Open Access Article
Open Access ArticleControlled degradation of polycaprolactone-based micropillar arrays†
Niamh
Geoghegan‡
 ab,
Mark
O'Loughlin‡
a,
Colm
Delaney
ab,
Mark
O'Loughlin‡
a,
Colm
Delaney
 c,
Keith D.
Rochfort
d,
Meabh
Kennedy
e,
Srikanth
Kolagatla
cf,
Lucia
Podhorska
c,
Keith D.
Rochfort
d,
Meabh
Kennedy
e,
Srikanth
Kolagatla
cf,
Lucia
Podhorska
 a,
Brian J.
Rodriguez
a,
Brian J.
Rodriguez
 f,
Larisa
Florea
f,
Larisa
Florea
 c and
Susan M.
Kelleher
*abe
c and
Susan M.
Kelleher
*abe
aSchool of Chemistry, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: susan.kelleher@dcu.ie
bCURAM, Science Foundation Ireland Centre for Research in Medical Devices, National University of Ireland Galway, Galway, Ireland
cSchool of Chemistry and AMBER, the SFI Research Centre for Advanced Materials and BioEngineering Research, Trinity College Dublin, the University of Dublin, College Green, Dublin 2, Ireland
dSchool of Nursing, Psychotherapy, and Community Health, Dublin City University, Glasnevin, Dublin 9, Ireland
eSchool of Chemical Sciences, Dublin City University, Glasnevin, Dublin 9, Ireland
fConway Institute of Biomolecular and Biomedical Research, University College Dublin, Belfield, Dublin 4, Ireland
First published on 24th February 2023
Abstract
Herein we demonstrate the fabrication of arrays of micropillars, achieved through the combination of direct laser writing and nanoimprint lithography. By combining two diacrylate monomers, polycaprolactone dimethacrylate (PCLDMA) and 1,6-hexanediol diacrylate (HDDA), two copolymer formulations that, owing to the varying ratios of the hydrolysable ester functionalities present in the polycaprolactone moiety, can be degraded in the presence of base in a controllable manner. As such, the degradation of the micropillars can be tuned over several days as a function of PCLDMA concentration within the copolymer formulations, and the topography greatly varied over a short space of time, as visualised using scanning electron microscopy and atomic force microscopy. Crosslinked neat HDDA was used as a control material, demonstrating that the presence of the PCL was responsible for the ability of the microstructures to degrade in the controlled manner. In addition, the mass loss of the crosslinked materials was minimal, demonstrating the degradation of microstructured surfaces without loss of bulk properties was possible. Moreover, the compatibility of these crosslinked materials with mammalian cells was explored. The influence of both indirect and direct contact of the materials with A549 cells was assessed by profiling indices reflective of cytotoxicity such as morphology, adhesion, metabolic activity, oxidative balance, and release of injury markers. No significant changes in the aforementioned profile were observed in the cells cultured under these conditions for up to 72 h, with the cell–material interaction suggesting these materials may have potential in microfabrication contexts towards biomedical application purposes.
1 Introduction
Engineered surface structures have been shown to influence mammalian cell growth and behaviour, aid in the delivery of drugs and biological payloads, enable sensors in wearable electronics, exhibit antibacterial properties, and define surface wettability.1–3 Nano and micropillars in particular have garnered much interest in the cell biology realm, owing to their high surface area and high aspect ratio. Nanoneedles, nanotubes, nanostraws, and nanowires have been interfaced with mammalian cells to investigate cell behaviour, cell growth and function, deliver biological payloads, monitor enzymatic activity, and probe nuclear mechanics.4–13 Several orders of magnitude larger, microneedles have been developed for controlled transdermal drug and vaccine delivery.14–17 Given the success of transcutaneous application, microneedle drug delivery has rapidly extended into other non – dermal tissues and organs such as the vascular tissues, oral mucosa, the gastric mucosa and the eye.18,19While both hard and soft material structures have demonstrated significant potential in biological applications, the importance of creating structures in soft polymers has been highlighted by recent advances in tissue engineering and medical device development, where creating biocompatible, degradable structures with tuneable chemical and physical properties has been shown to promote interactions with both tissue and cellular environments.20,21 Degradable biocompatible polymers are widely employed in biomedical in vivo applications as they break down into non-cytotoxic by-products such as water and carbon dioxide.22 Aliphatic polyesters such as polylactic acid (PLA), polyglycolic acid (PGA) and their copolymers are the most widely used degradable polymers in biomedical applications, having been used as resorbable sutures and implanted fixtures such as rods, pins, and screws, for decades.23,24 Such polyesters are susceptible to hydrolytic or enzymatic chain cleavage into α-hydroxyacids which are metabolized in the body.25 Polycaprolactone (PCL) has also found much use in the manufacture of long term implants and injectables for prolonged drug delivery applications due to its bioresorbable and hydrophobic nature, and its high drug permeability.26–28 PCL has a low tensile strength (∼23 MPa), but very high elongation at breakage (>700%).23 Therefore, to maintain structure integrity, PCL composite materials are often preferred.23,29 Such composites have been used as scaffolds for bone tissue engineering and regeneration, cardiac patches, and pelvic floor repair.30–34 Nano and micro-structures have been key to these applications, with electrospun fibrous membranes mimicking the extracellular matrix and promoting increased cell adhesion and proliferation, and electrospun nanofibers simulating the complex multiscale architectures of cardiac tissue.33,35 Vertical structures incorporating PCL have been employed for drug delivery applications, including microneedles which take advantage of the low melting point (55–60 °C) of PCL to thermally induce the melting of the needles which results in loaded drug release.29,36,37 Ko et al. have reported the fabrication of blended PCL/PEG (polyethylene glycol) composite microneedles with aspect ratios ranging from 2.25 to 2.47 through a solution casting and thermal pressing protocol. The structures were effective in the transdermal delivery of hydrophilic drugs like rhodamine 6G and FITC-collagen.38 Recently, similar work by Lee et al. demonstrated that PCL/PLA blends of varying concentrations could achieve microneedles with great mechanical strengths (up to 51.26 MPa) for the transdermal delivery of hispidin.39
Understanding the degradation, solubility and biocompatibility of such structured materials is essential for wide ranging applications in controlled drug release and delivery and implantable medical devices. The presence of specific degradable functionalities within the polymer chain is crucial for degradation to occur and by varying the concentration of such functional groups, control over degradability and the rate of degradation can be achieved. In 2017, Lee et al. demonstrated the fabrication of polymeric nanopillar arrays in poly(lactic-co-glycolic acid) (PLGA) through block copolymer micelle lithography. They then demonstrated the biodegradation of these nanopillars as a function of time exposed to an esterase solution with 85% biodegradation achieved after 7 days.40 Jung et al. showed the fabrication of dissolvable micropillars using a polymethyl methacrylate (PMMA) and hyaluronic acid blend. The average heights of the micropillars were 500 ± 63 μm with base diameters of 300 ± 21 μm. They then demonstrated the successful in vitro transdermal delivery of Rhodamine B using these loaded micropillars.41 In 2019, Milionis et al. presented the fabrication of fully organic cellulose-based micropillars with aspect ratios ranging from 0.5 to 12. Biodegradation tests then showed that the micropillars could be fully degraded in a seawater environment within 3 months.42 Some earlier work by Netti et al. also showcased the fabrication of PLGA micropillars through an innovative electro-drawing method. Their results reported that these micropillars were effective degradable devices for the penetration and delivery of model drugs (Rhodamine 6G, Nile Red, and human serum albumin) through the stratum corneum of pigs.43
PCL is a degradable polyester that has found much use in biomedical science due to its bioresorbable and hydrophobic nature, and its high drug permeability.26–28 PCL degrades via hydrolysis of the ester bond, into non-toxic products following implantation.44,45 The hydrolysis pathway can be catalysed by both an acid and a base. This work focuses on use of a strongly alkaline environment to accelerate PCL degradation, as a means to speed up long-term degradation studies, an approach often taken in the literature.46,47 Lam et al. showed that the degradation rate of PCL could be greatly increased in an alkaline environment. They demonstrated the fabrication of β-tricalcium phosphate (β-TCP) (20 wt%) modified PCL (Mn 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) scaffolds by fused deposition modelling.48 They then showed that the mass of neat PCL scaffolds decreased by 96% after 4 weeks in 5 M NaOH, and the PCL-β-TCP modified scaffolds degraded by the same amount after 48 hours. They attributed this rate increase for the PCL-β-TCP samples due to an increased water diffusion within the samples due to decreased hydrophobicity.
000 g mol−1) scaffolds by fused deposition modelling.48 They then showed that the mass of neat PCL scaffolds decreased by 96% after 4 weeks in 5 M NaOH, and the PCL-β-TCP modified scaffolds degraded by the same amount after 48 hours. They attributed this rate increase for the PCL-β-TCP samples due to an increased water diffusion within the samples due to decreased hydrophobicity.
