Open Access Article
Open Access ArticleMagnetically controlled cyclic microscale deformation of in vitro cancer invasion models†
Daphne O.
Asgeirsson
a,
Avni
Mehta
a,
Anna
Scheeder
ab,
Fan
Li
 a,
Xiang
Wang
a,
Xiang
Wang
 a,
Michael G.
Christiansen
a,
Michael G.
Christiansen
 a,
Nicolas
Hesse
a,
Rachel
Ward
a,
Andrea J.
De Micheli
ac,
Ece Su
Ildiz
a,
Nicolas
Hesse
a,
Rachel
Ward
a,
Andrea J.
De Micheli
ac,
Ece Su
Ildiz
 d,
Stefano
Menghini
d,
Stefano
Menghini
 a,
Nicola
Aceto
d and
Simone
Schuerle
a,
Nicola
Aceto
d and
Simone
Schuerle
 *a
*a
aDepartment of Health Sciences and Technology, Responsive Biomedical Systems Laboratory, ETH Zurich, 8093 Zurich, Switzerland. E-mail: simone.schuerle@hest.ethz.ch
bDepartment of Chemical Engineering and Biotechnology, University of Cambridge, Cambridge CB3 0AS, U.K
cDepartment of Oncology, Children's Research Center, University Children's Hospital Zurich, Zurich 8032, Switzerland
dDepartment of Biology, Institute of Molecular Health Sciences, ETH Zurich, 8093 Zurich, Switzerland
First published on 16th October 2023
Abstract
Mechanical cues play an important role in the metastatic cascade of cancer. Three-dimensional (3D) tissue matrices with tunable stiffness have been extensively used as model systems of the tumor microenvironment for physiologically relevant studies. Tumor-associated cells actively deform these matrices, providing mechanical cues to other cancer cells residing in the tissue. Mimicking such dynamic deformation in the surrounding tumor matrix may help clarify the effect of local strain on cancer cell invasion. Remotely controlled microscale magnetic actuation of such 3D in vitro systems is a promising approach, offering a non-invasive means for in situ interrogation. Here, we investigate the influence of cyclic deformation on tumor spheroids embedded in matrices, continuously exerted for days by cell-sized anisotropic magnetic probes, referred to as μRods. Particle velocimetry analysis revealed the spatial extent of matrix deformation produced in response to a magnetic field, which was found to be on the order of 200 μm, resembling strain fields reported to originate from contracting cells. Intracellular calcium influx was observed in response to cyclic actuation, as well as an influence on cancer cell invasion from 3D spheroids, as compared to unactuated controls. Furthermore, RNA sequencing revealed subtle upregulation of certain genes associated with migration and stress, such as induced through mechanical deformation, for spheroids exposed to actuation vs. controls. Localized actuation at one side of a tumor spheroid tended to result in anisotropic invasion toward the μRods causing the deformation. In summary, our approach offers a strategy to test and control the influence of non-invasive micromechanical cues on cancer cell invasion and metastasis.
Introduction
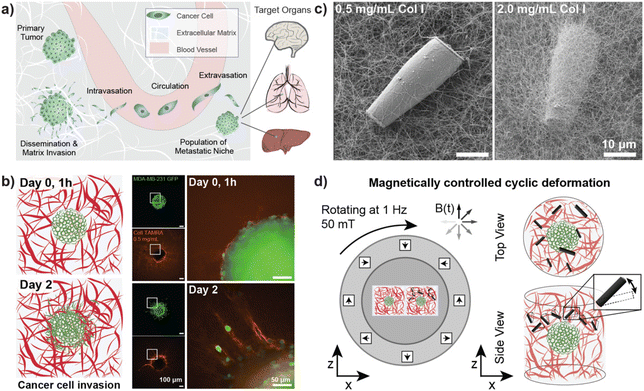
Metastasis is the process of cancer cells disseminating from primary or metastatic tumors, followed by the invasion of other organs or tissues and the subsequent formation of secondary tumor sites.1 The first key event in the metastasis cascade is the infiltration of single cells or cell clusters into the surrounding extracellular matrix (ECM).2,3 As this invasion is strongly influenced by the tumor microenvironment, research has concentrated on understanding the local mechanisms driving the metastatic transformation with the aim to find strategies for inhibition.4 While biochemical traits have been a long-standing focus of analysis, mechanical cues are increasingly considered a critical contributor to malignant cancer transformation and metastasis. The mechanical properties of the tumor and individual cells, however, are not the only factors affecting metastasis. The surrounding tumor microenvironment also provides essential biophysical cues such as matrix stiffness and tensile forces that critically modulate the metastatic process.5–7 These and other local physical properties are affected by the dynamic relationship between cancer cells and their surrounding microenvironment, which is characterized by alteration of tissue composition and architecture in a manner that supports malignant transformation and metastasis.6,8,9 The tumor stroma plays here a key role as it is populated by stromal cells such as fibroblasts and immune cells. Fibroblasts in particular have been shown to dynamically remodel the tissue microenvironment by exerting contractile forces of several tens of nN within the surrounding ECM. Furthermore, both cancer cells and fibroblasts can cause strain fields in their surrounding matrix exceeding 100 μm in diameter.10,11To study the effect of biophysical forces on malignant progression, cancer models are increasingly orienting towards three-dimensional (3D) culture matrices and away from historic approaches of two-dimensional (2D) cell culture performed on rigid plastic substrates. Composed of synthetic or biopolymers, these 3D models more accurately recapitulate the ECM environment and can be designed with specific stiffnesses, architectures, and the presence of surface ligands.12,13 Studies performed on stiffness-tuned cell culture substrates of tissue-derived or laboratory-generated materials include work presented by Cavo et al.14 where matrix elasticity has been shown to strongly regulate the rate of cell proliferation, cell morphology and the ability of cancer cells to occur in larger clusters upon exposure to softer hydrogels. Moreover, considerable progress has been made in the reconstitution of biomimetic cell culture models, such as through advances in 3D tumor models, implementation of engineered models for the surrounding tumor microenvironment, and the reorganization of in vitro models from patient-derived samples for personalized medicine approaches.15,16 Utilizing these 3D matrices to investigate cancer cell invasion, and the interaction of individual cancer cells or cell clusters with the surrounding matrix is an important step for physiologically relevant studies. As shown by Kim et al.,17 the application of tissue-derived material can improve the physiological relevance of cultured models compatible with fabrication using bioinks. The combination of laboratory-derived materials and the extraction of tissue components, whether from healthy or diseased organisms, allows for the introduction of disease specific markers that, in combination with controlled mechanical factors, support disease-specific cellular phenotypes. For instance, studies conducted in a 3D in vitro environment have demonstrated the influence of matrix stiffness on cancer cell migration and matrix invasion, with single-cell invasiveness exhibiting a biphasic response to increasing Collagen I concentrations.18,19 While such models have greatly increased the physiological relevance of applied cell culture systems, the high number of resistances against chemotherapeutic responses suggests that ex vivo model systems might profit from going beyond solely adapting the composition and architecture of in vitro tissue models.20 The active role of tumor-associated cells in the deformation of living tissue suggests that triggering a dynamic deformation to the surrounding tumor matrix can critically contribute to understanding the microenvironment experienced by cancer cells. To investigate the effect of matrix deformation on cancer cell migration, different approaches have been implemented.21,22 A particularly interesting approach is the application of remotely controlled microscale actuation of in vitro systems. One promising strategy is magnetically controlled micro- or nanoscale actuators embedded in samples with the capability to induce microscale deformations of the ECM. The remote control offers a non-invasive method and is compatible with sterile cell culture environments. A study performed by Menon et al. employed microparticles embedded in a 3D matrix, which were actuated using a permanent magnet to achieve a transient tugging force that was shown to enhance cancer cell invasion into 3D collagen matrices in a fibronectin–dependent manner.23 The same research group reported a cell-line-specific response to different types of mechanical stimuli such as matrix deformation and rigidity. They used a combination of substrates with varying degrees of compliance and triggered a cyclic deformation of the matrix using a single magnetic bead.24 Aiming to test the effect of local deformation of the ECM on cells residing there, further approaches have been developed but are often focused on individual cells and are not compatible with established 3D cancer invasion assays.25 In a recent study by Shou et al., 3D magnetic hydrogels were developed to study the effect of stiffness changes on embedded cancer spheroids. Their findings revealed that matrix stiffening promoted malignancies, while matrix softening downregulated the expression of mechano-transductors.26 While, in that study, the whole matrix stiffness could be rapidly altered to measure downstream effects, the influence of dynamic mechanical compression and strain of the network generated from individual point sources, like individual cells, remains to be studied.
This work applies magnetically controlled microscale actuation of 3D tumor invasion assays over several days of culture. Surface functionalized anisotropic rod-shaped iron microparticles (μRods) passivated with an outer layer of silica were covalently linked to a fibrous 3D Collagen I (Col I) network, which was employed as an ECM model in cancer spheroid invasion assays. Magnetically controlled cyclic actuation of the sample-embedded μRods in the vicinity of the tumor spheroids allowed for local deformation of the tumor microenvironment. Using particle velocimetry analysis, we determined the range of Col I deformation in response to the magnetic field which spanned several times the length of a single μRod—values comparable with strain fields reported to originate from contracting cells.10 Moreover, the patterns of Col I deformation resemble findings published for cells residing in Col I hydrogels.10,11 Changes in cellular calcium levels were observed in response to sustained actuation of the cell-embedding Col I matrix, indicating that calcium signaling might be influenced by the mechanical dynamics of the cellular microenvironment. Sustained actuation of the ECM model surrounding spheroids of the invasive cancer cell line MDA-MB-231 at 1 Hz over two days resulted in increased dissemination and invasion of cancer cells into the surrounding Col I network. Locally concentrated application of the μRods in the vicinity of Col I-embedded spheroids was shown to cause local attraction of the adjacent cancer cells and patterns of higher cell densities in the direction of the applied mechanical stimuli. Overall, this study suggests that local micromechanical deformation can promote cancer cell invasion and provides a means to apply and test the influence of controllable non-invasive micromechanical cues in 3D in vitro systems.
Experimental section
Cell culture
GFP labelled cells from the invasive cancer cell line MDA-MB-231 were ordered from GenTarget Inc. Cells were cultured in complete medium (Gibco DMEM, high Glucose, GlutaMAX Supplement, pyruvate, Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Biowest) and 1% penicillin/streptomycin (P/S, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1, Thermo Scientific) at 37 °C and 5% CO2. Cells were passaged at 70%–80% confluency by washing cells once with Dulbecco's phosphate buffered saline (Gibco DPBS, no calcium, no magnesium, Thermo Fisher Scientific), incubation with Trypsin-EDTA (0.25%, phenol red Thermo Fisher Scientific) and subsequent neutralization of the trypsin using complete medium. Cells were then concentrated by centrifugation, counted, and used for experiments or directly reseeded for further culture.
000 U mL−1, Thermo Scientific) at 37 °C and 5% CO2. Cells were passaged at 70%–80% confluency by washing cells once with Dulbecco's phosphate buffered saline (Gibco DPBS, no calcium, no magnesium, Thermo Fisher Scientific), incubation with Trypsin-EDTA (0.25%, phenol red Thermo Fisher Scientific) and subsequent neutralization of the trypsin using complete medium. Cells were then concentrated by centrifugation, counted, and used for experiments or directly reseeded for further culture.
Col I hydrogel preparation
Col I hydrogels were prepared as described previously.27 Briefly, Col I hydrogels were prepared from high-concentration rat-tail Col I (Corning, #354249, 9.48 mg mL−1) following the manufacturer's instructions. Concentrated Col I was mixed with cell culture medium and 0.5 M NaOH to adjust the mix to pH 7 to allow for polymerization. The sample was thoroughly mixed and incubated at 37 °C for a minimum of 30 minutes. For the preparation of fluorescently labelled Col I hydrogels, 10% of the Col I stock was replaced by fluorescently labelled Col I stock, prepared according to the protocol specified in the following section.Fluorescent labeling of Col I hydrogels
Fluorescent labelling of Col I was performed according to a previously published protocol.27 Briefly, a stock solution of 5(6)-TAMRA N-succinimidyl ester (C0027, Chemodex) was prepared in dimethyl sulfoxide (DMSO) at a concentration of 10 mg mL−1, vortexed thoroughly and stored under exclusion of light at −20 °C. 2 mL of concentrated Col I was injected to a 3 mL 3.5k MWCO Slide-a-Lyzer Dialysis Cassette (Thermo Fisher) and dialyzed overnight against 2 L of sterile filtered aqueous labeling buffer (0.25 M NaHCO3, 0.4 M NaCl, pH 9.5) at 4 °C. All subsequent steps were performed under exclusion of light.The dialysis cassette was removed from the labeling buffer, 100 μL of TAMRA stock solution was mixed with 900 μL of labeling buffer and injected to the dialysis cassette. The cassette was wrapped in aluminum foil and incubated on a rocking shaker overnight at 4 °C. Next, to remove excess dye and reconstitute the acidic pH of the Col I stock, the dialysis cassette was thoroughly washed in acidic washing buffers. First, the cassette was placed in 1 L of 0.5 M acetic acid at pH 4 and dialyzed at 4 °C overnight. Next, the cassette was subjected to two subsequent dialysis washes in each 1 L of 0.02 M acetic acid at pH 4 overnight at 4 °C. The fluorescently labelled Col I was withdrawn from the dialysis cassette and stored at 4 °C under exclusion of light.
Fabrication, functionalization and characterization of silica-coated magnetic iron μRods
Soft ferromagnetic iron μRods were fabricated using template restricted electrodeposition. The method to prepare plating templates via photolithography was developed based on work described in the thesis of B. Özkale.28 Subsequently, the magnetic iron μRods are coated with a layer of SiOx, functionalized, and characterized, see details in ESI Text 1 and ESI Fig. S1–S4.†PDMS pool preparation
PDMS pools were prepared as previously published.27 Briefly, polydimethylsiloxane (PDMS) polymer (SYLGARDTM 184 Silicone Elastomer kit, Dow Corning #01673921) was mixed according to manufacturer's instructions and degassed for 1 hour. The degassed PDMS mix was poured onto a blank wafer to obtain a layer thickness of 4 mm, degassed again for 1 hour and cured at 80 °C for 3 hours or overnight. The cured PDMS disc was detached from the wafer and rings were punched out with an inner diameter of 8 mm. Using oxygen plasma treatment, PDMS pools were bonded to glass cover slips and sterilized by washing with 70% ethanol in deionized water and UV treatment for a minimum of 30 minutes.Surface functionalization of glass and plastic surfaces
To enhance adhesion of Col I hydrogels to commercially available and self-fabricated sample substrates made from glass, PDMS or plastic, a surface modification with dopamine was performed, according to a protocol published by Park et al.29 If not already delivered in sterile packaging, the sample to be surface-treated was sterilized by thorough washing using 70% ethanol in deionized water and subsequent exposure to UV-radiation for at least 30 minutes. Next, the surfaces of interest were incubated with a sterile filtered solution of 2 mg mL−1 dopamine (Dopamine 62-31-7, Sigma Aldrich) in 10 mM Tris-HCl (pH 8.5) at room temperature for 2 hours. During the incubation, a change of color from transparent to dark grey-brown could be observed, indicating the progression of pH-induced oxidation. After incubation, the reaction solution was aspirated, samples were gently rinsed with autoclaved deionized water and left to dry. Until further use, samples were stored under sterile conditions in sealed plastic dishes at 4 °C.Magnetic actuation using an electromagnetic field generator
To infer suitable magnetic field parameters and allow live confocal imaging during magnetic actuation, an eight-coil electromagnetic field generator (MFG 100-I, Magnebotix AG) was used that was mounted on a spinning disk confocal microscope. Field magnitudes exceeding 20 mT were achieved using conical field focusing core extensions, resulting in a spherical workspace of 1 mm radius. Deflections upon actuation were analyzed optically for sweeps of field magnitudes and frequencies.Long-term magnetic actuation using a custom-built Halbach array
For cyclic magnetic actuation of multiple samples in parallel over several days, an incubator-compatible cylindrical Halbach array was built (ESI Fig. S5†). This Halbach array comprised eight positions of permanent magnets (NdFeB, N42) with dimensions of 0.5 × 0.5 × 2.0 cm arranged in a circular array. To obtain a cylinder of 8 cm in length, four of the bar magnets were placed in series at the magnet positions along the longitudinal axis of the cylindrical array. The dimensions of the cylindrical magnet array were determined using Finite Element Method Magnetics (FEMM)30 and designed to result in a uniform magnetic field of 50 mT within a cylindrical workspace of 3 cm. The Halbach array actuator was 3D printed from polyethylene terephthalate glycol (PETG) using a filament extruder and assembled with a motor (P25UK12, PiBorg) and a speed controller (Global Specialities) inside a custom-designed acrylic casing (0.5 cm thickness). The components were stably connected using dichloromethane (Sigma-Aldrich) and protected from humidity using silicon paste and thermoresistant tape. The rotational frequency of the assembled setup was measured and adjusted to 1 Hz using a hand-held tachometer (PeakTech 2790). The magnetic field of approximately 50 mT was confirmed inside the Halbach cylinder using a Hall Probe (Metrolab 3D Magnetometer) that was positioned using a piezo controller mounted on a custom-made 3D printed holder. Measurements of various positions (performed in triplicate) are displayed in ESI Fig. S6.†Fluorescence and brightfield imaging
Fluorescence imaging was performed using a Nikon Eclipse Ti2 microscope that was equipped with a CSU-W1 Confocal Scanner Unit and a Hamamatsu C13440-20CU ORCA Flash 4.0 V3 Digital CMOS camera. The Nikon NIS-Elements Advanced Research 5.02 (Build 1266) software was used for operation of the microscopy and image acquisition. Images were acquired with Nikon CFI Apochromat objectives (10×, 20×, 40×). See ESI Text 2† for image processing analysis.Collagen deformation with strain rate mapping
For Col I deformation studies, 700 μL of Col I hydrogel were prepared at a final concentration of 2 mg mL−1 as specified earlier. During preparation, 10 μL of the complete medium were substituted with a suspension of fluorescent microparticles (Fluoromax, 0.2 μm, R200, Thermo Scientific) prior to addition of the remaining components. Collagen hydrogels were transferred to PDMS wells with 4 mm diameter and left to polymerize for 45 minutes at 37 °C, 5% CO2 in a humidified Petri dish. To track collagen deformation, actuation experiments were conducted during live imaging using the magnetic field generator31 mentioned above. Actuation was performed at 50 mT field magnitude with fields rotating at a frequency of 1 Hz either in in-plane (xy), or out-of-plane (xz). Video sequences of 30 seconds were acquired with a 40× objective using confocal mode imaging in the TEXAS red channel (580 nm), see ESI Video V1 and V2.†Col I hydrogel deformation was recorded at a frame rate of 100 ms and the displacement of the fluorescent tracer particles was analysed via Particle Image Velocimetry using the PIVlab plugin for MATLAB.32–35 Image sequences were preprocessed by enabling the CLAHE (Contrast Limited Adaptive Histogram Equalization) function. PIV analysis was then performed using the FFT window deformation algorithm with an initial interrogation area of 128 pixels followed by two additional passes with decreased window size (halving the pixel number in every consecutive pass). The vectors indicating local displacement in m s−1 were scaled to 10 and superimposed with a display of the simple strain rate [1 s−1] presented in a color map. As the simple strain rate depicts the velocity gradient relative to the local velocity, their value indicates the direction of particle displacement, resulting in both positive and negative values within the obtained set of data.36
Ca2+ imaging experiments
3D Col I matrices that were populated with MDA-MB-231 cancer cells and enriched with surface-functionalized iron μRods for Ca2+ imaging in the presence and absence of magnetic actuation. 50 μL of Col I at a concentration of 2 mg mL−1 were added per PDMS well and incubated in a humidified chamber at 37 °C. Next, MDA-MB-231 cells were dispersed in 2 mg mL−1 Col I matrices at a concentration of 105 cells mL−1, 5 μL of iron μRods were added per 200 μL of Col I–cell mixture and 70 μL of the Col I-cell mixture were applied on the samples. Samples were incubated for 1 h, then covered with cell culture medium and further incubated for 3 days at standard incubator conditions. 1.5 h prior to the experiment, Col I samples were once gently washed with PBS and incubated with 10 μM solution of Calbryte 520 AM (AAT Bioquest), diluted in a labeling buffer prepared from Modified Hanks Buffer with Calcium and Magnesium, supplemented with 0.04% Pluronic F-127 and 20 mM HEPES. Samples were incubated at 37 °C, 5% CO2 for 1 h and gently washed twice with the labeling buffer prior to imaging.Live imaging was performed while samples were actuated using the magnetic field generator described earlier (ESI Video V3†). During the experiment, image sequences were acquired using the GFP channel with a frame rate of 200 ms to capture the Ca2+ influx while the deflection of the iron μRods was inferred separately via the brightfield channel. Images were analyzed using FIJI and raw integrated density was measured in regions of interest (ROI), representing the overall signal intensity within the selected ROI. The results were subsequently transformed into ΔF/F0 values, where F0 represents the average raw integrated density during the initial non-actuated period, and ΔF is the difference between the instantaneous raw integrated density and F0.
Spheroid preparation
Spheroids were prepared using low adhesion round bottom 96 well plates (Corning, Costar Ultra-Low Attachment 96-well plate). 5 × 103 cells were seeded per well in 100 μL of complete medium and plates were centrifuged at 500g for 5 minutes. After one day of culture, 100 μL of media containing 10 μg mL−1 Col I were added per well to obtain a final concentration of 5 μg mL−1. The plate was centrifuged again at 500g for five minutes and incubated for two more days.Spheroid embedding
Spheroids were embedded in Col I hydrogels after three days of culture. To enhance adhesion of Col I hydrogels, 8-well culture slides (μ-Slide 8 well, ibiTreat, IBIDI) were surface functionalized with dopamine as described above and briefly incubated with complete medium prior to use. 150 μL of Col I hydrogels prepared at the desired concentration were added per well. Spheroids were collected with a 200 μL pipette and transferred to Col I hydrogels, releasing them gently from the pipette without disruption of the collagen network and aiming to keep the amount of released medium to a minimum. Col I hydrogels were left to polymerize for approximately 30 minutes in a cell culture incubator at 37 °C and 5% CO2. Next, an additional layer of 100 μL of Col I was added on top of the previously prepared samples, either enriched with functionalized μRods or not. Samples were left to polymerize for 30 more minutes at 37 °C. To prevent dehydration of the hydrogels, samples were incubated in humidified chambers within the cell culture incubator.RNA isolation
RNA was extracted from Col I hydrogel-embedded spheroids after long-term magnetic actuation. All samples were transferred to 1.5 mL microcentrifuge tubes. TRIzol reagent (15596026, Thermo Scientific) was mixed with the samples and the mixtures were homogenized mechanically. Chloroform was subsequently added for phase separation purposes. The tubes were centrifugated, and the aqueous phases containing RNA were transferred to fresh tubes. Further steps including DNase treatment, RNA purification, and sample elution were carried out utilizing a Qiagen RNeasy Micro Kit (Qiagen 74004) according to manufacturer's instructions. Final RNA concentrations were measured using a NanoDrop Spectrophotometer (701-058112, Thermo Scientific).RNA sequencing and analysis
RNA analysis was performed at the Functional Genomics Center Zurich (University of Zurich). The library preparation was performed with Takara Clontech SMARTer Stranded Total RNA-Seq Kit v2 – Pico Input Mammalian using 1 ng of total RNA. The resulting libraries were sequenced on a NovaSeq 6000 (Illumina) in paired-end 250 bp mode. The raw reads were first cleaned by removing adapter sequences, trimming low quality ends, and filtering reads with low quality (phred quality <20) using Fastp (version 0.20).37 Sequence pseudo alignment of the resulting high-quality reads to the Human reference genome (build GRCh38.p13 and GENCODE gene models release 42) and quantification of gene level expression was carried out using Kallisto (version 0.46.1).38 Differential expression was computed using the generalized linear model implemented in the Bioconductor package DESeq2 (R version: 4.3.0, DESeq2 version: 1.40.1).39 Genes showing altered expression with adjusted (Benjamini and Hochberg method) p-value <0.05 were considered differentially expressed.Results
Analysis of tumor cell invasion into 3D Col I hydrogels
Cancer cell invasion marks the first step of the metastatic cascade (Fig. 1a). To model cancer cell invasion in vitro, 3D tumor spheroids from the invasive breast cancer cell line MDA-MB-231 were embedded in 3D Col I matrices and imaged over three days (Fig. 1b). As previously reported, our results confirmed that the invasion of tumor cells into the surrounding matrix is related to Col I concentration in a biphasic manner.40 Cancer cell invasion was observed to increase with increasing concentration of Col I hydrogels between 0.5 mg mL−1 to 3.0 mg mL−1 and a peak of invasive behavior was identified for Col I matrices at a concentration of 3 mg mL−1. Higher concentrations of Col I hydrogels, however, were found to reduce cancer cell invasion (ESI Fig. S7†).Fluorescent labelling of Col I hydrogels used for cancer invasion studies showed densification and remodeling of the Col I hydrogel network surrounding the invading tumor cells. Reorganisation of the matrix was more pronounced for hydrogels with lower concentrations of Col I, potentially because the embedded cells were able to more rapidly remodel the sparser matrix. Alternatively, changes in the matrix may have been easier to detect at lower Col I concentrations. Representative examples of tumor spheroids cultured in different concentrations of fluorescently labelled Col I matrices are displayed in ESI Fig. S7b–d.†
For the following studies on the influence of cyclic deformation of Col I hydrogel matrices on the invasive behavior of embedded tumor spheroids, a Col I hydrogel concentration of 2 mg mL−1 was used. Experimental data displayed in ESI Fig. S7b† show that a concentration of 3 mg mL−1 was found to result in the most pronounced spreading of cancer cells to the surrounding matrix (as displayed in ESI Fig. S7b†). In comparison, a concentration of 2 mg mL−1 showed similar behavior of matrix invasion while not reaching maximum values. Hence, a system using 2 mg mL−1 Col I was selected for experiments designed to test whether other stimuli increase or reduce the degree of cancer cell invasion, while allowing for the reconstitution of an invasive phenotype. This ensured that either positive or negative changes in invasiveness in response to local ECM deformation could be observable.
