DOI:
10.1039/D3BM01115A
(Paper)
Biomater. Sci., 2023,
11, 7169-7178
Delivery of siRNA using cationic rosette nanotubes for gene silencing†
Received
3rd July 2023
, Accepted 10th September 2023
First published on 21st September 2023
Abstract
The quest for new therapeutic treatments for hereditary diseases has led to many advances in RNA interference (RNAi) and gene silencing. While this technique has the potential to address many problems, the key to its continued use is the development of effective delivery strategies that would reduce cellular toxicity and increase silencing efficiency. Rosette nanotubes (RNTs) are biomimetic supramolecular nanostructures formed through the self-assembly of hybrid guanine–cytosine (G∧C) DNA bases. Here, we used bioactive RNTs for siRNA delivery and gene silencing. Fifteen lysine-functionalized twin-G∧C motifs (KnT, n = 1 to 15) were synthesized using solid phase peptide synthesis to produce building blocks that self-assembled to produce cationic RNTs under physiological conditions. The intracellular uptake of siRNA delivered by the oligo-L-lysine RNTs was examined and it was found that the complexation of siRNA was affected by the cationic charges from the lysine residues and the length of RNTs formed, with the higher charged KnT RNTs delivering siRNA to the cells at a faster rate. In addition, by protecting siRNA from serum degradation, KnT RNTs were shown to deliver their cargo to the cells effectively via the endocytic pathway. A reduction in the expression (∼70%) of the target stat3 protein was observed during gene expression analysis in HCT116 and A549 cell lines.
1. Introduction
The discovery of RNA interference1–3 has attracted a large number of researchers utilizing this tool for treatment of genetic diseases.4–7 The presence of double-stranded RNA (dsRNA) such as micro-RNA (miRNA) and small-interfering RNA (siRNA) in the cytoplasm of the cells triggers the RNAi pathway by incorporating with RNA-induced silencing complex (RISC).8–10 Only one strand of the dsRNA is kept by RISC as a guide strand to find the complementary messenger RNA (mRNA). After binding of RISC and mRNA, the Argonaute2 protein in RISC cleaves the mRNA, leading to a reduction or silencing of the corresponding protein.8,11 Thus, RNAi is touted as the future therapeutic treatment for genetic disease by knocking down selected gene expression.5,6,12 The perfect complementarity of siRNA and target mRNA makes the former remarkably specific for gene inactivation and hence, a highly focused area for current RNAi research.5,13 Naked siRNA molecules are however prone to serum degradation. In addition, their high molecular weight and negative charge prevent cellular uptake.5,14 As such, researchers have been developing new nanomaterials to capture, protect and deliver siRNA to the cells for gene silencing.5,6,15,16 Nanocarriers for siRNA delivery range from metal nanoparticles, cationic polymers, lipid-like materials such as liposomes or micelles, carbon nanotubes, which can either interact covalently or via electrostatic interaction with the siRNA. Liposomes are the most developed nanocarriers but the delivery needs to be controlled to prevent premature release of the siRNA cargo due to the interaction of the highly cationic liposomes with the molecules in the serum.17 Cationic polymers have also been commonly used to carry siRNA cargo,18 but cellular toxicity remains a concern due to their high molecular weight and low biodegradability.6,14 Although carbon nanotubes have been successfully used to deliver siRNA to cells,19–21 there have been concerns about the biodegradability and toxicity of these material.22–24
Rosette nanotubes (RNTs) are discrete nanotubular structures formed through the self-assembly of a hybrid guanine–cytosine (G∧C) module, having self-complementary hydrogen-bonding patterns (acceptor–donor–donor sites of guanine and donor–acceptor–acceptor sites of cytosine).25 In solution, these G∧C motifs lead to hexameric rosettes, which can then π–π stack to produce RNTs. The building blocks can have a single or two G∧C units, resulting into single or twin rosettes that are maintained by 18 or 36 hydrogen bonds, respectively.26,27 We have previously shown that the twin G∧C system produces more stable RNTs. Our group has worked extensively on the synthesis, characterization and applications of these biologically inspired nanomaterials.25–31 Previous studies have demonstrated favorable cytocompatibility of RNTs for various tissue-engineering applications due to their low toxicity and cargo-carrying capabilities.32,33 For instance, single G∧C-RNTs functionalized with the RGDSK peptide promoted osteoblast adhesion when coated on titanium substrates at low concentrations.34–36 RNTs, assembled from twin G∧C bases functionalized with the KRSR peptide, selectively improved osteoblast relative to fibroblast and endothelial cell adhesion.30 Similar observations were reported for composites made of poly(2-hydroxyethyl methacrylate) (pHEMA), hydroxyapatite and the twin-G∧C TBL RNTs.37 These biocompatible nanomaterials could potentially serve as implant coating materials that could increase the lifetime of implants in the human body. Since RNTs resemble the naturally occurring collagen and keratin in the skin, composites of pHEMA and TBL RNTs were investigated for wound healing applications and an increase in keratinocyte and fibroblast cell proliferation was observed.38 RNTs from the twin G∧C-RGDSK module enhanced human bone marrow mesenchymal stem cell (hMSC) chondrogenic differentiation that could make it an interesting candidate for cartilage regeneration.31 We have recently shown that cationic and fluorescent RNTs can capture plasmid DNA, penetrate the plant cell walls to deliver the cargo into live wheat microspores under mild conditions and in the absence of an external force.39
Herein, we present the synthesis of fifteen lysine-functionalized RNTs (KnT, n = 1–15), which were used to bind siRNA. The binding was examined at various molar ratios (KnT RNTs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA) by gel retardation assay and the resulting KnT RNTs/siRNA complexes were subjected to nucleases in bovine serum to determine the protective ability of KnT RNTs against serum degradation. Fluorescence and confocal microscopies were used to visualize the trafficking of KnT RNTs/FAM-labelled siRNA to the cells. The ability of the siRNA delivered to knockdown the targeted gene expression was investigated using firefly luciferase and stat3 protein assays.
siRNA) by gel retardation assay and the resulting KnT RNTs/siRNA complexes were subjected to nucleases in bovine serum to determine the protective ability of KnT RNTs against serum degradation. Fluorescence and confocal microscopies were used to visualize the trafficking of KnT RNTs/FAM-labelled siRNA to the cells. The ability of the siRNA delivered to knockdown the targeted gene expression was investigated using firefly luciferase and stat3 protein assays.
2. Experimental
Materials and methods
Reagent grade solvents and commercial reagents were purchased from Sigma Aldrich. Reagent grade CH2Cl2 and CH3OH were purified on an MBraun solvent purification system. All other solvents and reagents were used without further purification for the reactions. Standard Fmoc solid-phase peptide synthesis (SPPS) was used to prepare the target molecules on Wang resin having a loading capacity of 0.65 mmol g−1.27,39,40 The NMR data is presented as follows: chemical shift δ (ppm), multiplicity, coupling constant and integration. The following abbreviations were used to explain the multiplicities: s = singlet, d = doublet, t = triplet, m = multiplet, bs = broad singlet. Both 1H-NMR and 13C-NMR were carried out in 90% H2O in D2O at 600 MHz and 150 MHz, respectively. Cell lines A549 and HCT116 were purchased from ATCC. INTERFERin was obtained from Polyplus. McCoy's 5A, F-12K media, siRNAs (FAM, pGL3 and STAT3), Lipofectamine, Renilla–Firefly Luciferase Assay Kit, BCA Protein Assay Kit and antibodies (STAT3, GAPDH, goat anti-mouse IgG HRP) were procured from Thermo Fisher Scientific.
Synthetic procedures
Synthesis of Wang-Lys(Fmoc)-Boc (3).
Wang resin (1 g, 0.65 mmol g−1) was treated with a solution of Boc-L-Lys(Fmoc)-OH (1.28 g, 2.73 mmol) and p-dimethylaminopyridine (DMAP) (66 mg, 0.54 mmol) in DMF (8 mL). After activating the resin for 20 min, diisopropylcarbodiimide (DIC) (423 μL, 2.73 mmol) was added and the coupling reaction was maintained for 6 h. The resin was then filtered under vacuum, washed with (3 × 10 mL) DMF, DCM, and DMF and treated with 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 acetic anhydride/pyridine (5 mL, 1 × 10 min and 2 × 20 min) to cap the unreactive hydroxyl groups. The resin was subsequently filtered and washed with (3 × 10 mL) DMF, DCM and MeOH and dried under vacuum. The substitution degree (0.52 mmol g−1) was determined by spectroscopic quantification of the fulvene-piperidine adduct at 301 nm.