Herein, we extend on our previous work, which combined direct laser writing using 2 photon polymerisation (2PP) with nanoimprint lithography (NIL) to generate 3D microstructures.49 Our previous work studied the writing parameters which we needed to generate stable pore templates from which high aspect ratio micropillar arrays could be molded using photocurable polymers. We have advanced on this by producing large array templates, as well as dual height pore templates in this work. We have used these templates to make PCL-based copolymer arrays of micropillars, as well as a 1,6-hexanediol diacrylate (HDDA) control, to study local degradation of microstructures. By optimising a photocurable solvent-free photoresist comprising polycaprolactone dimethacrylate (PCLDMA) and HDDA we employ this technique to generate arrays of micropillars that can be sequentially degraded in basic media, resulting in a change in size and structure profile, and ultimately in complete pillar degradation. We show that surface structuring in degradable materials can be used to introduce a dynamic response of surface morphology, over time, even in the absence of measurable bulk degradation. Using this approach we can scale NIL templates to the macroscale, and exploit them for repeated use, to generate microstructured surfaces with controllable, time dependent degradation. These types of dynamic surfaces, where controlled release is possible, when paired with the mechanical strength of a robust bulk material, could have potential application in the delivery of therapeutics in vivo over prolonged periods.
The first step in investigating biocompatible, degradable polymeric pillar arrays for biological applications, is to interface them with cells in vitro. This work aims to carry out pilot cell studies for the development of cell screening protocols for investigating cytotoxicity, cell viability, and cell behaviour on the materials. With a view to applying the structured surfaces as implantable device coatings and in targeted drug delivery in the future, it was decided to focus on interfacing them with adherent cells. Apart from hematopoietic cells, almost every cell type found in the body are adherent or anchorage dependent, meaning they require a surface on which to grow.50 Adherent cells require polarity or an asymmetry to function optimally, with a cell orientating such that it has a distinct top apical membrane, and a bottom basal membrane.51 The external physical, biochemical, and mechanical cues of their environment can greatly influence the polarity of a cell, and in turn, the translocation of cellular machinery, proteins, organelles, etc. relative to such.52
Our focus with these materials is that they will be ultimately used for the slow release of therapeutics. As such, exploration of the influence the materials have on mammalian cells behaviour and health upon coming into contact with one another was of utmost interest, and we decided to utilise adherent cells over suspension counterparts as adherent cells would allow us to evaluate the materials effects from both an ‘indirect’ and ‘direct’ perspective as outlined in our experimental design. We therefore chose to study the interaction of A549 cells with the materials, and completed experiments to understand cell adhesion, viability, metabolic activity, oxidative balance, and injury marker release as part of this study.
2 Results and discussion
The fabrication method employed to produce micropillars involved the fabrication of pores using 2PP to form NIL templates (Fig. 1). Templates of cuboidal pores 2 × 2 × 2 μm were fabricated by using a laser power of 15 mW and a writing speed of 6000 μm s−1. These laser parameters were optimised to produce defined square pores at a relatively fast pace (successful printing of 390 × 390 μm area was completed in 65 minutes by stitching of smaller printed areas) to enable efficient production of templates for the study.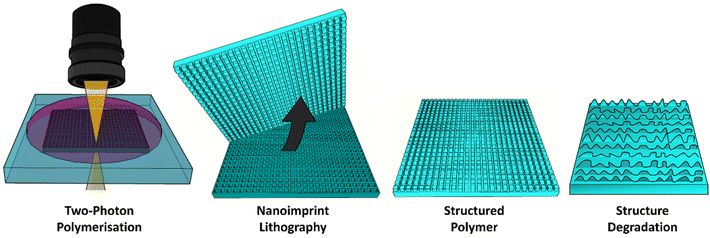 | ||
| Fig. 1 Schematic of direct laser writing and soft lithography techniques used to fabricate micropillars using novel copolymers. | ||
Two molecules containing terminal methacrylate groups, HDDA and PCLDMA, were mixed in 2 ratios and crosslinked to form a polymer network (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 and 20
90 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 PCLDMA
80 PCLDMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HDDA w/w%, hereafter referred to as 10%PH and 20%PH). In addition, neat HDDA was crosslinked as control. These 3 materials were crosslinked both on unstructured surfaces and microstructured template surfaces using nanoimprint lithography (NIL), both of which are discussed below. PCLMDA was prepared from a PCL diol precursor and methacryloyl chloride to produce a waxy solid with yields of 83–97% using a method adapted from Tian et al. (NMR and FTIR spectra of the methacrylated PCL are presented in Fig. S1 and S2†).53 The viscosity of the blend prior to replication was an important consideration for NIL.49,54 As the ratio of HDDA increased and PCLDMA decreased in the liquid monomer, the viscosities were reduced (15.5 cP and 10.8 cP for 20%PH and 10%PH, respectively). After photopolymerisation under white light, FTIR analysis was performed to confirm complete polymerisation (Fig. S3–5†), where for all 3 samples, as the curing progressed over 0–120 min, an incremental increase in transmission for the peaks at 1620 cm−1 and 1638 cm−1 was observed, which related to a reduction in C
HDDA w/w%, hereafter referred to as 10%PH and 20%PH). In addition, neat HDDA was crosslinked as control. These 3 materials were crosslinked both on unstructured surfaces and microstructured template surfaces using nanoimprint lithography (NIL), both of which are discussed below. PCLMDA was prepared from a PCL diol precursor and methacryloyl chloride to produce a waxy solid with yields of 83–97% using a method adapted from Tian et al. (NMR and FTIR spectra of the methacrylated PCL are presented in Fig. S1 and S2†).53 The viscosity of the blend prior to replication was an important consideration for NIL.49,54 As the ratio of HDDA increased and PCLDMA decreased in the liquid monomer, the viscosities were reduced (15.5 cP and 10.8 cP for 20%PH and 10%PH, respectively). After photopolymerisation under white light, FTIR analysis was performed to confirm complete polymerisation (Fig. S3–5†), where for all 3 samples, as the curing progressed over 0–120 min, an incremental increase in transmission for the peaks at 1620 cm−1 and 1638 cm−1 was observed, which related to a reduction in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C acrylate bond stretching as the polymerisation occurred.
C acrylate bond stretching as the polymerisation occurred.
Young's modulus of unpatterned, crosslinked bulk polymer samples was measured for each material using a Hysitron TI Premier Nanoindenter. The HDDA control sample recorded a Young's modulus of 1.91 GPa. For the co-polymers, the bulk elasticity decreased with increasing PCLDMA concentration; 10%PH had a value of 1.52 GPa, and 20%PH had a value of 1.15 GPa. A decrease in stiffness as a function of PCLDMA concentration can be explained by the decreased crosslink density within the material when the longer chain PCLDMA pre-polymer is added to the HDDA. This phenomenon is well understood with other systems, such as the curing of polyethylene glycol diacrylate pre-polymers as reported by Al-Nasassrah et al.55
Optimisation of the white light induced co-polymerisation of PCLDMA and HDDA polymers via NIL techniques from 2PP templates enabled production of high-fidelity replicas of square pillars with minimal loss of resolution between arrays of HDDA, 10%PH and 20%PH (Fig. 2). The NIL procedure indicated in the Experimental section details the solution composition of each polymer photoresist with 1 w/w% phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide (PBPO). These solutions were drawn onto the structured template via capillary forces between the substrate and a cover slide separated by PDMS spacers, 1 mm in height. For the control HDDA sample, and both the 10%PH and 20%PH solutions, arrays of micron sized pillars were characterised by SEM and AFM. Examples of these pillars are shown in Fig. 2, demonstrating the uniformity in shape and height. This was confirmed through AFM topographical imaging, an example of which is seen in Fig. 2(A), showing an average height of 1.72 ± 0.004 μm (n = 10). Also noteworthy is the surface topography changes evident on increasing the amount of PCLDMA in the pre-polymer solution (Fig. 2B–D). SEM images of the entire 390 × 390 μm 2PP templates and arrays from which these pillars were replicated can be seen in Fig. S6.† Fig. S7† shows SEM images of large arrays of pillars replicated using 10%PH from the large 390 × 390 μm template via NIL for reference. These images show the stitching of the smaller areas together and the long-range patterning achieved relative to the print area.
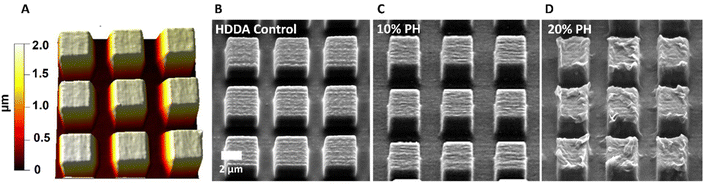 | ||
| Fig. 2 AFM topography image and SEM micrographs for 3 × 3 × 2 μm pillars; (A) AFM height trace and (B) SEM image of HDDA control; (C) SEM image of 10%PH; (D) SEM image of 20%PH. | ||
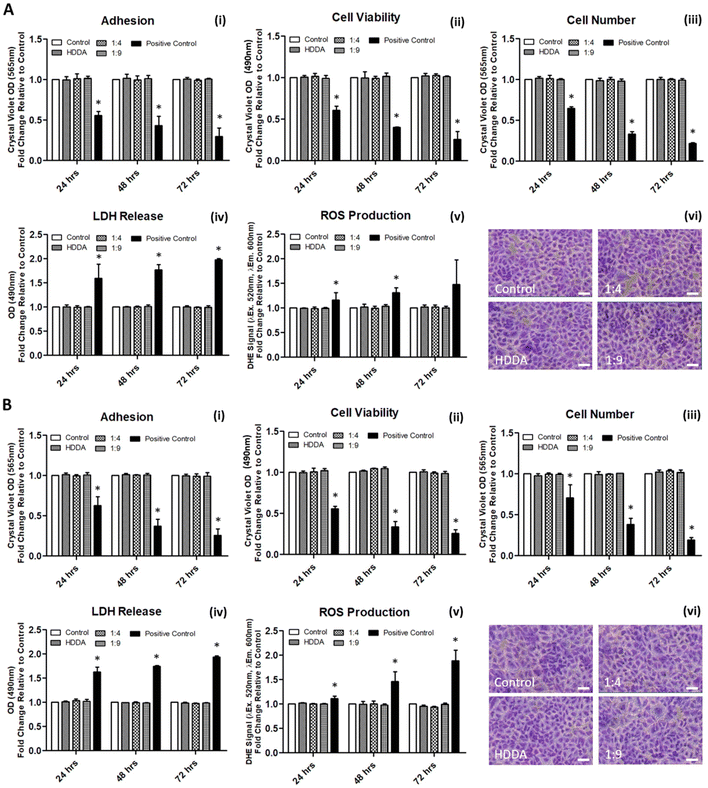
The cytotoxicity of the bulk materials towards mammalian cells was investigated using A549 cells; an epithelial cell line routinely used in toxicology studies and material science research.56–59 As an adherent cell line, the nature of the A549 cells would not only allow for the investigation of the effects of the material and its potential leachables on cell health, but also allow for an examination of whether the material could support culture of the cells through direct contact of the two. Initially, the influence of leachables from the materials was examined, with each material pre-incubated in cell culture media prior to application in A549 cultures. In contrast to the control cultures of cells grown on routine polystyrene plasticware, there was no significant difference in indices for adhesion, viability, cell number, LDH release, or ROS levels in cells grown on any of the materials tested (Fig. 3A, i–v). These trends were preserved from 24–72 h of incubation. Fig. 3(A, vi) visually shows no determinable differences in cultures grown in the presence of the material-conditioned media for 72 h, as compared to those treated with culture media conditioned in routine polystyrene. Similarly, assessment of the effects of the materials on indices of cell health when grown directly on the materials was also examined, with no significant differences observed when compared to the polystyrene control (Fig. 3B). The results are in agreement with previous research whereby cell–material interaction studies utilising HDDA or PCL, alone or as part of a material, exerted no significant effects on cell viability.60–62
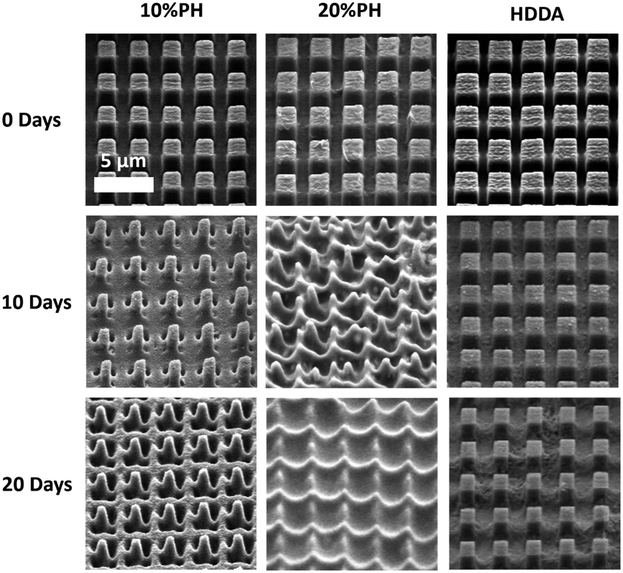
Next, the degradation of the 3 crosslinked materials was studied. Previous work has shown that the hydrolytic degradation of aliphatic ester linkages, such as that in PCL, occurs very slowly (2–3 years) due to its hydrophobic nature.26,27,63 For this reason, an accelerated degradation study was conducted using 5 M NaOH as the solution that would provide an alkaline environment. It was expected, based on the known literature, that hydrolysis of the ester bond in the HDDA and PCL chains would be the primary mode of degradation of the crosslinked materials. 4 mm diameter samples of unpatterned crosslinked HDDA, 10%PH and 20%PH were prepared and submerged in 5 M NaOH for up to 20 days. The mass loss of these samples was determined after this short time, and there was no significant change to the mass of the material after 5, 10 or 20 days. These samples were then left for longer-term studies and showed that there was minimal mass loss in the bulk over 160 days (see Fig. S8†). FTIR analysis studies on the surface of the HDDA, 10%PH and 20%PH unpatterned samples showed that hydrolysis of the ester was occurring, with greater loss in the ester signal in the material with the highest percentage of PCL (see Fig. S9–11†). This indicated that the materials were degrading via a surface degradation pathway rather than through the bulk of the crosslinked copolymers. To further understand the surface degradation, a set of microstructured arrays fabricated from the same master template were designed and produced. By removing samples from the 5 M NaOH solution at 5-day intervals over 20 days, and characterising using SEM and AFM, it was possible to monitor any changes in the topography over time (Fig. 4). The crosslinked HDDA pillars showed minimal degradation over time. For the 10%PH and 20%PH samples, the cubic arrays transitioned to truncated pillars, and ultimately to complete degradation of the surface structures. For the 10%PH samples, initial degradation of pillar height and width was followed by degradation of the pillar base. The exaggerated degradation at the base of the pores could be the result of a lower crosslinking density at the base of the pillars. Previous work has shown that the Young's modulus at the base of NIL-generated pillars was found to be half that of the planar bulk of the polymer, which may indicate a lower crosslinking density than the bulk.44 Sun et al. showed that Young's modulus of microstructures was reliant on the surface to volume ratio and the crosslinking density for a 2PP resin, SCR 500.64 They demonstrated that the polymerisation at the microstructures was confined to a small volume where the surface to volume area was much greater than the bulk. Therefore, there was an abundance of ‘dangling ends’ near the surface that were not attached to other polymer chains. The degree of crosslinking at these regions was reduced, decreasing the elastic strength at these confined areas. For the 20%PH samples, which possessed a higher concentration of PCLDMA in the prepolymer, complete degradation of the structures was the dominant response, which resulted in a surface-relief pattern on the polymer film. These results further indicated that this copolymer of PCLDMA and HDDA degraded via a surface degradation pathway, which is often observed for poly(α-hydroxy)esters of this nature.65 This occurs when the rate of hydrolysis exceeds the rate of water penetration into the bulk of the polymer. In this pathway, hydrolytic chain scission of the polymer backbone only occurs at the surface of the polymer.66 This surface erosion was visualised in the narrowing of the pillars and erosion at the pillar bases over time. This was supported by analysis of the bulk behaviour as described above, which showed minimal degradation over a period of 160 days.
Quantitative analysis of the change in topography across all samples using the tilted SEM images was not completed as the rate of variety in the degradation at the base of the pillars meant that the pillars themselves appeared to be getting larger as degradation occurred in certain samples.
However, side-on SEM analysis and AFM height profiles of samples after the 5-day degradation time point enables us to best demonstrate the 3-dimensional nature of degradation and change in geometry for 10%PH and 20%PH samples. Before degradation, the HDDA cuboidal pillars were measured using AFM to have a height of 1.72 ± 0.01 μm (n = 10). 5-Day accelerated degradation showed that this is reduced to 1.30 ± 0.02 μm (n = 10) for the 10%PH samples, with pillars adopting a conical shape. In the same time regime, the 20%PH sample showed a much more pronounced response, with the height of the resulting truncated pillar reduced to 0.74 ± 0.01 μm (n = 10).
The pitch (centre to centre distance) of the micropillars remained largely unchanged throughout the degradation process. The 10%PH and 20%PH pillars had a measured pitch of 2.93 ± 0.07 μm and 3.01 ± 0.12 μm respectively prior to undergoing basic degradation. After 20 days the pitch of the 10%PH pillars was measured to be 2.98 ± 0.13 μm, while the 20%PH pillars had fully degraded. The final pitch measurement of the 20%PH pillars was 3.13 ± 0.17 μm on day 15 before the pillars completely degraded.
While degradation in the structures was readily visualised using SEM and AFM, as described above, there was no noticeable degradation in the bulk of the sample. This led us to assume that the structured area must be degrading at a much faster rate than the bulk material, with the bulk material remaining intact. Mass loss studies were also carried out on samples where there was a microstructured array in the centre of the sample, and no significant mass loss was observed either. It should be noted that in a 4 mm diameter sample, the microstructured area accounts for only 1.2% of the surface area of the sample, and so the significance of the mass loss from the structured area would be negligible over the entire sample. The advantage of increased surface degradation at the structures is having specific release vehicles from which loaded cargo can be predictably released. The degradation of the material and, therefore, the release profile of a cargo could be improved by the addition of a structured surface to the hydrolysable material. The bulk remaining unchanged is characteristic of the surface degradation pathway often seen in polyester degradation. This could be of benefit when implanted biomaterial matrices were required to maintain their bulk stability and mechanical strength, while also enabling slow and controlled release of a loaded cargo.
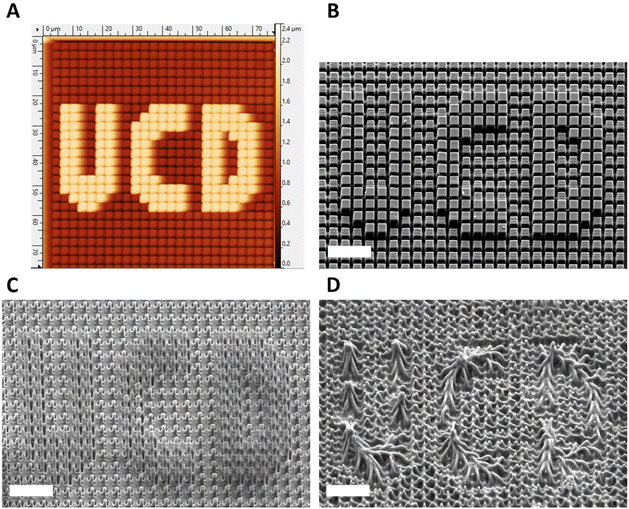
The potential to create changeable topographies via degradation was investigated by designing templates to produce pillars with two distinct heights. A second template was produced consisting of an array of pores that were printed to be 2 × 2 μm in width and 2 μm in depth, in which a deeper pore forming the letters “UCD” (University College Dublin) which were printed to be 2 × 2 in width but 3.5 μm deep were distributed. When used as a template, this produced arrays of crosslinked HDDA, 10%PH, and 20%PH pillars approx. 2 × 2 × 2 μm in size, with an arrangement of 2 × 2 × 3.5 μm, taller pillars forming UCD in the centre. Fig. 6A shows the AFM height image of the HDDA copolymer molded from the template. Fig. 6B shows a SEM micrograph of the same sample; height measurements as determined by manual measurements on 45° tilt images using ImageJ gave a height measurement of 1.89 ± 0.18 μm for the smaller pillars and 3.44 ± 0.13 μm for the taller pillars. This indicated that the fidelity of the moulding was high, with the pre-polymer solution reaching the bottom of the pore. For the 10%PH mold and the 20%PH mold, these values were 2.16 ± 0.14 μm and 3.64 ± 0.13 μm, and 2.00 ± 0.23 μm and 3.44 ± 0.15 μm, respectively. The 10%PH and 20%PH pillars can then be seen to degrade to form further dynamic topographies as time progresses. To quantify the degradation of the structures, two types of pillar measurements were determined to be most representative of the structure change over time in a 5 M NaOH alkaline environment. The first being manual measurements of the width of the middle of the pillar measured from FESEM images taken at a 45° tilt. The second was measuring the surface area of the tops of the pillars measured from SEM images taken top down. Table 1 shows the progression of the width and surface area of the pillars in the HDDA, 10%PH and 20%PH samples. There was a much larger decrease in both pillar width and pillar top surface area from the HDDA samples than was expected. The pillar widths decreased by 19% and the surface area by 45% after 20 days in 5 M NaOH. The 10%PH pillar width decreased by 42% and the surface area had a percentage loss of 58% by day 20. The 20%PH sample had no pillars left by day 20. Fig. 6C and D show representative SEM images of the 10%PH sample and the 20%PH sample after 10 days respectively. It can be seen how altering the PCLDMA concentration within the polymer formulation affects the degradation profile of the microstructures. This result further showcases the tunability of the degradation profile as a function PCLDMA concentration within the material.
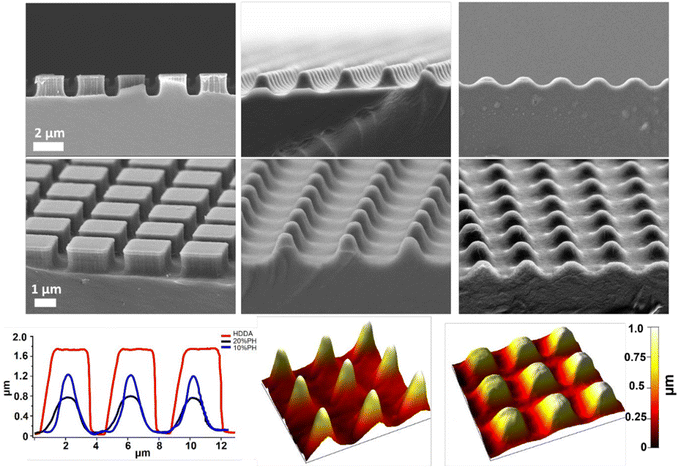 | ||
| Fig. 5 Top row: side on SEM images of HDDA pillars fabricated using NIL (left); 10%PH pillars submerged in 5 M NaOH for 5 days (middle); 20%PH pillars submerged in 5 M NaOH for 5 days (right); Middle row: tilted SEM images of HDDA pillars fabricated using NIL (left); 10%PH pillars submerged in 5 M NaOH for 5 days (middle); 20%PH pillars submerged in 5 M NaOH for 5 days (right); Bottom row: AFM line profiles (images from which these were taken can be seen in Fig. S12†) of 10%PH, 20%PH submerged in 5 M NaOH for 5 days, and HDDA control (left); AFM 3D renders of 10%PH submerged in 5 M NaOH for 5 days (middle); and 20%PH submerged in 5 M NaOH for 5 days (right). | ||
| Days in 5 M NaOH | HDDA | 10%PH | 20%PH | |||
|---|---|---|---|---|---|---|
| Pillar width (μm) | Pillar top surface area (μm2) | Pillar width (μm) | Pillar top surface area (μm2) | Pillar width (μm) | Pillar top surface area (μm2) | |
| 0 | 2.14 ± 0.07 | 4.09 ± 0.16 | 2.26 ± 0.11 | 4.17 ± 0.23 | 2.07 ± 0.07 | 3.27 ± 0.14 |
| 5 | 1.79 ± 0.07 | 2.94 ± 0.19 | 1.61 ± 0.08 | 2.31 ± 0.12 | 0.77 ± 0.04 | 1.02 ± 0.08 |
| 10 | 1.85 ± 0.05 | 3.22 ± 0.09 | 1.15 ± 0.09 | 1.28 ± 0.07 | 0.67 ± 0.10 | 0.37 ± 0.07 |
| 15 | 1.54 ± 0.07 | 2.24 ± 0.06 | 1.04 ± 0.07 | 0.95 ± 0.07 | No pillar | No pillar |
| 20 | 1.46 ± 0.06 | 1.91 ± 0.07 | 0.85 ± 0.14 | 0.74 ± 0.08 | No pillar | No pillar |
Dynamic surface topographies such as these could find applications in the area of drug delivery, whereby drug payloads could be controllably released from the degrading pillars on the polymer surface, while the bulk of the polymer remains intact, as part of an implantable device. The interaction of such degradable structured surfaces with adherent cell lines such as endothelial or epithelial cells may also influence cell growth and behaviour over time as the structures degrade and the topography changes. A number of research groups have also demonstrated that stem cells differentiate depending on the geometry and pitch of surface pillar structures.67–69 In addition to the changing topography, the difference in Young's modulus between the pillars and the surrounding planar areas that is thought to cause this degradation regime may also lead to varying stem cell differentiation e.g. Discher et al. have shown that stem cells are extremely sensitive to matrix mechanical properties and differentiate into certain cell lineages accordingly.55,70
3 Conclusions
Templates of square pores in a variety of dimensions were fabricated using 2PP. Optimised white light induced NIL of these 2PP templates using PCLDMA and HDDA formulations enabled production of high-fidelity arrays of micropillars. A series of experiments using A549 cells demonstrated that these materials produce no unwanted cytotoxic effect or stress response when cells were interfaced with them in a direct or indirect manner. Moreover, the adhesive A549 cells adhered to the materials, and experienced no ill effects. It was shown that the micropillar degradation was as a result of PCLDMA concentration within the copolymer formulations and time exposure to a 5 M NaOH environment. PCLDMA-containing micropillars generally followed a transition from cubic arrays, to truncated pillars, and ultimately to complete degradation of the surface structures. Dynamic surface topographies also featured whereby a greater degree of degradation was observed concerning the structured area of the sample compared to the surrounding planar bulk material. Further studies will be conducted to study long term degradation in environments closer to that of the human body, to determine the accurate rate of degradation of structured surface. One advantage of these materials is that controlled and enhanced degradation at the structured surface is possible, while the bulk material remains intact. This could open up corridors to increased rates of release of therapeutics from in vivo PCL-based materials. Loading and release studies will be investigated in future work to assess the ability of degradable structures to uptake and release drug molecules and even nanoparticles from the degrading structures in a controlled manner into surrounding cells and tissue for drug delivery applications.4 Experimental
4.1 Materials
Ethanol (≥99%), acetic acid, acetone (≥99%), 1,6-hexanediol diacrylate (HDDA) (technical grade, 80%), phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide (PBPO) (97%, powder), 3-(trimethoxysilyl)propyl methacrylate, propylene glycol methyl ether acetate, Sylgard 184 poly(dimethoxy) silane (PDMS) silicone elastomer and curing agent, sodium hydroxide, dimethyl sulfoxide (DMSO), industrial methylated spirits (IMS), 2-propanol, dichloromethane (DCM) (puriss. p.a., ACS reagent, reag. ISO, ≥99.9% (GC), contains 25 mg mL−1 amylene as a stabilizer), hexane, triethylamine (≥99%), methacryloyl chloride (97%, contains 200 ppm monomethyl ether hydroquinone as a stabilizer), sodium bicarbonate, crystal violet, dihydroethidium, phosphate buffered saline, Dulbecco's Modified Eagle Medium, fetal bovine serum, penicillin–streptomycin, formaldehyde, trypsin-EDTA, were purchased from Sigma-Aldrich, now known as Merck Life Sciences, Wicklow, Ireland. Tetrahydrofuran (99.5%, Extra Dry over Molecular Sieve, Stabilized) was bought from Thermo Scientific, Dublin, Ireland. Polycaprolactone diol (Mn 1250 g mol−1) was purchased from Polysciences Inc., Heddesheim, Germany. Borosilicate substrates (25 × 25 mm2; thickness 0.1 mm), dip-in laser lithography (DiLL) fused silica substrates (25 × 25 mm2; thickness 0.7 mm), and IP-Dip photoresist were purchased from Nanoscribe Gmbh, Karlsruhe, Germany. TAP300AI-G silicon probes and Pt/Ir-coated conductive tips for topographical imaging were purchased from BudgetSensors® Innovative Solutions Ltd, Bulgaria. Digital UV-ozone system PSD Pro Series was purchased from Novascan Technologies, Iowa, USA. The white light source was provided by Edmund Optics, MI 150 Illuminator, York, UK. A549 cells were purchased from ATCC, CCL-185, Virginia, USA.4.2 Fabrication of square pore arrays
Two photon polymerisation of square pore arrays was carried out using a commercial DLW apparatus, Photonic Professional (Nanoscribe Gmbh, Karlsruhe, Germany). The dip-in configuration was used, employing a 63× objective. Prior to laser writing the DiLL fused silica substrates were sonicated in ethanol for 30 min, followed by UV Ozone treatment for 15 min to activate the substrate surface. The surface of the DiLL substrates was modified by immersion in a solution of 3% v/v 3-(trimethoxysilyl)propyl methacrylate/0.1% v/v acetic acid in ethanol for 1 h. After rinsing with ethanol and drying under a stream of nitrogen, the substrates were cured for 10 min in an oven at 40 °C. The DiLL substrate was secured into the DiLL holder of the Photonic Professional. A single drop of the monomeric photoresist, IP-DIP, was deposited onto the centre of the substrate before the holder was placed into the system. The stereolithography structure file (.stl) of the desired array of square pores was loaded into the software, with distances of 100 nm used for both slicing and hatching. For pore array fabrication, the laser power and scan speed used were 15 mW and 6000 μm s−1. After laser writing, the structures were developed by immersing in propylene glycol methyl ether acetate for 20 min followed by 2-propanol (IPA) for 2 min. The structures were then rinsed with IPA, dried under a stream of nitrogen, and coated with a 10 nm layer of gold.4.3 Synthesis of polycaprolactone dimethacrylate
The synthesis was adapted from the method outlined by Tian et al. in 2019.53Fig. 7 shows the scheme for the reaction.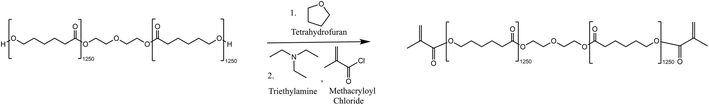 | ||
| Fig. 7 Reaction scheme for the methacrylation of PCL-diol using methacryloyl chloride and triethylamine. | ||
Tetrahydrofuran (THF) was dried over a 5 Å molecular sieve over 2 days. Polycaprolactone diol (7.97 g, 6.37 mmol, 1 eq.) was dissolved in the dry THF (80 mL) in a 2-neck 500 mL round bottom flask. Triethylamine (1.78 mL, 12.77 mmol, 2 eq.) and methacryloyl chloride (1.37 mL, 14.02 mmol, 2.2 eq.) were then added dropwise to the round bottomed flask and the reaction was covered with tin foil and left to stir at room temperature for 5 days. The resulting mixture was gravity filtered using an excess of hexane to remove salts precipitated during the reaction. THF and hexane were then removed by rotary evaporation and the isolated product was further purified by a washing and extraction step. The methacrylated polymer was dissolved in a minimal amount of dichloromethane (DCM) and then washed with 1 M hydrochloric acid three times. The isolated DCM layer was evaporated by rotary evaporation and the product was dried under vacuum for 2 days. The product was isolated as a white, waxy solid (7.72 g, 6.17 mmol, 96.9%). The 1H NMR was completed and used to determine that the degree of acrylation was 71.8%. The degree of acrylate capping was calculated as per the method described in the procedure outlined by Tian et al.531H NMR (500 MHz, CDCl3) δ 6.10 (s, 2H, CH), 5.54 (s, 2H, CH), 4.32–4.20 (m, 4H, CH2), 4.10–4.00 (m, 30H, CH2), 3.65–3.73 (m, 4H, CH2), 2.25–2.33 (m, 30H, CH2), 1.94 (s, 6H, CH3), 1.55–1.71 (m, 136H, CH2, CH2), 1.35–1.46 (m, 40H, CH2). FTIR-ATR (cm−1): 2945, 2862, 1720, 1639, 1471, 1419, 1396, 1364, 1323, 1292, 1237, 1161, 1106, 1044, 962, 815, 730.
4.4 Preparation of monomeric solutions
Three monomeric solutions were made by mixing varying amounts of PCLDMA and HDDA by weight in different ratios, neat HDDA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 PCLDMA
9 PCLDMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HDDA (10%PH), and 1
HDDA (10%PH), and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 PCLDMA
4 PCLDMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HDDA (20%PH) with each containing 1% w/w of the photoinitiator phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide (PBPO). The solutions were sonicated and vortexed thoroughly to ensure a homogeneous mixture was achieved.
HDDA (20%PH) with each containing 1% w/w of the photoinitiator phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide (PBPO). The solutions were sonicated and vortexed thoroughly to ensure a homogeneous mixture was achieved.
4.5 Fabrication of square micropillars
Nanoimprint lithography from a 2PP master of cylindrical pore arrays was carried out using three monomer solutions described above. Two PDMS spacers (thickness 600 μm) were placed firmly onto the DiLL substrates, positioned either side of the 2PP structures. A cover glass (22 × 22 mm) was placed firmly onto both PDMS spacers, positioned over the 2PP structures. The monomeric solution was pipetted into the space beneath the sealed cover glass and between the PDMS spacers until the space was filled and consequently the 2PP structures were fully covered. The monomeric solution was cured under white light for 2 h in total; 30 min at 25% power, 30 min at 50% power, 30 min at 75% power, and 30 min at 100% power before being separated from the 2PP master using tweezers.4.6 Degradation study in alkaline medium
A template 100 × 100 μm in width of 2 × 2 μm wide square pores with pore depths of both 2 and 4 μm were used in this degradation study. From the 2PP template, six replicas were made from each of the three solutions: HDDA, 10%PH and 20%PH. One replica of each polymer was left dry, unexposed to 5 M NaOH as the control sample. A single replica of each polymer was placed in a sealed vial of 20 mL 5 M NaOH for 5, 10, 15, 20 and 25 days. After each timepoint, the samples were removed from the vial of 5 M NaOH, rinsed thoroughly with deionised water and left in a desiccator to dry for 24 h. The samples were then sputtered with 10 nm of Au/Pd in preparation for SEM analysis.4.7 Mass loss in alkaline medium
To study the mass loss, unpatterned cured samples of the 3 monomeric solutions above were prepared. For each sample, the mixture (100 μL) was pipetted into uniform PDMS gaskets so that they were consistent in size, shape and volume. The solutions were cured for 2 h under white light as above. Each sample was then rinsed with deionised water and allowed to dry under vacuum in a desiccator for 24 h. The three cured samples (unpatterned HDDA, unpatterned 10%PH, and unpatterned 20%PH) were then weighed to obtain an average and standard deviation for the dry measurement. The samples were then immersed in 20 mL of 5 M NaOH in a sealed vial for 5, 10, 15, 20 and 25 days. For each timepoint, the samples were removed from the vial of 5 M NaOH, rinsed with deionised water, dried under vacuum for 24 h and weighed. The samples were then immersed in the 5 M NaOH again until the next time point at which time the samples were reweighed as above. This process was repeated three times for the HDDA, the 10%PH and the 20%PH. The average weight of the three repeats and the standard deviation was obtained for each time point. Data obtained was plotted using OriginPro8.5 software.4.8 SEM imaging and analysis
SEM imaging of the 2PP square pore templates and nanoimprinted micropillars was carried out. An accelerating voltage of 5 kV was used under SE2 or in-lens mode to acquire all images. Prior to SEM imaging, the structures were coated with a 10 nm Au–Pd layer using a Cressington Sputter Coater 208HR. A 57 × 0.1 mm Au–Pd target was used to coat the structures under an inert atmosphere of Argon for 10 s. Analysis of the acquired SEM images was carried out using ImageJ software.4.9 AFM imaging and analysis
Atomic force microscopy (AFM) was carried out using an Asylum Research MFP3D AFM (Santa Barbara, USA). For image in Fig. 6, amplitude modulation mode or tapping mode was employed for all scans. The AFM probe was a silicon tip with an aluminium reflex coating, a resonance frequency of 300 kHz and a nominal cantilever stiffness of 40 N m−1. Prior to imaging the substrate was secured onto a glass microscope slide using double sided tape. AFM images were post-processed and analysed using Gwyddion (Czech Metrology Institute, Jihlava, Czech Republic, version 2.60). Line profile data was obtained following image postprocessing and plotted using OriginPro8.5. For images in Fig. 2 and 5, imaging was performed in amplitude modulation mode and height, amplitude, and phase images were recorded. Data were recorded with 256 lines per scan direction and with a scan rate of 0.4 Hz and scan angle of 90°. A Pt/Ir-coated conductive tip (PPP-EFM-Nanosensor, with tip radius <15 nm, resonant frequency of 75 Hz, and a spring constant of 2 N m−1) was used for topography. Prior to imaging the substrate was secured onto a glass microscope slide. 2D and 3D representation of height images were produced using Gwyddion AFM software, all images were 1st order plane fit flattened. Line profile data was obtained following image post processing and plotted using Igor pro 6.38.4.10 Cell culture
A549 cells were used as a representative adherent cell model to examine indices of cell health in response to indirect and direct contact with the PCL-based materials. Briefly, cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% Fetal Bovine Serum, penicillin (100 U mL−1) and streptomycin (100 μg mL−1). Cells were seeded in 56 cm2 dishes at a density of 2000 cell per cm2 and maintained in 5% CO2/95% humidity at 37 °C. Culture media exchanged every 2–3 days, and cells were passaged at 80% confluency with all experimental procedures carried out in 96-well plates at a density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well.
000 cells per well.
4.11 Cell treatment
For indirect treatments, PCL-based materials were sterilised by soaking the materials in 70% ethanol for 20 minutes. Following sterilisation, the materials were then washed in sterile PBS to remove any residual ethanol. The materials were then transferred to individual wells of a 6-well dish and submerged in 2 mL of culture medium. A control well containing no materials and 2 mL of culture media was also prepared. Conditioned media exposed to materials for 24 h was then harvested and utilised as samples of ‘indirect contact’ for investigating the impact of potential leachable molecules from the PCL-based material and culture environment. Cells were then seeded on 96-well dishes (transparent for all assays apart from the ROS Assay which used an opaque white plate), in the presence of the conditioned media samples for 24, 48, and 72 h, before the respective assay was carried out.For direct treatments, circular disks of the PCL-based materials were prepared using a 5 mm hollow punch, and then sterilised by soaking the materials in 70% ethanol for 20 minutes. Following sterilisation, the disks were then washed in sterile PBS to remove any residual ethanol. Control disks of similar dimensions using cell culture grade plastic were also prepared. The materials were then transferred to the relevant 96-well plate (transparent for all assays apart from the ROS Assay which used an opaque white plate), and cells seeded at a density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per wells with respective assay measurements carried out 24-, 48-, and 72 h post-seeding.
000 cells per wells with respective assay measurements carried out 24-, 48-, and 72 h post-seeding.
In all experiments, a positive control of cells treated with TNF-α (200 ng mL−1) was included for each experimental timepoint.
4.12 Adhesion assay
To examine the influence of ‘indirect’ and ‘direct’ contact of the PCL materials on the adhesion profile of the cells, cells were seeded at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well into PCL material conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), all culture media was removed, and the wells were washed gently with PBS. 100 μL of 3.75% formaldehyde was then added to each well and left to incubate for 15 minutes. The cells were then gently washed with PBS before 100 μL of a 5 mg mL−1 solution of crystal violet was added to each well. The plate was left to incubate for 5 minutes before being washed with deionised water to remove excess crystal violet stain. The plate was then inverted and left to air dry overnight. 50 μL of a 2% SDS solution was added to each well, and the plate was gently agitated by placing it on a shaker to assist in cell lysis for 30 minutes. The absorbance was then read at 562 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
000 cells per well into PCL material conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), all culture media was removed, and the wells were washed gently with PBS. 100 μL of 3.75% formaldehyde was then added to each well and left to incubate for 15 minutes. The cells were then gently washed with PBS before 100 μL of a 5 mg mL−1 solution of crystal violet was added to each well. The plate was left to incubate for 5 minutes before being washed with deionised water to remove excess crystal violet stain. The plate was then inverted and left to air dry overnight. 50 μL of a 2% SDS solution was added to each well, and the plate was gently agitated by placing it on a shaker to assist in cell lysis for 30 minutes. The absorbance was then read at 562 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
4.13 MTS assay
To measure the impact of ‘indirect’ and ‘direct’ contact of the PCL materials on the viability of the cells, cells were seeded at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), 20 μl of CellTiter 96 AQueous Assay reagent was added to each well, and the plate was then incubated for 4 h at 37 °C in a humidified 5% CO2 incubator. The absorbance was then read at 490 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), 20 μl of CellTiter 96 AQueous Assay reagent was added to each well, and the plate was then incubated for 4 h at 37 °C in a humidified 5% CO2 incubator. The absorbance was then read at 490 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
4.14 Cell density
For cell density, the ‘Adhesion Assay’ protocol was utilised with the exception that for examining the effect of the PCL-conditioned media samples, cells were initially seeded in unconditioned culture media before this was exchanged for PCL-conditioned media once the cells had adhered. Prior to being lysed with the 2% SDS solution, images of the cells were captured using a Nikon Eclipse TS100 phase contrast microscope. Following acquisition of images, the cells were then lysed, and the absorbance was then read at 562 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.4.15 LDH release
To measure the impact of ‘indirect’ and ‘direct’ contact of the PCL materials on the release of stress-related biomarker lactate dehydrogenase, a cytotoxicity detection kit was used as per the manufacturer's instructions. Briefly, cells were seeded at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), 100 μL of culture media from each well was transferred into corresponding wells of a new 96-well plate. 100 μL of Reaction mixture was then added to each well, and the plate was left to incubate for 30 minutes. The absorbance was then read at 490 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), 100 μL of culture media from each well was transferred into corresponding wells of a new 96-well plate. 100 μL of Reaction mixture was then added to each well, and the plate was left to incubate for 30 minutes. The absorbance was then read at 490 nm on a Tecan Infinite M200 Pro Microplate Reader and readings compared against control cells as fold change.
4.16 ROS assay
To measure the impact of ‘indirect’ and ‘direct’ contact of the PCL materials on the oxidative balance of the cells, cells were seeded at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), dihydroethidium was added at a final concentration of 10 μM to each well and the plate was left to incubate for 30 minutes. The fluorescence was then read at excitation and emission wavelengths of 520 nm and 600 nm on a Tecan Safire 2 fluorospectrophotometer. Readings were normalised to cell number obtained from viability readings, and the resultant levels compared against control cells as fold change.
000 cells per well into PCL material-conditioned culture media, or directly onto the PCL materials. Following incubation for the designated amount of time (24–72 h), dihydroethidium was added at a final concentration of 10 μM to each well and the plate was left to incubate for 30 minutes. The fluorescence was then read at excitation and emission wavelengths of 520 nm and 600 nm on a Tecan Safire 2 fluorospectrophotometer. Readings were normalised to cell number obtained from viability readings, and the resultant levels compared against control cells as fold change.
4.17 Statistical analysis
Results are expressed as mean ± standard deviation. Experimental groups were performed in triplicate, with a minimum of three independent experiments (n = 3). Statistical comparison between control and experimental groups were by Student’ t-test for pairwise comparisons. A value of P ≤ 0.05 was considered significant.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Science Foundation Ireland (SFI), through the Starting Investigator Research Grant (SIRG) Scheme (Grant Number 15/SIRG/3429). S. K. and N. G. also acknowledge the SFI and European Regional Development Fund (ERDF) (Grant Number 13/RC/2073 and 17/RC-PhD/3480) through CúRAM, the SFI Research Centre for Medical Devices. S. K. and M. O. L. also acknowledge the Irish Research Council (IRC) (Grant Number GOIPG/2017/1037). L. F. acknowledges the European Research Council (ERC) Starting Grant (Project Number 802929-ChemLife) and SFI–ERDF (Grant Number 12/RC/2278_P2). B. R. also acknowledges SFI (SFI/17/CDA/4637). The 2PP-DLW fabrication and SEM imaging for this project were carried out at the Additive Research Laboratory (AR-Lab) and the Advanced Microscopy Laboratory (AML), Trinity College Dublin, Ireland. The AR-Lab and AML are SFI-supported centres, part of the CRANN Institute and affiliated to the AMBER centre.References
- Y. Wan, Z. Qiu, Y. Hong, Y. Wang, J. Zhang, Q. Liu, Z. Wu and C. F. Guo, A Highly Sensitive Flexible Capacitive Tactile Sensor with Sparse and High-Aspect-Ratio Microstructures, Adv. Electron. Mater., 2018, 4, 1700586 CrossRef.
- S. Wu, B. Zhang, Y. Liu, X. Suo and H. Li, Influence of surface topography on bacterial adhesion: A review (Review), Biointerphases, 2018, 13, 060801 CrossRef CAS PubMed.
- X. Li, Y. Li, I. Muzammil and M. Lei, Antireflection and Antiwetting Functionalities of Plasma-Nanotextured Polymer Surfaces with Biomimetic Nanopillars, Plasma Processes Polym., 2020, 17, 2000050 CrossRef.
- C. S. Hansel, S. W. Crowder, S. Cooper, S. Gopal, M. Cruz, L. de Oliveira Martins, D. Keller, S. Rothery, M. Becce, A. E. G. Cass, C. Bakal, C. Chiappini and M. M. Stevens, Nanoneedle-Mediated Stimulation of Cell Mechanotransduction Machinery, ACS Nano, 2019, 13, 2913–2926 CrossRef CAS PubMed.
- S. Gopal, C. Chiappini, J. Penders, V. Leonardo, H. Seong, S. Rothery, Y. Korchev, A. Shevchuk and M. M. Stevens, Porous Silicon Nanoneedles Modulate Endocytosis to Deliver Biological Payloads, Adv. Mater., 2019, 31, 1806788 CrossRef PubMed.
- Y. Chen, S. Aslanoglou, T. Murayama, G. Gervinskas, L. I. Fitzgerald, S. Sriram, J. Tian, A. P. R. Johnston, Y. Morikawa, K. Suu, R. Elnathan and N. H. Voelcker, Silicon-Nanotube-Mediated Intracellular Delivery Enables Ex Vivo Gene Editing, Adv. Mater., 2020, 32, 2000036 CrossRef CAS PubMed.
- A. R. Shokouhi, S. Aslanoglou, D. Nisbet, N. H. Voelcker and R. Elnathan, Vertically Configured Nanostructure-Mediated Electroporation: A Promising Route for Intracellular Regulations and Interrogations, Mater. Horiz., 2020, 2810–2831 RSC.
- R. Elnathan, B. Delalat, D. Brodoceanu, H. Alhmoud, F. J. Harding, K. Buehler, A. Nelson, L. Isa, T. Kraus and N. H. Voelcker, Maximizing Transfection Efficiency of Vertically Aligned Silicon Nanowire Arrays, Adv. Funct. Mater., 2015, 25, 7215–7225 CrossRef CAS.
- H. Y. Lou, W. Zhao, Y. Zeng and B. Cui, The Role of Membrane Curvature in Nanoscale Topography-Induced Intracellular Signaling, Acc. Chem. Res., 2018, 51, 1046–1053 CrossRef CAS PubMed.
- S. Park, H.-H. Park, K. Sun, Y. Gwon, M. Seong, S. Kim, T.-E. Park, H. Hyun, Y.-H. Choung, J. Kim and H. E. Jeong, Hydrogel Nanospike Patch as a Flexible Anti-Pathogenic Scaffold for Regulating Stem Cell Behavior, ACS Nano, 2019, 13, 11181–11193 CrossRef CAS PubMed.
- L. Schmiderer, A. Subramaniam, K. Žemaitis, A. Bäckström, D. Yudovich, S. Soboleva, R. Galeev, C. N. Prinz, J. Larsson and M. Hjort, Efficient and Nontoxic Biomolecule Delivery to Primary Human Hematopoietic Stem Cells Using Nanostraws, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 21267–21273 CrossRef CAS PubMed.
- Y. R. Na, S. Y. Kim, J. T. Gaublomme, A. K. Shalek, M. Jorgolli, H. Park and E. G. Yang, Probing Enzymatic Activity inside Living Cells Using a Nanowire-Cell “Sandwich” Assay, Nano Lett., 2013, 13, 153–158 CrossRef CAS PubMed.
- L. Hanson, W. Zhao, H. Y. Lou, Z. C. Lin, S. W. Lee, P. Chowdary, Y. Cui and B. Cui, Vertical Nanopillars for in Situ Probing of Nuclear Mechanics in Adherent Cells, Nat. Nanotechnol., 2015, 10, 554–562 CrossRef CAS PubMed.
- Y. C. Kim, J. H. Park and M. R. Prausnitz, Microneedles for Drug and Vaccine Delivery, Adv. Drug Delivery Rev., 2012, 1547–1568 CrossRef CAS PubMed.
- K. Ita, Ceramic Microneedles and Hollow Microneedles for Transdermal Drug Delivery: Two Decades of Research, J. Drug Delivery Sci. Technol., 2018, 314–322 CrossRef CAS.
- R. F. Donnelly, T. R. Raj Singh and A. D. Woolfson, Microneedle-Based Drug Delivery Systems: Microfabrication, Drug Delivery, and Safety, Drug Delivery, 2010, 187–207 CrossRef CAS PubMed.
- T. M. Tuan-Mahmood, M. T. C. McCrudden, B. M. Torrisi, E. McAlister, M. J. Garland, T. R. R. Singh and R. F. Donnelly, Microneedles for Intradermal and Transdermal Drug Delivery, Eur. J. Pharm. Sci., 2013, 623–637 CrossRef CAS PubMed.
- A. Panda, V. A. Matadh, S. Suresh, H. N. Shivakumar and S. N. Murthy, Non-Dermal Applications of Microneedle Drug Delivery Systems, Drug Delivery Transl. Res., 2022, 67–78 CrossRef CAS PubMed.
- S. R. Patel, A. S. P. Lin, H. F. Edelhauser and M. R. Prausnitz, Suprachoroidal Drug Delivery to the Back of the Eye Using Hollow Microneedles, Pharm. Res., 2011, 28, 166–176 CrossRef CAS PubMed.
- D. J. Lipomi, R. V. Martinez, L. Cademartiri and G. M. Whitesides, Soft Lithographic Approaches to Nanofabrication, Elsevier B.V, 2012, vol. 7 Search PubMed.
- G. M. Whitesides, E. Ostuni, S. Takayama, X. Jiang and D. E. Ingber, Soft Lithography in Biology and Biochemistry, Annu. Rev. Biomed. Eng., 2001, 3, 335–373 CrossRef CAS PubMed.
- R. Song, M. Murphy, C. Li, K. Ting, C. Soo and Z. Zheng, Current Development of Biodegradable Polymeric Materials for Biomedical Applications, Drug Des., Dev. Ther., 2018, 3117–3145 CrossRef CAS PubMed.
- P. Gunatillake, R. Mayadunne and R. Adhikari, Recent Developments in Biodegradable Synthetic Polymers, Biotechnol. Annu. Rev., 2006, 301–347 CAS.
- W. Amass, A. Amass and B. Tighe, A Review of Biodegradable Polymers: Uses, Current Developments in the Synthesis and Characterization of Biodegradable Polyesters, Blends of Biodegradable Polymers and Recent Advances in Biodegradation Studies, Polym. Int., 1998, 47, 89–144 CrossRef CAS.
- A. C. Vieira, J. C. Vieira, J. M. Ferra, F. D. Magalhães, R. M. Guedes and A. T. Marques, Mechanical Study of PLA-PCL Fibers during in Vitro Degradation, J. Mech. Behav. Biomed. Mater., 2011, 4, 451–460 CrossRef CAS PubMed.
- A. Heimowska, M. Morawska and A. Bocho-Janiszewska, Biodegradation of Poly(ε-Caprolactone) in Natural Water Environments, Pol. J. Chem. Technol., 2017, 19, 120–126 CrossRef CAS.
- R. Chandra and R. Rustgi, Biodegradable polymers, Prog. Polym. Sci., 1998, 23, 1273–1335 CrossRef CAS.
- O. S. Manoukian, M. R. Arul, N. Sardashti, T. Stedman, R. James, S. Rudraiah and S. G. Kumbar, Biodegradable Polymeric Injectable Implants for Long-Term Delivery of Contraceptive Drugs, J. Appl. Polym. Sci., 2017, 135, 46068 CrossRef PubMed.
- L. S. Nair and C. T. Laurencin, Biodegradable polymers as biomaterials, Prog. Polym. Sci., 2007, 32, 762–798 CrossRef CAS.
- J. M. Williams, A. Adewunmi, R. M. Schek, C. L. Flanagan, P. H. Krebsbach, S. E. Feinberg, S. J. Hollister and S. Das, Bone Tissue Engineering Using Polycaprolactone Scaffolds Fabricated via Selective Laser Sintering, Biomaterials, 2005, 26, 4817–4827 CrossRef CAS PubMed.
- B. S. Kim, S. S. Yang and C. S. Kim, Incorporation of BMP-2 Nanoparticles on the Surface of a 3D-Printed Hydroxyapatite Scaffold Using an ε-Polycaprolactone Polymer Emulsion Coating Method for Bone Tissue Engineering, Colloids Surf., B, 2018, 170, 421–429 CrossRef CAS PubMed.
- R. A. Surmenev, S. Shkarina, D. S. Syromotina, E. V. Melnik, R. Shkarin, I. I. Selezneva, A. M. Ermakov, S. I. Ivlev, A. Cecilia, V. Weinhardt, T. Baumbach, T. Rijavec, A. Lapanje, M. V. Chaikina and M. A. Surmeneva, Characterization of Biomimetic Silicate- and Strontium-Containing Hydroxyapatite Microparticles Embedded in Biodegradable Electrospun Polycaprolactone Scaffolds for Bone Regeneration, Eur. Polym. J., 2019, 113, 67–77 CrossRef CAS.
- M. Tallawi, D. Dippold, R. Rai, D. D'Atri, J. A. Roether, D. W. Schubert, E. Rosellini, F. B. Engel and A. R. Boccaccini, Novel PGS/PCL Electrospun Fiber Mats with Patterned Topographical Features for Cardiac Patch Applications, Mater. Sci. Eng., C, 2016, 69, 569–576 CrossRef CAS PubMed.
- S. Mukherjee, S. Darzi, A. Rosamilia, V. Kadam, Y. Truong, J. A. Werkmeister and C. E. Gargett, Blended Nanostructured Degradable Mesh with Endometrial Mesenchymal Stem Cells Promotes Tissue Integration and Anti-Inflammatory Response in Vivo for Pelvic Floor Application, Biomacromolecules, 2019, 20, 454–468 CrossRef CAS.
- P. Gentile, V. Chiono, C. Tonda-Turo, A. M. Ferreira and G. Ciardelli, Polymeric Membranes for Guided Bone Regeneration, Biotechnol. J., 2011, 1187–1197 CrossRef CAS.
- W. Yu, G. Jiang, Y. Zhang, D. Liu, B. Xu and J. Zhou, Near-Infrared Light Triggered and Separable Microneedles for Transdermal Delivery of Metformin in Diabetic Rats, J. Mater. Chem. B, 2017, 5, 9507–9513 RSC.
- Y. Zhang, D. Chai, M. Gao, B. Xu and G. Jiang, Thermal Ablation of Separable Microneedles for Transdermal Delivery of Metformin on Diabetic Rats, Int. J. Polym. Mater. Polym. Biomater., 2019, 68, 850–858 CrossRef CAS.
- P. T. Ko, I. C. Lee, M. C. Chen and S. W. Tsai, Polymer Microneedles Fabricated from PCL and PCL/PEG Blends for Transdermal Delivery of Hydrophilic Compounds, J. Taiwan Inst. Chem. Eng., 2015, 51, 1–8 CrossRef CAS.
- S. Lee, J. Lee, K. Choi, H. Kim, Y. Park, J. Yoon, J. Kim and S. Ryu, Polylactic Acid and Polycaprolactone Blended Cosmetic Microneedle for Transdermal Hispidin Delivery System, Appl. Sci., 2021, 11, 2774 CrossRef CAS.
- G. W. Lee, S. Lee, J. H. Kim, S. G. Yim, J. Ryu, E. Lee, J. Lee, S. I. Yoo and S. Y. Yang, Density-Controlled Freestanding Biodegradable Nanopillar Arrays Patterned via Block Copolymer Micelle Lithography, Macromol. Mater. Eng., 2017, 302, 1600361 CrossRef.
- C. R. Jung, S. F. Lahiji, H. Jung, Y. Kim and H. Kim, Rapidly Separable Micropillar Integrated Dissolving Microneedles, Pharmaceutics, 2020, 12, 1–11 Search PubMed.
- A. Milionis, C. S. Sharma, R. Hopf, M. Uggowitzer, I. S. Bayer and D. Poulikakos, Engineering Fully Organic and Biodegradable Superhydrophobic Materials, Adv. Mater. Interfaces, 2019, 6, 1801202 CrossRef.
- R. Vecchione, S. Coppola, E. Esposito, C. Casale, V. Vespini, S. Grilli, P. Ferraro and P. A. Netti, Electro-Drawn Drug-Loaded Biodegradable Polymer Microneedles as a Viable Route to Hypodermic Injection, Adv. Funct. Mater., 2014, 24, 3515–3523 CrossRef CAS.
- S. H. Oh, I. K. Park, J. M. Kim and J. H. Lee, In vitro and in vivo characteristics of PCL scaffolds with pore size gradient fabricated by a centrifugation method, Biomaterials, 2007, 28, 1664–1671 CrossRef CAS PubMed.
- M. Bartnikowski, T. R. Dargaville, S. Ivanovski and D. W. Hutmacher, Degradation mechanisms of polycaprolactone in the context of chemistry, geometry and environment, Prog. Polym. Sci., 2019, 96, 1–20 CrossRef CAS.
- A. Haryńska, J. Kucinska-Lipka, A. Sulowska, I. Gubanska, M. Kostrzewa and H. Janik, Medical-Grade PCL Based Polyurethane System for FDM 3D Printing—Characterization and Fabrication, Materials, 2019, 12, 887 CrossRef PubMed.
- Z.-X. Zhou, Y.-R. Chen, J.-Y. Zhang, D. Jiang, F.-Z. Yuan, Z.-M. Mao, F. Yang, W.-B. Jiang, X. Wang and J.-K. Yu, Facile Strategy on Hydrophilic Modification of Poly(ε-caprolactone) Scaffolds for Assisting Tissue-Engineered Meniscus Constructs In Vitro, Front. Pharmacol., 2020, 11, 471 CrossRef CAS PubMed.
- C. X. Lam, S. H. Teoh and D. W. Hutmacher, Comparison of the degradation of polycaprolactone and polycaprolactone–(β-tricalcium phosphate) scaffolds in alkaline medium, Polym. Int., 2007, 56, 718–728 CrossRef CAS.
- C. Delaney, N. Geoghegan, H. Ibrahim, M. O'Loughlin, B. J. Rodriguez, L. Florea and S. M. Kelleher, Direct Laser Writing to Generate Molds for Polymer Nanopillar Replication, ACS Appl. Polym. Mater., 2020, 2, 3632–3641 CrossRef CAS.
- O.-W. Merten, Advances in cell culture: anchorage dependence, Philos. Trans. R. Soc., B, 2015, 370, 20140040 CrossRef PubMed.
- D. St Johnston and J. Ahringer, Cell Polarity in Eggs and Epithelia: Parallels and Diversity, Cell, 2010, 141, 757–774 CrossRef CAS PubMed.
- E. Assémat, E. Bazellières, E. Pallesi-Pocachard, A. Le Bivic and D. Massey-Harroche, Polarity complex proteins, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 614–630 CrossRef PubMed.
- G. Tian, G. Zhu, T. Ren, Y. Liu, K. Wei and Y. X. Liu, The effects of PCL diol molecular weight on properties of shape memory poly(ε-caprolactone) networks, J. Appl. Polym. Sci., 2019, 136, 47055 CrossRef.
- S. Liparoti, V. Speranza and R. Pantani, Replication of Micro- and Nanofeatures in Injection Molding of Two PLA Grades with Rapid Surface-Temperature Modulation, Materials, 2018, 11, 1442 CrossRef PubMed.
- M. A. Al-Nasassrah, F. Podczeck and J. M. Newton, The effect of an increase in chain length on the mechanical properties of polyethylene glycols, Eur. J. Pharm. Biopharm., 1998, 46, 31 CrossRef CAS PubMed.
- L. Zhou, H. J. Forman, Y. Ge and J. Lunec, Multi-walled carbon nanotubes: A cytotoxicity study in relation to functionalization, dose and dispersion, Toxicol. in Vitro, 2017, 42, 292–298 CrossRef CAS PubMed.
- R. Ekambaram, S. Saravanan, V. P. S. Babu and S. Dharmalingam, Fabrication and evaluation of Docetaxel doped ZnO nanoparticles incorporated PCL nanofibers for its hemocompatibility, cytotoxicity and apoptotic effects against A549, Materialia, 2022, 21, 101278 CrossRef CAS.
- Y. Liu, J. Nguyen, T. Steele, O. Merkel and T. Kissel, A new synthesis method and degradation of hyper-branched polyethylenimine grafted polycaprolactone block mono-methoxyl poly (ethylene glycol) copolymers (hy-PEI-g-PCL-b-mPEG) as potential DNA delivery vectors, Polymer, 2009, 50, 3895–3904 CrossRef CAS.
- L.-K. Ma, X. Wang, X.-L. Xu, J. Zhou, Y. Liao, J.-G. Feng and L.-L. Tang, Micropillar-arrayed surfaces promote transforming growth factor beta 1 induced epithelial to mesenchymal transition by focal adhesion kinase-related signaling in A549 cells, Chin. Med. J. Search PubMed.
- G. González, D. Baruffaldi, C. Martinengo, A. Angelini, A. Chiappone, I. Roppolo, C. F. Pirri and F. Frascella, Materials Testing for the Development of Biocompatible Devices through Vat-Polymerization 3D Printing, Nanomaterials, 2020, 10, 1788 CrossRef PubMed.
- S. Atzet, S. Curtin, P. Trinh, S. Bryant and B. Ratner, Degradable Poly(2-hydroxyethyl methacrylate)-co-polycaprolactone Hydrogels for Tissue Engineering Scaffolds, Biomacromolecules, 2008, 9, 3370–3377 CrossRef CAS PubMed.
- B. J. Green, K. S. Worthington, J. R. Thompson, S. J. Bunn, M. Rethwisch, E. E. Kaalberg, C. Jiao, L. A. Wiley, R. F. Mullins, E. M. Stone, E. H. Sohn, B. A. Tucker and C. A. Guymon, Effect of Molecular Weight and Functionality on Acrylated Poly(caprolactone) for Stereolithography and Biomedical Applications, Biomacromolecules, 2018, 19, 3682–3692 CrossRef CAS PubMed.
- P. A. Gunatillake, R. Adhikari and N. Gadegaard, Biodegradable Synthetic Polymers for Tissue Engineering, Eur. Cells Mater., 2003, 1–16 CrossRef CAS PubMed.
- H. B. Sun, K. Takada and S. Kawata, Elastic Force Analysis of Functional Polymer Submicron Oscillators, Appl. Phys. Lett., 2001, 79, 3173–3175 CrossRef CAS.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- R. M. Ginde and R. K. Gupta, In Vitro Chemical Degradation of Poly(Glycolic Acid) Pellets and Fibers, J. Appl. Polym. Sci., 1987, 33, 2411–2429 CrossRef CAS.
- D. Schipper, P. Babczyk, F. Elsayed, S. E. Klein, M. Schulze and E. Tobiasch, in Nanostructures for Novel Therapy, ed. D. Ficai and A. M. Grumezescu, Elsevier, 2017, pp. 567–589 Search PubMed.
- H. Seong, S. G. Higgins, J. Penders, J. P. K. Armstrong, S. W. Crowder, A. C. Moore, J. E. Sero, M. Becce and M. M. Stevens, Size-Tunable Nanoneedle Arrays for Influencing Stem Cell Morphology, Gene Expression, and Nuclear Membrane Curvature, ACS Nano, 2020, 14, 5371–5381 CrossRef CAS PubMed.
- S. Dobbenga, L. E. Fratila-Apachitei and A. A. Zadpoor, Nanopattern-induced osteogenic differentiation of stem cells – A systematic review, Acta Biomater., 2016, 46, 3–14 CrossRef CAS PubMed.
- D. E. Discher, P. Janmey and Y. Wang, Tissue Cells Feel and Respond to the Stiffness of Their Substrate, Science, 2005, 310, 1139–1143 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00165b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |