3,4,5)P3 binding proteins.64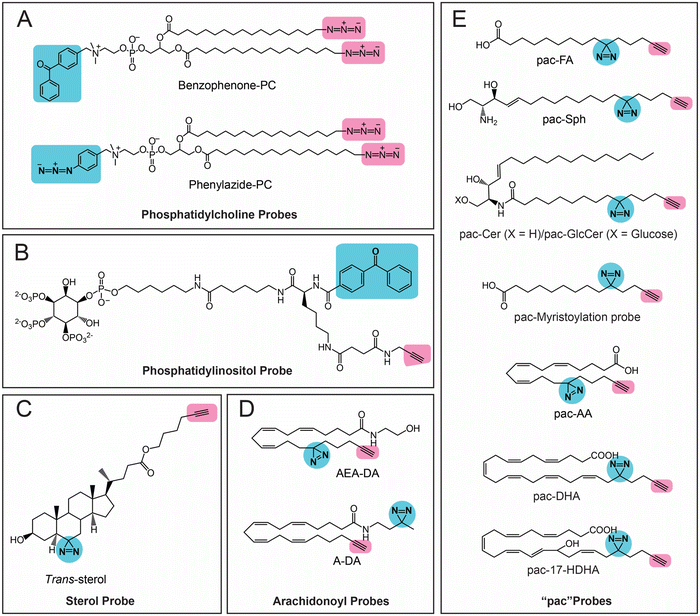 | ||
| Fig. 3 Bifunctional lipid probes containing a photo-crosslinking group (the moiety highlighted in blue) and a bioorthogonal handle (the moiety highlighted in pink) as described in the literature. | ||
Amongst the bifunctional lipid probes, two seminal studies by Cravatt and co-workers greatly expanded the repertoire of applications and our understanding of these probes. In the first study,65 they developed three sterol probes (cis-, trans- and epi-) that have different diastereomeric relationships between the C3-hydroxyl and the C5-hydrogen group of the cholesterol backbone, as well as containing the combination of a diazine and an alkyne group (Fig. 3C), in an effort to map the sterol interacting proteins in mammalian cells. The X-ray structures of these sterol probes revealed that only the trans-sterol probe had the correct overall stereochemistry and three-dimensional structure of cholesterol, and not surprisingly, from subsequent LC–MS/MS based chemoproteomics experiments, binding to known cholesterol interacting proteins and enzymes of the sterol biosynthesis pathway was also shown. Interestingly, this study also identified proteins with important biological functions that were previously not known to bind cholesterol. This study showed, for the first time, that the overall stereochemistry and structure of a lipid probe should closely resemble that of the parent lipid to minimize off-targets and enhance the enrichment of selective protein ligands. In their second study,66 Cravatt and co-workers synthesized various lipid probes (Fig. 3D), aimed at specifically mapping the protein ligands for different arachidonoyl-signaling lipids (e.g., arachidonic acid, anandamide), given the importance of endocannabinoids in human physiology and their association with diseases. By developing several advanced chemoproteomics platforms, they were able to show that several proteins pulled-down by these lipid probes (especially the arachidonoyl–lipid probes) were not part of the DrugBank database, and were for the first time shown to be pharmacologically liganded using these lipid probes. As an example, in this study, they identified the protein nucleobindin-1 as a novel metabolic regulator of arachidonoyl–lipid pools in mammalian cells, and showed that this protein is ligandable and has important functions in lipid metabolism. In the same study, using the synthesized lipid probes, Cravatt and co-workers also show through in situ profiling in mammalian cells that known clinically used drugs (especially those involved in regulating signaling lipids and their metabolism) have a high affinity for proteins that were previously thought to be “undruggable” and are in fact able to competitively inhibit protein–lipid interactions. Following up on this study, they have since shown that these lipid probes can also be used to map protein targets of biologically active natural products (e.g., KDT501, an anti-diabetic molecule derived from hops) and repurposed drugs of ill-defined mechanisms (e.g., miconazole, an anti-fungal agent used in the treatment of multiple sclerosis).67
Most of the aforementioned studies by Cravatt and co-workers on lipid probes were performed in vitro using lysates or in situ (ex vivo) in mammalian cells. Therefore, to visualize protein–lipid interactions in live cells, Schultz and co-workers developed “photoactivatable and clickable” (“pac”) probes for free fatty acid (pac-FA)68 and sphingosine (pac-Sph)69,70 (Fig. 3E), which consist of a diazirine and an alkyne handle, to systematically map the interactions of the two lipid classes in mammalian cells. Using both these probes, they demonstrated that, alongside the well-established