50 acetic anhydride/pyridine (5 mL, 1 × 10 min and 2 × 20 min) to cap the unreactive hydroxyl groups. The resin was subsequently filtered and washed with (3 × 10 mL) DMF, DCM and MeOH and dried under vacuum. The substitution degree (0.52 mmol g−1) was determined by spectroscopic quantification of the fulvene-piperidine adduct at 301 nm.
Synthesis of Wang-lysine oligomers (4).
The Fmoc protecting group was removed by incubating the resin in a 20% solution of piperidine in DMF (2 × 10 min). The resulting peptidyl resin was washed with DMF (4×), DCM (4×) and DMF (4×). The free amino group was then coupled to Boc-Lys(Fmoc)-OH (4 equiv. relative to resin loading) in the presence of N,N-diisopropylethylamine (DIPEA) (8 equiv.) and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyl-uroniumhexafluoro-phosphate (HBTU) (3.8 equiv.) in DMF for 2 h at 25 °C. The solution was then drained and the resin was washed with DMF (4×) and DCM (4×). Successful coupling was confirmed by the absence of free amino groups, as indicated by a yellow color in the Kaiser test. The above procedure was repeated for subsequent couplings.
Synthesis of protected twin-G∧C conjugated Wang-lysine oligomers (6).
tert-Butyl-4-(benzyloxy)-7-oxo-8-(2-oxoethyl)-7,8-dihydropyrimido[4,5-d]pyrimidine-2,5-diyltricarbamate (G∧C-aldehyde 5) (4 equiv.) and DIPEA (4 equiv.) was added to the peptidyl resin and shaken for 2 h in 1,2-DCE at 25 °C. NaBH(OAc)3 (4.8 equiv.) was then added and the mixture was shaken for an additional 72 h.
Synthesis of KnT.
Cleavage from the resin and deprotection was achieved by the treatment with 95% TFA in H2O for 2 h. The resin beads were filtered out over Celite and the resulting filtrate was concentrated to a viscous solution by rotary evaporation. Cold Et2O was then added to precipitate crude product. The precipitate was centrifuged, and the supernatant liquid was removed by decantation. After washing with ether (5×) to remove residual TFA, the precipitate was dried under vacuum for 12 h. The resulting materials were characterized by NMR, mass spectrometry and elemental analysis. The characterization data is included in the ESI (Table S1†).
Self-assembly of KnT into RNTs
Stock solutions of K1T–K15T RNTs (1 mg mL−1) were prepared by dissolving the monomers in deionized water, followed by sonication and heating in the oil bath at 90 °C for 15 min. Aliquots from the stock solutions were diluted and used for microscopy and complexation with siRNA. The molecular weight obtained from elemental analysis, molar concentration of each KnT RNTs solutions and net charges for the KnT compounds are summarized in Table S2.† The net charge of each KnT compound was calculated by the summation of all positive and negative charges per each KnT molecule. For instance, the K1T monomer has a net charge of +1 and K7T has a net charge of +7.
Agarose gel electrophoresis
Preparation of KnT RNTs/siRNA complexes.
Stock solutions of any siRNA sample (average MW ∼ 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) were dissolved in nuclease-free water (1 mg mL−1, 0.07 M). KnT RNTs were mixed with siRNA (0.1 nmol, 1.4 μL) at various molar ratios (0.5, 1, 2.5, 5, 10 and 20
000 g mol−1) were dissolved in nuclease-free water (1 mg mL−1, 0.07 M). KnT RNTs were mixed with siRNA (0.1 nmol, 1.4 μL) at various molar ratios (0.5, 1, 2.5, 5, 10 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, RNTs to siRNA respectively) in 10 μL of serum-free medium (SFM). The KnT RNTs/siRNA complexes were incubated for 30 min at room temperature prior to further experimental study.
1, RNTs to siRNA respectively) in 10 μL of serum-free medium (SFM). The KnT RNTs/siRNA complexes were incubated for 30 min at room temperature prior to further experimental study.
Gel retardation assay
The siRNA binding ability to KnT RNTs was studied by agarose gel retardation assay. The RNTs/siRNA complexes was mixed with 10× gel-loading buffer (1 μL) prior to loading the samples on 2% agarose gel, using Bio-Rad apparatus, running at 150 V for 20 min, followed by ethidium bromide (0.5 μg ml−1) staining for 20 min. Gel visualization was done using UVP BioDocIt BioImaging System. The signal of the unbound siRNA on the gel image was measured by ImageJ software. For the study of siRNA protection by KnT RNTs against serum degradation, the degree of siRNA degradation dose and time-dependency was first tested. Solutions of negative control siRNA (0.1 nmol) in 10 μL PBS (0, 10, 25, and 50% fetal bovine serum, FBS) were incubated in a 37 °C shaker (200 rpm) for 0, 1, 6, 12 and 24 h, followed by agarose gel electrophoresis. Next, complexes of K1T–K15T RNTs with siRNA (molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were prepared in PBS buffer. FBS was added to each solution to obtain final concentrations of 10% FBS. The RNTs/siRNA mixture was incubated in a 37 °C shaker for 24 h, followed by addition of heparin (5 μL, 20 μg μL−1) for 1 h at room temperature to displace siRNA from the RNTs. Agarose gel experiment was performed to detect the intact siRNA protected by RNTs from serum degradation.
1) were prepared in PBS buffer. FBS was added to each solution to obtain final concentrations of 10% FBS. The RNTs/siRNA mixture was incubated in a 37 °C shaker for 24 h, followed by addition of heparin (5 μL, 20 μg μL−1) for 1 h at room temperature to displace siRNA from the RNTs. Agarose gel experiment was performed to detect the intact siRNA protected by RNTs from serum degradation.
Cell culture
HCT116 (human colorectal carcinoma) and A549 (human lung carcinoma) cells were cultured in McCoy5A and F-12K media respectively (10% FBS, 100 units per mL penicillin, 100 μg mL−1 streptomycin) in a 5% CO2 atmosphere at 37 °C. Cells were detached for sub-culturing by trypsin solution.
Confocal and fluorescence imaging
The cells were seeded onto circular cover slips in a 12-well plate (1 mL per well, 50–60% cell confluence) and incubated for 24 h. The medium was then discarded and fresh SFM was added to the wells. The KnT RNTs/FAM siRNA complexes (molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were prepared in SFM (50 μL). Positive control INTERFERin/FAM siRNA complex was used (mixture of 4 μL INTERFERin solution and 1.4 μL FAM siRNA, 20 min incubation). The negative control sample was FAM-siRNA only (1.4 μL). The KnT RNTs/siRNA complexes and controls were added to the wells. After incubation, the cells were washed with PBS (3 × 30 s) and incubated with LysoTracker Red DND-99 in SFM (1 mL, 1 μM) for 20 min at 37 °C. The cells were washed with PBS (3 × 30 s) before fixation with 4% paraformaldehyde for 10 min. One drop of DAPI mounting medium was deposited on the microscope slide and the coverslip was mounted on the slide. The cells were imaged using fluorescence (Olympus IX81) and confocal laser scanning (Zeiss LSM 710) microscopes. Image processing was done using Metamorph software for fluorescence imaging, Zeiss LSM software (ZEN) for confocal imaging. FITC (green), DAPI (blue), Cy3 (red) channels were used to observe FAM-labelled siRNA, DAPI-labelled nuclei and DND99-labelled acidic compartments respectively. Sliced images of cells (in z-direction) were taken in selected area using z-stack experiment on ZEN software and processed by Imaris software to form 3D visualization of the imaged area. The setting of FITC channel was first set by imaging the positive control sample and was kept constant when imaging RNTs/siRNA-treated or negative control samples.
1) were prepared in SFM (50 μL). Positive control INTERFERin/FAM siRNA complex was used (mixture of 4 μL INTERFERin solution and 1.4 μL FAM siRNA, 20 min incubation). The negative control sample was FAM-siRNA only (1.4 μL). The KnT RNTs/siRNA complexes and controls were added to the wells. After incubation, the cells were washed with PBS (3 × 30 s) and incubated with LysoTracker Red DND-99 in SFM (1 mL, 1 μM) for 20 min at 37 °C. The cells were washed with PBS (3 × 30 s) before fixation with 4% paraformaldehyde for 10 min. One drop of DAPI mounting medium was deposited on the microscope slide and the coverslip was mounted on the slide. The cells were imaged using fluorescence (Olympus IX81) and confocal laser scanning (Zeiss LSM 710) microscopes. Image processing was done using Metamorph software for fluorescence imaging, Zeiss LSM software (ZEN) for confocal imaging. FITC (green), DAPI (blue), Cy3 (red) channels were used to observe FAM-labelled siRNA, DAPI-labelled nuclei and DND99-labelled acidic compartments respectively. Sliced images of cells (in z-direction) were taken in selected area using z-stack experiment on ZEN software and processed by Imaris software to form 3D visualization of the imaged area. The setting of FITC channel was first set by imaging the positive control sample and was kept constant when imaging RNTs/siRNA-treated or negative control samples.
