Open Access Article
Open Access Article3D printable acrylate polydimethylsiloxane resins for cell culture and drug testing†
Simona
Villata
 a,
Marta
Canta
a,
Désirée
Baruffaldi
a,
Marta
Canta
a,
Désirée
Baruffaldi
 a,
Alice
Pavan
a,
Annalisa
Chiappone
a,
Candido Fabrizio
Pirri
ab,
Francesca
Frascella
a,
Alice
Pavan
a,
Annalisa
Chiappone
a,
Candido Fabrizio
Pirri
ab,
Francesca
Frascella
 *a and
Ignazio
Roppolo
*a and
Ignazio
Roppolo
 a
a
aDipartimento di Scienza Applicata e Tecnologia, PolitoBIOMed Lab, Politecnico di Torino, C.so Duca degli Abruzzi 24, Turin 10129, Italy. E-mail: francesca.frascella@polito.it
bCenter for Sustainable Futures @PolitoIstituto Italiano di Tecnologia, Via Livorno 60, Turin 10144, Italy
First published on 13th March 2023
Abstract
Nowadays, most of the microfluidic devices for biological applications are fabricated with only few well-established materials. Among these, polydimethylsiloxane (PDMS) is the most used and known. However, it has many limitations, like the operator dependent and time-consuming manufacturing technique and the high molecule retention. TEGORad or Acrylate PDMS is an acrylate polydimethylsiloxane copolymer that can be 3D printed through Digital Light Processing (DLP), a technology that can boast reduction of waste products and the possibility of low cost and rapid manufacturing of complex components. Here, we developed 3D printed Acrylate PDMS-based devices for cell culture and drug testing. Our in vitro study shows that Acrylate PDMS can sustain cell growth of lung and skin epithelium, both of great interest for in vitro drug testing, without causing any genotoxic effect. Moreover, flow experiments with a drug-like solution (Rhodamine 6G) show that Acrylate PDMS drug retention is negligible unlike the high signal shown by PDMS. In conclusion, the study demonstrates that this acrylate resin can be an excellent alternative to PDMS to design stretchable platforms for cell culture and drug testing.
1 Introduction
In the biomedical field, there is an urgent need for preclinical testing models to predict drug response in human tissues or organs1,2 and to reduce the use of conventional 2D cell culture platforms and animal models.3,4 These devices can offer several advantages:5 lower volumes of reagents, fast response time, low fabrication costs, and high compactness and degree of scalability.3 Typical examples of such devices consist of microfluidic platforms, often obtained by replica molding of PDMS by means of soft lithography masters.6,7 PDMS is a silicone elastomer, optically transparent, mechanically flexible, chemically stable, biocompatible, non-cytotoxic, gas permeable, inert, and compatible with aqueous solutions.8 These properties make it appealing for biological application and microscopic observation.9,10 Unfortunately, this standard approach shows two highly disabling limitations:11 first, the manufacturing technique involves multiple steps,12 which makes it operator dependent, time consuming, material wasting and requires high-cost equipment.11 Furthermore, microfabrication allows to obtain 2.5 D devices, which need to be closed, with problems of material compatibility and bonding strategies.13 Recently, 3D printing was successfully applied for replica molding of PDMS-based devices, which allows consistent improvements in geometries achievable, avoiding the bonding step,14,15 but with the drawback of complicated removal of the master in 3D device.16 Alternatively, to obtain truly 3D structures, PDMS can be extruded17,18 by Direct Ink Writing (DIW) technique. Unlikely even DIW shows some drawbacks related to the inherent characteristics of this technology, in particular the presence of porosity and the limitation in geometries due to the need of support structures. At last, PDMS itself brings some criticism, since the absorption of small hydrophobic molecules, like most drugs, is really high.19,20 Consequently, in drug testing, the target molecule availability is lower than expected.19,21A promising solution17,18 can be to employ a material showing the same properties of PDMS22,23 in terms of transparency, mechanical flexibility and biocompatibility, but 3D printable with a different technique, in order to overcome the above mentioned drawbacks. In this context, Vat polymerization processes (SLA, DLP) could be good candidates. These additive manufacturing technologies, based on photopolymerization, enable high precision, direct 3D printing of complex geometries and, since they employ liquid formulations, ease of unreacted material removal.12 This may enhance reproducibility and speed of the process, avoiding waste of material or time.24 This approach was already investigated by some authors, obtaining excellent results.22,23
In this view, we recently investigated25 a photocurable silicone (TEGORad or Acrylate PDMS) 3D printable through Digital Light Processing (DLP) technology, as suitable candidate to replace PDMS in microfluidic platforms fabrication.25,26 Indeed, it gathers transparency, flexibility and peculiar chemical properties with good printability. On the other hand, it is well known that cells are influenced by the environment in which they live:27,28 mechanical, chemical and physical material properties can all have a deep impact on cell behavior.29 Consequently, each new material must be evaluated by cytotoxicity tests, which can manifest, for example, reduced cell viability, inflammatory response or DNA damage.30,31
Epithelial barriers are the body's natural defenses to regulate the passage of molecules from the external environment. Therefore, in vitro models of the outer epithelia of the human body (for instance, skin and lung) have found applications in both research and industrial settings in the effort to replace or partially substitute the use of animals in drug testing.32–35 For these reasons, the biological and chemical properties of 3D printed Acrylate PDMS have been evaluated, to assess the cytocompatibility of cell lines from two of the most important epithelial tissues: skin and lung.32–35 3D printed flexible devices for biological applications were fabricated, demonstrating the possible use as a drug testing platform. The investigation here performed aims at assessing the possibility to produce complex platforms for cell culture and drug testing, which gathers compatibility with advanced manufacturing (i.e. 3D printing) and flexibility. Knowing that cells are responsive to mechanical stimuli,36,37 the development of such flexible complex devices can be advantageous, since they can represent the building block for advanced multistimuli (mechanical, chemical, physical) biological testing platforms.