gel-based and LC–MS/MS based chemoproteomics techniques, such pac-lipid probes can also be used to visualize the intracellular localization and metabolic fate of the lipids in mammalian cells (albeit fixed), and even in organisms such as the nematode, C. elegans. Following up on these elegant studies with pac-lipid probes, Holthuis and co-workers have since developed pac-lipid probes for other sphingolipids, namely, ceramide (pac-Cer) and glucosylceramide (pac-GlcCer) in an effort to map the ceramide binding proteins in mammalian cells (Fig. 3E).71 Using the pac-Cer and pac-GlcCer probes, they showed that the steroidogenic acute regulatory protein D7 (StarD7) bound ceramide, and this binding was important in this protein's ability to import phospholipids, particularly PC, into mitochondria, and in turn to regulate the mitochondrial function, morphology and mitochondria-mediated cytotoxicity. In the same study, they also reported some non-canonical pac-Cer probes (e.g., 1-deoxy-pac-Cer, 3-deoxy-pac-Cer and 3-deoxy-N-methyl-pac-Cer), and used these to perform elegant structure–activity relationship studies against the ceramide transfer protein (CERT), a known ceramide binding protein.71 Interestingly, a recent preprint has also reported pac probes with lysosome targeting moieties, for sphingosine (Lyso-pac-Sph) and cholesterol (Lyso-pac-Chol), which together have shown that two known sterol transporters, NPC1 and LIMP-2, also act as sphingosine transporters.72 In addition, another preprint has also reported another pac-FA probe to study protein myristolyation (Fig. 3E) as well as lipid-mediated protein interactions and localization in different mammalian cells.73 Most recent amongst this series of lipid probes are those described by van der Stelt and co-workers, who report pac probes for omega-3 fatty acids, docosahexaenoic acid (DHA) and its oxidative metabolite 17-hydroxy-DHA (17-HDHA) to profile their metabolic pathways in human macrophages.74 Using the pac-DHA and pac-17-HDHA lipid probes (Fig. 3E), van der Stelt and co-workers showed for the first time that the enzyme prostaglandin reductase 1 (PTGR1), which is important in immune signaling cascades, binds to and uses 17-HDHA as a substrate. This exciting work has expanded the repertoire of oxidative lipid substrates for this key lipid metabolic enzyme beyond the eicosanoids.74
Trifunctional lipid probes
Building upon the work of the pac lipid probes, Schultz and co-workers have since pioneered the development of trifunctional lipid probes, which apart from the pac moieties, contain an orthogonal photocleavable caged fluorophore (e.g., coumarin) that enables better spatiotemporal resolution and localization of these lipid probes in mammalian cells.75,76 The presence of a photocleavable caging group protects the lipid probe from enzymatic degradation, keeping the probe intact and, in doing so, enabling it to accumulate in mammalian cells at desired sub-cellular locations (or in organelles) at near-physiological concentrations. Mammalian cells incubated with the probe are then exposed to an orthogonal wavelength of light (typically 400 nm), which cleaves the photocleavable caged group (but not the diazirine moiety), releasing the fluorophore (coumarin) that can possibly be used to report on the (sub)cellular localization of the lipid via live cell fluorescence imaging, and in doing so converts the trifunctional lipid probe into a bioactive bifunctional lipid probe that mimics the functions of the parent signaling lipid. The so-formed bifunctional lipid probe closely resembles the reported functions/effects of the parent signaling lipid, and is now amenable to gel and LC–MS/MS based chemoproteomics experiments described in the earlier section. Schultz and co-workers