Temperature-dependent endocytosis test
HCT116 cells were transfected with K15T RNTs/FAM siRNA for 2 h at 4 °C and 37 °C. The cells were fixed, stained with DAPI and imaged using fluorescence microscopy.
Cell uptake of KnT RNTs and siRNA complexes
The cells were treated with K1T–K15T RNTs/FAM siRNA for 24 h. The cells were fixed, stained with DAPI and imaged using fluorescence microscope.
Time-point study
HCT116 cells in 12 well-plates were treated with K5T, K10T and K15T RNTs/FAM siRNA for 3, 6, 12, 24, 36, 48, 72 and 96 h. Cells were labelled with DND99, fixed, stained with DAPI and imaged using fluorescence microscopies.
Cell viability study
HCT116 cells were seeded in 96-well plate (100 μL per well) for 24 h prior to transfection with RNTs. Cells were transfected with (A) K10T RNTs (0.25, 0.5, 1, 2 and 4 μM) for 24, 48, 72, 96 h, (B) K15T RNTs (0.002, 0.004, 0.008, 0.016, 0.031, 0.063, 0.125, 0.25, 0.5, 1 and 2 μM) for 48, 72 and 96 h and (C) K1T–K5T RNTs (0.75, 1.5, 3, 6 and 12 μM) RNTs for 24 h. At the end of the treatment, the medium from each well was discarded and cells were rinsed with PBS (2 × 30 s). Thiazolyl blue tetrazolium bromide in SFM (100 μL, 5 μg μL−1) was added to each well followed by 4 h of incubation at 37 °C. The media was then removed and DMSO (100 μL) was added to each well to dissolve the insoluble MTT formazan. Cell viability was determined by measuring the absorbance of each sample of the formazan product read at 570 nm using a microplate reader (AD-LD).
Proteins silencing by K3T RNTs
Luciferase experiment.
A549 and HCT116 cells were seeded in 6 or 12-well plate to 80–90% confluence for 24 h. The cells were then transfected for another 24 h with K3T RNTs/pGL3-siRNA complexes (molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) along with the positive control (INTERFERin/pGL3-siRNA) and negative control (pGL3-siRNA only). The medium was then discarded and all the cells were transfected with Lipofectamine/plasmids DNA complexes (1.6 μg of plasmids containing firefly pGL3 and Renilla-CMV in 10
1) along with the positive control (INTERFERin/pGL3-siRNA) and negative control (pGL3-siRNA only). The medium was then discarded and all the cells were transfected with Lipofectamine/plasmids DNA complexes (1.6 μg of plasmids containing firefly pGL3 and Renilla-CMV in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio w/w per well in 12-well plate) in SFM for an additional 24 h. Cells were washed with PBS (2 × 30 s) and lysed to collect proteins using Passive Lysis Buffer. Firefly, Renilla luminescent signals produced when exposing the lysed proteins to LAR II and Stop & Glow substrates respectively, were measured using FLUOstar Omega (BMG Labtech). Results were calculated as the ratio of signal produced by the targeted firefly luciferase to the control Renilla luciferase (FF/Renilla ratio).
1 ratio w/w per well in 12-well plate) in SFM for an additional 24 h. Cells were washed with PBS (2 × 30 s) and lysed to collect proteins using Passive Lysis Buffer. Firefly, Renilla luminescent signals produced when exposing the lysed proteins to LAR II and Stop & Glow substrates respectively, were measured using FLUOstar Omega (BMG Labtech). Results were calculated as the ratio of signal produced by the targeted firefly luciferase to the control Renilla luciferase (FF/Renilla ratio).
STAT3 silencing
After 24 h of seeding (50–60% confluence), cells were transfected with K3T RNTs/STAT3 siRNA (molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Cell lysis buffer (200 μL) was added to each well, followed by shaking of the plates on ice for 15 min. The lysed solutions were collected in the Eppendorf tubes and centrifuged (4 °C) for 15 min at 12
1). Cell lysis buffer (200 μL) was added to each well, followed by shaking of the plates on ice for 15 min. The lysed solutions were collected in the Eppendorf tubes and centrifuged (4 °C) for 15 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. BCA protein assay was performed to measure the concentration of collected proteins. The same amount of total protein for all lysed samples (20 to 25 μg) was resolved on SDS-PAGE (4% stacking gel, 12% running gel) at 150 V for 1 h. The gels were electro-blotted onto PVDF membranes. The membranes were blocked with 5% milk in TBST overnight at 4 °C, followed by washing with TBST (3 × 10 min). After incubation with primary antibody (STAT3 or GAPDH) at dilution of 1
000 rpm. BCA protein assay was performed to measure the concentration of collected proteins. The same amount of total protein for all lysed samples (20 to 25 μg) was resolved on SDS-PAGE (4% stacking gel, 12% running gel) at 150 V for 1 h. The gels were electro-blotted onto PVDF membranes. The membranes were blocked with 5% milk in TBST overnight at 4 °C, followed by washing with TBST (3 × 10 min). After incubation with primary antibody (STAT3 or GAPDH) at dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in 5% milk in TBST for 2 h at rt or overnight at 4 °C, the membranes were washed with TBST (3 × 10 min) and incubated with goat anti-mouse IgG HRP conjugated at a dilution of 1
1000 in 5% milk in TBST for 2 h at rt or overnight at 4 °C, the membranes were washed with TBST (3 × 10 min) and incubated with goat anti-mouse IgG HRP conjugated at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 for 1 h at rt or 2 h at 4 °C. Signals detected from the exposure to ECL Prime were collected onto CL-XPosure Film and measured with ImageJ software.
2000 for 1 h at rt or 2 h at 4 °C. Signals detected from the exposure to ECL Prime were collected onto CL-XPosure Film and measured with ImageJ software.
3. Results and discussion
Peptides containing up to 15 L-lysine residues were synthesized using solid-phase synthesis techniques and covalently attached to the G∧C aldehyde 5via a reductive amination reaction to produce the protecting group-free oligo-lysine twin G∧C compounds KnT (Fig. 1). The lysine containing twin-G∧C motifs self-assembled readily in solution to form RNTs through hydrogen bonding and π–π stacking interactions (Fig. 2). The presence of the lysine chains on the periphery of the RNTs resulted into cationic RNTs that were investigated as potential nanocarriers for siRNA delivery for gene silencing.
 |
| | Fig. 1 Synthetic scheme for preparation of KnT. | |
 |
| | Fig. 2 Lysine functionalized twin-G∧C motif K1T (A). Hierarchical self-assembly of K1T into double-stacked hexameric rosettes (B) which then further π–π stack to form RNTs (C). | |
The self-assembly of the KnT RNTs was investigated using scanning electron microscopy (SEM) (Fig. 3). While the K1T–K9T modules formed RNTs in unbuffered water, a decrease in the degree of self-assembly (reflected by shorter RNTs and lower abundance), was observed with an increase in the number of lysine residues per motif (Fig. S2†). It was therefore apparent that as the steric bulk and net-charged on the peptides was increased, formation of the nanostructures became more challenging and resulted into shorter and less abundant nanotubes. K10T–K15T did not produce RNTs in unbuffered water, but self-assembled at higher pH.27
 |
| | Fig. 3 SEM images of RNTs in unbuffered water: (A) K1T (1 × 10−4 M), (B) K4T (1.9 × 10−5 M) and (C) K5T (5.1 × 10−5 M). Scale bar = 500 nm. | |
Naked siRNA shows poor cellular uptake and degrades rapidly, due to which siRNA delivery systems are required to improve its stability, delivery to targeted site and cellular uptake. KnT RNTs have a cationic surface due to the presence of lysine side chains, which can bind non-covalently to siRNA via electrostatic interactions. Agarose gel retardation assay was used to measure the amount of unbound siRNA in SFM (Fig. 4) and it was found to have a higher binding ability as molar ratio of RNTs to siRNA was increased. In addition, with an increase in lysine oligomerization, the net charge on the RNTs increased, leading to a lower molar ratio of RNTs to siRNA for siRNA capture. A similar trend was observed with PBS buffer (Fig. S3†). These observations strongly indicate the role of the lysine residues and the corresponding cationic charges on G∧C molecules on electrostatic binding with siRNA.