2 Materials and methods
2.1 Materials
TEGO®Rad 2800 (TEGORad or Acrylate PDMS) is an acrylate polydimethylsiloxane copolymer kindly supplied by Evonik Industries AG (Essen, Germany).Phenyl bis(2,4,6-trimethylbenzoyl)-phosphine oxide (BAPO) is used as a photoinitiator. Since BAPO is not directly soluble in Acrylate PDMS, another liquid photoinitiator was used, 2-hydroxy-2 methylpropiophenone (HMP, Sigma-Aldrich), to disperse it, weight ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 between BAPO and HMP, respectively.
4 between BAPO and HMP, respectively.
Dansyl Chloride (purchased from Aldrich) was used as a light absorber dye to improve printing precision.38 Since Dansyl Chloride is not miscible in the Acrylate PDMS, a small amount of Methyl methacrylate (MMA, Aldrich Chemical Co) monomer was used to solvate the dye into the Acrylate PDMS oligomer with a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. All cellular experiments were performed using lung cancer epithelial cells (A549), kindly provided by Valentina Monica, of the Department of Oncology, University of Torino, AOU San Luigi Gonzaga and human keratinocytes (HaCaT) purchased from Antibody Research Corporation. A549 and HaCaT were maintained in BenchStable™ RPMI 1640 (Thermo Fisher scientific) or BenchStable™ DMEM (Thermo Fisher scientific), respectively, the first one supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin (all from Sigma Aldrich) and 1% L-glutamine (Biowest), while the second one supplemented with 15% fetal bovine serum, 1% penicillin/streptomycin, 1% sodium pyruvate (all from Sigma Aldrich), 2% L-glutamine (Biowest).
50. All cellular experiments were performed using lung cancer epithelial cells (A549), kindly provided by Valentina Monica, of the Department of Oncology, University of Torino, AOU San Luigi Gonzaga and human keratinocytes (HaCaT) purchased from Antibody Research Corporation. A549 and HaCaT were maintained in BenchStable™ RPMI 1640 (Thermo Fisher scientific) or BenchStable™ DMEM (Thermo Fisher scientific), respectively, the first one supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin (all from Sigma Aldrich) and 1% L-glutamine (Biowest), while the second one supplemented with 15% fetal bovine serum, 1% penicillin/streptomycin, 1% sodium pyruvate (all from Sigma Aldrich), 2% L-glutamine (Biowest).
2.2 PDMS replication molding
PDMS Sylgard® 184 is a heat curable PDMS supplied as a two-parts kit consisting of pre-polymer (base) and cross-linker (curing agent) components. Prepolymer and cross-linker were mixed at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio, respectively. In the preparation, the solution was stirred vigorously for 2 minutes. Before pouring, this mixture was placed in a vacuum oven for approximately 30 minutes. Once the mixture was degassed, it was poured onto the master. Finally, it was cured at 70 °C for 2 hours. After curing, the device was peeled off the master.
1 weight ratio, respectively. In the preparation, the solution was stirred vigorously for 2 minutes. Before pouring, this mixture was placed in a vacuum oven for approximately 30 minutes. Once the mixture was degassed, it was poured onto the master. Finally, it was cured at 70 °C for 2 hours. After curing, the device was peeled off the master.
2.3 3D printing
To prepare the photocurable formulations, first the photoinitiator solution (BAPO) was added to the Acrylate PDMS oligomer, to reach a final BAPO concentration of 0.8 wt%. Then, few drops of dye solution were added to the Acrylate PDMS formulation to obtain three different concentrations: 0.075 wt%, 0.05 wt%, 0.01 wt%. Afterwards, the mixtures were magnetic stirred at room temperature for 5 minutes, until the formulation became visibly homogeneous. Finally, the formulations were sonicated for 5 min at room temperature to degas.A DLP-3D printer (Asiga MAX X27 UV, Australia) was used for processing the Acrylate PDMS formulations. The light source of the printer is based on LEDs that emit at 385 nm. The structures were 3D printed by setting a printing slicing thickness of 50 μm and a light intensity at 48 mW cm−2. All CAD designs were produced with the FreeCAD program and exported in STL format to use them into Asiga Composer software. For the three different formulations the exposure time and separation velocity were optimized separately.
The freshly printed devices were placed in a covered beaker filled with acetone and left for 20 minutes. Then, the solvent was refreshed, and the samples were left overnight. The next day, a further change of the acetone was carried out and kept for another 20 minutes in immersion and dried. After washing, the sample were subjected to a post-curing treatment in a UV oven (Asiga Flash) to complete the crosslinking (5 minutes each side). Then, they were immersed in ultra-pure water and, sterilized by autoclave process (ML System, 20 L). Finally, they were left to dry at room temperature under sterile hood for 1 day. Before using, a sterilization protocol was followed. The 3D printed wells filled with ultra-pure water were inserted into a sterile multi well plate. The water was removed, and they were filled with PBS (200 μL each well), to avoid the cell osmotic stress. At this point they were sterilized for 30 minutes under a biological hood's UV light. Once sterilized, the PBS was removed right before cells seeding.
