Open Access Article
Open Access ArticleMatrisomal components involved in regenerative wound healing in axolotl and Acomys: implications for biomaterial development†
Nancy
Avila-Martinez‡
a,
Merel
Gansevoort‡
 a,
Juul
Verbakel§
a,
Haarshaadri
Jayaprakash
a,
Juul
Verbakel§
a,
Haarshaadri
Jayaprakash
 bc,
Ines Maria
Araujo
bc,
Ines Maria
Araujo
 ce,
Marta
Vitorino
bce,
Gustavo
Tiscornia
bd,
Toin H.
van Kuppevelt
a and
Willeke F.
Daamen
ce,
Marta
Vitorino
bce,
Gustavo
Tiscornia
bd,
Toin H.
van Kuppevelt
a and
Willeke F.
Daamen
 *a
*a
aDepartment of Medical BioSciences, Radboud Research Institute, Radboud university medical center, PO Box 9101, 6500 HB Nijmegen, The Netherlands. E-mail: Willeke.Daamen@radboudumc.nl
bCentre of Marine Sciences (CCMAR), University of Algarve, 8005-139, Faro, Portugal
cFaculty of Medicine and Biomedical Sciences (FMCB), University of Algarve, 8005-139, Faro, Portugal
dEugin Barcelona, Balmes, 236, 08006 Barcelona, Spain
eAlgarve Biomedical Center Research Institute (ABC-RI), University of Algarve, 8005-139, Faro, Portugal
First published on 1st August 2023
Abstract
Achieving regeneration in humans has been a long-standing goal of many researchers. Whereas amphibians like the axolotl (Ambystoma mexicanum) are capable of regenerating whole organs and even limbs, most mammals heal their wounds via fibrotic scarring. Recently, the African spiny mouse (Acomys sp.) has been shown to be injury resistant and capable of regenerating several tissue types. A major focal point of research with Acomys has been the identification of drivers of regeneration. In this search, the matrisome components related to the extracellular matrix (ECM) are often overlooked. In this review, we compare Acomys and axolotl skin wound healing and blastema-mediated regeneration by examining their wound healing responses and comparing the expression pattern of matrisome genes, including glycosaminoglycan (GAG) related genes. The goal of this review is to identify matrisome genes that are upregulated during regeneration and could be potential candidates for inclusion in pro-regenerative biomaterials. Research papers describing transcriptomic or proteomic coverage of either skin regeneration or blastema formation in Acomys and axolotl were selected. Matrisome and GAG related genes were extracted from each dataset and the resulting lists of genes were compared. In our analysis, we found several genes that were consistently upregulated, suggesting possible involvement in regenerative processes. Most of the components have been implicated in regulation of cell behavior, extracellular matrix remodeling and wound healing. Incorporation of such pro-regenerative factors into biomaterials may help to shift pro-fibrotic processes to regenerative responses in treated wounds.
1. Introduction
Throughout the animal kingdom, adult skin wounding can be resolved either by fibrotic scarring or tissue regeneration. With some exceptions such as ear punch closure in rabbits,1,2 antler regrowth in deer,3,4 digit tip regeneration in children and mice,5,6 most adult mammals repair their wounds by fibrotic scarring.7,8 In humans, fibrosis in response to the destruction of both the epidermis and dermis (full-thickness skin wounds), such as third degree burns and traumas, results in scars that can have a severe impact on the patients’ quality of life. Often, follow-up procedures, treatments, and medication are required to alleviate the functional impairment and discomfort caused by these scars.9 In contrast, some species of vertebrates (fish, amphibians, salamanders) are capable of skin regeneration. Why are some organisms capable of regeneration while others are not? Why is scarless regeneration in humans, with some notable exceptions, lost after early development? Both cell intrinsic and cell extrinsic factors are believed to play a role in the fibrosis vs. regeneration outcome, but the cellular and molecular mechanisms underlying fibrosis compared to regeneration remain poorly understood. Understanding the drivers of fibrotic scarring vs. regeneration will aid in developing approaches and therapies to induce skin wound regeneration in humans. These novel therapies would meet an important medical need. In this context the discovery of Acomys as an adult mammal capable of skin regeneration becomes significant.Fibrotic wound repair is a (patho)physiological mechanism where an exacerbated healing response occurs and a fibrous tissue is formed. In contrast, regeneration involves complete structural and functional reconstruction of a tissue or organ after wounding and is traditionally understood as a specialized re-enactment of development.10,11 Research into the underlying molecular and cellular mechanisms of both fibrosis and regeneration in the skin have often focused on cell-intrinsic behavior and underlying genetic pathways or molecular players. In contrast, the role and contribution of the extracellular matrix (ECM) and its components during both fibrosis and regeneration is still poorly understood. Initially regarded as a somewhat inert structural scaffold, it is now accepted that the ECM is a major determinant of cell behavior in all tissues, mediated by its structure, composition and modifications. For example, increasing the stiffness of 3D collagen hydrogels by introducing intra- and interfibrillar crosslinks activated the expression of pro-fibrotic genes by adipocytes.12 The ECM can regulate the capture and activity of growth factors, which in turn influences local cellular processes, as is seen in the maintenance of stem cell niches.13,14 It stands to reason that the ECM plays a role during skin regeneration and that the “regenerative matrix” is a highly specialized environment unique to regenerative species. The ECM and all its associated (glyco)proteins are collectively known as the matrisome. The Matrisome Project has assembled lists of all ECM-related genes, divided into two groups; core matrisome genes, (composed of collagens, glycoproteins and proteoglycans), and matrisome associated genes, including regulators, secreted factors and ECM-affiliated proteins.15,16 In particular, proteoglycans are underrepresented in the Matrisome Project. Proteoglycans consist of a core protein with glycosaminoglycan (GAG) side chains, the latter of which convey important biological properties. Proteoglycans are an important component of the ECM, but the high variability of the GAG side chains makes it difficult to assess their involvement. Proteoglycans (and thus GAGs) may play a significant role during regeneration. The metabolism and regulation of proteoglycans, in particular the subset of genes pertaining to GAG metabolism, are a focus of this review.
While many publications have focused on comparing regeneration competent to regeneration incompetent animals, two exceptional models of regeneration provide unique opportunities to explore the phenomenon of scarless regeneration: the axolotl (Ambystoma mexicanum) and the African spiny mouse (Acomys sp.). We will refer to Ambystoma mexicanum simply as ‘axolotl’. This is in line with other scientific reports and the Ambystoma genus also encompasses other salamanders that are not investigated here. On the other hand, we will address the African spiny mouse by its genus name ‘Acomys’. Various species of Acomys are investigated in this review and the genus name is most commonly used in relevant scientific literature. Acomys is unique since it is a remarkable exception to the rule that adult mammals cannot regenerate their tissues, with individuals displaying remarkable wound healing responses throughout all life stages. Some mouse strains have acquired regenerative characteristics through selective breeding or genetic manipulation (MRL mouse, p21−/− mouse), but the natural regenerative potential of Acomys is far superior.17 Every species of the Acomys genus examined so far has shown the ability to regenerate various tissues. The regenerative capacities of the axolotl are unparalleled, with individuals being able to regrow entire limbs. In contrast, frogs (Xenopus sp.) complete regeneration of amputated limbs with the formation of a cartilaginous spike.18 Zebrafish, another well characterized model for regeneration, shows limitations to fin regeneration based on the amputation plane and adult zebrafish are unable to regenerate skin.19,20 These limitations prompted us to focus our comparison on axolotl and Acomys. In this review, we investigate the role of matrisome components during wound healing in two regeneration competent animals by analyzing existing datasets from previous publications. We aim to compare the healing responses in terms of the ECM-related gene and protein profiles, with a focus on skin wound healing. We postulate that identification of matrisome-related targets involved in the regenerative process may lead to novel approaches in human wound healing, especially with regards to biomaterial development for skin wound healing. It is not our intention to compare or relate Acomys to humans. Instead, by comparing a regenerative competent amphibian (axolotl) and a regenerative competent mammal (Acomys) we may identify drivers of regeneration that transcend species. We will first briefly touch upon the process of regular wound healing ending in fibrosis and the process of regeneration via blastema as seen in some vertebrates.
2. Wound healing responses
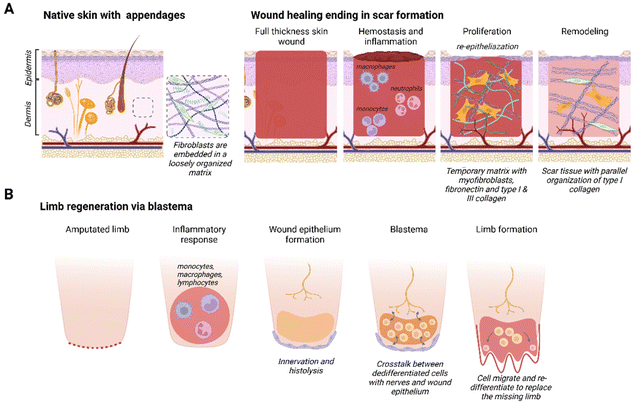
The mammalian skin wound healing response, culminating in scar formation, can be divided into three distinct phases: inflammation, proliferation and remodeling21 (Fig. 1A). The inflammatory phase occurs directly after wounding and is characterized by blood clot formation (hemostasis) and the invasion of neutrophils and monocytes.22 Macrophages play an important role by phagocytizing cell debris and bacteria, as well as secreting various growth factors and chemo-attractants. The proliferation phase, also known as the granulation tissue formation phase, encompasses both wound re-epithelialization and the formation of a dense network consisting of fibroblasts and neovasculature in a collagen and fibronectin rich matrix.23 This is a temporary matrix that is often highly disorganized compared to the original tissue and prone to rupturing. During the remodeling stage, the granulation tissue is remodeled into a smooth textured scar: rich in type I collagen fibers and lacking secondary skin appendages, such as hair follicles and sebaceous glands.24Some species resolve complete limb amputation through blastema-mediated regeneration. A blastema, also known as a regeneration bud, is an autonomous structure composed of a heterogeneous mass of dedifferentiated stem/progenitor cells that goes through morphogenesis, thereby creating a multitude of cell types to replace the missing organ or limb25,26 (Fig. 1B). The blastema is located under a layer of immature wound epithelium and it is created after the initial inflammatory response is resolved. Histolysis, re-innervation and ECM production will induce the migration and accumulation of cells under the wound epithelium that will differentiate into mesenchymal and ectodermal cell types.27 A requirement for blastema formation is the molecular and cellular interaction between the wound epithelium, nerves, dermal cells and the ECM at the wound site.28 In the final stage, cells inside the blastema differentiate and eventually form the missing organ, tissue or appendage.27 Blastema formation has been identified in different species, such as zebrafish,29 flatworms,30 urodeles26 and even some mammals (Acomys full-thickness ear wounds31 and mouse digit tip5). Intriguingly, under the right circumstances the fingertips of children up to 10 years of age heal similarly to the mouse digit tip.6
While the fibrotic response is the norm in most mammals, some exceptions exist as seen in the regeneration capacity of fetal wounds and digit tip regeneration in mice and children. Human skin wounds regenerate without scarring up to 24 weeks of gestation, thereafter the healing response will gravitate towards scar tissue formation.32 Research in fetal sheep indicated that, aside from gestational age, wound size also affects the regenerative capacity; with wounds exceeding 4 mm in diameter demonstrating an increased tendency to scar.33 A unique feature in fetal wounds is the ‘cable’ of actin filaments that develops around the wound edge. Wound closure through contraction of this actin ring has been compared to a drawstring closure.34–36 Recently, it has been postulated that myofibroblast-mediated wound contraction in adult mammals inhibits regeneration and drives scar formation.37 In addition, a distinct immune response has been recorded in fetal wound regeneration that contributes to a non-inflammatory ECM environment.32,38 The composition of fetal ECM, with an abundance of type III collagen and hyaluronan, is important in this aspect.32 Children up to ∼10 years of age are even able to fully regenerate a digit tip, as long as the wound is distal to the upper interphalangeal joint with sufficient nail bed39,40 and little to no intervention.41,42 Storer et al. showed that the ECM determines the blastema state and plasticity of mesenchymal cells in non-regenerative fingertip amputations,43 suggesting the importance of focusing further on the role of the matrix in regeneration.
3. Acomys, African spiny mouse, a mammal capable of regeneration
Recently, the African spiny mouse (Acomys) has emerged as a remarkable exception to the rule that adult mammals are incapable of regeneration.44 This rodent genus is found throughout Africa and the Middle East. Belonging to the subfamily Deomyinae, they are part of the Muroidea family and share a common ancestor with the Muridae, diverging about 23 million years ago.45 The genus comprises over 20 different species, but most studies have focused on A. cahirinus.Following up anecdotal reports of ‘skin jumping’ in Acomys, Seifert and colleagues reported A. kempi and A. percivali46 exhibited remarkable wound healing properties after extensive skin damage caused by mild handling of wild-caught animals. Following this initial report several groups have found that depending on type of organ and injury, Acomys shows remarkable responses to injury: extensive regeneration has been observed in skin, ear, muscle, digit tip and spinal cord47 (Fig. 2). Acute ischemia wound models of heart and kidney do not seem to regenerate but show resistance to wounding and diminished fibrosis.48 In contrast, Mus musculus shows extensive fibrotic scarring to similar injuries. Evolutionarily separated by only 23 million years, Acomys and Mus provide a powerful comparative framework to identify the cellular and molecular differences between regeneration and fibrotic scarring.
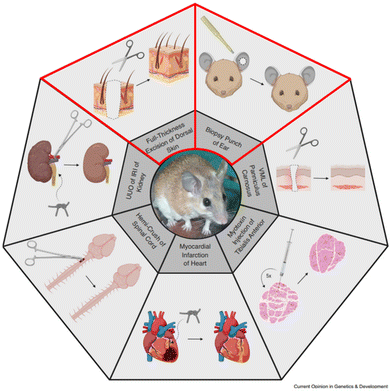 | ||
| Fig. 2 Overview of tissues/organs with a remarkable healing response in Acomys. This review aims to elucidate the involvement of the matrisome during regeneration of the skin and ear (lined in red). VML = volumetric tissue loss, UUO = unilateral obstruction, IRI = ischemia reperfusion injury. Reproduced from Sandoval & Maden (2020)48 with permission from Elsevier, 2020, under the STM permissions guidelines. | ||
In all regenerative systems, the immune system is thought to play an important role through its ability to modulate inflammation, as severe inflammation can be detrimental to the healing process. Several groups have extensively described the immune response observed in Acomys.48–52 In general, Acomys wounds demonstrated a distinct role for macrophage activity when compared to Mus.50 Mature macrophages (F4/80+) were absent in Acomys full-thickness skin wound beds, along with a distinct lack of pro-inflammatory cytokines.51 In Acomys ear wounds, CD86+ macrophages (classically activated M1) were absent from the blastema, being confined to the wound edges in the connective tissue distal to cartilage. CD206+ macrophages (M2, pro-regenerative) were restricted to the region beneath the wound epidermis and practically absent from the rest of the blastema.48 On the other hand, in Mus, both M1 and M2 macrophages are present throughout all connective tissues of the injured area.53 Interestingly, the depletion of macrophages through clodronate liposome injections severely delayed ear hole closure in Acomys.50 The neutrophil response in Acomys is delayed compared to Mus and differences were found in the number of neutrophils in various compartments, such as blood and bone marrow, when comparing several species of Acomys to Mus.52
A second factor of particular significance for skin regeneration is the skin biology of Acomys. Acomys dorsal skin is weak and tears easily when subjected to a mean tensile strength of 0.11 MPa, a force approximately 20 times weaker than that required to tear skin of Mus.46 Using atomic force microscopy to measure spatial tissue stiffness, intact Acomys skin did not exceed 15 kPa and wound centers measured no more than 5 kPa. Putting these findings into context, skin of C57Bl/6 mice measures 28 kPa, with wound beds of 10.5 kPa, further demonstrating the unique biomechanical properties of Acomys skin.54 Following fast re-epithelialization after skin injury, Acomys granulation matrix was loosely organized with less collagenous deposition and increased gene expression of the matrix metalloproteinases Mmp2 and Mmp9 compared to Mus.51,55 Overall, Acomys showed a distinct profile for protein remodeling and protein synthesis.56,57 Upon completion of regeneration, Acomys skin developed hair follicles, sebaceous glands and an intact panniculus carnosus.46,51 After wounding, enriched proteins belonged to pathways related to tight junction formation, endocytosis, ribosomes, proteasomes, Wnt signaling, MAPK signaling and vasopressin-regulated water reabsorption. Components of the ubiquitin-proteasome degradation pathway were strongly upregulated in Acomys and proteins in the ribosome related pathways were also implicated.56,57 Overall, enhanced protein turnover seems an important characteristic of skin wound healing in Acomys. The unique characteristics of Acomys’ skin may benefit wound healing, but the fact that Acomys is capable of regenerating other organs indicates that its skin biology may not be the only factor contributing to its regenerative potential.
Full-thickness skin wounds in Acomys regenerate without the presence of a blastema whereas ear punch wound closure in Acomys has been reported to occur through the formation of a blastema.27,31,46 Research using an ear hole punch model in A. cahirinus was reported by Matias Santos and colleagues,58 in response to the initial ear punch study.46 In these studies, reports on muscle regeneration were inconsistent, with the original study (Seifert 2012)46 mentioning the absence of muscle fibers in contrast to the study of Matias Santos (2016),58 who reported presence of muscle fibers within the regenerating region. Additionally, all regular skin appendages, such as hair follicles and sebaceous glands, had regenerated and nerve fibers were present.58 Gawriluk and colleagues demonstrated that the wound matrix of A. cahirinus exhibits several hallmarks of blastema-mediated regeneration.31 The Acomys ear wound beds showed several signs of the blastema-like wound epidermis, such as restriction of proliferating cells to the wound borders, lack of basal-apical polarity in basal keratinocytes and presence of keratin 17 in the wound matrix. Evidence of reinnervation, an essential process in epimorphic regeneration, was obtained as enrichment of genes involved in axon guidance, neuroactivity and growth was observed. Re-entry of mesenchymal cells into the cell cycle, another defining characteristic of the blastema, was also observed. Taken together these observations provide strong evidence for the formation of blastema in Acomys ear punch wounds and cements the importance of Acomys as a model system capable of tissue regeneration. In this review, we compare its wound healing responses to those of the axolotl, an animal well known for its regenerative abilities.
4. Axolotl, salamanders capable of full organ regeneration
The axolotl (Ambystoma mexicanum) is a paedomorphic (or ‘neotenic’) salamander belonging to the Urodele order of the Amphibia. The axolotl grows through a process of juvenilization, meaning it delays somatic development and retains physiologically juvenile features during its development to sexual maturity.59 Therefore, the axolotl is an animal that reaches sexual maturity without going through metamorphosis. Metamorphosis occurs only when they are in contact with an external and artificial stimulus, which is uncommon in the wild.60 The axolotl represents an important model organism in the field of regenerative medicine due to its highly efficient regeneration capacity. Regeneration has been observed in limbs and several organs such as the skin, heart, brain, lungs, and eyes61,62 (Fig. 3).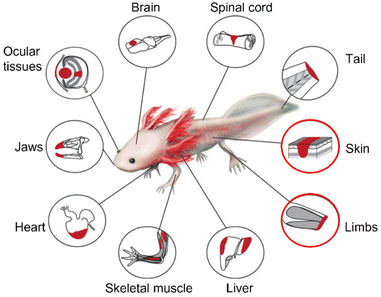 | ||
| Fig. 3 Summary of tissues/organs that can be regenerated by axolotls (Ambystoma mexicanum). The regeneration of skin and limbs, outlined in red, and the involvement of matrisome components is further explored in this review. Reproduced from Debuque & Godwin62 with permission from Springer International Publishing Switzerland, 2016, under the STM permissions guidelines. | ||
Limb regeneration has been extensively investigated in axolotl. Following limb amputation, wound closure continues to blastema formation and limb development.26 Wound closure takes several hours, and full regeneration of the limb typically occurs between 40 and 50 days.63 The formation of a blastema is essential to the regeneration of a limb. Research using the Accessory Limb Model has uncovered three requirements for successful blastema-mediated limb regeneration: the presence of a wound epithelium, the occurrence of nerve signaling and the availability of positional cues provided by pattern forming cells.64 Absence of any of these factors abrogates limb regeneration.65 Formation of a closed wound epithelium involves migrating keratinocytes that cover the wound surface.26 Under the influence of signals emanating from local nerve fibers, proliferation of this wound epithelium into a multilayered epithelium result in the apical epithelial cap.66 This epithelial cap is characterized by specific molecular markers and by an immature basement membrane beneath the epidermis.67 Nerve signaling causes basal epithelial cells to exit the cell cycle. This step is essential for the development of the blastema as cells of the apical epithelial cap will start secreting growth factors and enzymes that degrade ECM and facilitate migration.28 Next, fibroblasts originating from the limb circumference gather underneath the apical epithelial cap. The interaction between these cells with different positional identities is crucial for maintaining the blastema and ensuring regeneration as the ‘positional memory’ dictates the pattern of the missing limb.26
Transcriptomic analysis has revealed the importance of the immune system in axolotl wound healing and regeneration. There is a gradual increase of macrophages and fibroblast-like cells post amputation, whereas neutrophil populations decreased after wounding.68 Neutrophils evoke anti-inflammatory macrophages, which leads to a reduction in pro-inflammatory cytokines.69 Macrophages with anti-inflammatory and pro-resolving phenotypes were dominant in the first seven days of axolotl limb regeneration, thereby contributing to repressing the inflammatory response.70 When macrophages were systemically depleted, limb regeneration failed. A fibrotic ECM was observed with increased type I collagen deposition and the presence of αSMA-positive cells71 that could also inhibit apical epithelial cap formation and blastema-induction.72 ECM remodeling is of key importance to rapidly reconstruct the matrix70 and the ECM contains positional information required for limb pattern formation through location-specific differences in heparan sulfate sulfation. This was shown by grafting decellularized axolotl skin or decellularized mouse skin onto axolotl limb wounds.73 Depending on the developmental stage, positional origin and heparan sulfate sulfation pattern of the graft, limb patterning was either supported or inhibited. These results demonstrated that the ECM contains information, conveyed via heparan sulfate sulfation patterns, that can influence limb regeneration and this information is also present in the ECM of mice.73
Axolotl skin, which regenerates without blastema formation, provides a platform to uncover drivers of regeneration in the absence of blastema. The skin of juvenile axolotls consists of a pseudostratified epithelium which contains epithelial and Leydig cells, and the dermis presents a loosely organized network of thin collagen fibrils in which fibroblasts and mucus glands are embedded.74 A layer of compressed collagen fibers separates the hypodermis from the underlying muscle layer. Scarless skin wound healing has been reported in adult axolotls after making excisional wounds on the flanks. Wounds did not form scabs, instead epithelization was completed within 24 h and full skin healing including all skin appendages was completed within 80 days.67 Overall, the regenerating tissue in axolotl was characterized by high levels of matrix remodeling enzymes, relatively low abundance of fibronectin and persistently high levels of tenascin C throughout the wound bed. Type III collagen deposition was followed by type I collagen deposition and maturation from day 14 onwards. Following a full-thickness wound, the collagen architecture in the dermis did not return to the normal lattice-like pattern, but was disorganized, even though the healed skin appeared identical to unwounded skin. However, after blastema-mediated skin regeneration following limb amputation, the dermis, including collagen architecture, does fully regenerate.75
The healing capacity of the axolotl is unique and while directly translating regeneration in salamanders to mammals is difficult, some clues may be found in the regenerative response of axolotls. Axolotls display a minor inflammatory response with macrophages of a mostly anti-inflammatory phenotype. The fibrotic response is notably absent, ECM deposition is regulated, and matrix remodeling enzymes are prevalent (Fig. 4A). These hallmarks are also observed in Acomys regeneration, suggesting regeneration shares similar mechanisms across both models. However, there are differences in intensity and duration of the different events in regenerative and non-regenerative mammals, indicating the importance of timing of the various phases (Fig. 4B). The various wound healing responses described for the animals are summarized in Table 1 (full thickness skin wound healing) and Table 2 (blastema-mediated regeneration).
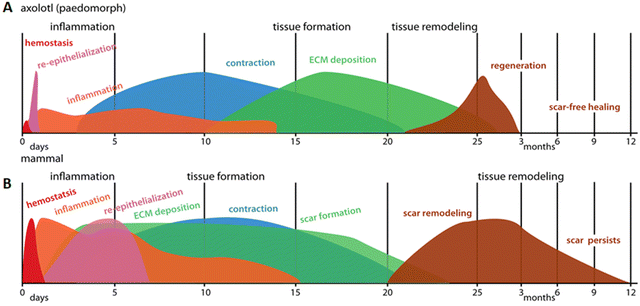 | ||
| Fig. 4 Schematic representation of the wound healing response in (A) paedomorphic axolotls and (B) mammals. Axolotl wound healing shows minimal hemostasis and very rapid re-epithelization followed by low levels of inflammation. ECM deposition occurs after wound contraction and tissue remodeling does not lead to a scar, but results in normal skin appearance. Contrary to that, in mammals, a strong inflammatory response is maintained even after re-epithelization is complete. Deposition of new ECM and wound contraction occur at the same time and scar tissue persists after completion of the months-long remodeling phase. Reproduced from Seifert et al. (2012)67 with permission from © 2012 Seifert et al., under the Creative Commons Attribution License. | ||
| Full-thickness skinwound healing | Mus musculus (mammal) | Acomys sp. (mammal) | Axolotl (amphibian) |
|---|---|---|---|
| Re-epithelialization | Slow (5–7 days) with a scab | Fast (3 days) with a scab | Fast (1 day) without a scab |
| Inflammation | High | Low | Low |
| Fast neutrophil response | Delayed neutrophil response | Low neutrophil response | |
| High expression of M1 macrophages | High expression of M2 macrophages | High expression of pro-resolving macrophages | |
| Pro-inflammatory cytokines ++ | Pro-inflammatory cytokines −− | Pro-inflammatory cytokines −− | |
| Proliferation | Early matrix deposition | Delayed matrix deposition | Delayed matrix deposition |
| Dense granulation matrix | Loose granulation matrix | Loose granulation tissue | |
| Collagen deposition+ | Collagen depostion − | Matrix degradation+ | |
| Remodeling | Scar remodeling | Scar-free remodeling | Scar-free remodeling |
| Type I collagen with parallel orientation | Type I collagen with basket weave orientation | Type I collagen did not return to normal lattice organization | |
| No regeneration of skin appendages | Regeneration of hair follicles and sebaceous glands | Regeneration of mucous glands |
| Acomys sp. (mammal) | Axolotl (amphibian) | |
|---|---|---|
| Blastema-mediated regeneration | Regeneration after ear punch via blastema-like process | Limb regeneration via blastema |
| Wound epithelium | Loss of basal-apical polarity in keratinocytes | Migrating keratinocytes close the wound |
| Presence of keratin 17 (blastema marker) | Multi-layered epithelium forms (apical epithelial cap) | |
| Recruitment of mesenchymal stem cells | Immature basement membrane | |
| Innervation | Upregulation of genes associated with axon guidance, neuron growth and neuroactivity | Apical epithelial cap (AEC) is innervated |
| Cell proliferation is restricted to wound borders | Cells exit cell cycle | |
| Blastema formation | Cell cycle re-entry, cell proliferation and cell division | Pro-migratory environment is formed (ECM degradation, growth factor secretion) |
| Increase of matrix degradation enzymes | Fibroblasts with various positional identities migrate to the AEC | |
| Cartilage pattern re-established the ear | Limb pattern is established by ECM and cells | |
| Differentiation | Regeneration of missing tissues | Cells redifferentiate to form missing limb |
5. Identification of matrisome-related genes and proteins during regeneration using existing datasets
Comparing the matrisome and GAG-related components present during regeneration in Acomys and axolotl will aid in uncovering similarities between these two animals. Through this approach, we aim to identify shared pro-regenerative factors between species, which may be translated to other species (i.e. humans).5.1 Selection of datasets
To select the databases that will be analyzed, papers that document regenerative wound healing in Acomys or axolotl were retrieved, with a focus on skin and blastema. Publications with either a proteomics or transcriptomics approach were extracted. For Acomys publications on both skin regeneration and ear hole closure were included in the search. A total of four papers were identified, of which three focused on the regeneration of full-thickness skin wounds55–57,76 and one publication described ear hole closure.31 The datasets of Brant et al. (2015)55 and Brant et al. (2019)76 were compared for overlap, as these publications investigate similar samples with different techniques. All genes identified by the 2015 publication were present in the 2019 publication. As the 2019 publication offered more data only this set was included for the comparison. Details of the selected papers are presented in Table 3.| Publication | Title | Tissue | Timepoints | Technique | Dataset |
|---|---|---|---|---|---|
| Brant et al. 2019 (Acomys)76 | Comparative transcriptome analysis of dermal wound healing reveals de novo skeletal muscle regeneration in Acomys cahirinus | Dorsal skin, 8 mm skin punch | Day 0, 7, 14 | De novo transcriptome assembly and comparative transcriptomics | Supplementary files of ref. 76: S7 data and S8 data |
| Gawriluk et al. 2016 (Acomys)31 | Comparative analysis of ear-hole closure identifies epimorphic regeneration as a discrete trait in mammals | Ear punch, 4 mm | Day 0, 5, 10, 15, 20 | De novo transcriptome assembly and comparative transcriptomics | Supplementary file of ref. 31: dataset 1 |
| Yoon et al. 2020 (Acomys)56,57 | Comparative proteomic analysis in scar-free skin regeneration in Acomys cahirinus and scarring Mus musculus | Dorsal skin, 8 mm skin punch | Day 0, 7, 14 | Shotgun proteomics using LC-MS/MS | Table 1 of ref. 56 and 57 |
| Monaghan et al., 2012 (axolotl)79 | Gene expression patterns specific to the regenerating limb of the Mexican axolotl | Flank skin outside of limb range, 4 mm skin punch | Day 0, 1, 3, 7 | Microarray by Affymetrix GeneChips | Supplementary data of ref. 79 in NCBI: GSE37198_RAW.tar |
| Stewart et al. 2013 (axolotl)80 | Comparative RNA-seq analysis in the unsequenced axolotl: The oncogene burst highlights early gene expression in the blastema | Right forelimbs at the mid-stylopod level | Hour 0, 3, 6, 12 and day 1, 3, 5, 7, 10, 14, 21, 28 | De novo assembly of axolotl transcript, RNA-seq | Supplementary data of ref. 80 In NCBI: GSE34394_RAW.tar |
| Rao et al. 2009 (axolotl)63 | Proteomic analysis of blastema formation in regenerating axolotl limbs | Bilateral hind limbs regenerating tissue and 1 mm of stump tissue | Day 0, 1, 4, 7 | Proteomics by LC- MS/MS | Supplementary files of ref. 63: Table 2 |
Several papers focusing on axolotl were reviewed such as Monaghan et al. (2009)77 and Wu et al. (2013).78 The limited availability of public transcriptomic datasets of Acomys led us to select publications on axolotl that best matched the timepoints and tissues analyzed in the selected Acomys publications. Proteomic and transcriptomic studies of the axolotl were selected based on their description of the regenerating limb blastema and/or non-blastema mediated skin regeneration, which resulted in the selection of three papers (Table 3). The paper by Monaghan et al. (2012)79 describes both limb regeneration (via blastema) and regeneration of a full-thickness skin wound. Both Rao et al. (2009)63 and Stewart et al. (2013)80 exclusively describe hind-limb regeneration via blastema at the proteomic and transcriptomic level, respectively.
5.2 Identification of the matrisome and GAG-related components
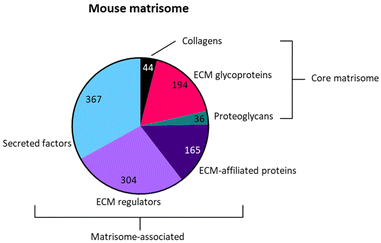
We next addressed the differences in gene annotations of the selected datasets. The publications focusing on Acomys used mouse gene symbols to annotate data, whereas the axolotl datasets were annotated using human gene symbols. Detailed lists of matrisome related genes of various species are available through the Matrisome Project.15 The mouse and human matrisome master lists contain 1110 and 1027 genes, respectively. As human-mouse orthologs are comparable the more extensive mouse matrisome was used for the selection of matrisome genes from all datasets.The genes of the matrisome are divided into core matrisome genes and matrisome-associated genes.15 The former is further subdivided into ECM glycoproteins, collagens and proteoglycans while the latter is subdivided into ECM-affiliated proteins, ECM regulators and secreted factors (Fig. 5, full gene list is available in Table S1†). Closely associated with the matrisome, but not fully included in the matrisome lists, are glycosaminoglycans (GAGs). These polysaccharides are generally present in the ECM and mostly attached to a core protein, together known as proteoglycans.81 Although the specialized role of GAGs in a multitude of biological processes is widely acknowledged, they are technically difficult to assess and are consequently often disregarded. After biosynthesis of the repeated disaccharide units, GAGs are extensively modified by enzymes that epimerize certain saccharides, remove acetyl groups and add sulfate groups at specific locations on the building blocks, resulting in highly heterogenous molecules. Methods such as a colorimetric staining or ELISA allow for the basic quantification of GAGs in various sample types.82 Methods that assess GAGs on the disaccharide level, e.g. RP-HPLC, are suited for identifying the various disaccharides but lose positional information in the chain. Raman spectroscopy may struggle to separate the signals of complex samples.83 Mass spectrometry has made significant progress with novel ionisation techniques, but is not yet a standard method for GAG analysis.84 Given the importance of GAGs in many biological processes, we performed an additional investigation into the presence of GAG related enzymes during the regenerative processes in axolotl and Acomys. Currently there are no methods available to directly assess GAG biosynthesis and modification. Instead, the expression of genes involved in GAG biosynthesis provides information on the regulation of GAGs during biological processes. A paper published in 2018 by Uijtdewilligen et al. provides a curated list of genes involved in proteoglycan/glycosaminoglycan metabolism,85 dividing GAG-related genes into various categories based on their role in GAG homeostasis. The following four categories were selected for use in our comparison analysis: linkage region formation (8 genes), GAG chain polymerization (13 genes), GAG chain modification (32 genes), and GAG chain degradation (19 genes) (the complete gene list is available in Table S2†). Several of these genes were also present in the greater matrisome list as part of the ECM regulators. These were GAG chain modificators sulfatase 1 and sulfatase 2, heparanase and hyaluronidase 1, 2 and 3. The matrisome and GAG-related genes were extracted from each dataset. Any adaptions made to a dataset prior to gene extraction are covered in the results section for each publication. The next section focuses on the datasets that were compared.
5.3 Comparison of the matrisome and GAG components
The same biological processes (full-thickness skin wound regeneration or blastema-mediated regeneration) in Acomys and axolotl were compared at the transcriptome level. On the proteomic level this comparison could not be made due to absence of proteomics studies on blastema-mediated regeneration in Acomys and skin regeneration in axolotl. Although the processes are different, we propose that comparing skin regeneration and blastema on the proteomic level could still yield beneficial information. Thus, the matrisome and GAG-related genes of the following datasets were compared:1. Transcriptomics of full-thickness skin wound regeneration in Acomys (Brant et al. 2019)76versus in axolotl (Monaghan et al. 2012).79
2. Transcriptomics of the (mammalian) blastema in Acomys (Gawriluk et al. 2016)31versus the (amphibian) blastema in axolotl (Stewart et al. 2013).80
3. Proteomics data of Acomys full-thickness skin wound healing (Yoon et al. 2020)56,57versus proteomics data of axolotl blastema-mediated limb regeneration (Rao et al. 2009).63
6. Results of the matrisome-GAG component analysis
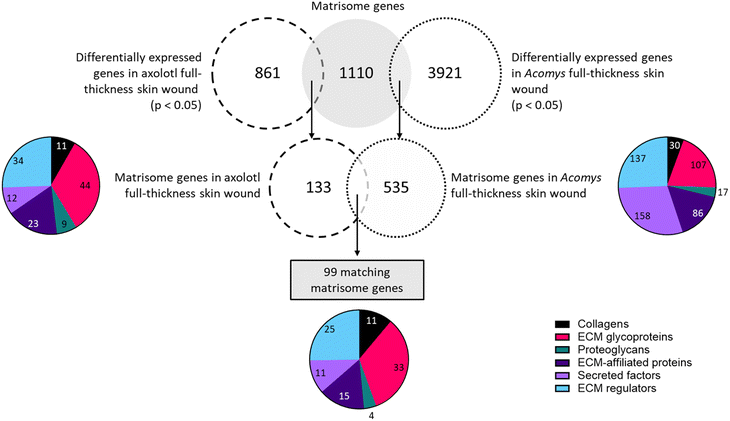
In the following sections we describe the results of the matrisome and GAG gene comparisons. Each comparison is divided into three sections. Section 1 describes the preparation of the datasets in order to perform the comparison, section 2 highlights the matrisome components identified in the comparisons and section 3 focuses on the GAG-related components.6.1. Comparison of the transcriptome profiles of regenerating full-thickness skin wounds on day 7 in Acomys (Brant et al. 2019)76versus axolotl (Monaghan et al. 2012)79
The only matching timepoint in both publications was day 7 after skin injury. Descriptive studies on skin wound healing in Acomys and axolotl reported complete re-epithelialization within the first 3–5 days in both species.46,79 Moreover, after wounding both animals exhibited a limited and low inflammatory response. After day 7 both animals start wound contraction, ECM deposition and new tissue formation.67,86 Therefore, Acomys and axolotl should be in a similar regeneration stage on day 7 after wounding and a comparison at this time point may highlight the matrisome and GAG genes involved during early regeneration. Thus, only the datasets of day 7 in Acomys and axolotl were compared. First, differentially expressed matrisome and GAG-related genes were identified in Acomys (Table S4†) and axolotl (Table S5†). Finally, the differentially expressed matrisome and GAG-related genes that were present in both Acomys and axolotl were identified (Table S6†).
A comparison of the collagens expressed in the two species revealed 11 matching genes. Among the top upregulated genes in axolotl were Col11a1, Col7a1, Col28a1, Col4a5 and Col12a1. In Acomys, the highest fold changes belonged to Col12a1, Col24a1, Col5a2, Col5a1 and Col7a1. All genes were upregulated in axolotl but in Acomys four genes were downregulated: Col4a5, Col4a6, Col17a1, and Col28a1. Amongst the glycoproteins a total of 33 matches were found. The most upregulated genes in both species were tenascin C (Tnc), fibronectin (Fn1), laminin alpha 1 (Lama1), thrombospondin 1 and 2 (Thbs1, Thbs2), peroxidasin homolog (Pxdn) and collagen triple helix repeat containing 1 (Cthrc1). The genes with the most negative fold changes were adiponectin C1q collagen domain containing (Adipoq), spondin 2 (Spon2) and multimerin 1 (Mmrn1). Only four proteoglycans were present in both datasets, of which three were upregulated in both species: proteoglycan 4 (Prg4), serglycin (Srgn) and decorin (Dcn). Osteoglycin (Ogn) was downregulated in both axolotl and Acomys. Fifteen genes matched in the compartment of ECM affiliated proteins. Several of these genes were upregulated in both species, with the highest fold changes found in: C1q tumor necrosis factor related protein 1 (C1qtnf1), lectin galactose binding soluble 9 (Lgals9), syndecan 2 (Sdc2), complement component 1 q subcomponent B (C1qb), and C chain (C1qc). Only C1qtnf2 was downregulated in both species. Notably, Lgals8 and Lgals2 were greatly downregulated in axolotl whereas their expression was barely affected in Acomys. Eleven secreted factors were identified in both datasets. Several genes were upregulated in both species, with the largest fold changes belonging to secreted frizzled-related protein 4 (Sfrp4), Srfp2, follistatin-like 1 (Fstl1) and angiopoietin-like 2 (Angptl2). Two genes were downregulated in both animals: chordin-like 1 (Chrdl1) and fibroblast growth factor 12 (Fgf12). A few notable differences in expression levels were present, the most notable being interleukin 1 beta (Ilb) which was the most upregulated gene in Acomys but showed a downregulation in axolotl. Among the ECM regulators 25 matches were found. The top upregulated genes in both species were matrix metallopeptidase 2 (Mmp2), Mmp3, Mmp13, procollagen lysine 2-oxoglutarate 5-dioxygenase 2 (Plod2), proprotein convertase subtilisin/kexin type 5 (Pcsk5), tissue inhibitor of metalloproteinase 1 (Timp1), peptidase domain containing associated with muscle regeneration 1 (Pamr1) and lysyl oxidase-like 2 (Loxl2). Only one out of 25 genes was downregulated in axolotl: cathepsin S (Ctss). In Acomys, three out of 25 matching genes were downregulated: Mmp28, tolloid-like 1 (Tll1) and Kazal-type serine peptidase inhibitor domain 1 (Kazald1). Interestingly, Kazald1 was the most upregulated gene in axolotl, but the most downregulated gene in Acomys.
In summary, of the 99 matrisome genes that were present in both species 77 genes (77%) had a similar expression pattern, where a gene was either upregulated (69 genes/70%) or downregulated (8 genes/8.1%) in both Acomys and axolotl. Twenty-two genes (22%) showed opposing expression patterns where a gene would be upregulated in one species but downregulated in the other species.
In Acomys, two genes involved with linkage region preparation were present: beta-1,4-galactosyltransferase 7 (B4galt7) and B4galt2, both were upregulated. Eleven genes related to GAG chain polymerization were found in Acomys, with the highest fold changes seen in hyaluronan synthase 1 (Has1), Has2 and Has3. Only two genes were downregulated: exostoses (multiple)-like 1 (Extl1) and Extl3. In axolotl, three genes relating to GAG chain modification were found, these were all upregulated: carbohydrate sulfotransferase 2 (Chst2), Chst11 and sulfatase 1 (Sulf1). In Acomys a total of 11 genes matched in this compartment, among which were the three genes identified in axolotl. Together with Sulf2 these were amongst the most upregulated genes in Acomys. Only three genes were downregulated: sulfatase modifying factor 1 and 2 (Sumf1, Sumf2) and carbohydrate sulfotransferase 15 (Chst15). Finally, 11 genes involved with GAG chain degradation were present in Acomys. The most highly upregulated genes were glucuronidase beta (Gusb), hexosaminidase A (Hexa), arylsulfatase B and J (Arsb, Arsj). No genes were downregulated.
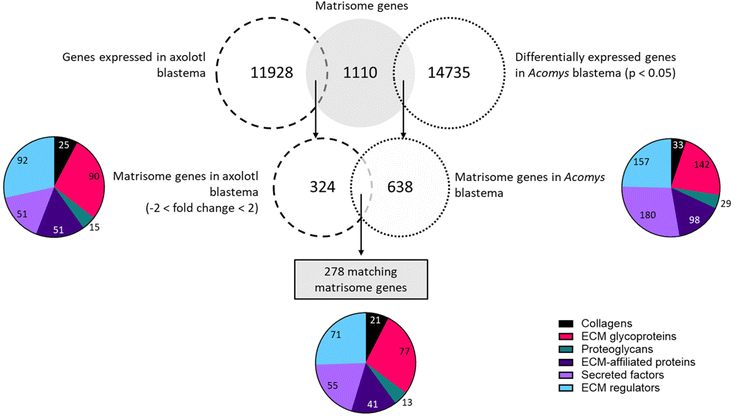
6.2 Comparison of the transcriptome profiles during blastemal regeneration in Acomys ear pinnae closure (Gawriluk et al. 2016)31vs. axolotl stylopod amputation (Stewart et al. 2013)80
In this comparison we focused only on the blastema stage. In axolotl this stage lasts approximately from day 3–4 up to day 21.71 A similar timeframe is described by Gawriluk et al. (2016)31 when investigating Acomys ear blastema. For this reason, we compare days 5, 10 and 14 in axolotl to days 5, 10 and 15 in Acomys. Later time points were excluded as they are not in the blastema stage in axolotl. The full dataset containing all matching matrisome and GAG-related genes of both species genes is available in the ESI (Table S9†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735 differentially expressed genes identified in Acomys ear pinna of which 638 genes (4.3%) were part of the matrisome. In axolotl, a total of 11
735 differentially expressed genes identified in Acomys ear pinna of which 638 genes (4.3%) were part of the matrisome. In axolotl, a total of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 928 genes were expressed in the regenerating limb. Following matching to the matrisome and application of the fold change threshold, 324 genes (2.7%) were extracted. Between Acomys and axolotl 278 matching matrisome genes were found.
928 genes were expressed in the regenerating limb. Following matching to the matrisome and application of the fold change threshold, 324 genes (2.7%) were extracted. Between Acomys and axolotl 278 matching matrisome genes were found.
Twenty-one genes that code for collagen proteins were present in both species during various points in the blastema formation. Of note, only three genes were upregulated in both models at all time points: Col18a1, Col6a2 and Col3a1. One gene was downregulated on all time points: Col22a1. The remaining collagen genes showed opposite expression patterns in each species. For example, Col4a6 was upregulated at all timepoints in axolotl but downregulated at all time points in Acomys; the reverse pattern was observed for Col4a1. In total 77 glycoproteins were identified in both species. Of the 17 genes that were upregulated in both species at all time points the top-upregulated genes were: collagen triple helix repeat containing 1 (Cthrc1), laminin alpha 1 (Lama1), tenascin C (Tnc), transforming growth factor beta induced (Tgfbi), thrombospondin 2 (Thbs2), peroxidasin homolog (Pxdn), fibronectin 1 (Fn1), and elastin microfibril interfacer 1 (Emilin1). A total of 10 genes were downregulated in both axolotl and Acomys, with the lowest fold changes seen in: adiponectin, C1Q and collagen domain containing (Adipoq), tenascin XB (Tnxb), leucine-rich repeat LGI family member 1 (Lgi1) and gliomedin (Gldn). One gene, Tenascin N (Tnn), displayed a similar expression profile in axolotl and Acomys. This gene was downregulated on day 5, but positive fold changes were observed on day 10 and day 14/15. The remaining genes showed differences in expression patterns such as matrilin 4 (Matn4), cartilage oligomeric matrix protein (Comp), Fraser syndrome 1 homolog (Fras1) and nephronectin (Npnt), all of which were upregulated in axolotl but downregulated in Acomys. In contrast, matrilin 2 (Matn2) and connective tissue growth factor (Ctgf) were upregulated in Acomys but downregulated in axolotl. Among the proteoglycan genes, 13 matches were identified. Three of these genes were upregulated at all time points in both animals: proteoglycan 4 (Prg4), versican (Vcan) and serglycin (Srgn). Similar expression profiles were seen in osteoglycin (Ogn), which was downregulated on day 5 and 10 in both animals, a positive fold change was seen on day 14 in axolotl and on day 15 in Acomys. A contradicting profile was observed for neurocan (Ncan): this gene was highly upregulated on day 5 in axolotl after which fold changes < 2 were seen, whereas in Acomys Ncan was absent on day 5 but displayed high fold changes on day 10 and 15. Among the ECM-affiliated matrisome 41 genes matched between axolotl and Acomys. A total of 15 genes were upregulated in both animals: C-type lectin domain family 4 member e (Clec4e), lectin galactose binding soluble 1 (Lgals1), complement component q subcomponent alpha polypeptide (C1qa), plexin D1 and C1 (Plxnd1, Plxnc1) and syndecan 1 (Sdc1). Only four genes were downregulated in both animals and during all timepoints, of which the most downregulated genes were: C-type lectin domain family 3 member a (Clec3a) and family 2 member d (Clec2d). The remaining genes in this compartment had opposing expression patterns. For example, Fras1 related extracellular matrix protein 2 (Frem2) was highly upregulated in axolotl but downregulated in Acomys. Surfactant associated protein C (Sftpc) was highly upregulated in Acomys but downregulated in axolotl. A total of 55 genes were found in the secreted factors subset. Seventeen genes were upregulated in both species at all timepoints, with the highest fold changes observed in chemokine C–X–C motif ligand 14 (Cxcl14), multiple EGF-like-domains 11 (Megf11), platelet-derived growth factor C polypeptide (Pdgfc), wingless-related MMTV integration site 5A (Wnt5a), sonic hedgehog (Shh), transforming growth factor beta 3 (Tgfb3) and secreted frizzled-related protein 2 (Sfrp2). Three genes were downregulated in all datasets: chemokine C–X–C motif ligand 12 (Cxcl12), follistatin (Fst) and bone morphogenic protein 5 (Bmp5). The remaining genes often showed opposing expression patterns. Heparin-binding EGF-like growth factor (Hbegf) and chemokine C–C motif ligand 5 (Ccl5), were downregulated in axolotl but upregulated in Acomys. The other way around: leptin (Lep) and macrophage stimulating 1 (Mst1) were upregulated in axolotl and downregulated in Acomys. Finally, 71 regulator genes matched between Acomys and axolotl of which 40 genes were upregulated at all timepoints in both datasets. The highest fold changes belonged to several matrix metallopeptidases (Mmp3, Mmp9, Mmp12, Mmp13, Mmp19), transglutaminase 3 E polypeptide (Tgm3), tolloid-like 2 (Tll2), tissue inhibitor of metalloproteinase 1 (Timp1), serine (or cysteine) peptidase inhibitor clade E member 1 (Serpine1), and cathepsin Z (Ctsz). Only one gene, tissue inhibitor of metalloproteinase 3 (Timp3), was downregulated in both animals across all days. Once again, the remaining genes displayed opposite expression patterns. This was the case for Kazal-type serine peptidase inhibitor domain 1 (Kazald1) and coagulation factor XIII A1 subunit (F13a1), which were among the most upregulated genes in axolotl but displayed downregulations in Acomys. The opposite was also seen: cathepsin H and S (Ctsh, Ctss) were downregulated in axolotl but upregulated in Acomys.
To summarize this comparison demonstrated that many matrisome genes are present in the blastema of both Acomys and axolotl. A total of 278 matching matrisome genes were identified and 115 of these genes (41%) showed similar expression patterns, being either upregulated (96 genes/35%) or downregulated (19 genes/7%) in both species.
6.3 Comparison of the proteomic profiles of skin regeneration in Acomys (Rao et al. 2009)63vs. blastema-mediated limb regeneration in axolotl (Yoon et al. 2020)56,57
In this section a comparison was performed of skin regeneration (absence of blastema) and limb regeneration (blastema mediated) on the proteomic level. The previous comparisons of transcriptome profiles each generated an extensive list of matrisome-related genes that were present in both species. However, up or downregulation on the transcriptional level does not necessarily translate to an increase or decrease in protein abundance, thus investigating regeneration at the proteomic level is important.Comparing the matrisome-related proteins from the proteomics studies on Acomys skin regeneration (Yoon et al. 202056,57) and axolotl limb regeneration (Rao et al. 200963) revealed only four proteins that were present in both species. The data from Yoon does not provide a fold change for the variations in protein abundance. To be able to compare trends a simple fold change for the Yoon data was calculated: fold change = [protein QV on day X]/[protein QV on day 0]. Following this, values of 0 > fold change < 1 indicate a decrease in the protein and these values were converted to negative fold changes by calculating −1/fold change. Values of fold change >1 indicate an increase in protein abundance, but this difference could not be statistically tested for a differential increase or decrease. Data are represented in Table 4.
| Collagens | Acomys (fold change) | Axolotl (fold change) | |||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Prot. description | Day 3 | Day 5 | Day 7 | Day 1 | Day 4 | Day 7 |
| Col12a1 | Collagen alpha-1(XII) chain | −1.29 | 1.90 | 1.80 | 1.32 | 1.05 | −1.12 |
| Col1a1 | Collagen alpha-1(1) chain | −1.32 | 1.87 | 1.62 | 1.43 | 1.44 | 1.92 |
| ECM glycoproteins | Acomys (fold change) | Axolotl (fold change) | |||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Prot. description | Day 3 | Day 5 | Day 7 | Day 1 | Day 4 | Day 7 |
| Fgb | Fibrinogen beta chain | 1.83 | 1.97 | 2.10 | 3.39 | 1.63 | 1.14 |
| Fgg | Fibrinogen gamma chain | 1.13 | 1.21 | 1.36 | 4.64 | 2.17 | 1.24 |
Of the matrisome collagens, only collagen type XII alpha 1 (Col12a1) and Col1a1 were found in both species. In Acomys the abundance of Col12a1 was lower on day 3 compared to day 0, but abundance had risen on day 5 and 7 as demonstrated by a positive fold change. On the other hand, Col12a1 abundance was increased in axolotl on day 1 and 4 but protein levels dropped with a negative fold change observed on day 7. The protein form of Col1a1 was again less abundant in Acomys on day 3, but the protein abundance increased with positive fold changes observed on day 5 and 7. In axolotl the abundance of Col1a1 was always increased. Among the ECM glycoproteins, only fibrinogen beta and gamma chain (Fgb and Fgg) matched between the datasets. No negative fold changes were observed in either animal. In Acomys the protein abundance of both Fgb and Fgg remained steady over all days, whereas in axolotl the fold change for both Fgb and Fgg was highest on day 1 with a decrease in protein abundance over the following days.
To conclude, only four matching matrisome proteins were identified and no matches were found among the GAG specific lists. This could be the result of comparing two distinct biological processes. While the axolotl hind limb regeneration is mediated by the blastema, the process of full-thickness skin regeneration in the spiny mouse has not been hallmarked as a blastema-mediated process.46 The blastema is a very specialized tissue, thus the proteins present during this process may not be present during skin regeneration in Acomys. An alternative explanation is that sample preparation methods resulted in biased protein sets in each species. A recent study that discusses methods to obtain ECM molecules from tissue samples states that the sample extraction method should be optimized to also obtain insoluble ECM,87 which was not done for the publications of Yoon et al. (2020)56,57 and Rao et al. (2009).63 Proteins from Acomys tissue were extracted using a buffer made with Tris-Cl, NaCl, ethylenediaminetetraacetic acid (EDTA), phosphatase inhibitors and protease inhibitors. Soluble proteins were then separated from undissolved tissue using centrifugation. Tissue derived from axolotl was homogenized in a lysis buffer containing urea and dithiothreitol (DTT) followed by further peptide extraction from cell lysates using triethylphosphine, iodoethanol and finally trypsin digestion. To determine the engagement of the matrisome on the proteomics scale it will be necessary to conduct carefully designed experiments that are tailored to the analysis of ECM.
7. Discussion
Identifying key regulatory factors of regeneration is crucial to develop biomaterials that are capable of inducing regeneration in systems that respond to wounding with fibrosis. Research on Acomys demonstrates that regeneration in mammals is possible. Here, we focused on the extracellular matrix (ECM) by performing a direct comparison of the matrisome components present during two distinct regenerative processes in two regenerative species. These were blastema formation and skin wound healing in axolotl and Acomys. This approach identified the common ECM-related denominators in the regenerative wound healing processes that occur in both species. Neither axolotl nor Acomys can be directly compared to humans due to obvious differences in their (skin) biology. However, the identification of ECM-related factors that were present in both an amphibian and mammal support the existence of shared drivers of regeneration that exceed the differences in species. We propose that these matrisome factors play a vital role in regeneration and that specific matrisome factors should be incorporated into novel biomaterials to improve skin healing in non-regenerative species.27,88Recent research provides evidence that regeneration can be induced using only a small selection of components. In axolotl the process of innervation is essential to achieve limb regeneration. In the absence of a nerve, the application of a combination of fibroblast growth factors (FGFs) and bone morphogenetic proteins (BMPs) was enough to rescue limb regeneration.89 Frogs (Xenopus laevis) lose the ability to replace amputated hind limbs through blastema after metamorphosis. Instead, adult individuals resolve amputated limbs through the formation of a cartilaginous spike. Murugan and colleagues reported on the ability of a functionalized device that induced hindlimb regeneration in adult Xenopus leavis.90 A wearable dome-shaped bioreactor (‘biodome’) constructed of a silicone sleeve and containing a silk-based hydrogel loaded with five drugs was attached to the amputation site during the first 24 hours. Animals were followed for 18 months, during which functional hindlimbs regenerated that resembled wildtype hindlimbs. The five drugs in question had been selected for their individual pro-regenerative effects. 1,4-Dihydrophenonthrolin-4-ene-3 carboxylic acid (1,4-DPCA), an inhibitor of prolyl 4-hydroxylase, is an enzyme involved in collagen formation.91 Brain-derived neurotrophic factor (BDNF) is associated with neuron development and axon regeneration.92,93 Growth hormone (GH) has many functions and a recent study on the side effects of GH as an anti-aging therapeutic identified its role as an inhibitor of TGF-β1-mediated myofibroblast activation.94 Resolvin D5, an oxidized lipid mediator, has anti-inflammatory properties by mediating immune cell behavior and recent work indicated this agent has a direct role during re-epithelialization.95,96 Finally, retinoic acid (a vitamin A metabolite) has been found in regenerating zebrafish tissues97 and it was shown to enhance the production of ECM components during wound healing.98 In another study, full-thickness skin wounds of 12 mm diameter were inflicted on the backs of fetal sheep.99 At this wound size and gestational age (day 79 of 140–147 days) fetal sheep are not able to regenerate the skin and instead a fibrotic scar forms.33 However, the regeneration of full-thickness skin wounds in fetal sheep was induced using type I collagen scaffolds functionalized with heparin, vascular endothelial growth factor (VEGF) and fibroblast growth factor 2 (FGF2).99 After postnatal analysis, there was an increase in the skin surface area, a reduction in myofibroblast numbers and evidence of hair follicle formation. In the regenerating ear tissue of Acomys sustained ERK activity was identified as a crucial pro-regenerative factor.100 The application of ERK activators (FGF2 and neuregulin-1) to Mus ear punch wounds induced a pro-regenerative matrisome profile that was characterized by the presence of matrix metalloproteinase 9 and fibronectin 1 and in addition hair follicle neogenesis was stimulated. Together these studies emphasize the feasibility of driving a biological system towards regeneration using only a few selected components.
Before we elucidate on the matrisome components that were extracted in our study the limitations of our study should be addressed. A single study which compares differentially expressed genes in human partial-thickness skin wounds to axolotl full-thickness skin wounds, using pre-existing datasets, is available. This study was seeking for genes that were upregulated in humans and downregulated in axolotl, and vice versa.101 This work identified genes involved in collagen formation, biosynthesis and modification, as well as ECM organization, ECM-receptor interactions and connective tissue development. To date, no transcriptomic or proteomic studies have been published regarding healing of adult human skin following a full-thickness skin wound. Despite the widely acknowledged caveats of extrapolating animal data to humans, comparing datasets obtained from regenerating animal models may be useful to gain knowledge that can be applied to human wound healing. We found a compelling overlap between the matrisome transcriptomes of axolotl and Acomys. This suggests there is a possibility of shared ECM/matrisome based regenerative mechanisms between amphibians and mammals that could potentially also be present, albeit inactive, in humans.102,103 While there was a clear overlap found between the datasets, a direct comparison of numerical data was challenging due to differences in sample harvesting, experimental methodologies and data analyses. Each step from sample extraction to data normalization can affect the final dataset.104–106 Therefore, the expression level of one gene cannot be directly compared between two datasets. For these reasons, our analysis will only show general trends of gene expression. Our approach also leaves out the effects of enzyme activity on protein synthesis, since an enzyme's gene expression does not need to be increased for the enzyme activity to be increased. An example of this may be found in the hyaluronan synthase genes, which were identified in only one comparison, even though hyaluronic acid is known to have positive effects on regeneration.107 Second, different publications use different sets of temporal data points which did not always coincide. In particular, the early timepoints of blastema formation (day 0–4) have not been characterized in Acomys. Comparing only the later time points means early contributors to regeneration are left unexplored. Third, the age range of the animals used in the experiments should be briefly considered. The publications focusing on axolotl all used juvenile animals of 7–11 cm in length.63,79,80 Axolotls reach sexual maturity after 9 to 12 months, when the animal is on average 15 cm long.89,108 The publications on Acomys used animals of varying ages; 6 weeks – 6 months,76 6 months only,31 or ‘sexually mature’.56,57 Both male and female Acomys reach sexually maturity at 2–3 months of age,44 thus the majority of the individuals in the experiments would have been mature. Comparing the life phases of axolotl and Acomys is negligible as axolotls naturally retain their juvenile characteristics even after obtaining sexual maturity. Lastly, the proteomic analyses of the incorporated papers are limited to the soluble fractions of the ECM. For example, only four proteins were found in the proteomics datasets and two of these (fibrinogen beta (Fgb) and gamma chain (Fgg)) are known soluble proteins. Fibrinogens have a well-known role in the healing response: after conversion to fibrin it assembles into a temporary support network. Fibrinogen-deficient mice have highlighted this role in wound stabilization and matrix organization.102 Although many important ECM proteins are soluble, identification of insoluble components of the ECM remains elusive.
Our analysis found multiple matrisome related genes that had similar expression patterns in both models and these genes could therefore potentially be essential to regeneration. In this discussion we will focus solely on the genes that were upregulated in both skin regeneration and blastema, as these could be candidates to be included in pro-regenerative biomaterials.
Glycoproteins are essential molecules in the ECM and are known to be involved in wound healing.109 The widespread functions of glycoproteins explain the presence of many highly upregulated glycoproteins during the regenerative processes. Glycoproteins that were highly upregulated in both axolotl and Acomys during skin regeneration and in the blastema were: tenascin C, fibronectin, laminin alpha 1, thrombospondin 1 and 2, peroxidasin homolog and collagen triple helix repeat containing 1. In general, tenascins are involved with cell adhesion modulation; thereby influencing cell proliferation and migration.110,111Tenascin C is activated after injury in axolotl and is deposited in the wound margins.67 Due to its anti-adhesive behavior and promotion of a softer matrix,112 Tenascin C is thought to lead to proliferation and migration of keratinocytes, fibroblasts, and endothelial cells.113 Tenascin C has been indicated as a component of a “transitional matrix” in regeneration-competent species such as newts and zebrafish.114–116 In humans, tenascin C is highly expressed during wound healing, inflammation and injury. A study using mice demonstrated that tenascin C polypeptides increased type I collagen expression, contributing to ECM strength in young skin.117Fibronectin 1, another component of the transitional matrix, was found in both Acomys and axolotl during skin regeneration and blastema-formation. Fibronectin is crosslinked to the fibrin matrix during hemostasis and enables fibroblast migration.118 Mouse full-thickness skin wounds sealed with a combination of plasma fibrinogen and fibronectin demonstrated enhanced wound healing.119 Similarly, pre-coating fibronectin on full-thickness skin wounds in mice before the application of autologous basal cells improved wound healing.120 Furthermore, it has been shown that fibronectin regulates collagen assembly.121,122 An in depth review on the role of fibronectin and its potential in regeneration was recently published.123Laminin alpha 1, a protein that interacts with fibronectin, was present and upregulated in all datasets. Laminin alpha 1 encodes the alpha I chain of the trimeric laminin protein and is a major component of the basement membrane, with an important role in re-epithelization and angiogenesis during wound healing.124 Laminins are also important in growth factor regulation: the heparin-binding domains on laminin can bind growth factors and fibrin matrices functionalized with laminin improved wound healing in diabetic mice.125 Another consistently upregulated gene during regeneration was the relatively unknown peroxidasin homolog. A study into the role of peroxidasin in the ECM demonstrated that human dermal fibroblasts treated with TGF-β1 to induce myofibroblasts secrete peroxidasin to the ECM where it may co-localize with fibronectin in thick bundles.126 Peroxidasin may also contribute to ECM stabilization via tyrosine crosslinking of proteins127 and it was identified as a catalyst in type IV collagen crosslinking through sulfilimine.128,129 Inhibition of peroxidasin reduced endothelial cell attachment and growth, and without peroxidasin the organization of type IV collagen, fibronectin and laminin into fibrillar networks was diminished.130 The potential role of peroxidasin in ECM formation coupled with its consistent presence in the regenerating matrix indicates this peroxidasin may be an important player in wound healing. The consistent presence of both fibrin(ogen) and fibronectin, as well as their associated proteins, demonstrates their importance during regeneration in regenerative and non-regenerative systems alike. Thrombospondin 1, a large trimeric glycoprotein secreted by a plethora of cell types, has been implicated in many (patho)physiological processes.131 Researchers demonstrated that the inhibition of thrombospondin 1 with antisense oligonucleotides in full-thickness skin wounds of mice markedly impaired the wound healing process.132 Recently thrombospondin 1 has been implicated in fibrillar collagen organization through its inhibitory effect on lysyl oxidase, which is an enzyme responsible for collagen crosslinking in the ECM.133 Thrombospondin 1 also indirectly inhibited myofibroblast differentiation through its effects on the organization of collagenous ECM.133Thrombospondin 2, like thrombospondin 1, is involved in ECM organization and it is secreted primarily by fibroblasts and smooth muscle cells.134 In thrombospondin 2 knock-out mice the skin displayed disorganized collagen fibers, contrasting the parallel orientated fibers found in wild-type mice.135 Additionally, the researchers demonstrated that dermal fibroblasts obtained from these mice produced more matrix metalloproteinase 2, indicating thrombospondin 2 may affect cell–matrix interactions. Taken together, the increased expression levels of thrombospondin 1 and 2 during regeneration underline the impact of ECM organization. Collagen triple helix repeat containing 1 (Cthrc1) is a secreted glycosylated protein that is usually present in embryonic and neonatal tissues. Cthrc1 has shown the ability to inhibit type I and III collagen synthesis.136,137 In skin wounds, it has been localized at sites of collagen deposition and in myofibroblast clusters.137 Moreover, Cthrc1 regulates and inhibits TGF-β1 production which may be a potential anti-fibrotic treatment in the skin.138,139 The consistent upregulation of Cthrc1 during regeneration is indicative of a need for controlled collagen synthesis.
Several genes involved in matrix degradation were upregulated in all transcriptomics datasets: matrix metalloproteinase 3, matrix metalloproteinase 13 and tissue inhibitor of metallopeptidase 1 (Timp1). Matrix remodeling is essential in wound healing, as demonstrated by the total of 14 differentially expressed MMPs that were found during healing of human split-thickness skin wounds, which do not scar.140 It has been described that matrix metalloproteinase 3 degrades both type I and III collagen.67 Research in regenerating axolotl digits showed that the presence of matrix metalloproteinases was essential to effective digit regeneration.141Matrix metalloproteinase 13 is capable of cleaving collagen type I, II, and III and various other fibrillar ECM components, thereby modulating fibroblast-matrix interactions and it is even upregulated in fetal wounds.142Timp1 can inhibit all matrix metalloproteinase family members.143 It has been shown that increased levels of this gene were implicated as a poor outcome marker for increased fibrosis in burn patients and persistence of foot ulcers in diabetic patients.144 On the other hand, a study performed in newts indicated that increased levels of Timp1 during regeneration is required to maintain optimal concentrations of matrix metalloproteinases.145
Regarding proteoglycans associated with the matrisome, proteoglycan 4 and serglycin were present and upregulated in all datasets. Proteoglycans consist of a core protein and one or more covalently attached glycosaminoglycan (GAG) side chains. They make up a major part of the ECM and have a range of purposes. Proteoglycan 4, also known as lubricin, contains the GAGs chondroitin sulfate and keratan sulfate. The biosynthesis of keratan sulfate was highly increased in the regenerating spinal cord of Acomys and the expression of one related biosynthesis gene, beta-1,3-N-acetylglucosaminyltransferase 7 (B3gnt7), was identified as an axon growth enhancer.47 Proteoglycan 4 is more formally associated with blastema formation because of its role in cartilage-bone protection.146 In humans, proteoglycan 4 is associated with the regeneration of cartilage tissue between joint surfaces.147 A recent study elaborated on the anti-fibrotic properties of proteoglycan 4 by demonstrating the ability of this proteoglycan to decrease synovial fibroblast activation in vitro and reduce fibrosis in vivo.148 Its presence during skin regeneration in both Acomys and axolotl underscores the importance of proteoglycan 4 to regeneration in general. This finding is supported by the results of Krawetz et al. (2022), who demonstrated proteoglycan 4 is essential in ear wound healing in mice.149 They showed that proteoglycan 4 modulates macrophage polarization, increases vascularization and promotes cartilage regeneration. The proteoglycan serglycin was upregulated during both blastema and skin regeneration. Serglycin is a well-known intracellular proteoglycan and one of its main roles is the regulation of inflammatory mediators in the granules of mast cells.150 After secrection serglycin may act as a vehicle for the extracellular delivery of the molecules inside granules, such as cytokines, or act as a scavenger in the ECM.151 Thus, serglycin may have a role as an inflammation mediator. Moreover, serglycin is upregulated under the influence of TGF-β1 during epithelial–mesenchymal transition, this transition is an important process during wound healing.152 Serglycin is densely packed with various GAGs, leading to a high density of GAG-binding proteins, and one of these associated GAGs is heparin.151 Heparin is most commonly associated with anticoagulation and anti-inflammation and has widespread use in therapeutic applications.153 For example, treatment of chronic ulcers with low molecular weight heparin resulted in increased numbers of healed patients and reduced wound recurrence rates.154,155 These findings illustrate the beneficial effects of heparin on the healing of chronic wounds.
Considering the upregulation of various proteoglycans, it is to be expected that several genes related to the linkage and modification of GAG chains were also upregulated during regeneration: carbohydrate sulfotransferase 2, carbohydrate sulfotransferase 11 and sulfatase 1. Carbohydrate sulfotransferases are responsible for the transfer of sulfate groups onto carbohydrates. Our analysis returned two carbohydrate sulfotransferase genes that were always upregulated during regeneration. Carbohydrate sulfotransferase 2, also known as N-acetylglucosamine 6-O-sulfotransferase or GlcNAc6ST1, is associated with the biosynthesis of keratan sulfate.156 This enzyme may play a direct role in the inflammatory response as it is a part of L-selectin ligand synthesis: these ligands are responsible for leukocyte rolling and capture in high endothelial venules.157,158Carbohydrate sulfotransferase 11, or chondroitin-4-sulfotransferase 1, is an enzyme involved in the biosynthesis of chondroitin sulfate.159 In Costello syndrome, where overexpression of the HRAS oncogene leads to a reduction in carbohydrate sulfotransferase 11, skin fibroblasts do not produce elastic fibers. Forced expression of carbohydrate sulfotransferase 11 rescued the elastic fiber production.160 Chondroitin sulfate itself was shown to improve palatal wound healing by promoting fibroblast adhesion and proliferation.161Sulfatase 1 is an extracellular enzyme that removes 6-O-sulfates from heparan sulfate and augments Wnt/β-catenin signaling. Following wounding of the corneal epithelium in mice sulfatase 1 was found in the wound edges and knockdown of sulfatase 1 led to a decrease in cell migration and delayed wound healing.162 Similarly, during hind limb regeneration in Xenopus laevis high levels of sulfatase 1 were present during the first three days in the blastema, which demonstrates its the importance in regeneration.163 Moreover, it was found that sulfatase 1 is upregulated during mouse embryonic skin development and is likely involved in cellular signaling.85 Taken together, the three enzymes that were found in the datasets regulate sulfation patterns of GAGs, which in turn modulates the bioactivity. The GAG heparan sulfate (HS) binds many proteins, such as growth factors, that reside in the ECM and the interaction of HS-binding proteins and receptors is modulated by HS proteoglycans.164,165 Changes in sulfation patterns of GAGs may thus have far-reaching effects in modulating cell behavior.
Lastly, two collagens were identified in the proteomics datasets. These collagens, type I collagen type alpha 1 chain and type XII collagen alpha 1 chain, are important during regeneration albeit in different ways. As part of the core matrisome, collagens have a great influence on the overall structure of the ECM and cell behavior. Type I collagen, a member of the fibrillar collagens, contributes to over 90% of the collagen content in skin ECM.166 Scar tissue consists of a dense collagenous matrix organized in thick parallel orientated bundles, in contrast to healthy dermis where the collagen is organized into a basket weave-like pattern.7 In regenerating Acomys skin, type I collagen was loosely organized and less abundant as shown through trichrome staining, resembling undamaged skin.51 To achieve skin regeneration, the architecture of type I collagen should match that of unwounded skin, which requires guiding the deposition and remodeling of type I collagen during wound healing. Type XII collagen is a member of the Fibril Associated Collagens with Interrupted Triple helices (FACIT). This collagen has binding sites for type I collagen, to assist in type I collagen packing, and GAGs (such as heparan sulfate).167,168 Additionally, type XII collagen helps to maintain the structure of the ECM and responds to mechanical stretch by absorbing stress. Type XII collagen is expressed in the dermis during skin development in humans, however, after birth it is found only in the papillary dermis and surrounding hair follicles.169 Following skin injury, the scarring Mus musculus displays significant increases in type XII collagen compared to Acomys, which indicates that an abundance of type XII collagen could also be detrimental to a regenerative matrix.51,55 Similarly, timing the deposition of type I and XII collagen has shown to play a key role in skin regeneration in the axolotl.103
In summary, our analysis of matrisome factors revealed several genes that are upregulated during regeneration in both axolotl and Acomys. In some cases, they encode proteins which are known to have a positive influence on wound healing in mammals or are in some way related to the structural organization of the ECM. The fact that fibrotic skin is characterized by the thick parallel organization of type I collagen while in undamaged skin a basket-weave pattern is seen suggests that a regenerative biomaterial should focus on re-establishing the normal ECM architecture and organization (Fig. 8).
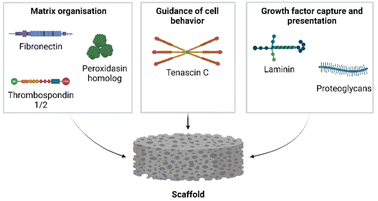 | ||
| Fig. 8 Components to be considered for incorporation in pro-regenerative biomaterials. Created with BioRender.com. | ||
Several of the components identified in our analysis may be considered for use in a regenerative biomaterial. The incorporation of fibronectin in a biomaterial would aid in the regulation of collagen fiber assembly. Several other components can be considered to strengthen the matrix organizing properties, such as thrombospondin 1 and/or 2, as well as peroxidasin homolog. Growth factors are an integral part of regeneration, and their capture should be facilitated. This may be achieved through the incorporation of molecules that have growth factor binding abilities, such as laminin alpha 1, or various proteoglycans which may bind growth factors through their GAG chains. The glycoprotein tenascin c is known for its positive influence on wound healing by enhancing cell migration. Its constant presence in our datasets supports the inclusion of tenascin c in a pro-regenerative biomaterial. The proteoglycans serglycin and proteoglycan 4 were an unexpected finding in our dataset, as their exact roles in skin wound healing are not well known. This warrants a more detailed investigation into the influence of the various proteoglycans on wound healing to determine which would be the most appropriate for incorporation in a pro-regenerative scaffold. Proteoglycan-associated GAGs, such as heparin in serglycin and chondroitin sulfate or keratan sulfate in proteoglycan 4, have shown potential in promoting wound healing and should be considered for incorporation into the pro-regenerative biomaterial. Phan et al. (2021) showed that heparan sulfates, together with fibroblast growth factor signaling, are required to provide the correct positional information during axolotl limb regeneration.170
There is a myriad of options available for the development and production of pro-regenerative biomaterials. Such materials may contain natural or synthetic components, or a combination of both. The application of natural-derived components is widespread. For example, decellularized tissues maintain the native tissue architecture and many of the signaling factors.171,172 A wide range of decellularized skin substitutes from human and animal sources are commercially available and have been used in clinical practice.173 Decellularized human nail beds have been used to stimulate bone regeneration in rats.174,175 These examples establish the ability of the native ECM to promote regenerative processes. In addition to naturally derived components, chemically or enzymatically produced components may be considered. Standardized processes greatly decrease variations between batches and the low immunogenicity makes synthetic compounds an interesting alternative.176 Besides the incorporation of proteinaceous compounds in biomaterials, carbohydrates – and GAGs in particular – encompass a class of molecules with diverse biological functions. Unlike proteins, they are more difficult to analyze due to the non-template driven biosynthesis and extensive structural heterogeneity of these long polysaccharides. In the context of biomaterial development their multitude of functions make them important to consider. GAGs are known for their role in organizing the extracellular matrix and for their ability to bind and present bioactive molecules, protecting these from proteolytic degradation, and regulating growth factor gradients.177 Many examples of the exploitation of such functions can be found in the literature, from hyaluronan injectables178 to collagen-based skin constructs containing dermatan sulfate and heparin.179 However, one GAG may have various domains in the sulfation pattern and its structure can vary during development and (patho)physiology. To avoid the use of heterogenous natural GAGs with potential batch-to-batch variations, novel strategies have been developed to obtain more defined preparations. One approach includes the use of degradation-resistant carboxymethyl-dextran sulfates: these heparan sulfate mimetics are capable of binding and protecting growth factors already present in the wound matrix, thereby allowing the tissue to regenerate.180 These ReGeneraTing Agents (RGTA®) have shown great potential during in vivo studies and were clinically used to treat ulcers.181,182 The basic research into defined saccharides offers an original approach to their application in biomaterials.183 In addition, chemo-enzymatic synthesis of well-characterized oligosaccharides with specific biological functions opens a whole new avenue in the field of regenerative medicine.184–186
Future efforts should be directed towards functionality-based studies in order to elucidate the function of the matrisome components identified here in the regenerating matrix. This may be aided by improving the proteomic analysis of insoluble matrisome components. For example, the insoluble ECM pellet can be digested to allow proteomic analysis of insoluble matrisome components.87 This can be achieved by digesting insoluble ECM pellets with hydroxylamine187 or with a photocleavable linker such as 4-hexylphenylazosulfonate.188,189 Both approaches yield a solubilized ECM fraction compatible with proteomic analysis pipelines and are relatively easy to implement. Efforts are already being made to improve the proteomic coverage of the matrisome. A universal tool titled ‘MatrisomeDB’ is available to all researchers, and it facilitates the reuse of proteomic datasets with a focus on the matrisome.190 To obtain robust insights in the trends in gene and protein expression patterns observed in both species, it would require a united effort to analyze all data via identical methods and with matching timepoints in the regenerative process. Furthermore, the investigation of early timepoints in Acomys is highly recommended to provide a detailed image of the processes that occur during the first phases of regeneration.
8. Conclusions
With the increasing knowledge on the complexity of regenerative environments, the next generation of biomaterials should focus on the rational incorporation of various pro-regenerative matrix molecules. By applying such knowledge to the development of biomaterials we open new avenues towards improving wound healing and obtaining regeneration in humans.Author contributions
Conceptualization, T. H. v. K. and W. F. D.; formal analysis N. A. M. and M. G.; methodology, N. A. M. and M. G.; funding acquisition, W. F. D.; project administration, W. F. D.; supervision, W. F. D. and T. H. v. K.; visualization, M. G. and N. A. M.; writing – original draft, N. A. M. and M. G., J. V.; writing – review and editing, W. F. D., T. H. v. K., G. T., M. V., I. M. A., H. J., J. V., M. G. and N. A. M.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Tahar van der Straaten, Field Applications Scientist, Microarray support from Thermo Fisher Scientific for his support in the extraction and analysis of the axolotl skin transcriptomics genes which were further used for the comparison with Acomys.This work has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement No. 955722 (SkinTERM, N. A. M. and H. J.) and from a Radboudumc RIMLS Young Investigator Grant (RIMLS, M. G.).
References
- P. K. Williams-Boyce and J. C. Daniel Jr., J. Exp. Zool., 1980, 212, 243–253 CrossRef CAS PubMed.
- P. G. ten Koppel, G. J. van Osch, C. D. Verwoerd and H. L. Verwoerd-Verhoef, Biomaterials, 2001, 22, 1407–1414 CrossRef CAS PubMed.
- J. Price and S. Allen, Philos. Trans. R. Soc., B, 2004, 359, 809–822 CrossRef CAS PubMed.
- U. Kierdorf and H. Kierdorf, Front. Biosci., Elite Ed., 2012, 4, 1606–1624 CrossRef PubMed.
- L. Yu, M. J. Han, M. Q. Yan, J. Lee and K. Muneoka, Dev. Biol., 2012, 372, 263–273 CrossRef CAS PubMed.
- J. Simkin, M. C. Sammarco, L. A. Dawson, P. P. Schanes, L. Yu and K. Muneoka, Regeneration, 2015, 2, 93–105 CrossRef CAS PubMed.
- P. Martin, Science, 1997, 276, 75–81 CrossRef CAS PubMed.
- J. H. W. Distler, A. H. Gyorfi, M. Ramanujam, M. L. Whitfield, M. Konigshoff and R. Lafyatis, Nat. Rev. Rheumatol., 2019, 15, 705–730 CrossRef CAS PubMed.
- P. C. Esselman, Arch. Phys. Med. Rehabil., 2007, 88, S3–S6 CrossRef PubMed.
- J. M. Reinke and H. Sorg, Eur. Surg. Res., 2012, 49, 35–43 CrossRef CAS PubMed.
- E. S. White and A. R. Mantovani, J. Pathol., 2013, 229, 141–144 CrossRef CAS PubMed.
- N. Di Caprio and E. Bellas, Adv. Biosyst., 2020, 4, e1900286 CrossRef PubMed.
- M. F. Brizzi, G. Tarone and P. Defilippi, Curr. Opin. Cell Biol., 2012, 24, 645–651 CrossRef CAS PubMed.
- F. Mercier, Cell. Mol. Life Sci., 2016, 73, 4661–4674 CrossRef CAS PubMed.
- A. Naba, K. R. Clauser, H. Ding, C. A. Whittaker, S. A. Carr and R. O. Hynes, Matrix Biol., 2016, 49, 10–24 CrossRef CAS PubMed.
- R. O. Hynes and A. Naba, Cold Spring Harbor Perspectives in Biology, 2012, vol. 4, p. a004903 Search PubMed.
- A. Heydemann, Front. Biol., 2012, 7, 522–538 CrossRef PubMed.
- M. Suzuki, N. Yakushiji, Y. Nakada, A. Satoh, H. Ide and K. Tamura, Sci. World J., 2006, 6 Suppl 1, 26–37 CrossRef PubMed.
- J. F. Denis, M. Levesque, S. D. Tran, A. J. Camarda and S. Roy, Adv. Wound Care, 2013, 2, 250–260 CrossRef PubMed.
- R. Richardson, K. Slanchev, C. Kraus, P. Knyphausen, S. Eming and M. Hammerschmidt, J. Invest. Dermatol., 2013, 133, 1655–1665 CrossRef CAS PubMed.
- S. E. Mutsaers, J. E. Bishop, G. McGrouther and G. J. Laurent, Int. J. Biochem. Cell Biol., 1997, 29, 5–17 CrossRef CAS PubMed.
- G. Henry and W. L. Garner, Surg. Clin. North Am., 2003, 83, 483–507 CrossRef PubMed.
- T. Velnar, T. Bailey and V. Smrkolj, J. Int. Med. Res., 2009, 37, 1528–1542 CrossRef CAS PubMed.
- H. P. Ehrlich, A. Desmouliere, R. F. Diegelmann, I. K. Cohen, C. C. Compton, W. L. Garner, Y. Kapanci and G. Gabbiani, Am. J. Pathol., 1994, 145, 105–113 CAS.
- M. C. Vogg, Y. Wenger and B. Galliot, Essays on Developmental Biology, Pt A, 2016, vol. 116, pp. 391–414 Search PubMed.
- C. McCusker, S. V. Bryant and D. M. Gardiner, Regeneration, 2015, 2, 54–71 CrossRef PubMed.
- A. W. Seifert and K. Muneoka, Dev. Biol., 2018, 433, 190–199 CrossRef CAS PubMed.
- A. Satoh, S. V. Bryant and D. M. Gardiner, Dev. Biol., 2012, 366, 374–381 CrossRef CAS PubMed.
- G. G. Whitehead, S. Makino, C. L. Lien and M. T. Keating, Science, 2005, 310, 1957–1960 CrossRef CAS PubMed.
- J. Girstmair, R. Schnegg, M. J. Telford and B. Egger, EvoDevo, 2014, 5, 37 CrossRef PubMed.
- T. R. Gawriluk, J. Simkin, K. L. Thompson, S. K. Biswas, Z. Clare-Salzler, J. M. Kimani, S. G. Kiama, J. J. Smith, V. O. Ezenwa and A. W. Seifert, Nat. Commun., 2016, 7, 11164 CrossRef CAS PubMed.
- B. J. Larson, M. T. Longaker and H. P. Lorenz, Plast. Reconstr. Surg., 2010, 126, 1172–1180 CrossRef CAS PubMed.
- D. L. Cass, K. M. Bullard, K. G. Sylvester, E. Y. Yang, M. T. Longaker and N. S. Adzick, J. Pediatr. Surg., 1997, 32, 411–415 CrossRef CAS PubMed.
- C. C. Yates, P. Hebda and A. Wells, Birth Defects Res., Part C, 2012, 96, 325–333 CrossRef CAS PubMed.
- P. Martin and J. Lewis, Nature, 1992, 360, 179–183 CrossRef CAS PubMed.
- J. Brock, K. Midwinter, J. Lewis and P. Martin, J. Cell Biol., 1996, 135, 1097–1107 CrossRef CAS PubMed.
- I. V. Yannas and D. S. Tzeranis, npj Regener. Med., 2021, 6, 39 CrossRef PubMed.
- M. M. W. Ulrich, in Textbook on Scar Management: State of the Art Management and Emerging Technologies, ed. L. Téot, T. A. Mustoe, E. Middelkoop and G. G. Gauglitz, Springer International Publishing, Cham, 2020, pp. 3–9, DOI:10.1007/978-3-030-44766-3_1.
- P. Vidal and M. G. Dickson, J. Hand Surg. Br., 1993, 18, 230–233 CrossRef CAS PubMed.
- R. B. Borgens, Science, 1982, 217, 747–750 CrossRef CAS PubMed.
- B. S. Douglas, Aust. Paediatr. J., 1972, 8, 86–89 CAS.
- S. K. Das and H. G. Brown, Hand, 1978, 10, 16–27 CrossRef CAS PubMed.
- M. A. Storer, N. Mahmud, K. Karamboulas, M. J. Borrett, S. A. Yuzwa, A. Gont, A. Androschuk, M. V. Sefton, D. R. Kaplan and F. D. Miller, Dev. Cell, 2020, 52, 509–524 CrossRef CAS PubMed.
- C. L. Haughton, T. R. Gawriluk and A. W. Seifert, J. Am. Assoc. Lab. Anim. Sci., 2016, 55, 9–17 Search PubMed.
- M. Maden and J. A. Varholick, Development, 2020, 147, dev167718 CrossRef CAS PubMed.
- A. W. Seifert, S. G. Kiama, M. G. Seifert, J. R. Goheen, T. M. Palmer and M. Maden, Nature, 2012, 489, 561–565 CrossRef CAS PubMed.
- J. Nogueira-Rodrigues, S. C. Leite, R. Pinto-Costa, S. C. Sousa, L. L. Luz, M. A. Sintra, R. Oliveira, A. C. Monteiro, G. G. Pinheiro, M. Vitorino, J. A. Silva, S. Simão, V. E. Fernandes, J. Provazník, V. Benes, C. D. Cruz, B. V. Safronov, A. Magalhães, C. A. Reis, J. Vieira, C. P. Vieira, G. Tiscórnia, I. M. Araújo and M. M. Sousa, Dev. Cell, 2022, 57, 440–450 CrossRef CAS PubMed.
- A. G. W. Sandoval and M. Maden, Curr. Opin. Genet. Dev., 2020, 64, 31–36 CrossRef CAS PubMed.
- M. Maden and J. O. Brant, Exp. Dermatol., 2019, 28, 436–441 CrossRef PubMed.
- J. Simkin, T. R. Gawriluk, J. C. Gensel and A. W. Seifert, eLife, 2017, 6, e24623 CrossRef PubMed.
- J. O. Brant, J. H. Yoon, T. Polvadore, W. B. Barbazuk and M. Maden, Wound Repair Regen., 2016, 24, 75–88 CrossRef PubMed.
- J. L. Cyr, T. R. Gawriluk, J. M. Kimani, B. Rada, W. T. Watford, S. G. Kiama, A. W. Seifert and V. O. Ezenwa, Integr. Comp. Biol., 2019, 59, 1138–1149 CrossRef CAS PubMed.
- Y. Kuninaka, Y. Ishida, A. Ishigami, M. Nosaka, J. Matsuki, H. Yasuda, A. Kofuna, A. Kimura, F. Furukawa and T. Kondo, Sci. Rep., 2022, 12, 20327 CrossRef CAS PubMed.
- H. I. Harn, S. P. Wang, Y. C. Lai, B. Van Handel, Y. C. Liang, S. Tsai, I. M. Schiessl, A. Sarkar, H. Xi, M. Hughes, S. Kaemmer, M. J. Tang, J. Peti-Peterdi, A. D. Pyle, T. E. Woolley, D. Evseenko, T. X. Jiang and C. M. Chuong, Nat. Commun., 2021, 12, 2595 CrossRef CAS PubMed.
- J. O. Brant, M. C. Lopez, H. V. Baker, W. B. Barbazuk and M. Maden, PLoS One, 2015, 10, e0142931 CrossRef PubMed.
- J. H. Yoon, K. Cho, T. J. Garrett, P. Finch and M. Maden, Sci. Rep., 2020, 10, 8641 CrossRef CAS PubMed.
- J. H. Yoon, K. Cho, T. J. Garrett, P. Finch and M. Maden, Sci. Rep., 2020, 10, 166 CrossRef CAS PubMed.
- D. Matias Santos, A. M. Rita, I. Casanellas, A. Brito Ova, I. M. Araújo, D. Power and G. Tiscornia, Regeneration, 2016, 3, 52–61 CrossRef PubMed.
- J. Gresens, Lab. Anim., 2004, 33, 41–47 CrossRef PubMed.
- A. Crowner, S. Khatri, D. Blichmann and S. R. Voss, Front. Endocrinol., 2019, 10, 237 CrossRef PubMed.
- S. R. Voss, H. H. Epperlein, E. M. Tanaka, S. Khattak, T. Richter, E. Nacu, D. Knapp, M. Kragl and R. B. Page and P. Cold Spring Harbor Laboratory, Emerging Model Organisms: Laboratory Manual, vol. 2, 2010, vol. 2009, 397–419 Search PubMed.
- R. J. Debuque and J. W. Godwin, Stem Cells Biol. Reg., 2016, 1–21, DOI:10.1007/978-3-319-44996-8_1.
- N. Rao, D. Jhamb, D. J. Milner, B. B. Li, F. Y. Song, M. Wang, S. R. Voss, M. Palakal, M. W. King, B. Saranjami, H. L. D. Nye, J. A. Cameron and D. L. Stocum, Bmc Biol., 2009, 7, 1–25 CrossRef PubMed.
- T. Endo, S. V. Bryant and D. M. Gardiner, Dev. Biol., 2004, 270, 135–145 CrossRef CAS PubMed.
- C. S. Thornton, J. Exp. Zool., 1957, 134, 357–381 CrossRef CAS PubMed.
- R. N. Christensen and R. A. Tassava, Dev. Dyn., 2000, 217, 216–224 CrossRef CAS PubMed.
- A. W. Seifert, J. R. Monaghan, S. R. Voss and M. Maden, PLoS One, 2012, 7, e32875 CrossRef CAS PubMed.
- A. K. Rodgers, J. J. Smith and S. R. Voss, Exp. Cell Res., 2020, 394, 112149 CrossRef CAS PubMed.
- J. A. Marwick, R. Mills, O. Kay, K. Michail, J. Stephen, A. G. Rossi, I. Dransfield and N. Hirani, Cell Death Dis., 2018, 9, 665 CrossRef PubMed.
- H. B. Li, X. Y. Wei, L. Zhou, W. Q. Zhang, C. Wang, Y. Guo, D. H. Li, J. Y. Chen, T. B. Liu, Y. Y. Zhang, S. Ma, C. Y. Wang, F. J. Tan, J. S. Xu, Y. Liu, Y. Yuan, L. Chen, Q. R. Wang, J. Qu, Y. Shen, S. S. Liu, G. Y. Fan, L. Q. Liu, X. Liu, Y. Hou, G. H. Liu, Y. Gu and X. Xu, Protein Cell, 2021, 12, 57–66 CrossRef PubMed.
- J. W. Godwin, A. R. Pinto and N. A. Rosenthal, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9415–9420 CrossRef CAS PubMed.
- A. Satoh, A. Hirata and A. Makanae, Zool. Sci., 2012, 29, 191–197 CrossRef CAS PubMed.
- A. Q. Phan, J. Lee, M. Oei, C. Flath, C. Hwe, R. Mariano, T. Vu, C. Shu, A. Dinh, J. Simkin, K. Muneoka, S. V. Bryant and D. M. Gardiner, Regeneration, 2015, 2, 182–201 CrossRef CAS PubMed.
- T. Demircan, A. E. Ilhan, N. Ayturk, B. Yildirim, G. Ozturk and I. Keskin, Acta Histochem., 2016, 118, 746–759 CrossRef PubMed.
- R. Kashimoto, S. Furukawa, S. Yamamoto, Y. Kamei, J. Sakamoto, S. Nonaka, T. M. Watanabe, T. Sakamoto, H. Sakamoto and A. Satoh, iScience, 2022, 25, 104524 CrossRef CAS PubMed.
- J. O. Brant, J. L. Boatwright, R. Davenport, A. G. W. Sandoval, M. Maden and W. B. Barbazuk, PLoS One, 2019, 14, e0216228 CrossRef CAS PubMed.
- J. R. Monaghan, L. G. Epp, S. Putta, R. B. Page, J. A. Walker, C. K. Beachy, W. Zhu, G. M. Pao, I. M. Verma, T. Hunter, S. V. Bryant, D. M. Gardiner, T. T. Harkins and S. R. Voss, BMC Biol., 2009, 7, 1–19 CrossRef PubMed.
- C. H. Wu, M. H. Tsai, C. C. Ho, C. Y. Chen and H. S. Lee, BMC Genomics, 2013, 14, 434 CrossRef CAS PubMed.
- J. R. Monaghan, A. Athippozhy, A. W. Seifert, S. Putta, A. J. Stromberg, M. Maden, D. M. Gardiner and S. R. Voss, Biol. Open, 2012, 1, 937–948 CrossRef PubMed.
- R. Stewart, C. A. Rascon, S. L. Tian, J. Nie, C. Barry, L. F. Chu, H. Ardalani, R. J. Wagner, M. D. Probasco, J. M. Bolin, N. Leng, S. Sengupta, M. Volkmer, B. Habermann, E. M. Tanaka, J. A. Thomson and C. N. Dewey, PLoS Comput. Biol., 2013, 9, e100293 Search PubMed.
- R. V. Iozzo and L. Schaefer, Matrix Biol., 2015, 42, 11–55 CrossRef CAS PubMed.
- F. Kubaski, H. Osago, R. W. Mason, S. Yamaguchi, H. Kobayashi, M. Tsuchiya, T. Orii and S. Tomatsu, Mol. Genet. Metab., 2017, 120, 67–77 CrossRef CAS PubMed.
- S. Heath, Y. Han, R. Hua, A. Roy, J. Jiang, J. S. Nyman and X. Wang, Bone, 2023, 171, 116751 CrossRef CAS PubMed.
- M. Grabarics, M. Lettow, C. Kirschbaum, K. Greis, C. Manz and K. Pagel, Chem. Rev., 2022, 122, 7840–7908 CrossRef CAS PubMed.
- P. J. E. Uijtdewilligen, E. M. Versteeg, E. M. A. van de Westerlo, J. van der Vlag, W. F. Daamen and T. H. van Kuppevelt, BioMed Res. Int., 2018, 2018, 9873471 CAS.
- T. R. Gawriluk, J. Simkin, C. K. Hacker, J. M. Kimani, S. G. Kiama, V. O. Ezenwa and A. W. Seifert, Front. Immunol., 2020, 11, 1695 CrossRef CAS PubMed.
- M. C. McCabe, L. R. Schmitt, R. C. Hill, M. Dzieciatkowska, M. Maslanka, W. F. Daamen, T. H. van Kuppevelt, D. J. Hof and K. C. Hansen, Mol. Cell. Proteomics, 2021, 20, 100079 CrossRef CAS PubMed.
- S. Owlarn, F. Klenner, D. Schmidt, F. Rabert, A. Tomasso, H. Reuter, M. A. Mulaw, S. Moritz, L. Gentile, G. Weidinger and K. Bartscherer, Nat. Commun., 2017, 8, 2282 CrossRef PubMed.
- W. A. Vieira, K. M. Wells and C. D. McCusker, Gerontology, 2020, 66, 212–222 CrossRef PubMed.
- N. J. Murugan, H. J. Vigran, K. A. Miller, A. Golding, Q. L. Pham, M. M. Sperry, C. Rasmussen-Ivey, A. W. Kane, D. L. Kaplan and M. Levin, Sci. Adv., 2022, 8, eabj2164 CrossRef CAS PubMed.
- T. Pihlajaniemi, R. Myllyla and K. I. Kivirikko, J. Hepatol., 1991, 13 Suppl 3, S2–S7 CrossRef CAS PubMed.
- S. Bathina and U. N. Das, Arch. Med. Sci., 2015, 11, 1164–1178 CrossRef CAS PubMed.
- C. E. McGregor and A. W. English, Front. Cell. Neurosci., 2018, 12, 522 CrossRef CAS PubMed.
- I. S. Thorey, B. Hinz, A. Hoeflich, S. Kaesler, P. Bugnon, M. Elmlinger, R. Wanke, E. Wolf and S. Werner, J. Biol. Chem., 2004, 279, 26674–26684 CrossRef CAS PubMed.
- H. W. Chun, J. Lee, T. H. Pham, J. Lee, J. H. Yoon, J. Lee, D. K. Oh, J. Oh and D. Y. Yoon, J. Microbiol. Biotechnol., 2020, 30, 85–92 CrossRef CAS PubMed.
- J. Hellmann, B. E. Sansbury, B. Wong, X. Li, M. Singh, K. Nuutila, N. Chiang, E. Eriksson, C. N. Serhan and M. Spite, J. Invest. Dermatol., 2018, 138, 2051–2060 CrossRef CAS PubMed.
- L. J. Gudas, Biochim. Biophys. Acta, 2012, 1821, 213–221 CrossRef CAS PubMed.
- M. E. Polcz and A. Barbul, Nutr. Clin. Pract., 2019, 34, 695–700 CrossRef CAS PubMed.
- C. Oostendorp, P. J. Geutjes, F. Smit, D. M. Tiemessen, S. Polman, A. Abbawi, K. M. Brouwer, A. J. Eggink, W. F. J. Feitz, W. F. Daamen and T. H. van Kuppevelt, Tissue Eng., Part A, 2020, 27, 10–25 CrossRef PubMed.
- A. Tomasso, T. Koopmans, P. Lijnzaad, K. Bartscherer and A. W. Seifert, Sci. Adv., 2023, 9, eadf2331 CrossRef CAS PubMed.
- E. K. Oktem, M. Yazar, G. Gulfidan and K. Y. Arga, OMICS, 2019, 23, 389–405 CrossRef PubMed.
- N. Suzuki and H. Ochi, Dev., Growth Differ., 2020, 62, 343–354 CrossRef PubMed.
- J. R. Erickson, M. D. Gearhart, D. D. Honson, T. A. Reid, M. K. Gardner, B. S. Moriarity and K. Echeverri, npj Regener. Med., 2016, 1, 16016 CrossRef PubMed.
- A. Conesa, P. Madrigal, S. Tarazona, D. Gomez-Cabrero, A. Cervera, A. McPherson, M. W. Szczesniak, D. J. Gaffney, L. L. Elo, X. Zhang and A. Mortazavi, Genome Biol., 2016, 17, 13 CrossRef PubMed.
- A. Conesa, P. Madrigal, S. Tarazona, D. Gomez-Cabrero, A. Cervera, A. McPherson, M. W. Szczesniak, D. J. Gaffney, L. L. Elo, X. Zhang and A. Mortazavi, Genome Biol., 2016, 17, 181 CrossRef PubMed.
- A. Stupnikov, C. E. McInerney, K. I. Savage, S. A. McIntosh, F. Emmert-Streib, R. Kennedy, M. Salto-Tellez, K. M. Prise and D. G. McArt, Comput. Struct. Biotechnol. J., 2021, 19, 3470–3481 CrossRef CAS PubMed.
- M. Litwiniuk, A. Krejner, M. S. Speyrer, A. R. Gauto and T. Grzela, Wounds, 2016, 28, 78–88 Search PubMed.
- L. Zambrano, E. Vega, L. G. Herrera, E. Prado and V. H. Reynoso, Anim. Conserv., 2007, 10, 297–303 CrossRef.
- F. X. Maquart and J. C. Monboisse, Pathol. Biol., 2014, 62, 91–95 CrossRef CAS PubMed.
- R. Chiquet-Ehrismann and R. P. Tucker, Cold Spring Harbor Perspect. Biol., 2011, 3, a004960 Search PubMed.
- D. J. Donaldson, J. T. Mahan, H. Yang and K. L. Crossin, Anat. Rec., 1991, 230, 451–459 CrossRef CAS PubMed.
- T. Y. Huang, C. H. Wu, M. H. Wang, B. S. Chen, L. L. Chiou and H. S. Lee, BioMed Res. Int., 2015, 2015, 1–10 Search PubMed.
- K. S. Midwood and G. Orend, J. Cell Commun. Signaling, 2009, 3, 287–310 CrossRef PubMed.
- S. Calve, S. J. Odelberg and H. G. Simon, Dev. Biol., 2010, 344, 259–271 CrossRef CAS PubMed.
- S. E. Mercer, C. H. Cheng, D. L. Atkinson, J. Krcmery, C. E. Guzman, D. T. Kent, K. Zukor, K. A. Marx, S. J. Odelberg and H. G. Simon, PLoS One, 2012, 7, e52375 CrossRef CAS PubMed.
- J. Govindan and M. K. Iovine, Gene Expression Patterns, 2015, 19, 21–29 CrossRef CAS PubMed.
- Y. E. Choi, M. J. Song, M. Hara, K. Imanaka-Yoshida, D. H. Lee, J. H. Chung and S. T. Lee, Int. J. Mol. Sci., 2020, 21, 8693 CrossRef CAS PubMed.
- E. Makogonenko, G. Tsurupa, K. Ingham and L. Medved, Biochemistry, 2002, 41, 7907–7913 CrossRef CAS PubMed.
- C. P. Jara, O. Wang, T. Paulino do Prado, A. Ismail, F. M. Fabian, H. Li, L. A. Velloso, M. A. Carlson, W. Burgess, Y. Lei, W. H. Velander and E. P. Araujo, Bioact. Mater., 2020, 5, 949–962 CrossRef PubMed.
- P. Wang, Z. Hu, X. Cao, S. Huang, Y. Dong, P. Cheng, H. Xu, B. Shu, J. Xie, J. Wu, B. Tang and J. Zhu, Stem Cell Res. Ther., 2019, 10, 154 CrossRef PubMed.
- J. A. Paten, C. L. Martin, J. T. Wanis, S. M. Siadat, A. M. Figueroa-Navedo, J. W. Ruberti and L. F. Deravi, Chem, 2019, 5, 2126–2145 CAS.
- K. E. Kubow, R. Vukmirovic, L. Zhe, E. Klotzsch, M. L. Smith, D. Gourdon, S. Luna and V. Vogel, Nat. Commun., 2015, 6, 8026 CrossRef CAS PubMed.
- J. Patten and K. Wang, Adv. Drug Delivery Rev., 2021, 170, 353–368 CrossRef CAS PubMed.
- V. Iorio, L. D. Troughton and K. J. Hamill, Adv. Wound Care, 2015, 4, 250–263 CrossRef PubMed.
- J. Ishihara, A. Ishihara, K. Fukunaga, K. Sasaki, M. J. V. White, P. S. Briquez and J. A. Hubbell, Nat. Commun., 2018, 9, 2163 CrossRef PubMed.
- Z. Peterfi, A. Donko, A. Orient, A. Sum, A. Prokai, B. Molnar, Z. Vereb, E. Rajnavolgyi, K. J. Kovacs, V. Muller, A. J. Szabo and M. Geiszt, Am. J. Pathol., 2009, 175, 725–735 CrossRef CAS PubMed.
- Z. Peterfi and M. Geiszt, Trends Biochem. Sci., 2014, 39, 305–307 CrossRef CAS PubMed.
- G. Bhave, C. F. Cummings, R. M. Vanacore, C. Kumagai-Cresse, I. A. Ero-Tolliver, M. Rafi, J. S. Kang, V. Pedchenko, L. I. Fessler, J. H. Fessler and B. G. Hudson, Nat. Chem. Biol., 2012, 8, 784–790 CrossRef CAS PubMed.
- E. Lazar, Z. Peterfi, G. Sirokmany, H. A. Kovacs, E. Klement, K. F. Medzihradszky and M. Geiszt, Free Radicals Biol. Med., 2015, 83, 273–282 CrossRef CAS PubMed.
- S. W. Lee, H. K. Kim, P. Naidansuren, K. A. Ham, H. S. Choi, H. Y. Ahn, M. Kim, D. H. Kang, S. W. Kang and Y. A. Joe, FASEB J., 2020, 34, 10228–10241 CrossRef CAS PubMed.
- N. Esemuede, T. Lee, D. Pierre-Paul, B. E. Sumpio and V. Gahtan, J. Surg. Res., 2004, 122, 135–142 CrossRef CAS PubMed.
- L. A. DiPietro, N. N. Nissen, R. L. Gamelli, A. E. Koch, J. M. Pyle and P. J. Polverini, Am. J. Pathol., 1996, 148, 1851–1860 CAS.
- S. Rosini, N. Pugh, A. M. Bonna, D. J. S. Hulmes, R. W. Farndale and J. C. Adams, Sci. Signaling, 2018, 11, eaar2566 CrossRef PubMed.
- N. E. Calabro, N. J. Kristofik and T. R. Kyriakides, Biochim. Biophys. Acta, 2014, 1840, 2396–2402 CrossRef CAS PubMed.
- P. Bornstein, T. R. Kyriakides, Z. Yang, L. C. Armstrong and D. E. Birk, J. Invest. Dermatol. Symp. Proc., 2000, 5, 61–66 CrossRef CAS PubMed.
- T. Durmus, R. J. LeClair, K. S. Park, A. Terzic, J. K. Yoon and V. Lindner, Gene Expression Patterns, 2006, 6, 935–940 CrossRef CAS PubMed.
- M. T. Shekhani, T. S. Forde, A. Adilbayeva, M. Ramez, A. Myngbay, Y. Bexeitov, V. Lindner and V. A. Adarichev, Arthritis Res. Ther., 2016, 18, 171 CrossRef PubMed.
- J. Li, J. Cao, M. Li, Y. Yu, Y. Yang, X. Xiao, Z. Wu, L. Wang, Y. Tu and H. Chen, Br. J. Dermatol., 2011, 164, 1030–1036 CrossRef CAS PubMed.
- Z. Shen, T. Su, J. Chen, Z. Xie and J. Li, Ann. Transl. Med., 2021, 9, 801 CrossRef CAS PubMed.
- K. Nuutila, A. Siltanen, M. Peura, J. Bizik, I. Kaartinen, H. Kuokkanen, T. Nieminen, A. Harjula, P. Aarnio, J. Vuola and E. Kankuri, Wound Repair Regen., 2012, 20, 830–839 CrossRef PubMed.
- T. Y. Huang, L. L. Zuo, S. K. Walczynska, M. Y. Zhu and Y. J. Liang, Cell Tissue Res., 2021, 385, 105–113 CrossRef CAS PubMed.
- M. J. Toriseva, R. Ala-aho, J. Karvinen, A. H. Baker, V. S. Marjomaki, J. Heino and V. M. Kahari, J. Invest. Dermatol., 2007, 127, 49–59 CrossRef CAS PubMed.
- M. P. Vincenti, Methods Mol. Biol., 2001, 151, 121–148 CAS.
- D. Ulrich, E. M. Noah, D. von Heimburg and N. Pallua, Plast. Reconstr. Surg., 2003, 111, 1423–1431 CrossRef PubMed.
- T. J. Stevenson, V. Vinarsky, D. L. Atkinson, M. T. Keating and S. J. Odelberg, Dev. Dyn., 2006, 235, 606–616 CrossRef CAS PubMed.
- H. Ogawa, E. Kozhemyakina, H. H. Hung, A. J. Grodzinsky and A. B. Lassar, Genes Dev., 2014, 28, 127–139 CrossRef CAS PubMed.
- Y. Lee, J. Choi and N. S. Hwang, Biomater. Res., 2018, 22, 9 CrossRef PubMed.
- M. Qadri, G. D. Jay, L. X. Zhang, H. Richendrfer, T. A. Schmidt and K. A. Elsaid, Arthritis Res. Ther., 2020, 22, 113 CrossRef CAS PubMed.
- R. J. Krawetz, S. Abubacker, C. Leonard, A. O. Masson, S. Shah, N. Narendran, P. Tailor, S. C. Regmi, E. Labit, N. Ninkovic, J. M. Corpuz, K. Ito, T. M. Underhill, P. T. Salo, T. A. Schmidt and J. A. Biernaskie, npj Regener. Med., 2022, 7, 32 CrossRef CAS PubMed.
- E. Ronnberg and G. Pejler, Proteoglycans, 2012, 836, 201–217 Search PubMed.
- S. O. Kolset and G. Pejler, J. Immunol., 2011, 187, 4927–4933 CrossRef CAS PubMed.
- M. Tellez-Gabriel, X. Tekpli, T. M. Reine, B. Hegge, S. R. Nielsen, M. Chen, L. Moi, L. S. Normann, L. R. Busund, G. A. Calin, G. M. Mælandsmo, M. Perander, A. D. Theocharis, S. O. Kolset and E. Knutsen, Front. Oncol., 2022, 12, 868868 CrossRef CAS PubMed.
- D. J. Tyrrell, A. P. Horne, K. R. Holme, J. M. Preuss and C. P. Page, Adv. Pharmacol., 1999, 46, 151–208 CAS.
- R. Serra, G. Buffone, V. Molinari, R. Montemurro, P. Perri, D. M. Stillitano, B. Amato and S. de Franciscis, Int. Wound J., 2015, 12, 150–153 CrossRef PubMed.
- C. Y. Huang and M. Y. Choong, Int. Wound J., 2017, 14, 589–590 CrossRef PubMed.
- B. Caterson and J. Melrose, Glycobiology, 2018, 28, 182–206 CrossRef CAS PubMed.
- B. N. Cook, S. Bhakta, T. Biegel, K. G. Bowman, J. I. Armstrong, S. Hemmerich and C. R. Bertozzi, J. Am. Chem. Soc., 2000, 122, 8612–8622 CrossRef CAS.
- X. Li, L. Tu, P. G. Murphy, T. Kadono, D. A. Steeber and T. F. Tedder, J. Leukocyte Biol., 2001, 69, 565–574 CrossRef CAS PubMed.
- M. Kluppel, Prog. Mol. Biol. Transl. Sci., 2010, 93, 113–132 CAS.
- M. Kluppel, P. Samavarchi-Tehrani, K. Liu, J. L. Wrana and A. Hinek, Eur. J. Hum. Genet., 2012, 20, 870–877 Search PubMed.
- X. H. Zou, W. C. Foong, T. Cao, B. H. Bay, H. W. Ouyang and G. W. Yip, J. Dent. Res., 2004, 83, 880–885 CrossRef CAS PubMed.
- I. Maltseva, M. Chan, I. Kalus, T. Dierks and S. D. Rosen, PLoS One, 2013, 8, e69642 CrossRef CAS PubMed.
- Y. H. Wang and C. Beck, Regeneration, 2015, 2, 19–25 CrossRef CAS PubMed.
- C. Lanzi and G. Cassinelli, Biochem. Pharmacol., 2020, 178, 114084 CrossRef CAS PubMed.
- D. Papy-Garcia and P. Albanese, Glycoconjugate J., 2017, 34, 377–391 CrossRef CAS PubMed.
- G. Theocharidis and J. T. Connelly, J. Anat., 2019, 235, 418–429 CrossRef PubMed.
- K. E. Keller, M. J. Kelley and T. S. Acott, Invest. Ophthalmol. Visual Sci., 2007, 48, 1164–1172 CrossRef PubMed.
- S. Delbaere, T. Dhooge, D. Syx, F. Petit, N. Goemans, A. Destree, O. Vanakker, R. De Rycke, S. Symoens and F. Malfait, Genet. Med., 2020, 22, 112–123 CrossRef CAS PubMed.
- K. Schonborn, S. Willenborg, J. N. Schulz, T. Imhof, S. A. Eming, F. Quondamatteo, J. Brinckmann, A. Niehoff, M. Paulsson, M. Koch, B. Eckes and T. Krieg, Matrix Biol., 2020, 94, 57–76 CrossRef PubMed.
- D. N. Phan, M. Q. Khan, N. T. Nguyen, T. T. Phan, A. Ullah, M. Khatri, N. N. Kien and I. S. Kim, Carbohydr. Polym., 2021, 252, 117175 CrossRef CAS PubMed.
- S. Ullah and X. Chen, Appl. Mater. Today, 2020, 20, 100656 CrossRef.
- M. Brovold, J. I. Almeida, I. Pla-Palacin, P. Sainz-Arnal, N. Sanchez-Romero, J. J. Rivas, H. Almeida, P. R. Dachary, T. Serrano-Aullo, S. Soker and P. M. Baptista, Adv. Exp. Med. Biol., 2018, 1077, 421–449 CrossRef CAS PubMed.
- J. Y. Park, T. G. Lee, J. Y. Kim, M. C. Lee, Y. K. Chung and W. J. Lee, Arch. Craniofac. Surg., 2014, 15, 14–21 CrossRef PubMed.
- Y. Yu, H. Cui, C. Zhang, D. Zhang, J. Yin, G. Wen and Y. Chai, J. Mater. Chem. B, 2020, 8, 4067–4079 RSC.
- Y. Yu, H. Cui, D. Zhang, B. Liang, Y. Chai and G. Wen, J. Tissue Eng. Regener. Med., 2019, 13, 1770–1778 CrossRef CAS PubMed.
- G. BaoLin and P. X. Ma, Sci. China: Chem., 2014, 57, 490–500 CrossRef PubMed.
- A. Varki, Glycobiology, 2017, 27, 3–49 CrossRef CAS PubMed.
- E. V. Maytin, Glycobiology, 2016, 26, 553–559 CrossRef CAS PubMed.
- S. T. Nillesen, G. Lammers, R. G. Wismans, M. M. Ulrich, E. Middelkoop, P. H. Spauwen, K. A. Faraj, J. Schalkwijk, W. F. Daamen and T. H. van Kuppevelt, Acta Biomater., 2011, 7, 1063–1071 CrossRef CAS PubMed.
- V. Rouet, A. Meddahi-Pelle, H. Q. Miao, I. Vlodavsky, J. P. Caruelle and D. Barritault, J. Biomed. Mater. Res., Part A, 2006, 78, 792–797 CrossRef PubMed.
- M. Tong, M. M. Zbinden, I. J. Hekking, M. Vermeij, D. Barritault and J. W. van Neck, Wound Repair Regen., 2008, 16, 294–299 CrossRef PubMed.
- M. Tong, B. Tuk, I. M. Hekking, M. Vermeij, D. Barritault and J. W. van Neck, Wound Repair Regen., 2009, 17, 840–852 CrossRef PubMed.
- P. Chopra, M. T. Logun, E. M. White, W. Lu, J. Locklin, L. Karumbaiah and G. J. Boons, ACS Chem. Biol., 2019, 14, 1921–1929 CrossRef CAS PubMed.
- W. Lu, C. Zong, P. Chopra, L. E. Pepi, Y. Xu, I. J. Amster, J. Liu and G. J. Boons, Angew. Chem., Int. Ed., 2018, 57, 5340–5344 CrossRef CAS PubMed.
- T. J. Boltje, T. Buskas and G. J. Boons, Nat. Chem., 2009, 1, 611–622 CrossRef CAS PubMed.
- D. Xu, K. Arnold and J. Liu, Curr. Opin. Struct. Biol., 2018, 50, 155–161 CrossRef CAS PubMed.
- A. S. Barrett, M. J. Wither, R. C. Hill, M. Dzieciatkowska, A. D'Alessandro, J. A. Reisz and K. C. Hansen, J. Proteome Res., 2017, 16, 4177–4184 CrossRef CAS PubMed.
- K. A. Brown, T. Tucholski, C. Eken, S. Knott, Y. Zhu, S. Jin and Y. Ge, Angew. Chem., Int. Ed., 2020, 59, 8406–8410 CrossRef CAS PubMed.
- S. J. Knott, K. A. Brown, H. Josyer, A. Carr, D. Inman, S. Jin, A. Friedl, S. M. Ponik and Y. Ge, Anal. Chem., 2020, 92, 15693–15698 CrossRef CAS PubMed.
- X. H. Shao, C. D. Gomez, N. Kapoor, J. M. Considine, C. Grams, Y. Gao and A. Naba, Nucleic Acids Res., 2023, 51(D1), D1519–D1530 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Tables S1–S9. See DOI: https://doi.org/10.1039/d3bm00835e |
| ‡ Both authors contributed equally to this work. |
| § Current address: Biointerface Science, Department of Biomedical Engineering, Eindhoven University of Technology, PO Box 513, 5600 MB Eindhoven, The Netherlands. |
| This journal is © The Royal Society of Chemistry 2023 |