 |
| | Fig. 4 Binding study of KnT RNTs and siRNA in SFM (molar ratio of 0.5, 1, 2.5, 5, 10, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) using agarose gel retardation assay, for KnT RNTs (n = 1–5 (A), n = 6–10 (B) and n = 11–15 (C)). 1) using agarose gel retardation assay, for KnT RNTs (n = 1–5 (A), n = 6–10 (B) and n = 11–15 (C)). | |
Protection of siRNA by KnT RNTs by serum degradation
In the physiological environment, unprotected siRNA undergoes serum degradation due to the presence of RNase. It is therefore important that RNTs bind strongly to the siRNA so that they are not displaced by polyanion competitors, thus preventing siRNA from being digested by RNase in the serum. We examined the protective ability of KnT RNTs by measuring the stability of the RNTs/siRNA complexes in fetal bovine serum (FBS) environment containing nuclease. The RNTs/siRNA complexes (molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were incubated in 10% FBS for 24 h at 37 °C (Fig. 5). It was found that K1T–K4T RNTs protected less than 40% of siRNA during the serum incubation. This could possibly be due to the low siRNA loading ability and the ease of siRNA displacement from K1T–K4T RNTs, leading to serum degradation of naked siRNA. On the other hand, K5T–K15T RNTs demonstrated good protective ability of siRNA against serum degradation with greater than 55% of siRNA recovered. For instance, K9T–K15T RNTs were able to protect more than 80% of siRNA during the 24 h of serum incubation. Improvement in protective ability from K1T–K15T RNTs against serum degradation of siRNA is likely due to the increase in electrostatic interaction in the RNTs/siRNA system. The high cationic charges on the RNTs create stable complexes with siRNA, thus preventing the displacement of the latter by negatively charged competitors and the nuclease access to the siRNA. The enhanced binding and protective ability of siRNA by KnT RNTs indicate that these RNTs can act as siRNA carrier, protecting them during delivery in the physiological environment. The condition chosen showed was fully capable of digesting the amount of siRNA used in this experiment (Fig. S4†).
1) were incubated in 10% FBS for 24 h at 37 °C (Fig. 5). It was found that K1T–K4T RNTs protected less than 40% of siRNA during the serum incubation. This could possibly be due to the low siRNA loading ability and the ease of siRNA displacement from K1T–K4T RNTs, leading to serum degradation of naked siRNA. On the other hand, K5T–K15T RNTs demonstrated good protective ability of siRNA against serum degradation with greater than 55% of siRNA recovered. For instance, K9T–K15T RNTs were able to protect more than 80% of siRNA during the 24 h of serum incubation. Improvement in protective ability from K1T–K15T RNTs against serum degradation of siRNA is likely due to the increase in electrostatic interaction in the RNTs/siRNA system. The high cationic charges on the RNTs create stable complexes with siRNA, thus preventing the displacement of the latter by negatively charged competitors and the nuclease access to the siRNA. The enhanced binding and protective ability of siRNA by KnT RNTs indicate that these RNTs can act as siRNA carrier, protecting them during delivery in the physiological environment. The condition chosen showed was fully capable of digesting the amount of siRNA used in this experiment (Fig. S4†).
 |
| | Fig. 5 Protective ability of siRNA by KnT RNTs (n = 1–15) in presence of FBS (+FBS) compared to the control of siRNA without FBS (−FBS). | |
Time-study on delivery of siRNA by RNTs
A continuous correlation in the binding, protection and cellular delivery of siRNA was observed with increasing number of lysine residues per monomer (Fig. 4, 5 and Fig. S5†). Since the performance of the adjacent KnT RNTs were fairly similar, only K5T, K10T and K15T RNTs were selected for the fluorescent (Fig. 6) and confocal imaging (Fig. 7) experiments. The cellular uptake rate of KnT RNTs/siRNA was investigated by delivery of RNTs/siRNA complexes to HCT116 cells at various time intervals (Fig. 6 and Fig. S5†). The siRNA was fluorescently labelled (FAM-siRNA) to allow visualization. Fig. 6 shows the fluorescence imaging on K5T, K10T and K15T RNTs/siRNA complexes at various time intervals (3, 12, 24, 48 and 96 h) and it was found that stronger green fluorescent signals due to siRNA in the cells were observed for K10T RNTs and K15T RNTs as compared to K5T RNTs. A similar trend was observed from K1T–K15T RNTs (Fig. S5†), reiterating the importance of cationic charges of KnT RNTs on the siRNA loading ability. This was in line with the binding study data where K10T and K15T RNTs captured 100% of siRNA compared to 60% of siRNA by K5T RNTs at molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 4). Moreover, increasing net charges among the KnT RNTs series demonstrated enhanced interaction with the negatively charged proteoglycans on the cellular membrane, resulting into the fastest cellular uptake for K15T RNTs/siRNA. While both K10T and K15T RNTs showed high siRNA fluorescent signal within 3 h of cell transfection and complete cell transfection within 24 h, K5T RNTs had a slower cell transfection rate. It was also observed that the green fluorescent signal of siRNA spread to the cytoplasm of the cells over time, indicating release or displacement of the siRNA from KnT RNTs/siRNA complexes. Based on these results, we propose that the length of the RNTs formed and the size of their complexes with siRNA impact the cellular uptake. As seen in Fig. S1,† the length of the RNTs formed decreases with an increase in the steric bulk due to oligomerization of lysine. Consequently, the complexes formed smaller and more dispersed aggregates, facilitating internalization.
1 (Fig. 4). Moreover, increasing net charges among the KnT RNTs series demonstrated enhanced interaction with the negatively charged proteoglycans on the cellular membrane, resulting into the fastest cellular uptake for K15T RNTs/siRNA. While both K10T and K15T RNTs showed high siRNA fluorescent signal within 3 h of cell transfection and complete cell transfection within 24 h, K5T RNTs had a slower cell transfection rate. It was also observed that the green fluorescent signal of siRNA spread to the cytoplasm of the cells over time, indicating release or displacement of the siRNA from KnT RNTs/siRNA complexes. Based on these results, we propose that the length of the RNTs formed and the size of their complexes with siRNA impact the cellular uptake. As seen in Fig. S1,† the length of the RNTs formed decreases with an increase in the steric bulk due to oligomerization of lysine. Consequently, the complexes formed smaller and more dispersed aggregates, facilitating internalization.
 |
| | Fig. 6 Fluorescence imaging for K5T, K10T, K15T RNTs complexes with siRNA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) at 3, 12, 24, 48 and 96 h. 1 molar ratio) at 3, 12, 24, 48 and 96 h. | |
 |
| | Fig. 7 Confocal image of HCT116 cells transfected with K5T, K10T and K15T RNTs/FAM-siRNA for 0.5, 2 and 6 h. | |
Confocal imaging was used to observe the trafficking of KnT RNTs/siRNA by the cells and monitor the uptake mechanism of the complexes. The confocal images (Fig. 7) were in agreement with fluorescent images, indicating the fastest intracellular delivery for K15T RNTs. The yellow fluorescent dots in the confocal images represent the co-localization of FAM-siRNA (green) and endosomes (red), which suggest that KnT RNTs/siRNA complexes were being internalized by the cells via the endocytosis pathway. As expected, cells transfected with KnT RNTs/siRNA showed higher in fluorescent signals compared to the positive and negative controls (Fig. S6 and S7†). Furthermore, the 3D image of HCT116 cells transfected with K10T RNTs/FAM-siRNA showed green fluorescent dots in the cytoplasm of the cells (Fig. S8†), supporting the endosomal escape of siRNA to the cytosol.
Endocytosis test
Temperature-dependent endocytosis assay was conducted to gain more insight on the uptake mechanism of KnT RNTs/siRNA complexes by HCT116 cells. Since K15T RNTs showed the fastest cellular uptake, it was selected for the endocytosis test. This also ensured that cell lysis was minimized for this temperature sensitive study. HCT116 cells were transfected with K15T RNTs/FAM-siRNA at low (4 °C) and physiological (37 °C) temperatures (Fig. 8). As expected, negligible fluorescent signal of siRNA was found at 4 °C in the cells since endocytosis processes are inhibited at low temperature. However, for cells incubated at 37 °C, a significant increase in intracellular uptake of the complexes was observed, which was reflected by the green region within the cells. The temperature-dependent assay strongly suggests that endocytosis pathway is the main mechanism for the uptake of KnT RNTs/siRNA complexes by the cells.
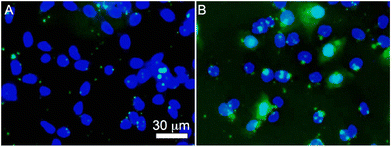 |
| | Fig. 8 Endocytosis assay of HCT116 cells treated with K15T RNTs/siRNA complexes at (A) 4 °C and (B) 37 °C for 2 h. | |
K1T–K5T RNTs/siRNA complexation
The higher molecular weight KnT RNTs with elevated charges (K10T–K15T) showed rapid cellular uptake at molar ratio of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 6 and 7). However, this may not be beneficial in vivo for siRNA due to their short circulation half-life. An increase in the circulation half-life of the delivery system in serum is desired to optimise the targeted cargo delivery and evade renal clearance and lower the toxicity of the carrier complexes.14 The higher molecular weight KnT RNTs could be better suited for the delivery of larger biomolecules such as mRNA or plasmid DNA. As such, the binding ability of K1T–K5T to siRNA was investigated using gel retardation assay and the binding interaction of siRNA was improved by increasing the molar ratio up to 50
1 (Fig. 6 and 7). However, this may not be beneficial in vivo for siRNA due to their short circulation half-life. An increase in the circulation half-life of the delivery system in serum is desired to optimise the targeted cargo delivery and evade renal clearance and lower the toxicity of the carrier complexes.14 The higher molecular weight KnT RNTs could be better suited for the delivery of larger biomolecules such as mRNA or plasmid DNA. As such, the binding ability of K1T–K5T to siRNA was investigated using gel retardation assay and the binding interaction of siRNA was improved by increasing the molar ratio up to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 due to their lower cationic charges on the RNTs (Fig. 9). Complete binding of siRNA at 50
1 due to their lower cationic charges on the RNTs (Fig. 9). Complete binding of siRNA at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio was observed for K3T, K4T and K5T RNTs. Based on these results, K3T was chosen for further investigation since it was the best potential nanocarrier for siRNA delivery due to its lower cationic charge (compared to K4T and K5T) and molar ratio used for complexation (compared to K1T and K2T).
1 molar ratio was observed for K3T, K4T and K5T RNTs. Based on these results, K3T was chosen for further investigation since it was the best potential nanocarrier for siRNA delivery due to its lower cationic charge (compared to K4T and K5T) and molar ratio used for complexation (compared to K1T and K2T).
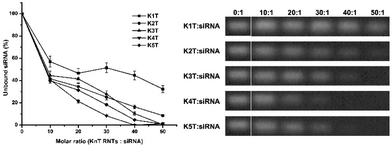 |
| | Fig. 9 Binding study of K1T–K5T RNTs to siRNA in SFM (molar ratio of 0.5, 10, 20, 30, 40, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) using agarose gel retardation assay. 1) using agarose gel retardation assay. | |
Characterization of K3T RNTs/siRNA complexation
While the data, so far, supported the complexation of the RNTs and siRNA, AFM and TEM were used to confirm the complexation and morphology of the bioconjugates at the nanoscale level using K3T–siRNA complex. AFM measurement showed the height profile of 4.1 ± 0.2 nm, 1.8 nm ± 0.2 nm and 5.6 ± 0.4 nm for K3T RNTs, siRNA and K3T RNTs/siRNA complex, respectively (Fig. 10). The morphology changes as well as the increased height clearly implied that negatively charged siRNA were adhering onto the surface of the RNTs. A similar trend was observed for TEM imaging with diameters of 4.3 ± 0.1 nm and 6.8 ± 0.8 nm for K3T RNTs and K3T RNTs/siRNA complex, respectively. The average height values from AFM corresponds to the diameter of the materials and these values were lower than the values obtained from the TEM measurements. RNTs are soft materials and when AFM measurements are carried out sample deformation from substrate compression and interactions with the AFM tip can lead to flattening of the tubes, thus resulting in an inferior value.41 In addition, the TEM images clearly reveal deposition of siRNA (dark spots) onto the nanotube's surface (Fig. 11).
 |
| | Fig. 10 AFM images of (A) K3T RNTs, (B) siRNA (C) K3T RNTs in SFM and (D) K3T–siRNA complex. | |
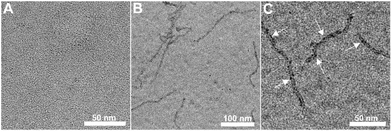 |
| | Fig. 11 TEM images: (A) siRNA only, (B) K3T RNTs/siRNA complexes at molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and (C) a magnified image of K3T RNTs/siRNA complexes at molar ratio of 50 1 and (C) a magnified image of K3T RNTs/siRNA complexes at molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, with arrows pointing at siRNA on the surface of the RNTs. 1, with arrows pointing at siRNA on the surface of the RNTs. | |
Transfection study by confocal microscopy for K3T RNTs/siRNA complexes
Confocal microscopy was used to visualize the uptake of K3T RNTs/FAM siRNA complexes by HCT116 cells at 5 and 10 h time-points. K3T RNTs/FAM-siRNA showed low cellular uptake within 5 h, which increased significantly in FAM fluorescent signal within 10 h (Fig. 12). The 3D images of K3T RNTs/FAM-siRNA complexes showed yellowish bright dots representing the overlapping between green and red fluorescence of siRNA and endosome, respectively. This further supported the proposed endocytic uptake pathway of KnT RNTs/siRNA by HCT116 cells.
 |
| | Fig. 12 Confocal images of HCT116 cells transfected with K3T RNTs/FAM-siRNA at (A) 5 h, (B) 10 h showing each channel separately and merged images overlay with differential interference contrast channel. (C) 3D image of cells at 10 h of transfection. | |
Gene silencing experiments
The ultimate goal for using RNTs as siRNA delivery system is to allow the delivered siRNA to enter the RNAi pathway for inhibition of targeted genes. Luciferase assay is a popular method that provides precise measurement of protein level based on the luminescent signals produced by luciferase enzyme activity. Dual luciferase assay was used where A549 and HCT116 cells were transfected with firefly (FF) and Renilla luciferase vectors by Lipofectamine-2000. The silencing was calculated by normalizing the enzyme activity of the targeted firefly luciferase to the activity of the control Renilla luciferase. For both cell lines, pGL3-siRNA delivered was shown to successfully function inside the cells as the signal produced by firefly luciferase was significantly reduced (Fig. 13A). The silencing effect from K3T RNTs/pGL3-siRNA treatment was compared to the negative control (naked pGL3-siRNA) and found to be 67 ± 20% for A549 cells and 72 ± 10% for HCT116 cells. The positive control of INTERFERin/pGL3-siRNA gave similar result of 74 ± 13% for A549 cells and 61 ± 10% for HCT116. This showed effective delivery of siRNA by K3T RNTs resulting in excellent silencing ability that was comparable to a commercial siRNA transfecting reagent (INTERFERin). In addition, a high toxicity of the positive control, INTERFERin, was observed compared to K3T RNTs system (Fig. S9 and S10†). The biocompatibility of KnT RNTs is essential for therapeutic application since the carrier system should not lead to non-specific cell death.
 |
| | Fig. 13 Cells transfected with K3T RNTs/siRNA (molar ratio 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, siRNA = 100 nM). (A) Luciferase assay of the K3T RNTs/pGL3 siRNA-treated samples compared with positive control (INTERFERin/pGL3 siRNA), and negative control (pGL3-siRNA only) in two cell lines A549 and HCT116. (B) Western blot detecting stat3 protein level in HCT116 cells after transfection with K3T RNTs/stat3-siRNA. 1, siRNA = 100 nM). (A) Luciferase assay of the K3T RNTs/pGL3 siRNA-treated samples compared with positive control (INTERFERin/pGL3 siRNA), and negative control (pGL3-siRNA only) in two cell lines A549 and HCT116. (B) Western blot detecting stat3 protein level in HCT116 cells after transfection with K3T RNTs/stat3-siRNA. | |
Stat3 protein silencing
Stat3 protein regulates gene expression in the cell nucleus and plays an important role in the growth and progression of cancer cells. The inhibition of stat3 gene is widely considered for cancer therapy, as this protein can upregulate other proteins expression and stimulate tumor angiogenesis and survival.42,43 Therefore, targeting endogenous protein such as stat3 protein is a preliminary step to examine the silencing efficiency of siRNA delivered by RNTs. HCT116 cells were treated with K3T RNTs/stat3 siRNA complexes at molar ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and INTERFERin/stat3 siRNA served as the positive control. Silencing from the siRNA delivered by K3T RNTs (35%) was comparable to the result obtained for the positive control (40%) (Fig. 13B). Similar silencing effect between K3T RNTs and INTERFERin was observed in firefly luciferase assay, which indicated the highest silencing effect was achieved in the treatment.
1 and INTERFERin/stat3 siRNA served as the positive control. Silencing from the siRNA delivered by K3T RNTs (35%) was comparable to the result obtained for the positive control (40%) (Fig. 13B). Similar silencing effect between K3T RNTs and INTERFERin was observed in firefly luciferase assay, which indicated the highest silencing effect was achieved in the treatment.
4. Conclusions
A series of fifteen lysine functionalized synthetic hybrid DNA bases (KnT) were synthesized which self-assembled into RNTs in physiological conditions. The KnT RNTs were able to form bioconjugates with siRNA via electrostatic interactions, which protected the siRNA against serum degradation. The number of lysine residues on the G∧C motif and the length of the RNTs formed highly impacted the uptake rate by the cells. This could be utilized to vary the circulation time of the bioconjugates by varying the KnT building block used. Along with the very low toxicity, the KnT nanomaterials have high delivery efficiency that can lead to significant inhibition of the target genes, and are therefore suitable for biological applications. We are currently exploring these materials for in vivo studies and mRNA delivery. The in vitro findings presented in this work would be used to fine tune the binding and delivery of siRNA, given that other biological factors can interfere with the circulation half-line and binding affinity of the RNTs and siRNA in in vivo testing.
Author contributions
The project was conceptualized by H. F. and supervised by H. F. and U. D. H. The compounds and RNTs were prepared by M. E.-B. All biological assays, bioconjugation experiments and confocal microscopy were conducted by U. H. and A. A. Electron microscopy and AFM was conducted by J.-Y. C. Molecular modeling was carried out by T. Y. The initial draft of the manuscript was written by U. H. and was reviewed and edited by all contributing authors.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgements
This work was funded by the National Research Council of Canada, a Federal Research and Development Organization of the Government of Canada, NSERC, and the University of Alberta.
References
- C. Napoli, C. Lemieux and R. Jorgensen, Plant Cell, 1990, 279–289 CAS.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- W. Ho, X.-Q. Zhang and X. Xu, Adv. Healthcare Mater., 2016, 5, 2715–2731 CrossRef CAS PubMed.
- D. Bumcrot, M. Manoharan, V. Koteliansky and D. W. Y. Sah, Nat. Chem. Biol., 2006, 2, 711–719 CrossRef CAS PubMed.
- A. de Fougerolles, H.-P. Vornlocher, J. Maraganore and J. Lieberman, Nat. Rev. Drug Discovery, 2007, 6, 443–453 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- R. L. Setten, J. J. Rossi and S.-p. Han, Nat. Rev. Drug Discovery, 2019, 18, 421–446 CrossRef CAS PubMed.
- P. D. Zamore, T. Tuschl, P. A. Sharp and D. P. Bartel, Cell, 2000, 101, 25–33 CrossRef CAS PubMed.
- G. Meister and T. Tuschl, Nature, 2004, 431, 343–349 CrossRef CAS PubMed.
- R. W. Carthew and E. J. Sontheimer, Cell, 2009, 136, 642–655 CrossRef CAS PubMed.
- J. Liu, M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J.-J. Song, S. M. Hammond, L. Joshua-Tor and G. J. Hannon, Science, 2004, 305, 1437–1441 CrossRef CAS PubMed.
- D. Haussecker, Mol. Ther. – Nucleic Acids, 2012, 2, e8 CrossRef PubMed.
- G. S. Mack, Nat. Biotechnol., 2007, 25, 631–638 CrossRef CAS PubMed.
- J. Wang, Z. Lu, M. G. Wientjes and J. L. S. Au, AAPS J., 2010, 12, 492–503 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- P. Ghosh, G. Han, M. De, C. Kim and V. Rotello, Adv. Drug Delivery Rev., 2008, 60, 1307–1315 CrossRef CAS PubMed.
- M. E. Gindy, A. M. Leone and J. J. Cunningham, Expert Opin. Drug Delivery, 2012, 9, 171–182 CrossRef CAS PubMed.
- S. Zhang, B. Zhao, H. Jiang, B. Wang and B. Ma, J. Controlled Release, 2007, 123, 1–10 CrossRef CAS PubMed.
- N. W. S. Kam, Z. Liu and H. Dai, J. Am. Chem. Soc., 2005, 127, 12492–12493 CrossRef CAS PubMed.
- R. Krajcik, A. Jung, A. Hirsch, W. Neuhuber and O. Zolk, Biochem. Biophys. Res. Commun., 2008, 369, 595–602 CrossRef CAS PubMed.
- D. L. Kirkpatrick, M. Weiss, A. Naumov, G. Bartholomeusz, R. B. Weisman and O. Gliko, Materials, 2012, 5, 278–301 CrossRef CAS PubMed.
- U. C. Nygaard, J. S. Hansen, M. Samuelsen, T. Alberg, C. D. Marioara and M. Løvik, Toxicol. Sci., 2009, 109, 113–123 CrossRef CAS PubMed.
- S.-T. Yang, J. Luo, Q. Zhou and H. Wang, Theranostics, 2012, 2, 271–282 CrossRef CAS PubMed.
- S. F. Hansen and A. Lennquist, Nat. Nanotechnol., 2020, 15, 3–4 CrossRef CAS PubMed.
- H. Fenniri, P. Mathivanan, K. L. Vidale, D. M. Sherman, K. Hallenga, K. V. Wood and J. G. Stowell, J. Am. Chem. Soc., 2001, 123, 3854–3855 CrossRef CAS PubMed.
- J. G. Moralez, J. Raez, T. Yamazaki, R. K. Motkuri, A. Kovalenko and H. Fenniri, J. Am. Chem. Soc., 2005, 127, 8307–8309 CrossRef CAS PubMed.
- U. D. Hemraz, M. El-Bakkari, T. Yamazaki, J.-Y. Cho, R. L. Beingessner and H. Fenniri, Nanoscale, 2014, 6, 9421–9427 RSC.
- R. S. Johnson, T. Yamazaki, A. Kovalenko and H. Fenniri, J. Am. Chem. Soc., 2007, 129, 5735–5743 CrossRef CAS PubMed.
- M. L. Puzan, B. Legesse, R. A. Koppes, H. Fenniri and A. N. Koppes, ACS Biomater. Sci. Eng., 2018, 4, 1630–1640 CAS.
- L. Zhang, U. D. Hemraz, H. Fenniri and T. J. Webster, J. Biomed. Mater. Res., Part A, 2010, 95A, 550–563 CrossRef CAS PubMed.
- A. Childs, U. D. Hemraz, N. J. Castro, H. Fenniri and L. G. Zhang, Biomed. Mater., 2013, 8(065003), 1–12 Search PubMed.
- W. S. Journeay, S. S. Suri, J. G. Moralez, H. Fenniri and B. Singh, Small, 2009, 5, 1446–1452 CrossRef CAS PubMed.
- W. S. Journeay, S. S. Suri, J. G. Moralez, H. Fenniri and B. Singh, Int. J. Nanomed., 2008, 3, 373–383 CAS.
- A. L. Chun, J. G. Moralez, T. J. Webster and H. Fenniri, Biomaterials, 2005, 26, 7304–7309 CrossRef CAS PubMed.
- A. L. Chun, J. G. Moralez, H. Fenniri and T. J. Webster, Nanotechnology, 2004, 15, S234–S239 CrossRef CAS.
- L. Zhang, F. Rakotondradany, A. J. Myles, H. Fenniri and T. J. Webster, Biomaterials, 2009, 30, 1309–1320 CrossRef CAS PubMed.
- L. Sun, L. Zhang, U. D. Hemraz, H. Fenniri and T. J. Webster, Tissue Eng., Part A, 2012, 18, 1741–1750 CrossRef CAS PubMed.
- L. Sun, D. Li, U. D. Hemraz, H. Fenniri and T. J. Webster, J. Biomed. Mater. Res., Part A, 2014, 102A, 3446–3451 CrossRef CAS PubMed.
- J.-Y. Cho, P. Bhowmik, P. L. Polowick, S. G. Dodard, M. El-Bakkari, G. Nowak, H. Fenniri and U. D. Hemraz, ACS Omega, 2020, 5, 24422–24433 CrossRef CAS PubMed.
- S. Hrapovic, C. F. Martinez-Farina, J. Sui, J.-D. Lavertu and U. D. Hemraz, Carbohydr. Polym., 2022, 298, 120108 CrossRef CAS PubMed.
- J. A. DeRose and J. P. Revel, Thin Solid Films, 1998, 331, 194–202 CrossRef CAS.
- K. Al Zaid Siddiquee and J. Turkson, Cell Res., 2008, 18, 254–267 CrossRef CAS PubMed.
- H. Yu, D. Pardoll and R. Jove, Nat. Rev. Cancer, 2009, 9, 798–809 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *bd and
Hicham
Fenniri
*abe
*bd and
Hicham
Fenniri
*abe
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA) by gel retardation assay and the resulting KnT RNTs/siRNA complexes were subjected to nucleases in bovine serum to determine the protective ability of KnT RNTs against serum degradation. Fluorescence and confocal microscopies were used to visualize the trafficking of KnT RNTs/FAM-labelled siRNA to the cells. The ability of the siRNA delivered to knockdown the targeted gene expression was investigated using firefly luciferase and stat3 protein assays.
siRNA) by gel retardation assay and the resulting KnT RNTs/siRNA complexes were subjected to nucleases in bovine serum to determine the protective ability of KnT RNTs against serum degradation. Fluorescence and confocal microscopies were used to visualize the trafficking of KnT RNTs/FAM-labelled siRNA to the cells. The ability of the siRNA delivered to knockdown the targeted gene expression was investigated using firefly luciferase and stat3 protein assays.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 acetic anhydride/pyridine (5 mL, 1 × 10 min and 2 × 20 min) to cap the unreactive hydroxyl groups. The resin was subsequently filtered and washed with (3 × 10 mL) DMF, DCM and MeOH and dried under vacuum. The substitution degree (0.52 mmol g−1) was determined by spectroscopic quantification of the fulvene-piperidine adduct at 301 nm.
50 acetic anhydride/pyridine (5 mL, 1 × 10 min and 2 × 20 min) to cap the unreactive hydroxyl groups. The resin was subsequently filtered and washed with (3 × 10 mL) DMF, DCM and MeOH and dried under vacuum. The substitution degree (0.52 mmol g−1) was determined by spectroscopic quantification of the fulvene-piperidine adduct at 301 nm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) were dissolved in nuclease-free water (1 mg mL−1, 0.07 M). KnT RNTs were mixed with siRNA (0.1 nmol, 1.4 μL) at various molar ratios (0.5, 1, 2.5, 5, 10 and 20
000 g mol−1) were dissolved in nuclease-free water (1 mg mL−1, 0.07 M). KnT RNTs were mixed with siRNA (0.1 nmol, 1.4 μL) at various molar ratios (0.5, 1, 2.5, 5, 10 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, RNTs to siRNA respectively) in 10 μL of serum-free medium (SFM). The KnT RNTs/siRNA complexes were incubated for 30 min at room temperature prior to further experimental study.
1, RNTs to siRNA respectively) in 10 μL of serum-free medium (SFM). The KnT RNTs/siRNA complexes were incubated for 30 min at room temperature prior to further experimental study.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were prepared in PBS buffer. FBS was added to each solution to obtain final concentrations of 10% FBS. The RNTs/siRNA mixture was incubated in a 37 °C shaker for 24 h, followed by addition of heparin (5 μL, 20 μg μL−1) for 1 h at room temperature to displace siRNA from the RNTs. Agarose gel experiment was performed to detect the intact siRNA protected by RNTs from serum degradation.
1) were prepared in PBS buffer. FBS was added to each solution to obtain final concentrations of 10% FBS. The RNTs/siRNA mixture was incubated in a 37 °C shaker for 24 h, followed by addition of heparin (5 μL, 20 μg μL−1) for 1 h at room temperature to displace siRNA from the RNTs. Agarose gel experiment was performed to detect the intact siRNA protected by RNTs from serum degradation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were prepared in SFM (50 μL). Positive control INTERFERin/FAM siRNA complex was used (mixture of 4 μL INTERFERin solution and 1.4 μL FAM siRNA, 20 min incubation). The negative control sample was FAM-siRNA only (1.4 μL). The KnT RNTs/siRNA complexes and controls were added to the wells. After incubation, the cells were washed with PBS (3 × 30 s) and incubated with LysoTracker Red DND-99 in SFM (1 mL, 1 μM) for 20 min at 37 °C. The cells were washed with PBS (3 × 30 s) before fixation with 4% paraformaldehyde for 10 min. One drop of DAPI mounting medium was deposited on the microscope slide and the coverslip was mounted on the slide. The cells were imaged using fluorescence (Olympus IX81) and confocal laser scanning (Zeiss LSM 710) microscopes. Image processing was done using Metamorph software for fluorescence imaging, Zeiss LSM software (ZEN) for confocal imaging. FITC (green), DAPI (blue), Cy3 (red) channels were used to observe FAM-labelled siRNA, DAPI-labelled nuclei and DND99-labelled acidic compartments respectively. Sliced images of cells (in z-direction) were taken in selected area using z-stack experiment on ZEN software and processed by Imaris software to form 3D visualization of the imaged area. The setting of FITC channel was first set by imaging the positive control sample and was kept constant when imaging RNTs/siRNA-treated or negative control samples.
1) were prepared in SFM (50 μL). Positive control INTERFERin/FAM siRNA complex was used (mixture of 4 μL INTERFERin solution and 1.4 μL FAM siRNA, 20 min incubation). The negative control sample was FAM-siRNA only (1.4 μL). The KnT RNTs/siRNA complexes and controls were added to the wells. After incubation, the cells were washed with PBS (3 × 30 s) and incubated with LysoTracker Red DND-99 in SFM (1 mL, 1 μM) for 20 min at 37 °C. The cells were washed with PBS (3 × 30 s) before fixation with 4% paraformaldehyde for 10 min. One drop of DAPI mounting medium was deposited on the microscope slide and the coverslip was mounted on the slide. The cells were imaged using fluorescence (Olympus IX81) and confocal laser scanning (Zeiss LSM 710) microscopes. Image processing was done using Metamorph software for fluorescence imaging, Zeiss LSM software (ZEN) for confocal imaging. FITC (green), DAPI (blue), Cy3 (red) channels were used to observe FAM-labelled siRNA, DAPI-labelled nuclei and DND99-labelled acidic compartments respectively. Sliced images of cells (in z-direction) were taken in selected area using z-stack experiment on ZEN software and processed by Imaris software to form 3D visualization of the imaged area. The setting of FITC channel was first set by imaging the positive control sample and was kept constant when imaging RNTs/siRNA-treated or negative control samples.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) along with the positive control (INTERFERin/pGL3-siRNA) and negative control (pGL3-siRNA only). The medium was then discarded and all the cells were transfected with Lipofectamine/plasmids DNA complexes (1.6 μg of plasmids containing firefly pGL3 and Renilla-CMV in 10
1) along with the positive control (INTERFERin/pGL3-siRNA) and negative control (pGL3-siRNA only). The medium was then discarded and all the cells were transfected with Lipofectamine/plasmids DNA complexes (1.6 μg of plasmids containing firefly pGL3 and Renilla-CMV in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio w/w per well in 12-well plate) in SFM for an additional 24 h. Cells were washed with PBS (2 × 30 s) and lysed to collect proteins using Passive Lysis Buffer. Firefly, Renilla luminescent signals produced when exposing the lysed proteins to LAR II and Stop & Glow substrates respectively, were measured using FLUOstar Omega (BMG Labtech). Results were calculated as the ratio of signal produced by the targeted firefly luciferase to the control Renilla luciferase (FF/Renilla ratio).