2.4 Characterization technique
Transmittance. 3D printed wells were put in a 12 well plate (TC treated, Greiner Bio-One) and the absorbance of the material was measured using Synergy™ HTX Fluorescence Multi-Mode Microplate Reader. The transmittance was calculated as follows:| T = e−A × 100 |
 , since Acrylate PDMS wells (A12 well) were put in a PS well plate (APS), the thickness of the bottom was 1 mm for the Acrylate PDMS wells and 0.250 mm for the PS well plate.
, since Acrylate PDMS wells (A12 well) were put in a PS well plate (APS), the thickness of the bottom was 1 mm for the Acrylate PDMS wells and 0.250 mm for the PS well plate.
Scanner 3D. 3D Scanner 3Shape E3 was used to scan the printed wells before and after the use of the mini-tray in order to be able to carry out a quantitative evaluation of the improvement of the printed objects.
Dynamic mechanical thermal analysis (DMTA). DMTA measurements were performed to evaluate the viscoelastic moduli as a function of temperature. They were carried out on PDMS and 0.075% dye Acrylate PDMS samples. The measurements were performed using a Triton Technology TTDMA. The tests were performed on 20 × 5 × 1.5 mm3 specimens in the range from −150 to 40 °C, with a heating rate of 3 °C min−1 (strain 20 μm, frequency 1 Hz).
Fourier Transform Infrared Spectroscopy. Attenuated total reflection (ATR) spectra were collected using a Thermo Scientific Nicolet iS50 FTIR spectrometer. 64 scans were collected for each sample in the range of 4000–400 cm−1, with a resolution of 4 cm−1.
Optical contact angle measurements (OCA). OCA was performed using an OCAH 200 Contact Angle System (Dataphysics Instruments, Germany) by the sessile drop technique. Briefly, each sample was put into contact with a 1.5 μL drop of deionised water (γd = 21.8 mN m−1, γ = 72.8 mN m−1) or with a 1.5 μL drop of diiodomethane (γd = 50.8 mN m−1, γ = 50.8 mN m−1, 99% purity, SIgma Aldrich) that were used as test liquids. After the determination of the drop profile, an ellipse fit was used in order to extrapolate the contact angle. Optical images of the samples were collected with a Leica DM2500 microscope at room temperature. Surface Energy was calculated using the OWRK method.
Surface profiler. A Tencor P-10 Surface Profiler was used to measure roughness. The area scan was 205 mm, with a vertical range = 160 μm and 1 Å vertical data resolution. The surface roughness (Ra) of each profiler image was determined as the average deviation of height values from the mean plane.
2.5 Oxygen plasma treatment
Samples were plasma treated in a Low-Pressure Plasma Polymerization System (IONVAC Process s.r.l.). A plasma treatment of 1 min, at a pressure of 30 mTorr, 50 W and O2 flow rate of 10 sccm was performed. After treatment, samples were immersed and kept in ultra-pure water.2.6 Cell proliferation assay
1 × 104 A549 or HaCaT cells were seeded onto a 96 well plate (TC treated, Greiner Bio-One) and on equivalent sterilized 3D printed Acrylate PDMS wells with 0.01, 0.05, 0.075 wt% dye (both treated with O2 plasma treatment or not) in complete medium (200 μL for each well) and cultured at 37 °C in 5% CO2. After 24 h and 72 h PrestoBlue™ Cell Viability Reagent was used to evaluate the viability of the seeded cells. The reagent was added at a concentration of 10% v/v and was incubated with cells for 1 hour at 37 °C. Then, the supernatant was transferred in a 96 Well white/clear bottom plate (TC treated, Thermofisher) and the change in the fluorescence of the test reagent (resazurin to resorufin35) was measured using Synergy™ HTX Fluorescence Multi-Mode Microplate Reader with the excitation/emission wavelengths set at 530/590 nm. The cell proliferation experiments were performed at least three times. Differences between groups were analyzed by three-way ANOVA.Conditioned medium assay. 1 × 104 A549 or HaCaT cells were seeded onto a 96 well plate (TC treated, Greiner Bio-One) in complete medium (200 μL for each well) that was previously incubated with sterilized Acrylate PDMS wells (one 48 like well for each 2 mL of medium) for 72 h at 37 °C at 5% CO2. Cells were then cultured at 37 °C in 5% CO2 and after 24 h and 72 h PrestoBlue™ Cell Viability Reagent was used to evaluate the cell viability. The reagent was added at a concentration of 10% v/v and was incubated with cells for 1 hour at 37 °C. Then the supernatant was transferred in a 96 well white/clear bottom plate and the change in the fluorescence of the test reagent (resazurin to resorufin39) was measured using Synergy™ HTX Fluorescence Multi-Mode Microplate Reader with the excitation/emission wavelengths set at 530/590 nm. The signal of the normal and conditioned medium without cells was used as background. The cell proliferation experiments were performed three times. Differences between groups were analyzed by two-way ANOVA.