have thus far reported and biochemically validated four such trifunctional lipid probes for free fatty acid, sphingosine, diacylglycerol and phosphatidylinositol lipids (Fig. 4). The studies reported using these trifunctional lipid probes have revealed considerable new mechanistic insights on the localization and trafficking of the aforementioned lipids in mammalian cells. It is of note that the trifunctional sphingosine probe has effectively been used to mechanistically probe and provides new cell biological insights into the pathology of Niemann–Pick type-C (NPC) disease, a sphingolipid metabolism disorder.In vivo synthesized lipid probes
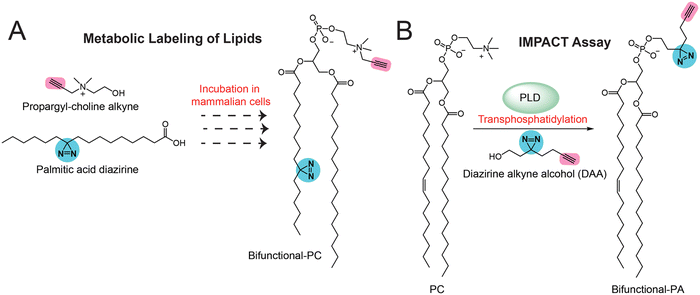
In all the studies discussed in earlier sections for the bifunctional and trifunctional lipid probes, these probes have been pre-made synthetically with all the functionalities. Although these studies worked very well, significant efforts are needed for synthetically making the lipid probes, which can prove challenging at times. Therefore, to overcome this particular setback, a few research groups have recently been leveraging the cellular machinery (mostly metabolic enzymes) to build various bifunctional lipids using building blocks that are much simpler to make synthetically and/or are commercially available. This approach of generating bifunctional lipid probes in situ in cells has popularly been referred to as the “metabolic labelling” of lipids. Initial studies attempting to prove this concept led to the development of different inositol derivatives carrying an azido group,77,78 which, when fed to mammalian cells, led to their successful uptake and the metabolic labeling of cellular phosphatidylinositol (PI) lipid pools and glycosyl-PI (GPI) anchored cell surface proteins. Besides this, numerous elegant studies have aimed at studying protein lipidation using diverse monofunctional lipid probes, and these have been summarized previously by Hang and co-workers.54 Following up on these studies, the first application of metabolic labeling to synthesize a complete bifunctional lipid probe in vivo was reported by Yao and co-workers in the design of bifunctional PCs in mammalian cells.79 In this study, they fed mammalian cells with two small molecules, i.e., an alkyne handle containing propargyl choline (the biosynthetic precursor of the PC head group) and a diazirine-containing palmitic acid (the lipid tail component of PC) (Fig. 5A). Using cellular lipidomics measurements, they confirmed that these two fragments resulted in the substantial formation of bifunctional PC (Fig. 5A) within cells, and that it could be used for gel, fluorescence microscopy and LC–MS/MS-based chemoproteomics applications in the mapping of protein–lipid interactions in mammalian cells.More recently, Baskin and co-workers pioneered the development of IMaging Phospholipase D (PLD) Activity with Clickable alcohols via Transphosphatidylation (IMPACT) assays80–83 for monitoring the activity of PLD (Fig. 5B), as well as assessing the production of different phospholipids (e.g., phosphatidic acid, phosphatidyl alcohols) in mammalian cells via PLD enzyme activity and studying the interactions of these phospholipids with diverse proteins in particular physiological contexts. In particular, they spatiotemporally mapped the activity of PLD isoforms in mammalian cells, showing its association with cellular signaling events, and extensively studied the protein interactors of phosphatidic acid (PA) in various physiological settings using diverse bioorthogonal reactions. In addition, using the same IMPACT technology, they developed photoswitchable PA analogs84 that served as useful tools in studying various PA-dependent cellular signaling pathways in mammalian cells.