1 ratio w/w per well in 12-well plate) in SFM for an additional 24 h. Cells were washed with PBS (2 × 30 s) and lysed to collect proteins using Passive Lysis Buffer. Firefly, Renilla luminescent signals produced when exposing the lysed proteins to LAR II and Stop & Glow substrates respectively, were measured using FLUOstar Omega (BMG Labtech). Results were calculated as the ratio of signal produced by the targeted firefly luciferase to the control Renilla luciferase (FF/Renilla ratio).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Cell lysis buffer (200 μL) was added to each well, followed by shaking of the plates on ice for 15 min. The lysed solutions were collected in the Eppendorf tubes and centrifuged (4 °C) for 15 min at 12
1). Cell lysis buffer (200 μL) was added to each well, followed by shaking of the plates on ice for 15 min. The lysed solutions were collected in the Eppendorf tubes and centrifuged (4 °C) for 15 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. BCA protein assay was performed to measure the concentration of collected proteins. The same amount of total protein for all lysed samples (20 to 25 μg) was resolved on SDS-PAGE (4% stacking gel, 12% running gel) at 150 V for 1 h. The gels were electro-blotted onto PVDF membranes. The membranes were blocked with 5% milk in TBST overnight at 4 °C, followed by washing with TBST (3 × 10 min). After incubation with primary antibody (STAT3 or GAPDH) at dilution of 1
000 rpm. BCA protein assay was performed to measure the concentration of collected proteins. The same amount of total protein for all lysed samples (20 to 25 μg) was resolved on SDS-PAGE (4% stacking gel, 12% running gel) at 150 V for 1 h. The gels were electro-blotted onto PVDF membranes. The membranes were blocked with 5% milk in TBST overnight at 4 °C, followed by washing with TBST (3 × 10 min). After incubation with primary antibody (STAT3 or GAPDH) at dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in 5% milk in TBST for 2 h at rt or overnight at 4 °C, the membranes were washed with TBST (3 × 10 min) and incubated with goat anti-mouse IgG HRP conjugated at a dilution of 1
1000 in 5% milk in TBST for 2 h at rt or overnight at 4 °C, the membranes were washed with TBST (3 × 10 min) and incubated with goat anti-mouse IgG HRP conjugated at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 for 1 h at rt or 2 h at 4 °C. Signals detected from the exposure to ECL Prime were collected onto CL-XPosure Film and measured with ImageJ software.
2000 for 1 h at rt or 2 h at 4 °C. Signals detected from the exposure to ECL Prime were collected onto CL-XPosure Film and measured with ImageJ software.



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) using agarose gel retardation assay, for KnT RNTs (n = 1–5 (A), n = 6–10 (B) and n = 11–15 (C)).
1) using agarose gel retardation assay, for KnT RNTs (n = 1–5 (A), n = 6–10 (B) and n = 11–15 (C)).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were incubated in 10% FBS for 24 h at 37 °C (Fig. 5). It was found that K1T–K4T RNTs protected less than 40% of siRNA during the serum incubation. This could possibly be due to the low siRNA loading ability and the ease of siRNA displacement from K1T–K4T RNTs, leading to serum degradation of naked siRNA. On the other hand, K5T–K15T RNTs demonstrated good protective ability of siRNA against serum degradation with greater than 55% of siRNA recovered. For instance, K9T–K15T RNTs were able to protect more than 80% of siRNA during the 24 h of serum incubation. Improvement in protective ability from K1T–K15T RNTs against serum degradation of siRNA is likely due to the increase in electrostatic interaction in the RNTs/siRNA system. The high cationic charges on the RNTs create stable complexes with siRNA, thus preventing the displacement of the latter by negatively charged competitors and the nuclease access to the siRNA. The enhanced binding and protective ability of siRNA by KnT RNTs indicate that these RNTs can act as siRNA carrier, protecting them during delivery in the physiological environment. The condition chosen showed was fully capable of digesting the amount of siRNA used in this experiment (Fig. S4†).
1) were incubated in 10% FBS for 24 h at 37 °C (Fig. 5). It was found that K1T–K4T RNTs protected less than 40% of siRNA during the serum incubation. This could possibly be due to the low siRNA loading ability and the ease of siRNA displacement from K1T–K4T RNTs, leading to serum degradation of naked siRNA. On the other hand, K5T–K15T RNTs demonstrated good protective ability of siRNA against serum degradation with greater than 55% of siRNA recovered. For instance, K9T–K15T RNTs were able to protect more than 80% of siRNA during the 24 h of serum incubation. Improvement in protective ability from K1T–K15T RNTs against serum degradation of siRNA is likely due to the increase in electrostatic interaction in the RNTs/siRNA system. The high cationic charges on the RNTs create stable complexes with siRNA, thus preventing the displacement of the latter by negatively charged competitors and the nuclease access to the siRNA. The enhanced binding and protective ability of siRNA by KnT RNTs indicate that these RNTs can act as siRNA carrier, protecting them during delivery in the physiological environment. The condition chosen showed was fully capable of digesting the amount of siRNA used in this experiment (Fig. S4†).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 4). Moreover, increasing net charges among the KnT RNTs series demonstrated enhanced interaction with the negatively charged proteoglycans on the cellular membrane, resulting into the fastest cellular uptake for K15T RNTs/siRNA. While both K10T and K15T RNTs showed high siRNA fluorescent signal within 3 h of cell transfection and complete cell transfection within 24 h, K5T RNTs had a slower cell transfection rate. It was also observed that the green fluorescent signal of siRNA spread to the cytoplasm of the cells over time, indicating release or displacement of the siRNA from KnT RNTs/siRNA complexes. Based on these results, we propose that the length of the RNTs formed and the size of their complexes with siRNA impact the cellular uptake. As seen in Fig. S1,† the length of the RNTs formed decreases with an increase in the steric bulk due to oligomerization of lysine. Consequently, the complexes formed smaller and more dispersed aggregates, facilitating internalization.
1 (Fig. 4). Moreover, increasing net charges among the KnT RNTs series demonstrated enhanced interaction with the negatively charged proteoglycans on the cellular membrane, resulting into the fastest cellular uptake for K15T RNTs/siRNA. While both K10T and K15T RNTs showed high siRNA fluorescent signal within 3 h of cell transfection and complete cell transfection within 24 h, K5T RNTs had a slower cell transfection rate. It was also observed that the green fluorescent signal of siRNA spread to the cytoplasm of the cells over time, indicating release or displacement of the siRNA from KnT RNTs/siRNA complexes. Based on these results, we propose that the length of the RNTs formed and the size of their complexes with siRNA impact the cellular uptake. As seen in Fig. S1,† the length of the RNTs formed decreases with an increase in the steric bulk due to oligomerization of lysine. Consequently, the complexes formed smaller and more dispersed aggregates, facilitating internalization.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) at 3, 12, 24, 48 and 96 h.
1 molar ratio) at 3, 12, 24, 48 and 96 h.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 6 and 7). However, this may not be beneficial in vivo for siRNA due to their short circulation half-life. An increase in the circulation half-life of the delivery system in serum is desired to optimise the targeted cargo delivery and evade renal clearance and lower the toxicity of the carrier complexes.14 The higher molecular weight KnT RNTs could be better suited for the delivery of larger biomolecules such as mRNA or plasmid DNA. As such, the binding ability of K1T–K5T to siRNA was investigated using gel retardation assay and the binding interaction of siRNA was improved by increasing the molar ratio up to 50
1 (Fig. 6 and 7). However, this may not be beneficial in vivo for siRNA due to their short circulation half-life. An increase in the circulation half-life of the delivery system in serum is desired to optimise the targeted cargo delivery and evade renal clearance and lower the toxicity of the carrier complexes.14 The higher molecular weight KnT RNTs could be better suited for the delivery of larger biomolecules such as mRNA or plasmid DNA. As such, the binding ability of K1T–K5T to siRNA was investigated using gel retardation assay and the binding interaction of siRNA was improved by increasing the molar ratio up to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 due to their lower cationic charges on the RNTs (Fig. 9). Complete binding of siRNA at 50
1 due to their lower cationic charges on the RNTs (Fig. 9). Complete binding of siRNA at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio was observed for K3T, K4T and K5T RNTs. Based on these results, K3T was chosen for further investigation since it was the best potential nanocarrier for siRNA delivery due to its lower cationic charge (compared to K4T and K5T) and molar ratio used for complexation (compared to K1T and K2T).
1 molar ratio was observed for K3T, K4T and K5T RNTs. Based on these results, K3T was chosen for further investigation since it was the best potential nanocarrier for siRNA delivery due to its lower cationic charge (compared to K4T and K5T) and molar ratio used for complexation (compared to K1T and K2T).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) using agarose gel retardation assay.
1) using agarose gel retardation assay.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and INTERFERin/stat3 siRNA served as the positive control. Silencing from the siRNA delivered by K3T RNTs (35%) was comparable to the result obtained for the positive control (40%) (Fig. 13B). Similar silencing effect between K3T RNTs and INTERFERin was observed in firefly luciferase assay, which indicated the highest silencing effect was achieved in the treatment.
1 and INTERFERin/stat3 siRNA served as the positive control. Silencing from the siRNA delivered by K3T RNTs (35%) was comparable to the result obtained for the positive control (40%) (Fig. 13B). Similar silencing effect between K3T RNTs and INTERFERin was observed in firefly luciferase assay, which indicated the highest silencing effect was achieved in the treatment.