Magnetically controlled actuation of in vitro ECM models
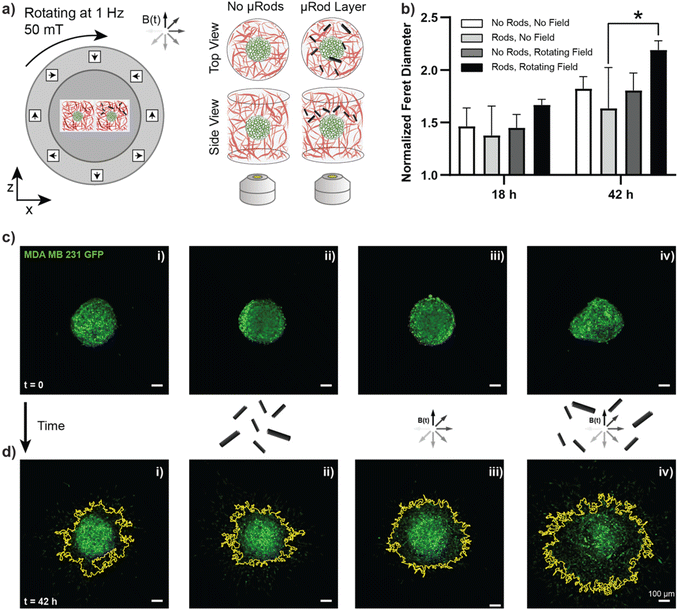
Tumor cells interact and invade their surrounding microenvironment, which is also populated by other cell types, such as highly contractile fibroblasts. Fig. 1b illustrates how individual cancer cells disseminate from their original location (here a tumor spheroid) and invade the surrounding Col I matrix while further reorganizing the surrounding fiber structure. Our study aimed to investigate how cancer cell invasion could be influenced by cyclic local deformation of the tumor microenvironment, mimicking the interaction of cells residing in the tissue with their surrounding ECM network.Magnetic μRods, approximately 25 μm in length and 3.5 μm in radius, covalently linked to the Col I hydrogels (Fig. 1c) were used in 3D tumor invasion models as ECM-embedded microscale mechanical actuators. Acute cytotoxicity of the iron μRods was excluded by an MTT assay, as displayed in ESI Fig. S4.† To determine the field magnitude used for the actuation experiments, Col I hydrogel samples were prepared at a concentration of 2 mg mL−1 and mixed with surface-functionalized μRods. Using a versatile electromagnetic field generator mounted on a confocal fluorescence microscope, rotating magnetic fields (RMF) of varying magnitude were tested to determine a field magnitude that would allow for readily detectable deflection of the Col I -embedded μRods without disrupting the surrounding Col I network. The experimental setup is depicted in ESI Fig. S8.† A working RMF magnitude of 50 mT was determined, and hereafter a RMF of 50 mT with 1 Hz frequency was adopted for collagen deflection and cell invasion studies (Fig. 1d). In addition, to enable exposure of multiple tumor spheroid samples to a uniform RMF over several days under standard cell culture conditions, a custom incubator-compatible sample actuation system based on a rotating Halbach cylinder was built. The device consists of permanent magnets arranged in a first order Halbach cylinder41 designed to produce a magnitude of 50 mT. It was mounted laterally on a ball bearing and coupled to a motor that drove its rotation around the x-axis with a frequency of 1 Hz.
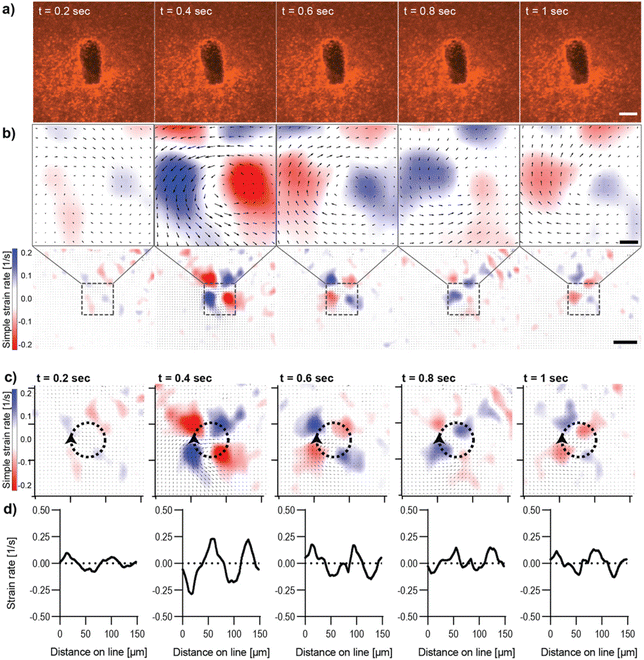
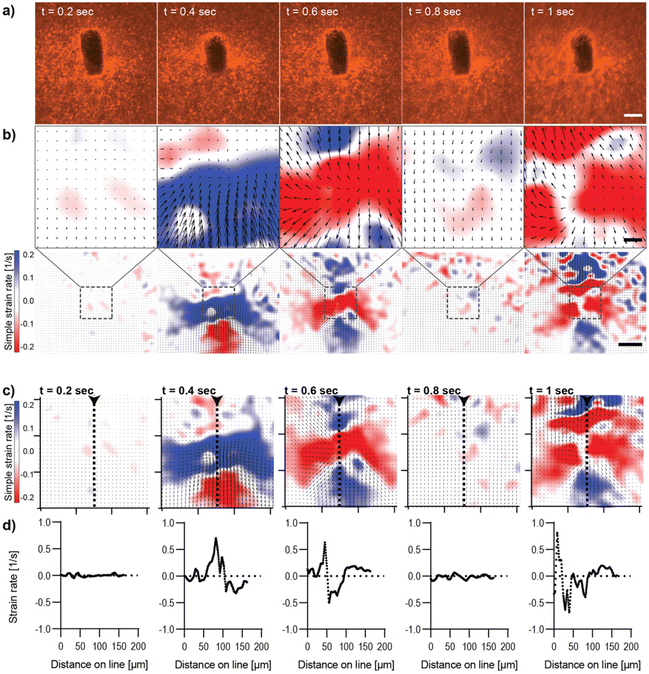
Characterization of magnetically controlled Col I hydrogel deformation
Live imaging of the deformation of Col I hydrogels through magnetic actuation of μRods embedded in samples was investigated using the microscope-mounted magnetic field generator. In all experiments, the same actuation parameters (50 mT, 1 Hz) were applied, as previously determined and used for the incubator-compatible Halbach actuation setup. To visualize Col I hydrogel deformation during magnetic actuation, hydrogel samples were supplemented with fluorescent polystyrene nanoparticles (200 nm diameter) and imaged via confocal fluorescence microscopy. The resulting deformation patterns of the Col I matrices by the embedded magnetic μRods were analyzed for both in-plane rotation (about the z-axis) and out-of-plane rotation (about the x axis) under the specified RMF conditions. We then quantified the strain rates resulting from cyclic deflection. Although it would, in principle, be possible to integrate these spatially dependent strain rates in time to map the resulting strain fields, strain rate maps are favored here because they emphasize changes from frame to frame and more closely reflects the underlying PIV data. Results for in-plane and out-of-plane RMF are displayed in Fig. 2 and 3 respectively. In both modes of actuation, the spatial extent of the strain field and sign of the strain rates was found to change over the course of one cycle, as visualized in Fig. 2 and 3. The torque applied in both modes was estimated to be on the order of 80 pNm for the μRods, with details of the torque calculation provided in ESI Text 3.† This magnitude is similar to previously reported cell contractile moments exhibited e.g. by fibroblasts.11,42–45In-plane rotation of the magnetic field was shown to cause a radially symmetric pattern of strain rate distribution around the center of rotation of the magnetic μRod (Fig. 2b–d, ESI Video V1†). Maximal values were found at 0.4 s after the onset of deflection and reached values of simple strain rates of approximately ±0.2 s−1. The extraction of strain rates along the circumference of a circle surrounding the center of μRod deflection with an assigned radius of 24 μm further displays the pattern of alternating positive and negative values of strain rates surrounding the center of μRod deflection in four quadrants (Fig. 2b and c). This behavior is associated with the direction of Col I network displacement during the rotation of the actuated μRod. Considerable strain is detectable within regions spanning 200 μm in diameter around the μRod, which is in line with reported strain fields generated by cells, as noted earlier.10,11
The analysis of image sequences acquired during out-of-plane actuation of magnetic μRods within the Col I hydrogel was performed analogously to the in-plane magnetic field rotation and is shown in Fig. 3 (ESI Video V2†). Here, as expected for any cyclic deformation, two directions of Col I displacement were detected. However, the strain rates measured during out-of-plane rotation were more than double the strain rate values observed for in-plane field rotations. The pattern of Col I deformation caused by out-of-plane deflection of a μRod displays regions expressing positive and negative strain rates, respectively, to extend laterally, perpendicular to the long axis of the μRod, hence along the axis of the field rotation (Fig. 3b and c). The values of the strain rates identified in the analysis are shown to vary cyclically, with representative values from one period of field rotation depicted in the image sequences of Fig. 3. In an originally relaxed state (Fig. 3b, 0.2 s) strain rates are low, reflecting only marginal particle displacement. During deflection, strain rates are increasing, as shown at timepoints t = 0.4 and t = 0.6 s, respectively, resulting from a larger displacement of tracer particles per time, and thus, increased particle velocity. At t = 0.8 s, a planar position of the μRod within the image plane is again observed, similar to the fluorescence micrograph shown for timepoint t = 0.2s.
The values extracted from the region of the center of deformation (Fig. 3c and d) indicate local strain rates that start close to zero and assume peak values of approximately 0.7 s−1 (for positive strain rates) and close to −1 s−1 for the negative strains rates, as observed at timepoint t = 1 s of the magnetic field rotation. The extraction of strain rates along a line perpendicular to the axis of rotation confirms the alternation of positive strain and negative strain rates within the area depicted in the strain rate maps (Fig. 3c and d). Furthermore, the strain rates extracted from image sequences during the actuation of a μRod caused by the out-of-plane rotation of a magnetic field show detectable strains developed over an area exceeding 200 μm in diameter, as indicated in the overview maps in Fig. 3b, bottom. The higher out-of-plane strain rate values may be attributable to the local mechanical environment created by the method of sample preparation, in which μRods were added in unpolymerized Col I atop an already polymerized Col I hydrogel. This is likely to lead to differences in crosslinking within and between the layers at the resulting interface and could account for the apparent difference in compliance for in-plane and out-of-plane rotation.
Moreover, despite higher values, neither out-of-plane actuation nor in-plane actuation seem to induce any damage or plastic deformation in the surrounding collagen matrix, as strain maps and the matrix pore size distributions remained unaltered over time (ESI Fig. S9–S12).† In summary, both modes provide means to generate physiologically relevant strain fields originating from individual localized sources of single cell dimensions.
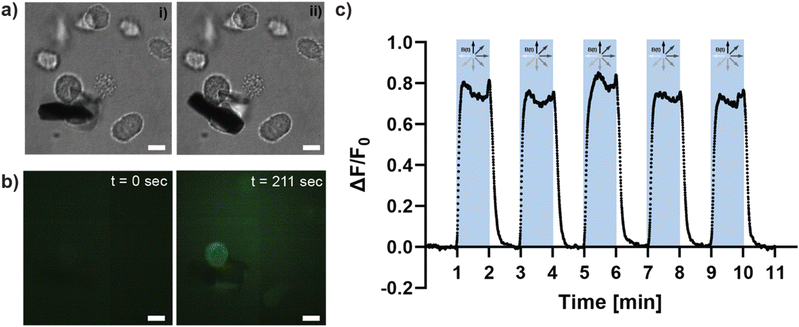
Magnetically induced mechanical deformation triggers increased calcium flux
Having examined the matrix deformations produced by the μRods, we next sought to test their impact on cancer cells. Mechanosensitive ion channels are a prime example of mechanical signal transduction in living cells. Embedded in the cellular membrane, they are activated by deformation of the membrane and allow, for example, the influx of calcium ions that trigger intracellular signaling processes. To test whether the mechanical forces resulting from magnetic actuation of Col I-embedded iron μRods provoke a change in Ca2+ levels within living cells, experiments were performed to investigate changes in cellular Ca2+ levels during actuation of Col I networks surrounding MDA-MB-231 cancer cells. Cancer cells were embedded with μRods in Col I hydrogels (at a concentration of 2 mg mL−1). By means of a fluorescent Ca2+ indicator probe, changes in intracellular Ca2+ levels were monitored during magnetic actuation of the embedded μRods (Fig. 4). An image sequence of samples containing cancer cells treated with the fluorescent Ca2+ indicator probe was analyzed for fluorescence intensity levels during cyclic magnetic actuation of the μRods embedded in their vicinity (Fig. 4a). Prior to actuation (Fig. 4a, i and Fig. 4b, t = 0 s), a low and relatively homogenous fluorescence signal is observed. Upon actuation of the sample with a magnetic field of 50 mT at 1 Hz frequency for 1 minute, the repeated exposure of the cells to a detectably deflected μRod (as depicted in Fig. 4a, ii) was accompanied by an increase of fluorescence intensity in the region of μRod-adjacent cancer cells (see also ESI Video V3†). This trend is detected as prominent peak in the fluorescence values that are plotted as function of time in Fig. 4c. Analyzing these changes in fluorescence intensity of the intracellular Ca2+ probe, our data suggests that repeated deformation of the cellular environment by adjacent μRods mechanically triggers the influx of Ca2+ ions into the cellular body.Micromechanical deformation of Col I hydrogels increases tumor cell invasion
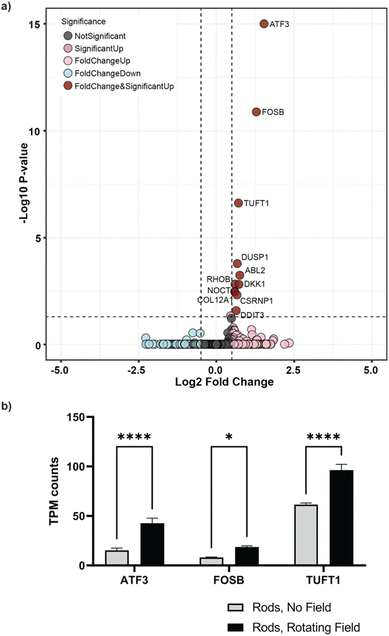
After establishing that actuated μRods can provoke observable cellular responses, we next sought to investigate the influence of long-term cyclic micromechanical deformation of the tumor microenvironment on the invasive behavior of MDA-MB-231 spheroids. Based on the previously described tumor invasion model, spheroids prepared from MDA-MB-231 GFP cells were embedded in Col I hydrogels of 2 mg mL−1 concentration. A second layer Col I hydrogel containing the surface-functionalized iron μRods was then applied and allowed to polymerize, followed by the addition of another layer of Col I hydrogel on top. Imaging was performed 1 h post spheroid embedding, before samples were exposed to RMF. Samples were imaged via confocal microscopy after 18 h and 42 h of incubation with or without magnetic actuation, respectively (Fig. 5a). As displayed in Fig. 5b, invasion of tumor cells into the surrounding Col I hydrogel matrix was significantly increased after 42 h of magnetic actuation compared to the control condition, where spheroids were cultured in the presence of μRods but without exposure to a rotating magnetic field. Furthermore, the invasive behavior of MDA-MB-231 cells was neither expected to be appreciably affected by the application of RMF alone, nor did our experiment reveal a statistically significant effect on samples without μRods that were exposed to RMF, as compared to unexposed control samples. Representative images of the spheroids imaged before and after incubation in the presence and absence of a RMF and μRods are displayed in Fig. 5c (before incubation) and Fig. 5d (after 42 h of incubation). The yellow outline indicates the boundary of spheroid spreading. As is apparent from the data obtained from the Feret diameter calculations displayed in Fig. 5b, the spread of tumor cells into the surrounding Col I matrix is readily detectable in the fluorescence images.Beyond these observed changes in cell migration and invasion upon mechanical actuation, we sought to investigate potential alterations on a gene level. To do so, spheroids embedded in collagen with dispersed μRods were dissociated after 48 h of actuation and mRNA was extracted for subsequent sequencing and transcriptomic analysis. Spheroids without exposure to cyclic actuation served as controls. Comparing actuated and non-actuated samples, 11 genes were identified to be significantly upregulated in samples exposed to cyclic deformation (Fig. 6a, ESI Fig. S13†). Among these, we found three genes of particular interest in the context of our study due to their association in cancer cell proliferation,46 migration,47–49 and cellular stress response that can arise from mechanical strain:50–52 the stress and cancer related Activating Transcription Factor 3 (ATF-3) gene, and the tuftelin 1 (TUFT1) gene, and the proto-oncogene of the Activator Protein-1 family FosB. For instance, knockdown of ATF-3 expression in MDA-MB-231 cells was shown to reduce cell migration and the expression levels of invasive and metastatic genes such as MMP-13 and Runx2.53
Furthermore, TUFT1 expression is known to be increased in breast cancer, and its downregulation has been shown to reduce proliferation in vitro and to restrict tumor growth in vivo.54 In a colon cancer study, upregulation of TUFT1 was correlated with increased migration and invasion of LoVo cells.55 Upregulated ATF-3 and TUFT1 expression in samples exposed to cyclic deformation thus seems consistent with the increased invasive behavior observed via analysis of confocal micrographs of the spheroid invasion. Both FosB and ATF-3 expression are related to stress response and their upregulation may be associated with the mechanical load applied during cyclic deformation.50,51,56 In particular, FosB is rapidly induced via mechanical stress.51 Interestingly, its knockout has been also studied as a target in triple negative breast cancer. Thus, its upregulation may also be linked to invasion.57 While overall the fold increase between actuated and unactuated samples may be subtle (Fig. 6b), it should be noted that only cells in the periphery of spheroids may contribute to the observed upregulated gene expressions leading to a rather small effect, while the majority of cells in deeper layers of the spheroids may be merely affected by mechanical stimuli of their immediate surroundings. Nevertheless, the observed changes in gene expression related to migration and stress seem to support the impact of the applied mechanical cues on a cellular level.
Local application of cyclic strain triggers anisotropic invasion
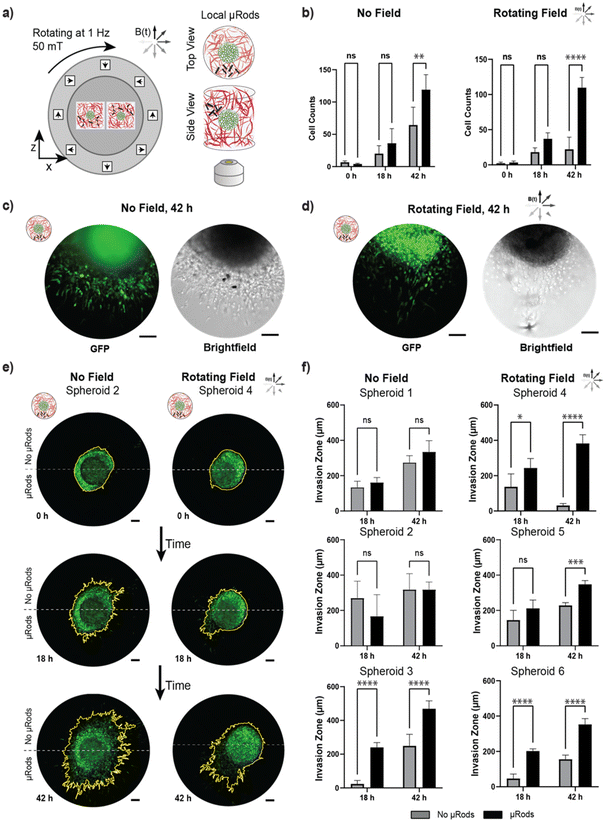
To test the localized effect of micromechanical deformation, an experiment was conducted analogously to the assay presented in the previous section with spatially confined μRods embedded at one side of a tumor spheroid at variable distances ranging from tens to hundreds of μm. Thus, instead of the global distribution of μRod-enriched Col I hydrogel volumes across the whole surface of the spheroid sample, the suspension was applied only locally in a region adjacent to a spheroid (Fig. 7a). Thus, mechanical actuation of the Col I matrix could be spatially focused to localized sources with a deformation spanning about 200 μm (see Fig. 3) and, accordingly, the cells in closest proximity to the applied μRods were exposed to the strongest cyclic micromechanical deformation. We hypothesized that the cells would respond in a spatially dependent manner to mechanical stimulation and acquired images before actuation at t = 0 h, 18 h, and 42 h. To quantify anisotropic response, a measure of invasion capturing directionality is required, which can be provided by considering the ‘invasion zone’ through the number of leading invading cells in a certain region.58 To do so, images of spheroids were divided into two regions: one half associated with the hemisphere with locally applied μRods and an opposite hemisphere without μRods, and thus, lesser or no exposure to mechanical deformation (see also ESI Fig. 9†). Next, we analyzed and compared the cell count within stimulated and unstimulated hemispheres (Fig. 7b). Less pronounced increase in cell invasion between the hemispheres was observed for spheroids that did not experience actuation, in contrast to spheroids exposed to RMF that showed a strong anisotropic increase in cell invasion after 42 h. Representative fluorescence and bright field images of tumor spheroids with locally applied μRods with and without actuation are shown in Fig. 7c and d.Overall, a trend of cell migration toward the location of the actuated μRods was observed. A particularly distinct example is shown Fig. 7e (left), where the spheroid was exposed to RMF and cell migration occurs most intensely in the direction of the μRods (all analyzed spheroids are shown in ESI Fig. S14†). The extent of the invasion zone of the tumor spheroid into the Col I matrix was more closely investigated by measuring the distance of the five leading invading cells from the tumor rim (Fig. 7f and ESI Fig. S9†). All spheroids exposed to RMF showed a significant increase in invasion zone distances in regions with μRods compared to regions without μRods after 42 h. We did, however, also observe one spheroid which had no exposure to RMF with apparent increase in invasion zone distance for the region with μRods versus without. The exact cause is uncertain, but this behavior could have been triggered through the structural influence of the μRods on the Col I matrix that polymerized around them. Furthermore, the exact placement of μRods adjacent to spheroids was challenging to robustly control, thus, in some instances the source of actuation was further away and less impactful on a spheroid than in others.
Discussion and conclusion
Cells and their surrounding microenvironment exist in a dynamic relationship, involving the exchange of both biochemical and biomechanical signals. In particular, the process of cancer cell invasion is marked by the close interaction between cells and the surrounding ECM network. This work presents the controlled application of long-term mechanical deformation within Col I hydrogels that were used as model systems for studies of cancer cell invasion. Controlled by RMF, iron μRods served as embedded microscale actuators to achieve local deformation of the surrounding fibrous matrix.Using PIV analysis, strain rates within the hydrogel networks were analyzed regarding magnitude and range of influence within the examined sample environments, showing regions of influence spanning approximately 200 μm or larger, which is consistent with cell-generated strain fields. In long-term actuation experiments over 42 h, we showed that invasion of MDA-MB-231 cells from spheroids embedded in Col I hydrogels increased upon cyclic deformation of the Col I matrix by globally distributed μRods. Local application and actuation of μRods on one side of the tumor spheroids showed a local influence on cell migration, indicating local attraction of the invasive cancer cells by the localized strain.
By means of magnetic micro-actuators that were stably attached to Col I hydrogel matrices and showed good compatibility with cell culture systems, cyclic strain was applied within a 3D Col I network for 42 h. The analysis of Col I displacement revealed the rates of strain expressed in the Col I network during one cycle of actuation. The patterns of positive and negative values reflect the direction of the strain and show alternation during one cycle of actuation, as expected. The strain rates recorded through particle velocimetry analysis were dependent on the mode of actuation (in-plane or out-of-plane). This anisotropy is likely accounted for by the specific method of sample preparation, which involved polymerizing a layer of Col I atop a preexisting layer of Col I. Certainly, the local structure of the Col I hydrogel might have also favored the higher compliance and higher rate of strain observed for out-of-plane actuation by μRods. As an additional but likely more minor effect, the displacement of fluorescent tracer particles during μRod deflection out-of-plane might have resulted in tracer particles passing in and out of the focal plane. This could result in higher apparent degrees of displacement for the out-of-plane rotation of the magnetic field, but the impact of this artifact should be mitigated by the fact that during any given timestep, most of the tracer particles remain within the focal plane.
Col I is a biomaterial that is characterized by strain stiffening. The exposure of mechanically sensitive cells to these cyclically strained materials during magnetic actuation is therefore particularly interesting with regards to the temporal aspect of mechanical alteration of the cell surrounding matrix. Indeed, Ca2+ imaging revealed increased Ca2+ levels within cells exposed to sustained actuation of the surrounding matrix and increased invasion was observed for tumor spheroids of the invasive cell line MDA-MB-231. This partially supports previous findings by Menon et al. in 2011, reporting on increased invasion of the invasive sarcoma cell line HT1080 in response to cyclic strain resulting from magnetically controlled displacement of magnetic beads.23 However, Menon et al. attributed this increase of invasive behavior to a mechanism dependent on the presence of fibronectin as additional ECM component for the reception of the involved mechanical signals. In contrast to these findings, the presented system of Col I hydrogels does not require the addition of fibronectin to mechanically provoke increased invasive behavior. Potential reasons might include the investigated cell line, different patterns and magnitudes of magnetic particle actuation or different quality of the applied Col I matrix. Alternatively, the mechanisms underlying the change of invasion might differ with regards to the magnitude of the strain caused by deflecting μRods and the “tugging” forces described by Manon et al. While the displacement of beads resulting in the described “tugging” forces that enhanced cancer cell invasion in an integrin-dependent manner, the strains caused during the deflection of μRods might evoke strain stiffening of the Col I network, targeting stiffness-dependent modes of migration.59
Additional insight into the mechanistic influence of localized cyclic deformation on the cancer cells was gained through RNA sequencing, which identified 11 upregulated genes, with at least three of these (ATF-3, TUFT1, and FosB) seeming to offer explanatory value for the observed changes in invasive behavior. We note that, in the future, these approaches could be refined by performing RNA sequencing with greater spatial resolution to directly probe the cells subjected to the most intense mechanical stimulus. Proteomic analysis could be another source of information about the intracellular processes triggered, amplified or suppressed by the cyclic micromechanical stimulus.
The potential effects of constant deformation over longer times might include a change in patterns of Col I architecture resulting from the altered interaction of cells with their surrounding matrix. Further, the ECM is known to exhibit local accumulation of metabolites and signaling factor reservoirs that can be released through mechanical deformation by cells. A potential consequence of long-term cyclic deformation of the ECM network might be to influence the accumulation of such reservoirs and, for instance, result in the release or interfere with the local accumulation of otherwise bound molecules, thereby causing changes in the biochemical environment of the tissue model.60
The presented work applies magnetic μRods that are stably attached to a Col I hydrogel matrix as sample-embedded microscale actuators. The design of a magnetic field generator that is suitable for use in a standard cell culture incubator allows for the actuation of several sample replicates under uniform conditions over several days of culture. Applied in the context of a 3D model for cancer invasion, the combination of techniques applied in this study is shown to influence the invasive behavior of cancer cells and to direct cell migration. While demonstrated on one cancer cell line, future studies could employ the presented techniques for the evaluation of mechanobiological influences in the context of other model systems and perform further readouts of biological markers.
Author contributions
Conceptualization: D. A., S. S.; data curation: D. A., A. M., N. H., F. L., X. W., S. M.; formal analysis: D. A., A. M., N. H., F. L., R. W., S. S.; funding acquisition S. S.; investigation: S. S.; methodology: D. A., M. C., S. S.; project administration: S. S.; resources: N. A., S. S.; software: D. A., A. M., A. S., A. D. M.; supervision: S. S.; validation: D. A.; visualization: D. A., F. L., X. W., R. W.; writing – original draft: D. A., S. S., A. S., R. W.; writing – review & editing: all authors.Conflicts of interest
S. S. is co-founder, member of the board, and scientific advisor of Magnebotix AG.Acknowledgements
The authors would like to thank Prof. Salvador Pané, Siyu Deng, Fabian Landers and Valentin Gantenbein for technical support regarding the μRod fabrication and access to their electroplating setup. They gratefully acknowledge the Functional Genomics Center Zurich (FGCZ) of University of Zurich and ETH Zurich, and in particular Lennart Opitz, for assistance with Genomics analyses. They also thank Dr. Claire Schirmer for support, Dr. Cameron Forbrigger for proof reading, and the staff of the FIRST clean room and of the Physics workshop at ETH Zurich for support.References
- A. Ring, B. D. Nguyen-Sträuli, A. Wicki and N. Aceto, Nat. Rev. Cancer, 2023, 23, 95–111 CrossRef CAS PubMed.
- I. J. Fidler, Nat. Rev. Cancer, 2003, 3, 453–458 CrossRef CAS PubMed.
- D. Wirtz, K. Konstantopoulos and P. C. Searson, Nat. Rev. Cancer, 2011, 11, 512–522 CrossRef CAS PubMed.
- X. Guan, Acta Pharm. Sin. B, 2015, 5, 402–418 CrossRef PubMed.
- B. Emon, J. Bauer, Y. Jain, B. Jung and T. Saif, Comput. Struct. Biotechnol. J., 2018, 16, 279–287 CrossRef CAS PubMed.
- K. S. Kopanska, Y. Alcheikh, R. Staneva, D. Vignjevic and T. Betz, PLoS One, 2016, 11, e0156442 CrossRef PubMed.
- F. Kai, A. P. Drain and V. M. Weaver, Dev. Cell, 2019, 49, 332–346 CrossRef CAS PubMed.
- J. Insua-Rodríguez and T. Oskarsson, Adv. Drug Delivery Rev., 2016, 97, 41–55 CrossRef PubMed.
- M. W. Conklin, J. C. Eickhoff, K. M. Riching, C. A. Pehlke, K. W. Eliceiri, P. P. Provenzano, A. Friedl and P. J. Keely, Am. J. Pathol., 2011, 178, 1221–1232 CrossRef PubMed.
- T. M. Koch, S. Münster, N. Bonakdar, J. P. Butler and B. Fabry, PLoS One, 2012, 7, e33476 CrossRef CAS PubMed.
- S. A. Maskarinec, C. Franck, D. A. Tirrell and G. Ravichandran, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 22108–22113 CrossRef CAS PubMed.
- G. Bahcecioglu, G. Basara, B. W. Ellis, X. Ren and P. Zorlutuna, Acta Biomater., 2020, 106, 1–21 CrossRef CAS PubMed.
- A. Micalet, E. Moeendarbary and U. Cheema, ACS Biomater. Sci. Eng., 2021, 9(7), 3729–3741 CrossRef PubMed.
- M. Cavo, M. Fato, L. Peñuela, F. Beltrame, R. Raiteri and S. Scaglione, Sci. Rep., 2016, 6, 35367 CrossRef CAS PubMed.
- E. R. Shamir and A. J. Ewald, Nat. Rev. Mol. Cell Biol., 2014, 15, 647–664 CrossRef CAS PubMed.
- D. P. Saraiva, A. T. Matias, S. Braga, A. Jacinto and M. G. Cabral, Front. Oncol., 2020, 10, 1543 CrossRef PubMed.
- J. Kim, J. Jang and D.-W. Cho, Front. Bioeng. Biotechnol., 2021, 9, 605819, DOI:10.3389/fbioe.2021.605819.
- N. R. Lang, K. Skodzek, S. Hurst, A. Mainka, J. Steinwachs, J. Schneider, K. E. Aifantis and B. Fabry, Acta Biomater., 2015, 13, 61–67 CrossRef CAS PubMed.
- H. Ahmadzadeh, M. R. Webster, R. Behera, A. M. Jimenez Valencia, D. Wirtz, A. T. Weeraratna and V. B. Shenoy, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(9), E1617–E1626 CrossRef CAS PubMed.
- A. Singh, I. Brito and J. Lammerding, Trends Cancer, 2018, 4, 281–291 CrossRef CAS PubMed.
- J. M. Tse, G. Cheng, J. A. Tyrrell, S. A. Wilcox-Adelman, Y. Boucher, R. K. Jain and L. L. Munn, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 911–916 CrossRef CAS PubMed.
- A. N. Gasparski, S. Ozarkar and K. A. Beningo, J. Cell Sci., 2017, 130(11), 1965–1978 CAS.
- S. Menon and K. A. Beningo, PLoS One, 2011, 6, e17277 CrossRef CAS PubMed.
- I. Indra, A. N. Gasparski and K. A. Beningo, PLoS One, 2018, 13, e0207490 CrossRef PubMed.
- F. E. Uslu, C. D. Davidson, E. Mailand, N. Bouklas, B. M. Baker and M. S. Sakar, Adv. Mater., 2021, 33, 2102641 CrossRef CAS PubMed.
- Y. Shou, X. Y. Teo, X. Li, L. Zhicheng, L. Liu, X. Sun, W. Jonhson, J. Ding, C. T. Lim and A. Tay, ACS Nano, 2023, 17, 2851–2867 CrossRef CAS PubMed.
- D. O. Asgeirsson, M. G. Christiansen, T. Valentin, L. Somm, N. Mirkhani, A. H. Nami, V. Hosseini and S. Schuerle, Lab Chip, 2021, 21, 3850–3862 RSC.
- B. Özkale, An electrochemical approach to the development of complex materials for nanorobotic applications, PhD Thesis, ETH Zürich, 2016.
- S. E. Park, A. Georgescu, J. M. Oh, K. W. Kwon and D. Huh, ACS Appl. Mater. Interfaces, 2019, 11, 23919–23925 CrossRef CAS PubMed.
- D. Meeker, Finite Element Method Magnetics (version 4.2), https://www.femm.info, 28feb2018 Build Search PubMed.
- S. Schuerle, S. Erni, M. Flink, B. E. Kratochvil and B. J. Nelson, IEEE Trans. Magn., 2013, 49, 321–330 Search PubMed.
- W. Thielicke and E. J. Stamhuis, J. Open Res. Softw., 2014, 2(1), e30 Search PubMed.
- W. Thielicke, The flapping flight of birds, PhD Thesis,University of Groningen, 2014.
- W. Thielicke and R. Sonntag, J. Open Res. Softw., 2021, 9, 12 CrossRef.
- D. Garcia, Exp. Fluids, 2011, 50, 1247–1259 CrossRef PubMed.
- E. J. Stamhuis, Aquat. Ecol., 2006, 40, 463–479 CrossRef.
- S. Chen, Y. Zhou, Y. Chen and J. Gu, Bioinformatics, 2018, 34, i884–i890 CrossRef PubMed.
- N. L. Bray, H. Pimentel, P. Melsted and L. Pachter, Nat. Biotechnol., 2016, 34, 525–527 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- A. J. Perumpanani and H. M. Byrne, Eur. J. Cancer, 1999, 35, 1274–1280 CrossRef CAS PubMed.
- P. Blümler, Cells, 2021, 10, 2708 CrossRef PubMed.
- T. Pompe, S. Glorius, T. Bischoff, I. Uhlmann, M. Kaufmann, S. Brenner and C. Werner, Biophys. J., 2009, 97(8), 2154–2163 CrossRef CAS PubMed.
- D. H. Kim, A. J. Ewald, J. S. Park, Kshitiz, M. Kwak, R. S. Gray, C. Y. Su, J. Seo, S. S. An and A. Levchenko, Sci. Rep., 2018, 8, 14210 CrossRef PubMed.
- S. S. An, J. Kim, K. Ahn, X. Trepat, K. J. Drake, S. Kumar, G. Ling, C. Purington, T. Rangasamy, T. W. Kensler, W. Mitzner, J. J. Fredberg and S. Biswal, Biochem. Biophys. Res. Commun., 2009, 382(4), 697–703 CrossRef CAS PubMed.
- A. R. Yoon, I. Stasinopoulos, J. H. Kim, H. M. Yong, O. Kilic, D. Wirtz, Z. M. Bhujwalla and S. S. An, Cancer Biol. Ther., 2015, 16(3), 430–437 CrossRef CAS PubMed.
- W. Liu, L. Zhang, Z. Jin, M. Zhao, Z. Li, G. Chen, L. Sun and B. Chen, Oncotarget, 2017, 8, 74962–74974 CrossRef PubMed.
- Y. Yang, T. Zhang and L. Wu, Biochem. Genet., 2021, 59, 1018–1032 CrossRef CAS PubMed.
- H. Lin, W. Zeng, Y. Lei, D. Chen and Z. Nie, Pathol. Oncol. Res., 2021, 27, 640936 CrossRef PubMed.
- S. Liu, J. Luan and Y. Ding, Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics, 2018, vol. 26, pp. 683–690 Search PubMed.
- D. Inoue, S. Kido and T. Matsumoto, J. Biol. Chem., 2004, 279, 49795–49803 CrossRef CAS PubMed.
- C. Haasper, M. Jagodzinski, M. Drescher, R. Meller, M. Wehmeier, C. Krettek and E. Hesse, Exp. Toxicol. Pathol., 2008, 59, 355–363 CrossRef CAS PubMed.
- X. Wang, J. Fan, M. Zhang, Z. Sun and G. Xu, PLoS One, 2013, 8, e63467 CrossRef CAS PubMed.
- M. Gokulnath, N. C. Partridge and N. Selvamurugan, Tumor Biol., 2015, 36, 1923–1931 CrossRef CAS PubMed.
- W. Liu, L. Zhang, Z. Jin, M. Zhao, Z. Li, G. Chen, L. Sun and B. Chen, Oncotarget, 2017, 8, 74962–74974 CrossRef PubMed.
- Y. Yang, T. Zhang and L. Wu, Biochem. Genet., 2021, 59, 1018–1032 CrossRef CAS PubMed.
- Y. Tanaka, A. Nakamura, M. S. Morioka, S. Inoue, M. Tamamori-Adachi, K. Yamada, K. Taketani, J. Kawauchi, M. Tanaka-Okamoto, J. Miyoshi, H. Tanaka and S. Kitajima, PLoS One, 2011, 6, e26848 CrossRef CAS PubMed.
- C.-H. Ting, Y.-C. Chen, C.-J. Wu and J.-Y. Chen, Oncotarget, 2016, 7, 40329–40347 CrossRef PubMed.
- F. Geiger, L. G. Schnitzler, M. S. Brugger, C. Westerhausen and H. Engelke, PLoS One, 2022, 17, e0264571 CrossRef CAS PubMed.
- F. Geiger, D. Rüdiger, S. Zahler and H. Engelke, PLoS One, 2019, 14, e0225215 CrossRef CAS PubMed.
- K. Gelse, Adv. Drug Delivery Rev., 2003, 55, 1531–1546 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00583f |
| This journal is © The Royal Society of Chemistry 2023 |