Open Access Article
Open Access ArticleA machine learning approach to predict cellular uptake of pBAE polyplexes†
Aparna
Loecher‡
a,
Michael
Bruyns-Haylett‡
 a,
Pedro J.
Ballester
a,
Salvador
Borros
a,
Pedro J.
Ballester
a,
Salvador
Borros
 b and
Nuria
Oliva
b and
Nuria
Oliva
 *ab
*ab
aDepartment of Bioengineering, Imperial College London, SW7 2AZ London, UK. E-mail: nuria.oliva@iqs.url.edu
bDepartment of Bioengineering, Institut Quimic de Sarria, Via Augusta 390, 08017 Barcelona, Spain
First published on 30th June 2023
Abstract
The delivery of genetic material (DNA and RNA) to cells can cure a wide range of diseases but is limited by the delivery efficiency of the carrier system. Poly β-amino esters (pBAEs) are promising polymer-based vectors that form polyplexes with negatively charged oligonucleotides, enabling cell membrane uptake and gene delivery. pBAE backbone polymer chemistry, as well as terminal oligopeptide modifications, define cellular uptake and transfection efficiency in a given cell line, along with nanoparticle size and polydispersity. Moreover, uptake and transfection efficiency of a given polyplex formulation also vary from cell type to cell type. Therefore, finding the optimal formulation leading to high uptake in a new cell line is dictated by trial and error, and requires time and resources. Machine learning (ML) is an ideal in silico screening tool to learn the non-linearities of complex data sets, like the one presented herein, with the aim of predicting cellular internalisation of pBAE polyplexes. A library of pBAE nanoparticles was fabricated and the uptake studied in 4 different cell lines, on which various ML models were successfully trained. The best performing models were found to be gradient-boosted trees and neural networks. The gradient-boosted trees model was then analysed using SHapley Additive exPlanations, to interpret the model and gain an understanding into the important features and their impact on the predicted outcome.
Introduction
Gene therapy (including DNA- and RNA-based therapies) is a promising strategy to treat a wide range of diseases through the transfer of nucleic acids into the cells of a patient,1 with the goal of modulating gene and protein expression. Non-viral delivery vectors have emerged as a safer, simpler, and more affordable approach to viral vectors, especially for the cytoplasmic delivery of nucleic acids. Moreover, they have no constraint on the size and number of nucleic acid inserts, making them an excellent alternative delivery vector.2 However, transfection efficiency is typically lower than that observed with viral vectors and is dependent on the physicochemical properties of the nanoparticles and the cell type. This implies that nanoparticle formulations need to be optimised on a case-by-case basis for each specific cell type.Poly-β-amino esters (pBAEs) are highly versatile polymers with amenable chemistry that enable facile tunability of their physicochemical properties, like polarity, molecular weight, and charge. Their cationic nature enables the electrostatic binding and condensation of negatively charged nucleic acids into nanoparticles.3 Furthermore, they are biodegradable and biocompatible. Initial high throughput screening of large pBAE libraries (over 2000 formulations) revealed promising polymer structures with efficient transfection in COS-7 cells, an easy-to-transfect cell system useful for high-throughput biological assays.4 Linear pBAEs with an amine/acrylate ratio of 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and terminal secondary amines were found to have much higher cellular uptake and transfection efficiency, as did also those pBAEs forming nanoparticles smaller than 200 nm and near neutral zeta (ζ) potential. While this combinatorial chemistry approach revealed key insights into pBAE-mediated gene-delivery, synthesis of over 2000 polymers is time-consuming and costly. Moreover, the data gathered had no prediction potential and was valid only for the cell line of study. Therefore, knowing which nanoparticle formulation will result in optimal cellular internalisation in each cell line before performing transfection experiments is almost impossible, therefore making it a process of largely trial and error and requiring high amounts of time and resources.5
1 and terminal secondary amines were found to have much higher cellular uptake and transfection efficiency, as did also those pBAEs forming nanoparticles smaller than 200 nm and near neutral zeta (ζ) potential. While this combinatorial chemistry approach revealed key insights into pBAE-mediated gene-delivery, synthesis of over 2000 polymers is time-consuming and costly. Moreover, the data gathered had no prediction potential and was valid only for the cell line of study. Therefore, knowing which nanoparticle formulation will result in optimal cellular internalisation in each cell line before performing transfection experiments is almost impossible, therefore making it a process of largely trial and error and requiring high amounts of time and resources.5
Artificial intelligence (AI), and more concretely its machine learning (ML) branch, can bypass trial and error and be utilised to optimise this process.5 This is achieved by building ML models that can find trends and predict outcomes by exploiting and learning from large, complex, and non-linear data sets. One example are those models built from nanoparticle uptake and transfection data, which are really effective tools for optimising nanomedicine. In fact, the utilisation of ML models to predict nanoparticle behaviour is becoming increasingly widespread. A recent study developed a ML model to predict cellular internalisation of carbon nanoparticles (CNP) in different breast cancer cells. Numerous physicochemical properties of the CNP's were used as inputs to the model, which returned the cellular internalisation as output, minimising the number of nanoparticles needed to be tested in vitro. In another study, Damiati et al. constructed an ML model to predict the insertion potential of cell-penetrating peptides as delivery vehicles, which could then predict the cellular insertion with high accuracy.6 A recently published study transfected 488 barcoded cancer cell lines with liposomes, poly(lactic-co-glycolic acid) (PLGA) or polystyrene (PS) nanoparticles and demonstrated that core composition is a key predictor of cell uptake.7 Moreover, ML revealed that the expression of solute carrier family 46 member 3 (SLC46A3) was inversely correlated with liposome cellular trafficking but had no effect on PLGA and PS uptake and downstream efficacy.
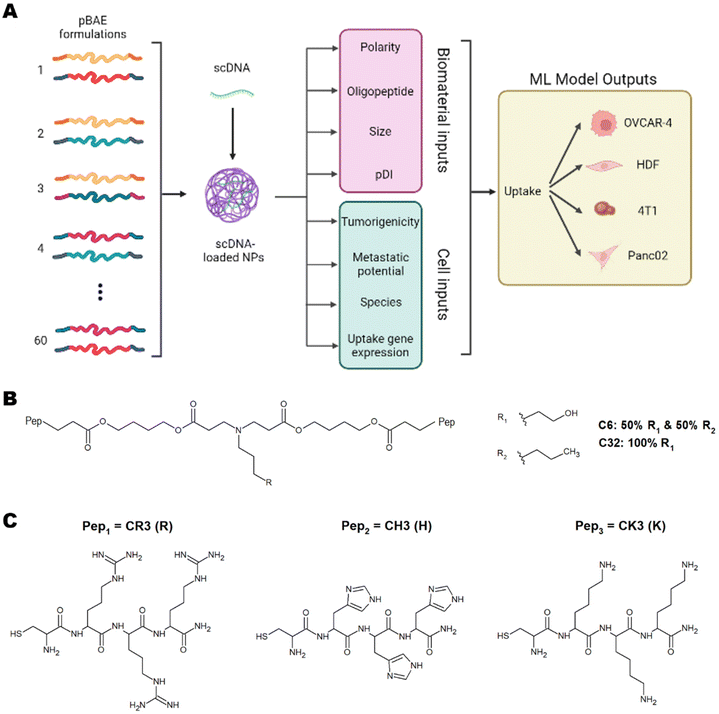
These studies have successfully implemented ML models on various types of nanoparticles and have highlighted the importance of using ML to understand nanoparticle interactions with cells to predict toxicity, uptake, and therapeutic efficiency.5,6,8 This understanding will pave the way for personalised medicine. To this day, however, there have been no studies using ML for predicting and understanding the cellular internalisation of pBAE nanoparticles. Of special relevance to tissue engineering and regenerative medicine, there is no previous data shedding light on the parameters dictating internalisation in non-cancerous cells. In this work, we have developed a library of pBAE nanoparticles of varying core chemistry, terminal oligonucleotides, and size, and have built and optimised a model of ML as a proof of concept that demonstrates accurate prediction of cellular uptake in a range of cell types (Fig. 1A). With respect to the nanoparticle-related model inputs, our previous expertise demonstrates that the polymer backbone has a large impact on the crossing of the cellular membrane due to variations in polarity caused by the pendant chemical groups: the C6 polymer is more hydrophobic than the C32 polymer3,9 (Fig. 1B). The addition of terminal oligopeptides composed of basic amino acids such as histidine (H), arginine (R) and lysine (K) (Fig. 1C) also creates different transfection efficiencies based on the type and ratio of oligopeptides.9,10 Size has also been shown to dictate the cellular uptake of pBAE polyplexes.4,9
We have built a proof-of-concept ML model using four distinct cell lines, three cancerous (OVCAR-4, Panc02 and 4T1) and one non-cancerous (Human Dermal Fibroblasts, HDFs). The cancerous cell lines have been chosen based on their characteristics and previous empirical observations. OVCAR-4 and 4T1 have been demonstrated to have overall high levels of uptake across most pBAE formulations. They are both metastatic cancer cell lines, with OVCAR-4 being an ovarian one of human origin, and 4T1 a breast cancer cell line of murine origin. Panc02 has been explored as 4T1 non-metastatic counterpart (murine pancreatic cancer cell line). Finally, HDFs have been used as a model of non-cancerous cells, which are notoriously harder to transfect using nanoparticles.3 The three main microscale endocytic pathways through which cells uptake foreign substances are clathrin-, caveolae-, and dynamin-mediated endocytosis,11 and the uptake route most responsible for transfection can change depending on the pBAE properties, such as size and charge.12 Additionally, the main endocytic mechanism can vary across different cell lines.11 Thus, determining the most prevalent mechanisms in each cell type and the preferred mechanism for each nanoparticle is very relevant. For pBAEs, clathrin- and caveolae-mediated endocytosis have been reported as the most prevalent uptake mechanisms.13 For this reason, we have chosen the normalised expression of genes involved in these pathways as cell-related inputs for the model. Nanoparticle- and cell-related inputs for 60 pBAE formulations and the 4 cell lines described above have been used to train various ML models to establish trends within these inputs and confer the ability to predict the uptake of pBAE polyplexes.
Materials and methods
Materials
Reagents and solvents were purchased from Sigma-Aldrich (Spain) and used as received unless otherwise stated. Catalogue number and suppliers are specified next to each chemical in this section. Oligopeptides were purchased from Ontores Biotechnologies Inc. Untagged and 3′-AlexaFluor488 tagged DNA sequences (5′-CCTCAAGTGGGACCATCATAA-[AlexaFluor488]-3′) were purchased from IDT (Custom made, UK). Human Dermal Fibroblasts (HDFs) were isolated from adult skin after abdominoplasty procedures, kindly provided by Dr Higgins from Imperial College London. Vials of cancerous cell lines OVCAR-4 (RRID:CVCL_1627), 4T1 (RRID:CVCL_0125) and Panc02 (RRID:CVCL_D627) were provided by Prof. McNeish, Dr Keshavarz and Dr Ishihara, respectively, all from Imperial College London (UK). Products for cell culture (DMEM, RPMI-1640, FBS, phosphate-buffered saline (PBS), glutamine and penicillin–streptomycin solutions, trypsin-EDTA 0.25%) were obtained from Thermo Fisher (UK).Cell culture conditions
Panc02 and 4T1 cell lines were cultured in RPMI 1640 Medium (A10491, Thermo Fisher) and OVCAR-4 and HDF cell lines in DMEM (11995, Thermo Fisher). Both media were supplemented with 10% FBS (26140079, Thermo Fisher) and 1% penicillin–streptomycin (15070063, Thermo Fisher). Cells were kept in an incubator at 37 °C and 5% CO2. Cells were thawed and passaged using established techniques.Synthesis of pBAE polymer backbones
Acrylate-terminated poly(β-aminoester)s C32 and C6 (Fig. 1B) were synthesised following a procedure previously described in the literature by Dosta et al.10 Specifically, the polymer formation occurs by addition reaction of primary amines with diacrylates. C32 polymer was obtained by stirring 5-amino-1-pentanol (7.7 g, 75 mmol; 123048 Sigma Aldrich) and 1,4-butanediol diacrylate (18 g, 82 mmol; 411744 Sigma Aldrich) together at 90 °C for 20 h. For C6 polymer, 5-amino-1-pentanol (3.9 g, 38 mmol; 123048 Sigma Aldrich) was firstly mixed with 1-hexylamine (3.8 g, 38 mmol; 219703 Sigma Aldrich). Then, 1,4-butanediol diacrylate (18 g, 82 mmol; 411744 Sigma Aldrich) was added to the mixture and heated at 90 °C for 20 h. 1H-NMR spectra were recorded in a 400 MHz Varian (Varian NMR Instruments, Claredon Hills, IL, USA) and methanol-d4 was used as solvent unless otherwise stated. Polymer backbones were characterised by 1H-NMR as described in our previous works,9,10,14,15 using MestReNova Software v14.3.2 (ESI Fig. 1†).Modification of acrylate-ended pBAEs with oligopeptides
Peptides were purchased as trifluoro acetic acid salts. The first step was the substitution of trifluoro acetic acid for hydrochloride as counterions. Generally, oligopeptides (100 mg) were dissolved in HCl 0.1 M (10 mL, 320331 Sigma Aldrich) and frozen at −80 °C for an hour. The solution was then freeze-dried. Oligopeptides used in the present work were Cys-Arg-Arg-Arg (CR3), Cys-His-His-His (CH3) and Cys-Lys-Lys-Lys (CK3) (Fig. 1C). Peptides hydrochlorides were reacted with acrylate-ended C32 or C6 polymers following a Michael-type addition at a pBAE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide molar ratio of 1
peptide molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5. PBAEs and peptides were dissolved separately in dimethyl sulfoxide (DMSO, 472301 Sigma Aldrich) at 100 mg mL−1 concentration. Then, polymer solution was added dropwise to the peptide solution. At this point, triethylamine (471283 Sigma Aldrich) was added to the solution in a peptide
2.5. PBAEs and peptides were dissolved separately in dimethyl sulfoxide (DMSO, 472301 Sigma Aldrich) at 100 mg mL−1 concentration. Then, polymer solution was added dropwise to the peptide solution. At this point, triethylamine (471283 Sigma Aldrich) was added to the solution in a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) triethylamine molar ratio of 1
triethylamine molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8. The mixture was allowed to react at room temperature for 48 h. 1H-NMR spectra were recorded in a 400 MHz Varian (Varian NMR Instruments, Claredon Hills, IL, USA) and methanol-d4 was used as solvent unless otherwise stated. OM-pBAEs were characterised by as described in our previous works,9,10,14,15 using MestReNova Software v14.3.2 (ESI Fig. 2–4†).
8. The mixture was allowed to react at room temperature for 48 h. 1H-NMR spectra were recorded in a 400 MHz Varian (Varian NMR Instruments, Claredon Hills, IL, USA) and methanol-d4 was used as solvent unless otherwise stated. OM-pBAEs were characterised by as described in our previous works,9,10,14,15 using MestReNova Software v14.3.2 (ESI Fig. 2–4†).
PBAE polyplexes formulation optimisation
Oligopeptide-modified C6 and C32 pBAE nanoparticles were prepared following protocols based on previous works.16–18 PBAEs and polynucleotides were kept in stocks at 100 mg mL−1 in DMSO or 1 mg mL−1 in nuclease-free water, respectively. First, the DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratio was optimised to ensure all DNA had been encapsulated without compromising cell viability. A model pBAE formulation previously used in the group, called C6RH (C6CR3
polymer ratio was optimised to ensure all DNA had been encapsulated without compromising cell viability. A model pBAE formulation previously used in the group, called C6RH (C6CR3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6CH3 in a 60
C6CH3 in a 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 ratio), was used for optimisation purposes.3 Polyplexes were formed using a fixed concentration of DNA (0.06 μg μL−1) and increasing concentrations of C6RH pBAE at DNA
40 ratio), was used for optimisation purposes.3 Polyplexes were formed using a fixed concentration of DNA (0.06 μg μL−1) and increasing concentrations of C6RH pBAE at DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratios of 1
polymer ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, 1
25, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 and 1
75 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. Encapsulation efficiency was analysed by agarose gel electrophoresis. Briefly, 10 μL of nanoparticle solution were mixed with 2 μL loading buffer (10816015 Thermo Fisher, UK), loaded onto a gel prepared with 2.5% agarose (AG002 Appleton Woods, UK) in 1× TBE buffer (15581044 Thermo Fisher UK), and run for 30 minutes at 80 V and 400 mA (Mini-Sub Cell GT, 1704406 Bio-Rad). Cell viability was measured with Presto Blue metabolic assay (A13262 Thermo Fisher UK), following established protocols. Fluorescence signal was recorded using a CLARIOstar Plus plate reader.
100. Encapsulation efficiency was analysed by agarose gel electrophoresis. Briefly, 10 μL of nanoparticle solution were mixed with 2 μL loading buffer (10816015 Thermo Fisher, UK), loaded onto a gel prepared with 2.5% agarose (AG002 Appleton Woods, UK) in 1× TBE buffer (15581044 Thermo Fisher UK), and run for 30 minutes at 80 V and 400 mA (Mini-Sub Cell GT, 1704406 Bio-Rad). Cell viability was measured with Presto Blue metabolic assay (A13262 Thermo Fisher UK), following established protocols. Fluorescence signal was recorded using a CLARIOstar Plus plate reader.
Library of PBAE polyplexes for ML model
A library of 60 different pBAE formulations was created by altering the ratios of both the polymer (C6 or C32) and the oligopeptide (R, H or K), as shown in Table 1 on the left. For each formulation, polyplexes were synthesised as previously described, following the optimal pBAE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratio of 50
DNA ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, determined as described above for this particular DNA structure. Briefly, 0.4 μL of pBAE stock solution (100 mg mL−1 in DMSO) and 0.8 μL scramble DNA solution (1 mg mL−1 in RNase/DNase free water) were diluted in 12.1 μL and 11.8 μL acetate buffer (12.5 mM, 4,8 pH), respectively. These two solutions were then mixed with a pipette for a few seconds and left at room temperature for 30 min. The resulting nanoparticles could then be used for transfecting cells, dynamic light scattering (DLS) or gel electrophoresis.
1, determined as described above for this particular DNA structure. Briefly, 0.4 μL of pBAE stock solution (100 mg mL−1 in DMSO) and 0.8 μL scramble DNA solution (1 mg mL−1 in RNase/DNase free water) were diluted in 12.1 μL and 11.8 μL acetate buffer (12.5 mM, 4,8 pH), respectively. These two solutions were then mixed with a pipette for a few seconds and left at room temperature for 30 min. The resulting nanoparticles could then be used for transfecting cells, dynamic light scattering (DLS) or gel electrophoresis.
| Formulation number | Ratio |
|---|---|
| 1 | 6R/6H – 80/20 |
| 2 | 6R/6H – 60/40 |
| 3 | 6R/6H – 20/80 |
| 4 | 6R/6K – 80/20 |
| 5 | 6R/6K – 60/40 |
| 6 | 6R/6K – 20/80 |
| 7 | 6H/6K – 80/20 |
| 8 | 6H/6K – 60/40 |
| 9 | 6H/6K – 20/80 |
| 10 | 6H – 100 |
| 11 | 6R – 100 |
| 12 | 6K −100 |
| 13 | 32R/32H – 80/20 |
| 14 | 32R/32H – 60/40 |
| 15 | 32R/32H – 20/80 |
| 16 | 32R/32K – 80/20 |
| 17 | 32R/32K – 60/40 |
| 18 | 32R/32K – 20/80 |
| 19 | 32H/32K – 80/20 |
| 20 | 32H/32K – 60/40 |
| 21 | 32H/32K – 20/80 |
| 22 | 32H – 100 |
| 23 | 32R – 100 |
| 24 | 32K − 100 |
| 25 | 32R/6H – 80/20 |
| 26 | 32R/6H – 60/40 |
| 27 | 32R/6H – 40/60 |
| 28 | 32R/6H – 20/80 |
| 29 | 32R/6R – 80/20 |
| 30 | 32R/6R – 60/40 |
| 31 | 32R/6R – 40/60 |
| 32 | 32R/6R – 20/80 |
| 33 | 32R/6K – 80/20 |
| 34 | 32R/6K – 60/40 |
| 35 | 32R/6K – 40/60 |
| 36 | 32R/6K – 20/80 |
| 37 | 32H/6H – 80/20 |
| 38 | 32H/6H – 60/40 |
| 39 | 32H/6H – 40/60 |
| 40 | 32H/6H – 20/80 |
| 41 | 32H/6R – 80/20 |
| 42 | 32H/6R – 60/40 |
| 43 | 32H/6R – 40/60 |
| 44 | 32H/6R – 20/80 |
| 45 | 32H/6K – 80/20 |
| 46 | 32H/6K – 60/40 |
| 47 | 32H/6K – 40/60 |
| 48 | 32H/6K – 20/80 |
| 49 | 32K/6H – 80/20 |
| 50 | 32K/6H – 60/40 |
| 51 | 32K/6H – 40/60 |
| 52 | 32K/6H – 20/80 |
| 53 | 32K/6R – 80/20 |
| 54 | 32K/6R – 60/40 |
| 55 | 32K/6R – 40/60 |
| 56 | 32K/6R – 20/80 |
| 57 | 32K/6K – 80/20 |
| 58 | 32K/6K – 60/40 |
| 59 | 32K/6K – 40/60 |
| 60 | 32K/6K – 20/80 |
Data collection for ML model
Along with polymer chemical characteristics, measured inputs for the model included size, polydispersity and gene expression, to predict the cellular uptake as model output. All experimental data for the model can be found in GitHub (https://github.com/mbhaylett23/pBAE-cellular-uptake-ML).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio, and encapsulated in the polyplexes to enable fluorescence tracking. Cells (OVCAR-4, 4T1, Panc02 or HDF) were seeded in a 96-well plate at a density of 10
10 ratio, and encapsulated in the polyplexes to enable fluorescence tracking. Cells (OVCAR-4, 4T1, Panc02 or HDF) were seeded in a 96-well plate at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well, and incubated for 24 hours at 37 °C. Polyplexes containing 10% fluorescent DNA were mixed with non-supplemented DMEM in a 1
000 cells per well, and incubated for 24 hours at 37 °C. Polyplexes containing 10% fluorescent DNA were mixed with non-supplemented DMEM in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio, to reach a final concentration of 0.003 μg μl−1 DNA in each well, and 100 μL of the nanoparticle medium was added into each well and incubated at 37 °C for 3 hours. The media was then exchanged to supplemented DMEM. In total, each cell line was transfected with 60 different formulations, performing duplicates for each one. The cells were then detached and the fluorescence intensity per cell was measured in duplicate for each well, using a Countess 3 FL fluorescence cell counter. This gave 4 uptake measurements to average for each formulation in every cell line. Gating conditions were determined using untreated controls, with intensities below 97 RFU being considered background (ESI Fig. 5A–C†).
10 ratio, to reach a final concentration of 0.003 μg μl−1 DNA in each well, and 100 μL of the nanoparticle medium was added into each well and incubated at 37 °C for 3 hours. The media was then exchanged to supplemented DMEM. In total, each cell line was transfected with 60 different formulations, performing duplicates for each one. The cells were then detached and the fluorescence intensity per cell was measured in duplicate for each well, using a Countess 3 FL fluorescence cell counter. This gave 4 uptake measurements to average for each formulation in every cell line. Gating conditions were determined using untreated controls, with intensities below 97 RFU being considered background (ESI Fig. 5A–C†).
Modelling
All modelling was performed in Python (v3.10.10) – the code can be found on GitHub: (https://github.com/mbhaylett23/pBAE-cellular-uptake-ML). Overall, the models that were created included multi-linear regression (LM), random forests (RF), gradient-boosted trees (GBT) and neural networks (NN). For all models, feature normalisation was performed on the data, and the data was split into training, validation and test sets in an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio using the scikit-learn (v1.2.2) library. The models were trained on the training set, tuned on the validation set and then evaluated on the test set. Performance of the models was evaluated by calculating the mean absolute error (MAE) between the model's uptake prediction and the actual uptake. The MAE is the average absolute difference between the predicted and observed outputs, and is useful for assessing the performance of a model on a particular dataset.19
10 ratio using the scikit-learn (v1.2.2) library. The models were trained on the training set, tuned on the validation set and then evaluated on the test set. Performance of the models was evaluated by calculating the mean absolute error (MAE) between the model's uptake prediction and the actual uptake. The MAE is the average absolute difference between the predicted and observed outputs, and is useful for assessing the performance of a model on a particular dataset.19
Statistical analysis
All statistical analyses of data were performed with the GraphPad Prism software (v9.5.1), using ANOVA (analysis of variance) tests (one-way and two-way) unless stated otherwise. Statistical significance was calculated based on p ≤ 0.05, where *, **, *** and **** represent p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001, respectively.Results and discussion
Optimisation of DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) polymer ratio
polymer ratio
Four different DNA-to-pBAE proportions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, 1
25, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 and 1
75 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) were synthesised and analysed for encapsulation efficiency and cytotoxicity. The DNA was completely encapsulated at DNA
100) were synthesised and analysed for encapsulation efficiency and cytotoxicity. The DNA was completely encapsulated at DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pBAE ratios of 1
pBAE ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and higher (ESI Fig. 6A†), as evident by the disappearance of the free DNA band in gel electrophoresis. No statistically significant cytotoxicity was observed for any formulation except the 1
50 and higher (ESI Fig. 6A†), as evident by the disappearance of the free DNA band in gel electrophoresis. No statistically significant cytotoxicity was observed for any formulation except the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio (ESI Fig. 6B†). Therefore, for the rest of this study, a DNA
100 ratio (ESI Fig. 6B†). Therefore, for the rest of this study, a DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pBAE ratio of 1
pBAE ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 was used.
50 was used.
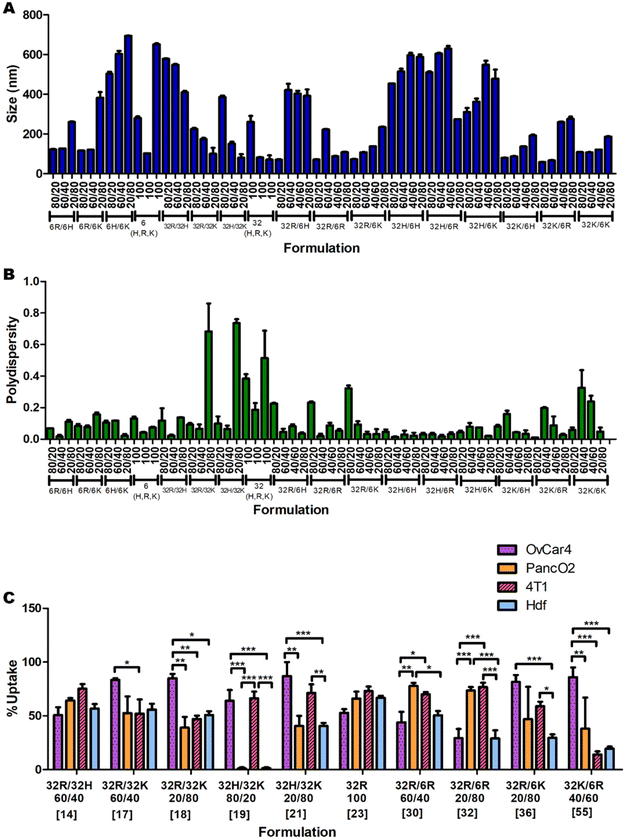
Nanoparticle size and polydispersity
The measurements of the sizes (Fig. 2A) and polydispersities (Fig. 2B) of all 60 formulations presented statistically significant variations (p < 0.0001 for both One-Way ANOVAs). The sizes ranged between 58 and 694 nm in diameter, and the polydispersities between 0.01 and 0.737. In general formulations with high amounts of C6K and C32H tend to result in larger sizes, and formulations with high amounts of C32K tend to result in higher polydispersity.Cellular uptake of nanoparticles
The uptake of the 60 nanoparticle formulations was measured in OVCAR-4, HDF, 4T1 and Panc-02 cell lines (ESI Fig. 7A, B and 8A, B†). A summary of a few significant formulations is shown in Fig. 2C to demonstrate the apparent unpredictability of the system and confirm the hypothesis that tuning the backbone polymer and the oligopeptide ratios affects the cellular uptake. Additionally, there is variation in the amount of uptake for specific nanoparticles between different cell lines. For instance, formulation 19 has medium-to-high transfection efficiency in OVCAR-4 and 4T1, but almost-zero uptake in Panc02 and HDF. However, formulation 32 has high uptake in Panc01 and 4T1 cells, and low cellular entry in OVCAR-4 and HDF. Each cell line was found to have a different formulation that resulted in the highest uptake: for OVCAR-4 it was formulation 21, with 87% uptake, for 4T1 it was formulation 31, with 83%, for Panc02 it was formulation 30, with 77% and for HDF it was formulation 23, with 66% uptake.Predicting which formulation would have resulted in the highest uptake in each cell line without a ML model would have been impossible, showing the need for a predictive model. Another interesting observation is that the cancer cell lines all had a higher average uptake than the non-cancerous cell line. The average uptakes of OVCAR-4, 4T1, Panc02 and HDF were 44%, 40%, 29% and 23% respectively. The trend that cancer cells have higher uptake of various types of nanoparticles has been reported in the literature.20 This is mostly because cancer cells consistently undergo endocytosis more rapidly than noncancerous cells, to provide themselves with more nutrients.21 This highlights the importance of ML models to maximise uptake and transfection in non-cancerous cells and enable this way the use of pBAE polyplexes in regenerative medicine. Within cancer cells, metastatic ones (OVCAR-4 and 4T1) present higher average uptake than non-metastatic Panc02 (44 and 40% versus 29%, respectively). To further understand the effect of the cell type on uptake and provide additional prediction capability to the model, we investigated the expression of key genes involved in polyplexes’ uptake and cell trafficking.
Gene expression
It has been previously described in the literature that there is a preferential uptake of polyplexes into cells through clathrin- and caveolae-mediated endocytosis.13 More interestingly, previous studies using pBAEs demonstrated that altering the polymer backbone chemistry and terminal groups preferentially triggered one pathway over the other.12,22 Therefore, incorporating the expression of key genes regulating these cellular trafficking pathways is a promising approach to improve the prediction capability of the ML model. The expression of genes involved in clathrin- and caveolae-mediated endocytosis (Fig. 3A) were extracted from publicly available microarray data and normalised to the total microarray intensity (Fig. 3B) prior to data input in the models. Interestingly, the expression of these genes is significantly different in the four cell lines of study, and overall there seems to be an overexpression of genes involved in clathrin-mediated endocytosis and underexpression of caveolae-related genes.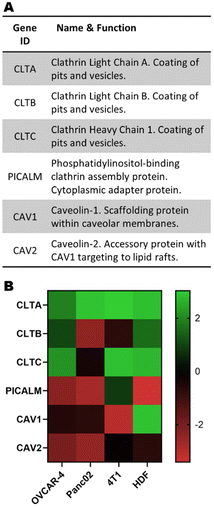 | ||
| Fig. 3 (A) Subset of genes linked to clathrin- and caveolae-mediated endocytosis and (B) their normalised expression. | ||
Initial data analysis
Some initial data analysis on the experimental uptake data was performed to discover trends. Pure formulations (only one type of polymer) were evaluated first (Fig. 4). In all cell lines, polymers C6H and C32H resulted in approximately zero uptake (Fig. 4A and B). Additionally, given the oligopeptide terminal R or K, the C32 backbone results in higher uptake than the C6 backbone across all cell lines. Lastly, OVCAR-4 seems to have the highest affinity to C32K, while 4T1, Panc02 and HDF have the highest affinities to C32R.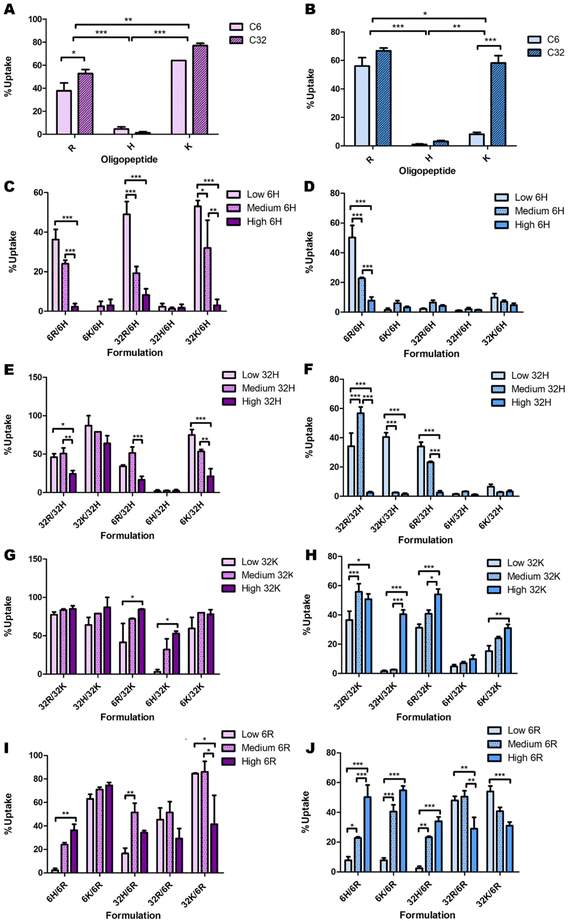 | ||
| Fig. 4 Initial analysis of the effects of backbone chemistry and terminal oligopeptide on cellular uptake in OVCAR-4 (A, C, E, G & I; purple bars) and HDF (B, D, F, H & J; blue bars), both for (A and B) pure formulations and low, medium and high ratios of (C and D) C6H, (E and F) C32H, (G and H) C32K and (I and J) C6R. The remaining combinations presented no clear trends (Fig. S4 and S5†). | ||
Delving further into these trends, the data point to a consistent decrease in nanoparticle uptake as the percentage of C6H and C32H increase (Fig. 4C–F & ESI Fig. 9†). However, there are a few exceptions in which 60% C32H (referred to as “medium” amounts in the corresponding graphs) increase the uptake (Fig. 4E and F). This is explained by the endocytic process leading to transfection. To efficiently deliver the genetic material, endosomal escape of the particles must occur after uptake. The process called ‘proton sponge effect’ facilitates endosomal escape, and is driven by terminal amines with high buffering capacity.9 Histidine has the highest buffering capacity, making it the best for inducing endosomal escape inside cells. However, this study focuses only on cellular uptake, without considering the ability to successfully transfect cells (which would include endosomal escape). Since high transfection has been found to be a result of high cellular uptake, rather than high endosomal escape,9 we focused on this first as a proof-of-concept study on cellular uptake only. Thus, while histidine decreases cellular uptake, and might seem dispensable in this system, including minimal amounts of histidine in the formulation will have an impact on overall transfection efficiency through increased endosomal escape.
A similar analysis on C6K revealed inconclusive trends (ESI Fig. 8C and D†). While C32K effects on Panc02 and 4T1 cell lines were also inconclusive (ESI Fig. 8E and F†), OVCAR-4 and HDF seem to follow a trend with higher uptake as the amount of C32K increases (Fig. 4G and H). Interestingly, HDFs had low affinity for pure C32K. This suggests that mixing polymers and oligopeptides results in completely different interactions with cells. Finally, there is no conclusive trend in C6R and C32R uptake (Fig. 4I and J & ESI Fig. 10A–F†). However, medium amounts of 6R result in higher uptake in most formulations in OVCAR-4, Panc02 and 4T1 cell lines, while uptake trends are inconclusive in HDF.
Overall, while a few trends can be observed, complex nonlinear relationships at play exist that are not obvious. This further highlights the need for a model to learn these complexities and accurately predict which formulations result in high uptake in a certain cell line.
Model results
After creating and tuning the four different models to have low MAE (ESI Fig. 10A†), and balance bias and variance, the optimal hyper-parameters are shown in Table 2. MAE values showed GBT was the best performing model, followed by the NN model and the RF model, with multi-linear regression (LM) performing the most poorly. Non-linear ML models GBT and NN have statistically significant decreases in MAE compared to multi-linear regression (ESI Fig. 11†). Both the GBT and NN models have statistically non-significantly different mean performances, with an MAE of 10.57 and 11.17, respectively, about 30% better than that of the multi-linear regression model. This shows that the uptake data and its corresponding features have complex non-linearities, from which ML models are able to learn from and better capture trends.| Model | Hyper-parameter 1 | Hyper-parameter 2 | Test MAE |
|---|---|---|---|
| Multi-linear regression | N/A | N/A | 14.09 |
| Random forest | Tree depth = 13 | Max. Num of trees = 75 | 13.09 |
| Gradient boosted trees | Tree depth = 5 | Max. Num of Trees = 50 | 10.57 |
| Neural network | Hidden layers = 2 | Num. of nodes = 41 | 11.17 |
SHAP (shapley additive exPlanations) analysis
A model that returns good predictions is useful, however, if there is no understanding as to how the model uses the inputs to make its predictions, it can be of limited use. The SHAP approach gives an understanding of both the contributions of the features globally, as well as contributions for individual observations.23 This renders the ‘black box’ ML model interpretable and is especially relevant here, because understanding the importance of each feature can help in the design process of the nanoparticles. Since the GBT and NN models had the best performance overall, and calculating SHAP-values for GBT was computationally more efficient than for the NN, SHAP analysis was implemented on the GBT model.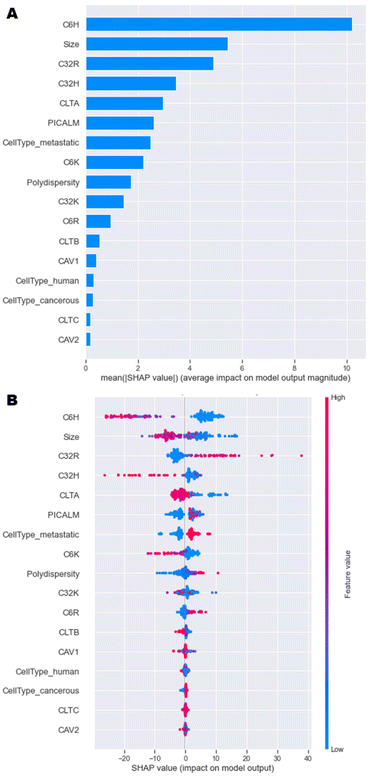 | ||
| Fig. 5 SHAP analysis of model data. (A) Overall importance of each feature on the model output, and (B) Influence of each feature value on the model output. | ||
A more in-depth explanation of how the specific value of each feature contributes to the model output is depicted in Fig. 5B. High negative SHAP-values mean that features greatly decrease the uptake, while highly positive SHAP-values mean that features strongly increase the uptake. For C6H, C32H and C6K, high percentages result in high negative SHAP-values and vice versa. This trend was observed in initial data analysis in Fig. 4. High amounts of C32R and C6R lead to higher model outputs and vice versa, while C32K displays no clear trend. Overall, as size values are lower, there is a high positive impact on the output of the model, which is in line with research having shown that optimal sizes of nanoparticles that enter most cells endocytically are between 100–200 nm.4,9
In terms of the cell type and phenotype, high expression of CLTA results in low model output, while high levels of PICALM lead to high model outputs. This seems contradictory, as both genes are part of the same clathrin-mediated endocytosis pathway. Interestingly, while high expression of CLTA (pink dots) has a consistent negative impact on the model output, low levels (blue dots) might have a positive or negative impact on uptake. Similarly, high PICALM expression consistently improves uptake, while low levels can lead to either high or low model output. These data suggest that the expression of clathrin-mediated genes is key for some formulations, but not for others, as previously described.12,22 A more in depth, partial dependence investigation delves into these findings in the next section. Lastly, metastatic cells display higher levels of uptake than non-metastatic cells, while tumorigenicity and species (human or murine) have no impact on the model outputs. Overall, this SHAP analysis shows that the model has consistently been able to learn the trends initially observed.
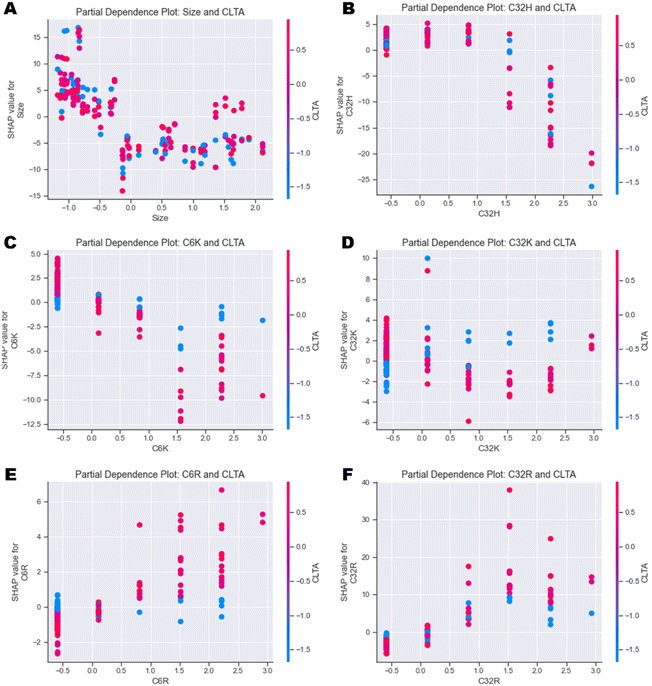 | ||
| Fig. 6 Partial dependence plots investigating the individual feature interactions between CLTA expression and (A) Size, (B) C32H, (C) C6K, (D) C32K, (E) C6R and (F) C32R. | ||
In general, the output of the model decreases as size increases independently of the cell type (Fig. 6A and ESI Fig. 12A†). Similarly, high values of C32H and C6H also have a negative impact on uptake independently of cell type (Fig. 6B and ESI Fig. 12B–D†). Partial dependence plots of arginine (R) and lysine (K) polymers versus CLTA and PICALM reveal very interesting correlations. Presence of C6K and C32K leads to higher uptake in those cells with lower expression of CLTA (Fig. 6C & D), while C6R and C32R triggered higher uptake in cells expressing high levels of CLTA (Fig. 6E & F). This suggests that polymers with terminal arginines potentially use clathrin-mediated endocytosis. PICALM dependence plots show similar trends: C6K and C32K display higher uptake in those cells with low PICALM expression (ESI Fig. 12E & F†) while C6R and C32R result in higher uptake in cells with higher PICALM levels (ESI Fig. 12G & H†). Finally, further feature interaction analysis shows that C32K polyplexes have higher affinity to metastatic cells, while C6K, C6R and C32R present no clear trend (ESI Fig. 13A–D†), which had already been identified in Fig. 4.
Conclusions
In summary, data relating to the size, polydispersity and uptake of 60 different nanoparticle formulations in 4 cell lines were collected. Biomaterial and cellular inputs were used to successfully train various ML models to predict the cellular uptake in these cell lines. It is important to highlight that despite the relatively low number of data points (240 uptake values compared to 1000s of points typically used for ML), the SHAP analysis carried out on the GBT model showed that it was successfully able to learn many trends seen in the data. In the future, new cells lines will be investigated to continue to grow the training and validation sets of the model, improving accuracy and reducing MAE.Aspects like polyplex size, backbone chemistry and terminal oligopeptides play distinct roles in cellular uptake, which often display divergent behaviour in different types of cell lines. Using an ML model approach, we have also identified two genes in the clathrin-mediated endocytosis pathway, CLTA and PICALM, which seem to play a key role controlling cellular trafficking as a function mainly of the identity of the terminal oligopeptides. The data suggests that high expression of these genes makes cells more receptive to the uptake arginine polymers (C6R and C32R), while low levels of these genes trigger the uptake of lysine polymers (C6K and C32K). Histidine is an important feature of the model because high percentages of histidine polymers abrogate the cellular uptake, which explains the lack of any partial dependence with CLTA or PICALM.
This proof-of-concept study demonstrates that ML is a key tool to gain in depth understanding of the complex non-linearities underlying pBAE cellular uptake. This work has been a step towards the ultimate goal of being able to use a model to scan across a number of nanoparticle formulations in a new cell line and predict those with the highest transfection efficiency.
Author contributions
AL curated the data and performed formal analysis, MBH carried out formal analysis and validation, PJB and SB provided supervision and project administration, NO conceptualised the project, acquired funding and carried out project administration and supervision. AL, MBH and NO wrote the original draft, all authors reviewed and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Prof. McNeish, Dr Keshavarz, Dr Ishihara and Dr Higgins for providing the cells for the study. NO acknowledges an Imperial College Research Fellowship, a Royal Society Research Grant (ID: RGS\R2\212038) and a ‘la Caixa’ foundation Junior Leader Fellowship (ID: LCF/BQ/PR22/11920009).References
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt and P. R. Cullis, et al., The current landscape of nucleic acid therapeutics, Nat. Nanotechnol. Nat. Res., 2021, 16, 630–643 CrossRef CAS PubMed.
- I. Roy, M. K. Stachowiak and E. J. Bergey, Nonviral gene transfection nanoparticles: function and applications in the brain, Nanomedicine, 2008, 4, 89–97 CrossRef CAS PubMed.
- J. A. Duran-Mota, J. Q. Yani, B. D. Almquist, S. Borrós and N. Oliva, Polyplex-Loaded Hydrogels for Local Gene Delivery to Human Dermal Fibroblasts, ACS Biomater. Sci. Eng., 2021, 7(9), 4347–4361 CrossRef CAS PubMed.
- J. J. Green, R. Langer and D. G. Anderson, A combinatorial polymer library approach yields insight into nonviral gene delivery, Acc. Chem. Res., 2008, 41, 749–759 CrossRef CAS PubMed.
- M. Alafeef, I. Srivastava and D. Pan, Machine Learning for Precision Breast Cancer Diagnosis and Prediction of the Nanoparticle Cellular Internalization, ACS Sens., 2020, 5(6), 1689–1698 CrossRef CAS PubMed.
- S. A. Damiati, A. L. Alaofi, P. Dhar and N. A. Alhakamy, Novel machine learning application for prediction of membrane insertion potential of cell-penetrating peptides, Int. J. Pharm., 2019, 567, 118453 CrossRef CAS PubMed.
- N. Boehnke, J. P. Straehla, H. C. Safford, M. Kocak, M. G. Rees and M. Ronan, et al., Massively parallel pooled screening reveals genomic determinants of nanoparticle delivery, Science, 2022, 377(6604), eabm5551 CrossRef CAS PubMed.
- N. Boehnke and P. T. Hammond, Power in Numbers: Harnessing Combinatorial and Integrated Screens to Advance Nanomedicine, JACS Au, 2022, 2(1), 12–21 CrossRef CAS PubMed.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization, Acta Biomater., 2014, 10(5), 2147–2158 CrossRef CAS PubMed.
- P. Dosta, V. Ramos and S. Borrós, Stable and efficient generation of poly(β-amino ester)s for RNAi delivery, Mol. Syst. Des. Eng., 2018, 3(4), 677–689 RSC.
- S. Kumari, S. Mg and S. Mayor, Endocytosis unplugged: Multiple ways to enter the cell, Cell Res., 2010, 20, 256–275 CrossRef CAS PubMed.
- J. Kim, J. C. Sunshine and J. J. Green, Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(β-amino ester) polyplexes in human breast cancer cells, Bioconjugate Chem., 2014, 25(1), 43–51 CrossRef CAS PubMed.
- J. Rejman, A. Bragonzi and M. Conese, Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes, Mol. Ther., 2005, 12(3), 468–474 CrossRef CAS PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed.
- N. Segovia, M. Pont, N. Oliva, V. Ramos, S. Borrós and N. Artzi, Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer, Adv. Healthcare Mater., 2014, 4(2), 271–280 CrossRef PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization, Acta Biomater., 2014, 10(5), 2147–2158 CrossRef CAS PubMed.
- P. Dosta, V. Ramos and S. Borrós, Stable and efficient generation of poly(β-amino ester)s for RNAi delivery, Mol. Syst. Des. Eng., 2018, 3(4), 677–689 RSC.
- H. H. Rashidi, S. Albahra, S. Robertson, N. K. Tran and B. Hu, Common statistical concepts in the supervised Machine Learning arena, Front. Oncol., 2023, 13, 1130229 CrossRef PubMed.
- K. Bromma, A. Bannister, A. Kowalewski, L. Cicon and D. B. Chithrani, Elucidating the fate of nanoparticles among key cell components of the tumor microenvironment for promoting cancer nanotechnology, Cancer Nanotechnol., 2020, 11(1), 8 CrossRef CAS PubMed.
- D. Wang, S. Liu and G. Wang, Establishment of an Endocytosis-Related Prognostic Signature for Patients With Low-Grade Glioma, Front. Genet., 2021, 12, 709666 CrossRef CAS PubMed.
- J. C. Sunshine, D. Y. Peng and J. J. Green, Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties, Mol. Pharm., 2012, 9(11), 3375–3383 CrossRef CAS PubMed.
- S. M. Lundberg, G. Erion, H. Chen, A. DeGrave, J. M. Prutkin and B. Nair, et al., From local explanations to global understanding with explainable AI for trees, Nat. Mach. Intell., 2020, 2(1), 56–67 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00741c |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2023 |