2.7 Fluorescence staining and microscopy
3 × 104 A549 or HaCaT cells were seeded onto a 48 well plate (TC treated, Greiner Bio-One) and on equivalent sterilized 3D printed Acrylate PDMS wells with 0.01, 0.05, 0.075% wt% dye O2 plasma treated in 400 μl per well of complete medium and grown at 37 °C in 5% CO2. After 72 h cells were fixed with 4% paraformaldehyde (PFA, Sigma) at room temperature for 15 min and washed twice with PBS. For the staining, cells were incubated with 0.4 μM DAPI in PBS and 0.25 μM FITC-conjugated phalloidin in PBS (200 μL) for 30 min at room temperature. The samples were imaged using the microscope Eclipse Ti2 Nikon (Tokyo, Japan) equipped with a Crest X-Light spinning disk.2.8 Flow cytometry analysis
1.2 × 105 A549 or HaCaT cells were seeded onto a 12 well plate (TC treated, Greiner Bio-One) and on equivalent 3D printed Acrylate PDMS wells with 0.01, 0.05, 0.075 wt% dye in complete medium (1.5 mL for each well) and grown at 37 °C in 5% CO2. After 72 hours, 1 × 106 cells for each condition were fixed with 2% paraformaldehyde (PFA, Alfa Aesar) on ice for 15 min, then washed two times with cold PBS and kept at −20 °C until the cytofluorimetric analysis. For the staining, cells were washed with cold TBS pH 7.4, permeabilized with TST (4% Fetal bovine serum, 0.1% Triton X-100 in TBS) on ice for 10 minutes and stained with phosphorylated Histone H2AX Alexa Fluor 647-conjugated Antibody (IC2288R, R&D) or Alexa Fluor 647-conjugated isotype ctrl (IC1051R, R&D) for 1 hour at 4 °C in the dark, then washed two times with cold TBS and analysed by flow cytometry. Flow cytometry was performed with a Guava Easycyte 6-2L flow cytometer (Merck Millipore), collecting for each sample 5 × 103 or 1 × 104 events, excluding cell debris. The pH2AX and the isotype fluorescence were detected using the red laser for the excitation and the Red in Red channel to collect the signal. Data were analyzed by FCS express 6 flow (De Novo Software).2.9 Drug-like absorption test
A solution of drug-like substance (Rhodamine 6G) was used to test PDMS and Acrylate PDMS material absorption. For this purpose, 3D printed microfluidics made of Acrylate PDMS with 0.01, 0.05, 0.075 wt% dye have been designed. Afterwards, the devices have been sealed by plasma bonding to a glass microscope slide. PDMS microfluidic were realized by reaplica molding. A solution of Rhodamine 6G (1 μM in PBS) was pumped through the devices and left in place for 1 hour. Then the microluidic were washed three times with PBS and fresh PBS solution was left in the microfluidics overnight. At last, three more washes with PBS were performed. The microfluidics were imaged using the microscope Eclipse Ti2 Nikon (Tokyo, Japan) equipped with a Crest X-Light spinning disk.3 Results and discussion
3.1 Acrylate PDMS characterization
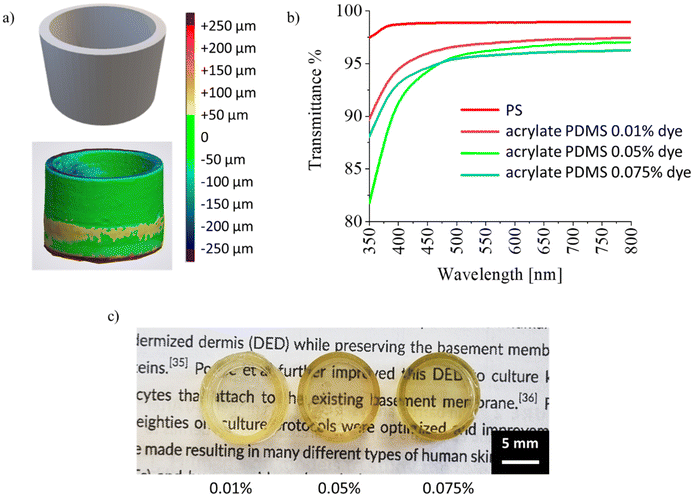
Starting from previous investigations25,26 that already assessed Acrylate PDMS formulation and printing parameters, in this study different geometries were 3D printed. Printing parameters and the employed digital models are reported in ESI, Table S1.† According to the purpose of this study, many well-like structures were 3D printed, to perform all the biological studies directly in 3D printed components. The printed structures were produced with good reproducibility and CAD fidelity of the 3D printed wells was also measured by 3D scanner, evidencing ±50 μm of precision (Fig. 1a).While controlled absorption is important in the emission range of the 3D printer (i.e. 385 nm), high transparency is instead required in the visible range, in order to be able to check cell culture experiments on going. Acrylate PDMS turned out to be an optically transparent material, in particular for wavelengths above 470 nm, even in presence of dyes (i.e. transmittance % ≥ 95% for of all the dye concentrations tested) (Fig. 1b and c). This suggests that the monitoring of the cell culture on Acrylate PDMS devices is possible, even for the formulation with the higher concentration of dye.
As mentioned, mechanical properties are important too. Here, the thermomechanical properties were evaluated by DMTA analysis, to establish the glass transition temperature (Tg) and the storage modulus (E′) of the 3D printed Acrylate PDMS, compared to PDMS (Fig. S1†). It is important to mention that PDMS mechanical properties can be adjusted by changing the ratio between resin and hardener.40 Here the properties were compared with the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (prepolymer–crosslinker) formulation, which is somehow a standard one. The analysis showed that, approaching room temperature, the elastic modulus E′ of Acrylate PDMS is very similar to the one of PDMS, resulting in similar flexibility of the two materials.
1 (prepolymer–crosslinker) formulation, which is somehow a standard one. The analysis showed that, approaching room temperature, the elastic modulus E′ of Acrylate PDMS is very similar to the one of PDMS, resulting in similar flexibility of the two materials.
3.2 Acrylate PDMS cytocompatibility
Once the use of Acrylate PDMS as ink for DLP-printing to obtain transparent and flexible objects was proved, we investigated its suitability to produce 3D printed devices for biological applications. Direct and indirect tests were performed to investigate cell adhesion and proliferation for both lung and skin epithelium, using lung adenocarcinoma cells (A549) and epidermal keratinocyte (HaCaT). Cells were seeded on all the 3D printed samples differing for the amount of dye dispersed. The viability was analyzed at 24 h and 72 h.41,42 Unfortunately, direct seeding on 3D printed objects demonstrated to be not suitable (Fig. 2a and b) due to low proliferation. Therefore, similarly to other plastic components conventionally used for cell culture, O2 plasma treatment was performed.As well-known, plasma O2 treatment modifies the surface properties of the materials, leading to partial surface oxidation and increase of hydrophilicity. This effect was evaluated by physical and chemical measurements, as detailed in ESI (Table S2 and Fig. S2),† evidencing a consistent increase of the polarity after plasma. From a morphological point of view, surface roughness was evaluated by means of a profiler. O2 Plasma treated Acrylate PDMS had a smoother surface than Acrylate PDMS, as detailed in ESI (Table S3).† This improvement in the surface profile may have been caused by a shallow etch process during plasma treatment with oxygen gas.
Furthermore, O2 plasma treatment resulted to be essential to ensure cell adhesion on 3D printed samples surfaces, sustaining cell growth for 72 hours, since cells reached confluence in most of the conditions.
Viability data show that Acrylate PDMS allows the growth of both cell lines A549 and HaCaT, even with a lower extent of HaCaT at 72 h, compared to the positive control (Fig. 2). Moreover, it is important to note that the growth of both cell lines from 24 h to 72 h is statistically significant, confirming the ability of Acrylate PDMS samples to become a good candidate for devices in which cell adhesion and proliferation are requested. These results indicate that cell culturing is possible only on O2 plasma treated Acrylate PDMS, and that this treatment is sufficient to obtain a significant proliferation of both cell lines, while it is not on the bare material. This property gives versatility to the material, exploitable in future devices, localizing biocompatibility properties by means of a localized treatment, for instance for having cell-cultured surfaces and not cultured microfluidics.
To better investigate the material cytotoxicity, the effect of conditioned medium for every Acrylate PDMS formulation on the viability of both cell lines was investigated. Cytotoxicity tests were performed to identify any possible toxic release of compounds from Acrylate PDMS samples. These was perfomed also in the not cultured microfluidic parts, where cells are not cultured and their metabolism does not affect the release of substances from the bulk material. Proliferation trends (Fig. 2c and d) results very similar to the previous adhesion and proliferation tests directly on Acrylate PDMS wells. This behaviour suggests that the bland cytotoxic effect revealed during the cell culture (especially for HaCaT cells) could be due to substance released by the material, as already witnessed in other printable resins.43 These results highlight that this effect has to be considered not only for the surfaces in which cell culture is performed, but also in the bare microfluidics.
Furthermore, the ability of the 3D printed materials to support cell colonization was confirmed through immunostaining. DAPI/Phalloidin fluorescence analysis were performed to observe the surface colonization of both cell lines (A549 and HaCaT) cultured for 72 h on wells of each Acrylate PDMS formulations, treated with O2 plasma treatment. The aim was to visualize the surfaces to understand if they were homogeneously colonized or cells were growing in clusters or aggregates. Cell nuclei were stained with DAPI (blue), while Phalloidin was used to stain actin and visualize the cytoskeleton of cells (green). Fluorescence images (Fig. 3) show that both cell lines survive and proliferate on all the Acrylate PDMS wells. The material surfaces were fully colonized with a confluent layer of well dispersed cells. This confirms that O2 plasma treated Acrylate PDMS can be a suitable surface for cell culture. It is important to highlight that O2 plasma treatment can be used both on plane surfaces (as in this case) but also in confined spaces like microfluidics.44
3.3 Acrylate PDMS genotoxicity
Possible DNA damages induced by the material were tested, evaluating the extent of histone H2AX phosphorylation on Ser-139 (double strands breaks, DSBs) by flow cytometry.45–47Fig. 4a and b show the comparison between the control on PS well plate and cells grown on Acrylate PDMS wells. The black horizontal line indicates the maximum intensity of pH2AX signal obtained for the control while the beginning of y axis is the limit of the aspecific signal (isotype control). It can be observed how the population of cells grown on Acrylate PDMS wells do not cross the black line, which indicates that the DNA damage induced by the material is not more relevant than in normal conditions. These results highlight the biocompatibility of all Acrylate PDMS formulations, confirming its potential application as a cell culture platform.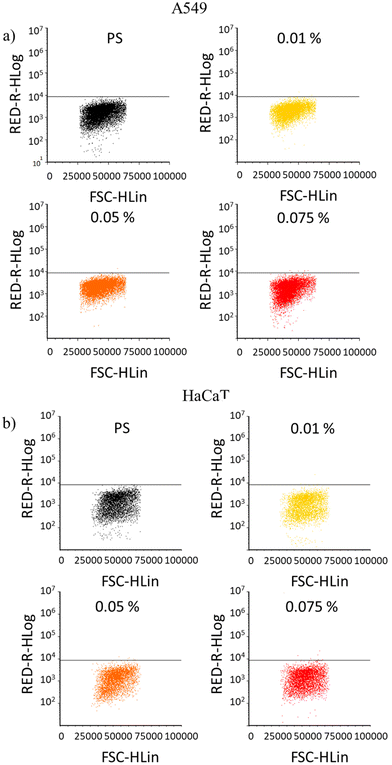 | ||
| Fig. 4 Genotoxicity of the Acrylate PDMS 0.01–0.05–0.075% dye samples on (a) A549 and (b) HaCaT cell lines. | ||
3.4 Drug-like absorption test
At last, 3D printed microfluidic structures were used to test the adsorption of the drug-like molecule (Rhodamine 6G).4 As mentioned, this is somehow problematic for PDMS standard devices since this material presents high absorption of small hydrophobic molecules, like most of the drugs, so it is important to propose a material that retains less.19,20Rhodamine 6G is a well-known dye and, like most drugs, a hydrophobic molecule.48,49 Its molecular weight is 479 g mol−1,50 very close to the average molecular weight of drugs marketed up during 2021, that has been reported 477 g mol−1.51 For these reasons, Rhodamine 6G has been selected as drug-like molecule to investigate the material absorption.
As reference, a PDMS microfluidic was fabricated through replica molding technique. Acrylate PDMS microfluidics were then printed leaving one side open, to compare directly with PDMS geometry. For both materials, after O2 plasma treatment of the exposed surface, PDMS and Acrylate PDMS were sealed with a glass microscope slide.
After perfusing 1 μM Rhodamine 6G solution through the microfluidic and rinsing the channel deeply with PBS, following the protocol described in the Experimental section, Acrylate PDMS and PDMS microfluidic devices were observed under a fluorescence microscope to evaluate the different contribution of the drug-like substance (Fig. 5a and b). A clear difference between the Acrylate PDMS formulations and PDMS was readily noticeable, with acrylate PDMS absorbing less than PDMS. A possible explanation can be that Acrylate PDMS has a higher cross-linking density than PDMS (as shown by values of rubbery plateau in DMTA analysis 2.3.52), which hinders the diffusion of Rhodamine 6G in the polymeric network. Moreover, it has to be considered that the different production processes (i.e. replica molding and DLP printing) can introduce different roughness or in general different physical characteristics at the surface level,53 that can be also responsible for the different drug-like molecule retention. This result suggests a higher suitability of Acrylate PDMS with respect of PMDS in drug testing applications.
4 Conclusions
In this study, it was demonstrated that 3D printable formulations based on an Acrylate PDMS (TEGORad) can be promising alternative materials to PDMS, in the development of biomedical devices. First of all, these formulations showed easy and fast printing, enabling the production of complex devices without having time-consuming steps or a great amount of waste. While this aspect may seem secondary for standard biological platforms (i.e. multiwell), it acquires great importance in view of developing complex multiplex platforms, in which cell cultures should be accompanied by stimulation and real-time observation. In this view the compatibility with light-induced 3D printing can simplify laboratory manufacturing, especially for integrated structures in which microfluidic is required. For what concerns the biological aspect, it was demonstrated that O2 plasma treatment is crucial to obtain cytocompatible and cell adhesive surfaces.41,42 Moreover, cell proliferation of both lung and skin epithelium after O2 plasma treatment of the surface, together with the absence of genotoxicity of Acrylate PDMS formulations are proofs that this material can be suitable for cell culture. At last, the low drug-like molecule retention, and the low cell adhesion on the material if not treated with O2 plasma treatment suggested a possible application in the fabrication of microfluidic devices. As stated before, one of the main problems of PDMS is its high adsorption, which led to misestimate the molecule dose delivered to the sample, making the in vivo translation difficult.For what concerns the material feasibility in terms of microfluidics, conditioned medium investigation revealed that the low cytotoxic effect could be related with the release of some molecule from the material, effect that has to be considered for drug testing experiments.
Another important aspect of Acrylate PDMS is its excellent stretchability,25 after mechanical stimulation. This means that cells can be cultured on a stretchable material, which can be of particular interest to better mimicry organ natural environment in terms of mechanical stimuli. This would be of great importance especially referring to epithelium,54–56 since alveolar epithelium is exposed to cyclic tensile strain during breathing54 and skin epithelium is costantly under mechanical stimuli.56 In future investigations, cell viability tests on Acrylate PDMS mechanically stimulated samples would be of interest for many applications.
To conclude, the versatility of the developed 3D printable formulation promotes it as a good candidate for fabricating microfluidic devices for multi-stimulus cell cultures, analytical and diagnostic tests in the laboratory or in vitro pharmacological experiments.
Author contributions
Simona Villata: methodology, validation, investigation, data, writing, visualization; Marta Canta: methodology, validation, investigation; Désirée Baruffaldi: conceptualization, methodology; Alice Pavan: investigation; Annalisa Chiappone: investigation; Candido Fabrizio Pirri: funding acquisition; Francesca Frascella: conceptualization, methodology, funding acquisition; Ignazio Roppolo: conceptualization, methodology.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The present work was developed in the framework of DEFLeCT project funded by Regione Piemonte POR-FESR 2014–2020 and PNRR MUR NODES and D3-4HEALTH funding.References
- V. Das, F. Bruzzese, P. Konečný, F. Iannelli, A. Budillon and M. Hajdúch, Drug Discovery Today, 2015, 20, 848–855 CrossRef CAS PubMed.
- C. W. Chi, A. R. Ahmed, Z. Dereli-Korkut and S. Wang, Bioanalysis, 2016, 8, 921–937 CrossRef CAS PubMed.
- A. Chiadò, G. Palmara, A. Chiappone, C. Tanzanu, C. F. Pirri, I. Roppolo and F. Frascella, Lab Chip, 2020, 20, 665–674 RSC.
- K. Domansky, D. C. Leslie, J. McKinney, J. P. Fraser, J. D. Sliz, T. Hamkins-Indik, G. A. Hamilton, A. Bahinski and D. E. Ingber, Lab Chip, 2013, 13, 3956–3964 RSC.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Nat. Rev. Mater., 2018, 3, 257–278 CrossRef.
- K. Ren, J. Zhou and H. Wu, Acc. Chem. Res., 2013, 46, 2396–2406 CrossRef CAS PubMed.
- M. Radisic and P. Loskill, ACS Biomater. Sci. Eng., 2021, 7, 2861–2863 CrossRef CAS PubMed.
- S. Torino, B. Corrado, M. Iodice and G. Coppola, Inventions, 2018, 3, 65 CrossRef.
- I. D. Johnston, D. K. McCluskey, C. K. L. Tan and M. C. Tracey, J. Micromech. Microeng., 2014, 24, 035017 CrossRef CAS.
- S. Halldorsson, E. Lucumi, R. Gómez-Sjöberg and R. M. T. Fleming, Biosens. Bioelectron., 2015, 63, 218–231 CrossRef CAS PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS.
- G. Gonzalez, I. Roppolo, C. F. Pirri and A. Chiappone, Addit. Manuf., 2022, 55, 102867 CAS.
- A. Lai, N. Altemose, J. A. White and A. M. Streets, J. Micromech. Microeng., 2019, 29, 107001 CrossRef CAS.
- A. Iuliano, E. van der Wal, C. W. B. Ruijmbeek, S. L. M. in ‘t Groen, W. W. M. P. Pijnappel, J. C. de Greef and V. Saggiomo, Adv. Mater. Technol., 2020, 5(9), 2000344 CrossRef CAS.
- M. del Rosario, H. S. Heil, A. Mendes, V. Saggiomo and R. Henriques, Adv. Biol., 2022, 6(4), 2100994 CrossRef PubMed.
- V. Saggiomo and A. H. Velders, Adv. Sci., 2015, 2(9), 1500125 CrossRef PubMed.
- V. Ozbolat, M. Dey, B. Ayan, A. Povilianskas, M. C. Demirel and I. T. Ozbolat, ACS Biomater. Sci. Eng., 2018, 4, 682–693 CrossRef CAS PubMed.
- H. Zhang, T. Qi, X. Zhu, L. Zhou, Z. Li, Y. F. Zhang, W. Yang, J. Yang, Z. Peng, G. Zhang, F. Wang, P. Guo and H. Lan, ACS Appl. Mater. Interfaces, 2021, 13, 36295–36306 CrossRef CAS PubMed.
- B. J. van Meer, H. de Vries, K. S. A. Firth, J. van Weerd, L. G. J. Tertoolen, H. B. J. Karperien, P. Jonkheijm, C. Denning, A. P. Ijzerman and C. L. Mummery, Biochem. Biophys. Res. Commun., 2017, 482, 323–328 CrossRef CAS PubMed.
- V. S. Shirure and S. C. George, Lab Chip, 2017, 17, 681–690 RSC.
- M. W. Toepke and D. J. Beebe, Lab Chip, 2006, 6, 1484–1486 RSC.
- C. Chen, B. T. Mehl, A. S. Munshi, A. D. Townsend, D. M. Spence and R. S. Martin, Anal. Methods, 2016, 8, 6005–6012 RSC.
- N. Bhattacharjee, C. Parra-Cabrera, Y. T. Kim, A. P. Kuo and A. Folch, Adv. Mater., 2018, 30(22), 1800001 CrossRef PubMed.
- J. Raczkowska, S. Prauzner-Bechcicki, J. Lukes, J. Sepitka, A. Bernasik, K. Awsiuk, C. Paluszkiewicz, J. Pabijan, M. Lekka and A. Budkowski, Appl. Surf. Sci., 2016, 389, 247–254 CrossRef CAS.
- G. Gonzalez, A. Chiappone, K. Dietliker, C. F. Pirri and I. Roppolo, Adv. Mater. Technol., 2020, 5(9), 2000374 CrossRef CAS.
- L. Pezzana, G. Riccucci, S. Spriano, D. Battegazzore, M. Sangermano and A. Chiappone, Nanomaterials, 2021, 11, 1–17 CrossRef PubMed.
- I. G. Siller, A. Enders, P. Gellermann, S. Winkler, A. Lavrentieva, T. Scheper and J. Bahnemann, Biomed. Mater., 2020, 15(5), 055007 CrossRef CAS PubMed.
- Z. Nejedlá, D. Poustka, R. Herma, M. Liegertová, M. Štofik, J. Smejkal, V. Šícha, P. Kaule and J. Malý, RSC Adv., 2021, 11, 16252–16267 RSC.
- D. Baruffaldi, G. Palmara, C. Pirri and F. Frascella, ACS Appl. Bio Mater., 2021, 4, 2233–2250 CrossRef CAS PubMed.
- D. Baruffaldi, C. F. Pirri and F. Frascella, Bioprinting, 2021, 22(3), e00135 CrossRef.
- M. Zanon, D. Baruffaldi, M. Sangermano, C. F. Pirri, F. Frascella and A. Chiappone, Biomed. Sci. Eng., 2021, 5, s1 CrossRef.
- N. Arumugasaamy, J. Navarro, J. Kent Leach, P. C. W. Kim and J. P. Fisher, Ann. Biomed. Eng., 2019, 47(1), 1–21 CrossRef PubMed.
- Y. Mohammed, A. Holmes, P. C. L. Kwok, T. Kumeria, S. Namjoshi, M. Imran, L. Matteucci, M. Ali, W. Tai, H. A. E. Benson and M. S. Roberts, Adv. Drug Delivery Rev., 2022, 186, 114293 CrossRef CAS PubMed.
- Y. Bin Choy and M. R. Prausnitz, Pharm. Res., 2011, 28, 943–948 CrossRef PubMed.
- S. Gordon, M. Daneshian, J. Bouwstra, F. Caloni, S. Constant, D. E. Davies, G. Dandekar, C. A. Guzman, E. Fabian, E. Haltner, T. Hartung, N. Hasiwa, P. Hayden, H. Kandarova, S. Khare, H. F. Krug, C. Kneuer, M. Leist, G. Lian, U. Marx, M. Metzger, K. Ott, P. Prieto, M. S. Roberts, E. L. Roggen, T. Tralau, C. van den Braak, H. Walles and C. M. Lehr, ALTEX, 2015, 32, 327–378 CrossRef PubMed.
- P. A. Janmey and C. A. McCulloch, Annu. Rev. Biomed. Eng., 2007, 9, 1–34 CrossRef CAS PubMed.
- R. Goetzke, A. Sechi, L. de Laporte, S. Neuss and W. Wagner, Cell. Mol. Life Sci., 2018, 75, 3297–3312 CrossRef CAS PubMed.
- M. Gastaldi, F. Cardano, M. Zanetti, G. Viscardi, C. Barolo, S. Bordiga, S. Magdassi, A. Fin and I. Roppolo, ACS Mater. Lett., 2021, 3, 1–17 CrossRef CAS.
- J. L. Chen, T. W. J. Steele and D. C. Stuckey, Biotechnol. Bioeng., 2018, 115, 351–358 CrossRef CAS PubMed.
- A. Santiago-Alvarado, A. S. Cruz-Félix, J. González-García, O. Sánchez-López, A. J. Mendoza-Jasso and I. Hernández-Castillo, Mater. Res. Express, 2020, 7, 045301 CrossRef CAS.
- M. Amerian, M. Amerian, M. Sameti and E. Seyedjafari, J. Biomed. Mater. Res., Part A, 2019, 107, 2806–2813 CrossRef CAS PubMed.
- E. D. Yildirim, D. Pappas, S. Güçeri and W. Sun, Plasma Processes Polym., 2011, 8, 256–267 CrossRef CAS.
- G. González, D. Baruffaldi, C. Martinengo, A. Angelini, A. Chiappone, I. Roppolo, C. F. Pirri and F. Frascella, Nanomaterials, 2020, 10, 1–13 Search PubMed.
- S. H. Tan, N. T. Nguyen, Y. C. Chua and T. G. Kang, Biomicrofluidics, 2010, 4, 032204 CrossRef PubMed.
- T. Toyooka, M. Ishihama and Y. Ibuki, J. Invest. Dermatol., 2011, 131, 1313–1321 CrossRef CAS PubMed.
- A. Kinner, W. Wu, C. Staudt and G. Iliakis, Nucleic Acids Res., 2008, 36, 5678–5694 CrossRef CAS PubMed.
- F. K. Noubissi, A. A. McBride, H. G. Leppert, L. J. Millet, X. Wang and S. M. Davern, Sci. Rep., 2021, 11, 8945 CrossRef CAS PubMed.
- J. Bouman, P. Belton, P. Venema, E. van der Linden, R. de Vries and S. Qi, Pharm. Res., 2016, 33, 673–685 CrossRef CAS PubMed.
- C. Lin, S. K. Katla and J. Perez-Mercader, J. Photochem. Photobiol., A, 2021, 406, 112992 CrossRef CAS.
- T. Rasheed, M. Bilal, H. M. N. Iqbal, H. Hu and X. Zhang, Water, Air, Soil Pollut., 2017, 228, 291 CrossRef.
- U. Fagerholm, bioRxiv, 2022, DOI:10.1101/2022.09.21.508888.
- J. Qian and C. Berkland, Adv. Healthc. Mater., 2021, 10(12), 2100015 CrossRef CAS PubMed.
- V. Mehta and S. N. Rath, Bio-Des. Manuf., 2021, 4, 311–343 CrossRef.
- C. L. Thompson, S. Fu, M. M. Knight and S. D. Thorpe, Front. Bioeng. Biotechnol., 2020, 8(10), 602646 CrossRef PubMed.
- A. Varone, J. K. Nguyen, L. Leng, R. Barrile, J. Sliz, C. Lucchesi, N. Wen, A. Gravanis, G. A. Hamilton, K. Karalis and C. D. Hinojosa, Biomaterials, 2021, 275, 120957 CrossRef CAS PubMed.
- N. Mori, Y. Morimoto and S. Takeuchi, Biofabrication, 2018, 11(1), 011001 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00152k |
| This journal is © The Royal Society of Chemistry 2023 |