Challenges with lipid probes
Although lipid affinity-based probes have received significant attention over the past decade and have been used in a variety of applications, they do possess some limitations that still prevent their wider use across the entire spectrum of lipid classes. These mostly include:(a) Synthesizing the probes
Given their overall inert chemical nature, natural lipids by themselves are rather difficult to chemically modify in order to develop such bioorthogonal probes discussed in earlier sections. Thus, such lipid probes have to be developed de novo from complex synthetic reactions, which can often be challenging, time consuming and rather expensive. In addition, the photoreactive groups are fairly labile and sensitive to light and certain solvents, and therefore such reactions often have lower yields and need extra precautions to ensure the purity of the final probes.(b) Ensuring comparable ligand properties
Since the lipid probes typically have at least two additional functional moieties (e.g., a photoreactive group and a bioorthogonal handle or reporter tag), during the development of such a probe for an as-of-yet unstudied lipid class, extensive structure–activity relationship studies through the development of various synthetic analogs are needed to ensure that the final lipid probe is indeed comparable to the natural lipid ligand, before proceeding with any chemoproteomics studies in mammalian cells and tissues. The effective placement of the various functionalities on the lipid probes is of prime importance to ensure the correct biological readouts.(c) Off-target activity
In an effort to map the “backgroundome” of a minimally photoreactive probe, Sieber and co-workers showed from a recent study that such a probe had a promiscuous propensity for binding a variety of proteins, even in the absence of a specificity group.85 Based on this study, and coupled with published studies on different multi-functional probes, it is evident that both the natural lipid and the various functionalities appended to it do in fact interact with non-specific “off-target” proteins,86 and therefore, contrary to the well-established activity-based probes,87,88 very stringent control (e.g., UV exposure) and competition (e.g., with natural lipid) chemoproteomics and biological validation experiments are needed to pull down “true” protein interactors of such lipid-affinity based probes.Future outlook
Over the past decade, ligand affinity-based proteomic profiling technologies have been instrumental in mapping the protein ligands and/or interactors of various small molecules and/or their metabolites, and in the context of lipid biology have greatly facilitated the annotation of (new) functions to poorly characterized proteins that have important physiological functions in mammals, including humans. In addition to this, bioorthogonal lipid probes have, for the first time, enabled the spatiotemporal visualization of lipids and the signaling pathways they modulate in real time in live mammalian cells. While tremendous progress has been made in developing such lipid probes, the associated technologies or chemical reactions, and an appreciable body of new knowledge has emerged in the field of lipid biology, it is important to note that lipid probes are available only for a handful of lipid classes. Prominently missing amongst these are signaling lysophospholipids (e.g., lysophosphatidylserine89) and oxidized lipids generated during ferroptosis,90–93 which are poorly characterized in terms of their known protein interactors, even though they have important physiological implications in several autoimmune human diseases. The development of probes for such signaling lipids will significantly add to our understanding of the pathologies associated with autoimmune diseases in humans, and will facilitate the development of new therapeutic paradigms for treating them. Finally, most of the studies until now with the aforementioned lipid probes have been performed in mammalian cells, and very limited studies have been possible in animal models. Hence, as recently demonstrated by Ye, Cravatt and co-workers with small molecules drugs,94 the next frontier in this field is perhaps the development of bioorthogonal lipid probes that will enable intricate anatomical mapping studies of lipids and the signaling pathways that they modulate in mammalian tissues and animal models.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a SwarnaJayanti Fellowship from the Science and Engineering Research Board (SERB), Government of India (grant number: SB/SJF/2021-22/01) to S. S. K.References
- E. A. Dennis, J. Biol. Chem., 2016, 291(47), 24431–24448 CrossRef CAS PubMed.
- E. A. Dennis, FASEB J., 2016, 30(S1), 114.3 Search PubMed.
- M. Sud, E. Fahy, D. Cotter, A. Brown, E. A. Dennis, C. K. Glass, A. H. Merrill, Jr., R. C. Murphy, C. R. Raetz, D. W. Russell and S. Subramaniam, Nucleic Acids Res., 2007, 35, D527–532 CrossRef CAS PubMed.
- S. M. Houten, S. Violante, F. V. Ventura and R. J. Wanders, Annu. Rev. Physiol., 2016, 78, 23–44 CrossRef CAS PubMed.
- P. J. Gonzalez-Cabrera, S. Brown, S. M. Studer and H. Rosen, F1000Prime Rep., 2014, 6, 109 Search PubMed.
- H. Rosen, P. J. Gonzalez-Cabrera, M. G. Sanna and S. Brown, Annu. Rev. Biochem., 2009, 78, 743–768 CrossRef CAS PubMed.
- J. L. Blankman and B. F. Cravatt, Pharmacol. Rev., 2013, 65, 849–871 CrossRef PubMed.
- D. Piomelli and O. Sasso, Nat. Neurosci., 2014, 17, 164–174 CrossRef CAS PubMed.
- E. Ricciotti and G. A. FitzGerald, Arterioscler., Thromb., Vasc. Biol., 2011, 31, 986–1000 CrossRef CAS PubMed.
- D. L. Simmons, R. M. Botting and T. Hla, Pharmacol. Rev., 2004, 56, 387–437 CrossRef CAS PubMed.
- I. Ishii, N. Fukushima, X. Ye and J. Chun, Annu. Rev. Biochem., 2004, 73, 321–354 CrossRef CAS PubMed.
- J. J. A. Contos, I. Ishii and J. Chun, Mol. Pharm., 2000, 58, 1188–1196 CrossRef CAS PubMed.
- K. Shanbhag, A. Mhetre, N. Khandelwal and S. S. Kamat, J. Membr. Biol., 2020, 253, 381–397 CrossRef CAS PubMed.
- J. Jin, X. Zhi, X. Wang and D. Meng, J. Cell. Physiol., 2021, 236, 3220–3233 CrossRef CAS PubMed.
- B. R. Martin and B. F. Cravatt, Nat. Methods, 2009, 6, 135–138 CrossRef CAS PubMed.
- S. B. Wortmann, M. Espeel, L. Almeida, A. Reimer, D. Bosboom, F. Roels, A. P. de Brouwer and R. A. Wevers, J. Inherited Metab. Dis., 2015, 38, 99–110 CrossRef CAS PubMed.
- D. K. Nomura and J. E. Casida, Chem. – Biol. Interact., 2016, 259, 211–222 CrossRef CAS PubMed.
- M. P. Wymann and R. Schneiter, Nat. Rev. Mol. Cell Biol., 2008, 9, 162–176 CrossRef CAS PubMed.
- J. Z. Long and B. F. Cravatt, Chem. Rev., 2011, 111, 6022–6063 CrossRef CAS.
- W. Yu and J. M. Baskin, Curr. Opin. Chem. Biol., 2022, 69, 102173 CrossRef CAS PubMed.
- R. Tei and J. M. Baskin, Curr. Opin. Chem. Biol., 2021, 65, 93–100 CrossRef CAS PubMed.
- S. Xie, C. Moya, B. Bilgin, A. Jayaraman and S. P. Walton, Expert Rev. Proteomics, 2009, 6, 573–583 CrossRef CAS PubMed.
- N. R. Burton, P. Kim and K. M. Backus, Org. Biomol. Chem., 2021, 19, 7792–7809 RSC.
- R. E. Galardy, L. C. Craig, J. D. Jamieson and M. P. Printz, J. Biol. Chem., 1974, 249, 3510–3518 CrossRef CAS PubMed.
- J. Gubbens, M. Slijper, B. de Kruijff and A. I. de Kroon, Biochim. Biophys. Acta, 2008, 1784, 2012–2018 CrossRef CAS PubMed.
- G. W. Fleet, J. R. Knowles and R. R. Porter, Biochem. J., 1972, 128, 499–508 CrossRef CAS PubMed.
- J. Gubbens, P. Vader, J. M. Damen, M. C. O'Flaherty, M. Slijper, B. de Kruijff and A. I. de Kroon, J. Proteome Res., 2007, 6, 1951–1962 CrossRef CAS PubMed.
- J. Brunner, Annu. Rev. Biochem., 1993, 62, 483–514 CrossRef CAS PubMed.
- J. Brunner, H. Senn and F. M. Richards, J. Biol. Chem., 1980, 255, 3313–3318 CrossRef CAS PubMed.
- A. L. Mackinnon and J. Taunton, Curr. Protoc. Chem. Biol., 2009, 1, 55–73 CrossRef PubMed.
- C. G. Parker, A. Galmozzi, Y. Wang, B. E. Correia, K. Sasaki, C. M. Joslyn, A. S. Kim, C. L. Cavallaro, R. M. Lawrence, S. R. Johnson, I. Narvaiza, E. Saez and B. F. Cravatt, Cell, 2017, 168, 527–541 CrossRef CAS PubMed.
- A. V. West, G. Muncipinto, H. Y. Wu, A. C. Huang, M. T. Labenski, L. H. Jones and C. M. Woo, J. Am. Chem. Soc., 2021, 143, 6691–6700 CrossRef CAS PubMed.
- X. Chen and Y. W. Wu, Org. Biomol. Chem., 2016, 14, 5417–5439 RSC.
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS PubMed.
- N. K. Devaraj, ACS Cent. Sci., 2018, 4, 952–959 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- A. E. Speers and B. F. Cravatt, Chem. Biol., 2004, 11, 535–546 CrossRef CAS PubMed.
- A. E. Speers, G. C. Adam and B. F. Cravatt, J. Am. Chem. Soc., 2003, 125, 4686–4687 CrossRef CAS PubMed.
- V. Hong, N. F. Steinmetz, M. Manchester and M. G. Finn, Bioconjugate Chem., 2010, 21, 1912–1916 CrossRef CAS PubMed.
- S. T. Laughlin, J. M. Baskin, S. L. Amacher and C. R. Bertozzi, Science, 2008, 320, 664–667 CrossRef CAS PubMed.
- J. M. Baskin, J. A. Prescher, S. T. Laughlin, N. J. Agard, P. V. Chang, I. A. Miller, A. Lo, J. A. Codelli and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16793–16797 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- J. C. Jewett and C. R. Bertozzi, Chem. Soc. Rev., 2010, 39, 1272–1279 RSC.
- J. C. Jewett, E. M. Sletten and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 3688–3690 CrossRef CAS PubMed.
- M. J. Hangauer and C. R. Bertozzi, Angew. Chem., Int. Ed., 2008, 47, 2394–2397 CrossRef CAS PubMed.
- F. L. Lin, H. M. Hoyt, H. van Halbeek, R. G. Bergman and C. R. Bertozzi, J. Am. Chem. Soc., 2005, 127, 2686–2695 CrossRef CAS PubMed.
- J. A. Prescher, D. H. Dube and C. R. Bertozzi, Nature, 2004, 430, 873–877 CrossRef CAS PubMed.
- K. L. Kiick, E. Saxon, D. A. Tirrell and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 19–24 CrossRef CAS PubMed.
- E. Saxon, J. I. Armstrong and C. R. Bertozzi, Org. Lett., 2000, 2, 2141–2143 CrossRef CAS PubMed.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, ACS Chem. Biol., 2014, 9, 16–20 CrossRef CAS PubMed.
- M. L. Blackman, M. Royzen and J. M. Fox, J. Am. Chem. Soc., 2008, 130, 13518–13519 CrossRef CAS PubMed.
- H. C. Hang, J. P. Wilson and G. Charron, Acc. Chem. Res., 2011, 44, 699–708 CrossRef CAS PubMed.
- T. W. Bumpus and J. M. Baskin, Trends Biochem. Sci., 2018, 43, 970–983 CrossRef CAS PubMed.
- Y. Tian and Q. Lin, ACS Chem. Biol., 2019, 14, 2489–2496 CrossRef CAS PubMed.
- Y. Xia and L. Peng, Chem. Rev., 2013, 113, 7880–7929 CrossRef CAS PubMed.
- C. Thiele, M. J. Hannah, F. Fahrenholz and W. B. Huttner, Nat. Cell Biol., 2000, 2, 42–49 CrossRef CAS PubMed.
- M. J. Janssen, F. van Voorst, G. E. Ploeger, P. M. Larsen, M. R. Larsen, A. I. de Kroon and B. de Kruijff, Biochemistry, 2002, 41, 5702–5711 CrossRef CAS PubMed.
- G. P. Jose and T. J. Pucadyil, Curr. Protoc. Protein Sci., 2020, 101, e110 CAS.
- G. P. Jose, S. Gopan, S. Bhattacharyya and T. J. Pucadyil, Traffic, 2020, 21, 297–305 CrossRef CAS PubMed.
- K. Bertheussen, M. van de Plassche, T. Bakkum, B. Gagestein, I. Ttofi, A. J. C. Sarris, H. S. Overkleeft, M. van der Stelt and S. I. van Kasteren, Angew. Chem., Int. Ed., 2022, 61, e202207640 CrossRef CAS PubMed.
- C. H. Knittel and N. K. Devaraj, Acc. Chem. Res., 2022, 55(21), 3099–3109 CrossRef CAS PubMed.
- M. M. Rowland, H. E. Bostic, D. Gong, A. E. Speers, N. Lucas, W. Cho, B. F. Cravatt and M. D. Best, Biochemistry, 2011, 50, 11143–11161 CrossRef CAS PubMed.
- J. J. Hulce, A. B. Cognetta, M. J. Niphakis, S. E. Tully and B. F. Cravatt, Nat. Methods, 2013, 10, 259–264 CrossRef PubMed.
- M. J. Niphakis, K. M. Lum, A. B. Cognetta 3rd, B. E. Correia, T. A. Ichu, J. Olucha, S. J. Brown, S. Kundu, F. Piscitelli, H. Rosen and B. F. Cravatt, Cell, 2015, 161, 1668–1680 CrossRef CAS PubMed.
- K. M. Lum, Y. Sato, B. A. Beyer, W. C. Plaisted, J. L. Anglin, L. L. Lairson and B. F. Cravatt, ACS Chem. Biol., 2017, 12, 2671–2681 CrossRef CAS PubMed.
- P. Haberkant, R. Raijmakers, M. Wildwater, T. Sachsenheimer, B. Brugger, K. Maeda, M. Houweling, A. C. Gavin, C. Schultz, G. van Meer, A. J. Heck and J. C. Holthuis, Angew. Chem., Int. Ed., 2013, 52, 4033–4038 CrossRef CAS PubMed.
- P. Haberkant, F. Stein, D. Hoglinger, M. J. Gerl, B. Brugger, P. P. Van Veldhoven, J. Krijgsveld, A. C. Gavin and C. Schultz, ACS Chem. Biol., 2016, 11, 222–230 CrossRef CAS PubMed.
- M. J. Gerl, V. Bittl, S. Kirchner, T. Sachsenheimer, H. L. Brunner, C. Luchtenborg, C. Ozbalci, H. Wiedemann, S. Wegehingel, W. Nickel, P. Haberkant, C. Schultz, M. Kruger and B. Brugger, PLoS One, 2016, 11, e0153009 CrossRef PubMed.
- S. Bockelmann, J. G. M. Mina, S. Korneev, D. G. Hassan, D. Muller, A. Hilderink, H. C. Vlieg, R. Raijmakers, A. J. R. Heck, P. Haberkant and J. C. M. Holthuis, J. Lipid Res., 2018, 59, 515–530 CrossRef CAS PubMed.
- J. Altuzar, J. Notbohm, F. Stein, P. Haberkant, S. Heybrock, J. Worsch, P. Saftig and D. Hoglinger, 2021, bioRxiv, 2021.11.10.468010.
- R. O. Fedoryshchak, A. Gorelik, M. Shen, M. M. Shchepinova, I. Pérez-Dorado and E. W. Tate, 2021, bioRxiv, 2021.12.15.472799.
- B. Gagestein, J. H. von Hegedus, J. C. Kwekkeboom, M. Heijink, N. Blomberg, T. van der Wel, B. I. Florea, H. van den Elst, K. Wals, H. S. Overkleeft, M. Giera, R. E. M. Toes, A. Ioan-Facsinay and M. van der Stelt, J. Am. Chem. Soc., 2022, 144(41), 18938–18947 CrossRef CAS PubMed.
- R. Muller, M. Citir, S. Hauke and C. Schultz, Chemistry, 2020, 26, 384–389 CrossRef PubMed.
- D. Hoglinger, A. Nadler, P. Haberkant, J. Kirkpatrick, M. Schifferer, F. Stein, S. Hauke, F. D. Porter and C. Schultz, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 1566–1571 CrossRef PubMed.
- L. Lu, J. Gao and Z. Guo, Angew. Chem., Int. Ed., 2015, 54, 9679–9682 CrossRef CAS PubMed.
- T. J. Ricks, C. D. Cassilly, A. J. Carr, D. S. Alves, S. Alam, K. Tscherch, T. W. Yokley, C. E. Workman, J. L. Morrell-Falvey, F. N. Barrera, T. B. Reynolds and M. D. Best, ChemBioChem, 2019, 20, 172–180 CrossRef CAS PubMed.
- D. Wang, S. Du, A. Cazenave-Gassiot, J. Ge, J. S. Lee, M. R. Wenk and S. Q. Yao, Angew. Chem., Int. Ed., 2017, 56, 5829–5833 CrossRef CAS PubMed.
- R. Tei and J. M. Baskin, J. Biol. Chem., 2022, 298, 101810 CrossRef CAS PubMed.
- D. Liang, R. W. Cheloha, T. Watanabe, T. J. Gardella and J. M. Baskin, Cell Chem. Biol., 2022, 29, 67–73 CrossRef CAS PubMed.
- W. Yu, Z. Lin, C. M. Woo and J. M. Baskin, ACS Chem. Biol., 2021 DOI:10.1021/acschembio.1c00584.
- T. W. Bumpus, D. Liang and J. M. Baskin, Methods Enzymol., 2020, 641, 75–94 CAS.
- R. Tei, J. Morstein, A. Shemet, D. Trauner and J. M. Baskin, ACS Cent. Sci., 2021, 7, 1205–1215 CrossRef CAS PubMed.
- P. Kleiner, W. Heydenreuter, M. Stahl, V. S. Korotkov and S. A. Sieber, Angew. Chem., Int. Ed., 2017, 56, 1396–1401 CrossRef CAS PubMed.
- D. Mellacheruvu, Z. Wright, A. L. Couzens, J. P. Lambert, N. A. St-Denis, T. Li, Y. V. Miteva, S. Hauri, M. E. Sardiu, T. Y. Low, V. A. Halim, R. D. Bagshaw, N. C. Hubner, A. Al-Hakim, A. Bouchard, D. Faubert, D. Fermin, W. H. Dunham, M. Goudreault, Z. Y. Lin, B. G. Badillo, T. Pawson, D. Durocher, B. Coulombe, R. Aebersold, G. Superti-Furga, J. Colinge, A. J. Heck, H. Choi, M. Gstaiger, S. Mohammed, I. M. Cristea, K. L. Bennett, M. P. Washburn, B. Raught, R. M. Ewing, A. C. Gingras and A. I. Nesvizhskii, Nat. Methods, 2013, 10, 730–736 CrossRef CAS PubMed.
- M. J. Niphakis and B. F. Cravatt, Annu. Rev. Biochem., 2014, 83, 341–377 CrossRef CAS PubMed.
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef CAS PubMed.
- N. Khandelwal, M. Shaikh, A. Mhetre, S. Singh, T. Sajeevan, A. Joshi, K. N. Balaji, H. Chakrapani and S. S. Kamat, Cell Chem. Biol., 2021, 28, 1169–1179 CrossRef CAS PubMed.
- X. Jiang, B. R. Stockwell and M. Conrad, Nat. Rev. Mol. Cell Biol., 2021, 22, 266–282 CrossRef PubMed.
- B. R. Stockwell, X. Jiang and W. Gu, Trends Cell Biol., 2020, 30, 478–490 CrossRef CAS PubMed.
- B. R. Stockwell and X. Jiang, Cell Chem. Biol., 2020, 27, 365–375 CrossRef CAS PubMed.
- D. S. Kelkar, G. Ravikumar, N. Mehendale, S. Singh, A. Joshi, A. K. Sharma, A. Mhetre, A. Rajendran, H. Chakrapani and S. S. Kamat, Nat. Chem. Biol., 2019, 15, 169–178 CrossRef CAS PubMed.
- Z. Pang, M. A. Schafroth, D. Ogasawara, Y. Wang, V. Nudell, N. K. Lal, D. Yang, K. Wang, D. M. Herbst, J. Ha, C. Guijas, J. L. Blankman, B. F. Cravatt and L. Ye, Cell, 2022, 185, 1793–1805 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |