Open Access Article
Open Access ArticleSolute transport in the brain tissue: what are the key biophysical parameters tying in vivo and in vitro studies together?
Daniel
Alcaide
a,
Jean
Cacheux
ab,
Aurélien
Bancaud
 abc,
Rieko
Muramatsu
d and
Yukiko T.
Matsunaga
abc,
Rieko
Muramatsu
d and
Yukiko T.
Matsunaga
 *ab
*ab
aInstitute of Industrial Science, The University of Tokyo, Tokyo 153-8505, Japan. E-mail: mat@iis.u-tokyo.ac.jp
bLIMMS, CNRS-IIS UMI 2820, The University of Tokyo, Tokyo 153-8505, Japan
cCNRS, LAAS, 7 Avenue Du Colonel Roche, F-31400, Toulouse, France
dDepartment of Molecular Pharmacology, National Institute of Neuroscience, National Center of Neurology and Psychiatry, 4-1-1 Ogawa-higashi, Kodaira, Tokyo 187-8502, Japan
First published on 4th April 2023
Abstract
The mechanisms of solute transport in brain tissues are still under debate. The medical relevance of this topic has put the blood–brain barrier and the mechanisms of solute transport through the brain parenchyma in the spotlight, notably in the context of brain clearance. In the last decade, the classical view of pure diffusive flow across the brain parenchyma was tested against the recent proposal of an active, convectional fluid flow model known as the glymphatic model. Experimental studies of brain transport on living humans and animals have temporal and spatial limitations to validate any of these models. Therefore, detailed microscopic observations, mostly ex vivo tissue and simplified in vitro brain models with the support from computational models, are necessary to understand transport mechanisms in brain tissues. However, standardization is lacking between these experimental approaches, which tends to limit the generality of conclusions. In this review, we provide an overview of the output and limitations of modern brain solute transport studies to search for key parameters comparable across experimental setups. We emphasize that in vitro models relying on physiological material and reproducing the biophysical setting of the brain, as well as computational/mathematical models constitute powerful solutions to understand the solute transport phenomena inside of the brain tissue. Finally, we suggest the blood–brain barrier permeability and the apparent diffusion coefficient through the brain parenchyma to be robust biophysical parameters for the extraction of cross-model conclusion.
Introduction
The brain vasculature is composed of tightly joint endothelial cells with specific transport systems that differ from the rest of the organism's vasculature. This interface is known as the blood–brain barrier (BBB), and it is responsible for brain homeostasis and protection against damaging circulating agents.1–3 The integrity of the BBB is critical for correct brain functioning.4 Several diseases are associated with BBB dysfunction, such as stroke, epilepsy, multiple sclerosis, Alzheimer's disease, and mental disorders.5 Given this multi-disease and pluri-field relevance, medical studies regarding transport mechanisms through the BBB and preservation of its adequate functioning have gained attention in the past decades.However, solute transport in the parenchyma after passing the BBB has not yet been fully elucidated. Because the brain was typically thought to be lacking lymphatic clearance, solute transport through the extracellular space (ECS) was classically attributed to diffusive movements.6,7 In the last decade, the evidence for a fluid clearance pathway in the nervous system (CNS), the so-called glymphatic system has been proposed. This system considers active water and ion transport from the periarterial spaces through the interstitial space to the perivenous space. This occurs thanks to polarized water channels, specifically aquaporin-4 (AQP-4), and ionic channels on the membrane of a type of glial cells, known as astrocytes, in a convective, directional manner with arterial pulsation cyclic movements as flow motor force.8,9 Studies have confirmed the presence of a meningeal lymphatic vessel network along the outer dura mater of the brain that leads to the lymphatic nodes in the skull base,10,11 showing that diffusion may not be all about solute movement through the brain's ECS and leaving space for models that also consider convective flow.
Dysfunction of the glymphatic system can reduce waste removal from the brain tissue, leading to neuroinflammation and CNS disorders such as Alzheimer's disease, associated with the accumulation of amyloid-β and tau. Fluid clearance in the glymphatic system is mostly active during sleep. However, this system degrades with age, suggesting a causal link between sleep disturbances and the progression of neurodegenerative dementia symptoms. Thus, fluid transport through glymphatic system has been considered as a potential therapeutic target for CNS diseases.12,13 Drugs can also affect the glymphatic function of the brain, mainly through the modulation of CSF–ISF exchange and AQP-4 channels. Special attention has been paid to anesthetic drugs, which induce unconsciousness and increase glymphatic function.13,14 However, the mechanisms of action of these drugs seem to impact glymphatic function modulation, suggesting that they affect brain transport more globally than only through anesthesia.14
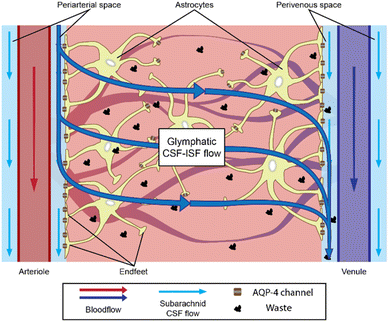
In the glymphatic context, astrocytes are thought to be relevant to fluid transport through the brain parenchyma. However, astrocytes also have various functions in the brain, depending on their surrounding neural tissues, ranging from molecular homeostasis maintenance by transporting major ions, protons, and neurotransmitter precursors, to defining the cytoarchitecture of the gray matter, working as structural guides with connections to the vasculature.15 Astrocytes located near the brain microvasculature revolve around it with their endfeet, where the expression of AQP-4 appears particularly relevant.16 Those endfeet constitute an intricate layer, normally referred to as the astrocytic endfeet network, which separates the perivascular spaces filled with cerebrospinal fluid (CSF)8,9,17 from the brain parenchyma full of interstitial fluid (ISF). CSF and ISF have a similar composition but serve different purposes. Waste from neural activities are secreted directly into the ISF, from where they are removed and leave the central nervous system (CNS) through the CSF.18 CSF also serves as the buoyancy cushion for the brain and compensates for the blood volume changes inside the skull. Astrocytes are thought to work as a regulator of the CSF–ISF exchange through the periarterial spaces into the brain ECS and out of the brain through the perivenous spaces, thanks to AQP-4 water channels19,20 (Fig. 1).
Nonetheless, conflicting experimental results regarding the glymphatic model have been found. Huge differences in microvasculature coverage by the endfeet network have been observed in ex vivo samples using different cell fixation methods before electron microscopy imaging,21 which raises doubts about the sieving function of astrocytes. Some experiments even suggest that basal membranes contribute or in some cases substitute the sieving function of the endfeet network.17 At the capillary scale, the basal membrane is fused between the endfeet network and the endothelial barrier,22,23 which makes differentiating the filtering contribution of each structure difficult. Albargothy et al. have also indicated the existence of pathways for the CSF influx through the basal membrane structures in the brain arterioles of mice.24
Other controversies come from the use of animal models, which have considerably progressed to allow the application of invasive observation techniques. Disagreement arises from key differences between animal and human brains. The astrocyte-to-neuron ratio is higher in humans than in rodents, and human astrocytes are approximately 2.6 times larger in diameter. In the rodent brain, pericytes around the microvasculature appear to guide AQP-4 polarization, but this conclusion is questionable in human.25,26 These disagreements may be avoided using in vitro models (normally referred to as microfluidic chips or brain-on-a-chip) employing human cell lines. Thanks to three-dimensional (3D) cocultures of relevant cell lines (astrocytes, endothelial cells, etc.), researchers recapitulate biological phenomena in a controlled environment and perform real-time and high-resolution observations. Transport across the BBB27 is one of the most common mechanism studied on in vitro platforms, whereas solute transport through the extracellular matrices (ECMs) appears to be quite out of the conversation probably due to the difficulty in recreating accurate brain ECM in vitro.
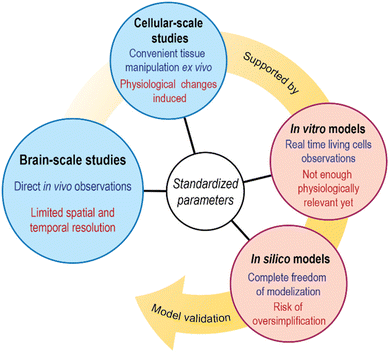
Many great reviews summarizing current brain solute transport models and concepts have been published in the last decade.18,28 However, not so many pursue the parametrical analysis of such models considering the standardization of measurements. By identifying parameters measurable in various experimental setups, cross-study conclusion extraction may become easier. Thus, this review summarizes recent results of experiments on solute transport through the brain parenchyma. We look at solute transport studies through the whole brain parenchyma and transport studies at the cellular scale, where in vitro models also provide interesting input. Then, we briefly overview the attempts of computational modeling to tie all previous studies together. Finally, we discuss relevant biophysical parameters that could be used for validation purposes across all those approaches (Fig. 2).
Brain-scale transport observations
To observe the fluid movement inside the brain, studies are mainly conducted in vivo. Since transport through the brain parenchyma was initially thought to be purely diffusive, experiments over two decades quantitatively measured the apparent diffusion coefficient (ADC) of tracers in the human brain to distinguish the gray matter, white matter, and CSF. ADC is commonly understood as the diffusion coefficient obtainable from measurements such as magnetic resonance images, and it was then used to indicate possible methodological uncertainties that may influence the pure diffusion coefficient measured value. Previously published values of ADC of water in living humans are (2.9–3) × 10−3 mm2 s−1 for CSF, (0.75–1) × 10−3 mm2 s−1 for the gray matter, and (0.2–1) × 10−3 mm2 s−1 depending on which part of the white matter is measured.29,30 At baseline, the self-diffusion coefficient of water at 35 °C is approximately 2.9 × 10−3 mm2 s−1,31,32 which is basically the same value as the water ADC in the CSF. In a more recent study, Valnes et al. used diffusion tensor images to determine the ADCs of water in both gray and white matters, obtaining 1.1 ± 0.3 × 10−3 mm2 s−1 and 0.8 ± 0.2 × 10−3 mm2 s−1, respectively, in the healthy human brain, which is still in the range of previously mentioned records.33In a more recent noninvasive study conducted in living humans, Wu et al. observed complete contrast agent clearance from the CSF in a span of 18 h maximum on 217 patients, based on sequential magnetic resonance images after intravenous administration of gadobutrol (Fig. 3A).34 The clearance from the periarterial spaces to the perivenous spaces and lastly to the meningeal lymphatics was observed, which follows the path predicted by the glymphatic theory. Other experiments in the same line revealed that drainage of intrathecally injected tracers to the lymphatic nodes in the skull base takes up to 24 h.35 These experiments are suitable for the evaluation of the overall movement of the CSF and ISF inside the brain; however, observations both in real time and at high resolution are lacking, inhibiting us from observing brain waste removal mechanisms in action.
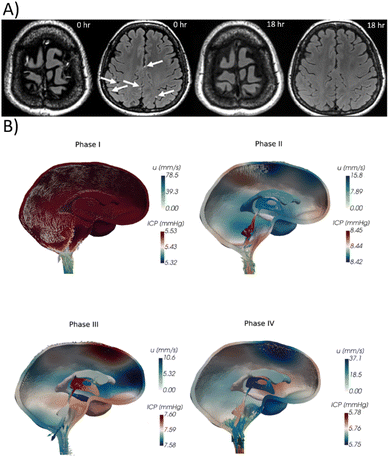 | ||
| Fig. 3 Whole brain flow visualization. (A) Contrast enhanced gadolinium (Gd) T2 FLAIR images. Perivenous enhancement and Gd leakage are visible (arrows). Follow-up images 18 h after initial images present no Gd signals.34 © 2020 American Neurological Association. (B) Computational calculation of ICP variation (colormap) and CSF velocity u (arrows) due to cardiac pulsations.50 The pulsation is divided in high net blood inflow (phase I), end of the net blood inflow (phase II), inflow and outflow equilibrium (phase III) and high blood net outflow (phase IV).51 | ||
Higher temporal and spatial resolutions can be achieved while remaining in vivo in animal models using more invasive observational procedures. A cranial opening is typically carved in the skull of the test subjects, allowing for detailed observations of the outer brain layers.36,37 So far, however, these studies have suffered from the critical issue associated to the change in the intracranial pressure (ICP) to the atmospheric pressure.38 Studies have proved that even small variations in the ICP can significantly change the results of ISF transport experiments.39,40 Notably, increased ICP after intrathecal injection in the closed skull of mice was one of the controversial points by which early glymphatic model experiments were criticized, although later low-flow injections in large brain cavities do not increase ICP significantly.40 Other factors, which contribute to change the ICP, including the stage of development and position of the subjects during the experiment, tend to affect the consistency of results across experimental setups.41,42 Thus, recently, other techniques that enable the monitoring of the ICP have been tested. Demeulenaere et al. mapped the whole mouse brain vasculature by non-invasive ultrafast ultrasound localization. This technique allowed the imaging of 0.3 mm × 0.3 mm2 volumes 750 times per s.43 These are incredibly promising results for large-volume, highly resolved, and non-invasive vasculature imaging that could be applied to humans; although these types of studies, which require acquisitions of several tens of seconds and a processing time of several hours, have not been published yet.
Experiments with living subjects also allow researchers to monitor effects of the whole organism in brain solute transport, such as the sleeping state, vascular pulsations, or disease. Studies on the natural necessity for sleeping and its implications on brain clearance show that during sleeping hours, brain permeability is enhanced and the interstitial space volume is increased by up to 60%.44 Demiral et al. studied variations in slow and fast ADC in the sleeping human brain, showing an increase in the overall CSF volume, but only localized changes in these coefficients in the sleeping brain. Astrocytes may help regulate sleep homeostasis45 through changes in their cell volume in favor of greater interstitial space volume in the sleeping brain, facilitating CSF–ISF flux throughout the brain.46,47 Regarding vascular pulsations,48 a study suggested the coexistence of different pulsations moving CSF along perivascular spaces.49 Data from such studies has recently motivated computational models that also show the relationship between CSF displacement and ICP variations during cardiac pulsation cycles (Fig. 3B).50,51
Related studies have shown general behaviors of brain fluid dynamics, mostly ignoring how microscopic elements such as the different cell barriers and astrocyte AQP-4 channels may influence CFS–ISF movements through the brain parenchyma. One way to identify the relevance on the fluid transport of those microscopic elements in the brain is to have them be altered artificially by drugs,14 due to disease or any other special conditions in the subjects, and then compare them with control subjects. This notably occurs in Alzheimer's disease, where AQP-4 depolarization and glymphatic impairment take place.52 Disease-like effects are also seen in genetically mutated mice with non-expression of AQP-4.53–56 To our knowledge, only a few noninvasive studies on AQP-4-deficient mice have been published, and some of them have reported a significant decrease in the water exchange rate between the brain vasculature and parenchyma through non-invasive MRI.54 Still, most of these studies conduct brain dissection for microscopic observations.
Cellular-scale observations
For microscopic observations, biopsied brain tissue is usually collected from animals, postmortem humans, or sometimes from living humans as part of another surgical process. The extracted tissue is then subjected to fixation that preserves it ex vivo, or when taken from a living subject, it can be kept alive inside a controlled in vitro microenvironment that provides it sufficient oxygen and nutrients for its survival. The latter technique is still not widely used; however, the coupling of in vitro devices with brain samples can produce very interesting platforms.57–60 Fixed brain tissue is much easier to handle, although the combination of terminal ischemia and the fixation process induce adverse effects on the cellular structure of the tissue, up to reducing the ECM space in half.36D'Arceuil et al. compared the ADC of primate (macaque) brains in vivo, postmortem ex vivo, and fixed samples. In vivo ADC values are in the same range as those in humans, as stated in the previous section. However, ADC values decrease in the white matter to 48% ± 20% and 20% ± 16% of the in vivo value in postmortem ex vivo and fixed brains, respectively. A similar degree of variation was observed for the gray matter.61 In another study, Thelwall et al. demonstrated that fixation processes, specifically aldehyde-based fixation, irreversibly reduce cell membrane permeation.62 A more recent study showed relevant morphological differences in mouse brain slices between chemical fixation and cryofixation. Specifically, astrocyte endfeet vasculature coverage appears to rise from 62.9% in cryofixation to 94.4% in chemical fixation (Fig. 4A), where the ECS has nearly completely collapsed (Fig. 4B).21 Cryofixation is thought to preserve better the in vivo-like tissue structure than chemical fixation, although the latter is the most commonly used. Surprisingly, D'Arceuil et al. also showed that diffusion anisotropy, i.e., the diffusion coefficient variation as a function of the direction inside the brain tissue, present in in vivo brains is well preserved in ex vivo fixed samples.61
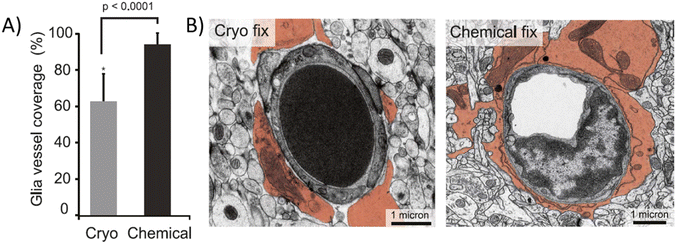 | ||
| Fig. 4 Chemically fixed and cryofixed brain tissue presents morphological differences. (A) Average vasculature coverage in cryo and chemically fixed mouse brain tissue. (B) Electro micrograph images of transversal sections of cryo and chemically fixed cerebral tissue (astrocytic endfeet colored). Chemically fixed samples present almost complete vasculature covering by the astrocytic endfeet while some uncovered areas can be seen in cryo fixed samples.21 | ||
Despite these alterations, we cannot disregard the useful information about the structure of the BBB and the parenchymal astrocyte endfeet network provided by microscopic observations in biopsied tissue. Nuriya and Yasui et al. used mice brain slices to show that astrocytic endfeet revolving around the brain vasculature constitute a slow diffusive barrier that may control solute concentrations around the BBB, whereas the rest of the astrocytic processes present higher diffusion rates. This local decrease in diffusion rates may allow astrocytes to regulate each region of the BBB.19,63 Similarly, Ezan et al. showed that the depletion of some gap junction proteins present in astrocytic endfeet, which occurs under several neuropathological conditions,64 weakens the BBB, making it leaky under stress.65 Also, brain disease mice models have proven to be clinically relevant tools, even showing perturbed patterns of AQP-4 expression throughout the brain tissue similar to post-mortem humans in Alzheimer's disease studies.66
Deep analysis of brain tissues reveals some differences in AQP-4 distribution between human and rodent brains. Higher contents of AQP-4 in human astrocyte's parenchymal membranes have been reported, whereas endfeet surrounding the vasculature appear to have similar AQP-4 densities in both cases.26 When working with AQP-4 null mice, Saadoun et al. did not detect abnormalities in brain morphology nor BBB integrity.67 Even so, under edema conditions, AQP-4 null mice experience greater brain water accumulation than wild-type mice,68 suggesting that AQP-4 not only participates in the inflow of water into the brain (even during brain edema formation),69 but also in the outflow of water from the brain. However, these experiments are extremely difficult to replicate in human brain tissues, leaving the door open for extrapolation and interpretation of these results in humans. An attempt to closing this gap is through engineered culture systems using human cell lines to approximate the brain anatomy in vitro or directly employing biopsied living human tissues, even though the latter is harder to come by.
In vitro models
Brain in vitro models attempted to recapitulate the basic biological and physical elements of the brain, also known as brain-on-a-chip (BoC). These models offer researchers controlled; high-throughput systems where precise measurements can be made. Standard 2D culture systems, which involve endothelial cells interacting with other cell types through porous membranes, are simple to reproduce and cost-effective but lack physiological relevance.70 Instead, 3D engineered cultures are built onto microfluidic devices with cell cultures of endothelial cells, typically cocultured with pericytes and/or astrocytes to recreate the structure of the 3D BBB and the brain tissue as close as possible to the pathophysiological conditions.71,72 The relevance of these systems would rely on the correct representation of key characteristics such as engineered ECM composition, endothelial BBB tightness, coculture cellular interactions, selective permeability and diffusion rates and pressure and stress responses, among others.73Unfortunately, the direct use of human brain cells or brain tissue is not always possible because the cellular materials are limited and endothelial brain cells tend to lose their phenotype during the cell handling process.74 Recent studies have addressed this limitation using human-induced pluripotent stem cells that differentiate into brain endothelial human cells.75 This technique have also been implemented in pericyte cells.76 It is also not uncommon to find in vitro assays using animal cell lines instead of human cell lines.73,77,78 In addition, as the cell lines of choice for BoC models might vary, BoC device design and measurement readout are also often different among models. This variety of BoC platforms complicates cross-model comparisons, which is the reason for the requests for standardization of measurements on microfluidic platforms in the last decade.27,79,80 van der Helm et al. suggested the use of the measured permeability coefficient or apparent permeability coefficient of multiple analytes, which can be calculated in each device, to compare model results with in vivo measurements. If the geometry of the device allows for it, they also suggest placing electrodes for measuring transendothelial electrical resistance (TEER) for barrier tightness assessment.27
The geometrical disposition of the BoC model directly affects BBB permeability measurements and requires calibration to confirm physical relevance of the model.27 Non-transwell microfluidic BoC devices (Fig. 5A) enable shear stress and hydrodynamic pressure effects over the BBB to be controlled by endothelial cells directly in contact with the ECM that contains supporting cellular coculture. Cylindrical geometries also influence the EC phenotype, morphology, and polarization of key proteins.81 However, unlike in 2D and 2.5D, TEER measurements in cylindrical 3D models are more difficult and normally substituted by tracer permeability studies modeled after radial diffusion of molecules from a cylindrical source.82 In addition, most 3D cylindrical models have much larger diameters (100–800 μm) than brain capillaries. Thus far, the only way to achieve diameters as low as 30 μm is through vasculogenesis; however, it is still in development, and the measurement of BBB permeability is very complicated.80
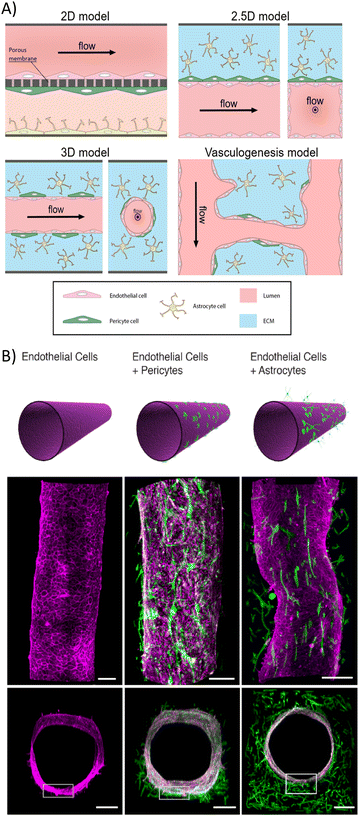 | ||
| Fig. 5 Common geometric and cellular compositions of BoC systems. (A) Schematic of typical setups of 2D, 2.5D, 3D, and vasculogenesis brain-on-a-chip models. 2D model: EC culture is separated from the supporting cell cocultures by a porous membrane. Shear stress effects are hard to maintain, but transport observations and TEER measurements are easy to perform. 2.5D model: non-cylindrical 3D EC culture in contact with an external ECM containing supporting cell coculture by one of its walls (2D cell interaction). Shear effects are maintained while still easing transport observations and TEER measurements. 3D cylindrical model: cylindrical vessel suspended in ECM containing coculture. Great shear flow and geometrical effects in ECs, but TEER measurements are more complex. 3D vasculogenesis model: ECs grow smaller capillaries between two larger ones. This method requires specific ECM compositions, but local measurements are hard to perform.80 (B) Confocal views of 3D BBB coculture models. Schematic, longitudinal and cross-section views are presented. ECs are colored in pink, whereas PC and astrocytes appear in green. Scale bar, 200 μm.86 | ||
An in vivo like BBB structure is achieved in BoC models when endothelial cells create a continuous layer and present junctions rich in tight junction proteins (occludin, claudin-5 and ZO-1).83,84 The cellular response to shear stress and pressure in a 3D matrix motivates the formation of these tight cellular junctions and lowers cell barrier permeability.27,85 Herland et al. also showed that the coculture of endothelial cylindrical BBB with pericytes or astrocytes (Fig. 5B)86 decreases the diffusive permeability of endothelial barriers, which relates to the diffusive flux through the barrier, against small molecules (3 kDa). The diffusive permeability values measured in monoculture conditions were 4 × 10−6 cm s−1, 3 × 10−6 cm s−1 in pericyte coculture and 2 × 10−6 cm s−1 in astrocyte coculture. This may result from an increase in tight junction proteins induced by the presence of astrocytes and/or pericytes. Some experiments use astrocyte-conditioned medium instead of astrocytes because of its lower cost and simple storage.87 However, results obtained in BoCs with astrocyte-conditioned medium are inconsistent, showing both an increase and a decrease in BBB permeability.84,88 Therefore, the use of astrocytes is preferred. Other factors that may influence the BBB permeability are disease-related biochemical compounds, such as pro-inflammatory cytokines, e.g., tumor necrosis factor alpha (TNF-α), which decreased severely the integrity of the cultured BBB.85,86
Similar to their in vivo counterparts, cases where in vitro astrocyte AQP-4 polarization on the endfeet facing the cultured endothelial barrier have been reported. This phenotype appears to be specific to 3D cultures.89 Indeed, 2D-cultured astrocytes become active, a process known as reactive gliosis, and they present less AQP-4 polarization than when they are cultured in a 3D hydrogel.90 Even more, the composition of the hydrogel matrix plays a major role on in vitro astrocyte development. Hydrogels such as collagen, commonly used in vitro culture are far from the real brain ECM, where collagen cannot be found.91 Astrocyte activation and morphology was different in collagen hydrogels than hydrogels mixing other components also present in the brain ECM, such as hyaluronic acid and/or proteoglycan compounds by adding Matrigel.92 Astrocytes branched much more in hydrogels containing those three materials and remained unactive, much more like they are found in vivo. These early approaches towards a more physiologically relevant in vitro ECM are motivating the design of glymphatic in vitro platforms focusing on the transport across astrocyte cultured ECM. Using a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of collagen type 1, Matrigel and hyaluronic acid, a very recent glymphatics-on-a-chip was proposed.93 The chip consisted in two parallel endothelial vessels passing through one common chamber containing the astrocyte seeded hydrogel. Transport of an amyloid-β solution from one vessel to the other was observed but no control over the hydrostatic pressure gradient between the vessels was incorporated in the design.
1 mixture of collagen type 1, Matrigel and hyaluronic acid, a very recent glymphatics-on-a-chip was proposed.93 The chip consisted in two parallel endothelial vessels passing through one common chamber containing the astrocyte seeded hydrogel. Transport of an amyloid-β solution from one vessel to the other was observed but no control over the hydrostatic pressure gradient between the vessels was incorporated in the design.
For future transport studies, as argued by Wan et al., the use of relevant ex vivo samples into microfluidic devices and the fine control of the hydrostatic pressure gradient across the ECM in glymphatics-on-a-chip would be necessary for accurate transport studies through the parenchyma simulation.94 Since this pressure gradient is likely to determine the mechanism of transport driven by diffusion or advection, recreating the same range of pressure gradients present inside the brain in vivo for accurate in vitro representation is strongly advised. Development in this direction is being made: for instance, Coloma et al. studied reverse transport in the perivascular spaces on in vitro platforms with quantified fluid transport velocity and different boundary conditions.95 Still, there is a severe lack of in vitro platforms where pressure gradient and its consequences onto the brain tissue deformation and permeability are taken into consideration. The employment of engineered hydrogels, whose composition, mechanical properties, and influence in cell cultures can be very well characterized and tuned,96 may ease the future design of these pressure-controlled glymphatics-on-a-chip devices. For now, one way to study such effects are computational models.
In silico models
To broaden our insight into brain solute transport, well known aspects of brain physiology can be coupled with mathematical calculations of the parenchymal flow in computational/mathematical models, commonly known as in silico models. In silico models are constructed to test the robustness of hypotheses extracted from experimental observations, and they may also serve to find a hierarchy between parameters that control the solute transport in the brain.97 Normally, in silico models can be highly detailed but focused in specific areas of the parenchymal flow phenomena or broader in their modeling area but sacrificing fine details in favor of computational viability.98Highly localized parenchymal transport in silico models are capable to use finite-element or even analytical methods in most cases. Under these settings, models isolate specific sections of the brain for study. Using a finite-element modeling of poroelastic brain tissue around a penetrating arteriole, Kedarasetti et al. found that arterial pulsations could drive convective flow through the ECM.99 Transport through the perivascular spaces is also a common topic of this kind of models, either on idealized or image-based geometries.100,101 In their perivascular flow model, Vinje et al. found that geometry differences between periarterial and perivenous spaces favor flow velocity in the first ones, which is in agreement with some previous experimental data.
On the other hand, more complete parenchymal transport in silico models brain fluid transport commonly consider the following basic elements: an influx source into the ECM (normally the periarterial space), different transport channels through the EC (AQP-4 channels, paracellular pathways, etc.), and an outflow source from the ECM (normally the perivenous space). Flow calculation may be performed by translating the fluid movement into an electric circuit based on hydraulic resistance paths102,103 or simulating the biological tissue based on electron microscopy images of brain tissue and then implementing Navier–Stokes104,105 and/or Darcy model106,107 calculations on them.
In these hydrodynamic or poromechanic models, parameters such as the scale, ECM geometry, flow-driving forces, and astrocyte contributions, among others, may be implemented differently,97 which leads to slight differences in their conclusions. Table 1 shows the parameters and general conclusions of four example parenchymal flow models. Regardless of small deviations, these four models agree that diffusion through the ECM is the principal mechanism of transport, as the majority of in silico brain clearance models do.102–105 However, only two of them pointed out that astrocytic AQP-4 channels may be a major facilitator of ISF flow.102,104 Meanwhile, more recent general models describe fluid movement in the brain as the composition of diffusion with net zero mean flow induced by the cyclic arterial pulsation, i.e., dispersion,107 which also substitutes the need for advective flow to explain brain clearance.108
| Model type | Ref. | Parameters and considerations | Conclusions |
|---|---|---|---|
| Hydraulic resistance model | 102 | ■ Distance between vessels = 300 μm | ■ Whole astrocytes facilitate water exchange through AQP-4 in all the CNS tissue. |
| ■ Astrocyte domain diameter = 50 μm | ■ Diffusion dominates in the parenchyma. Advection and diffusion both occur in the PVS. | ||
| ■ Flow pathways: | ■ AQP-4 depletion hinders the flow rate up to 46%. | ||
| (1) Endfeet AQP-4 only for water | |||
| (2) Inter-endfeet for solutes up to 20 nm | |||
| (3) Basement membrane contributions | |||
| ■ Gap junction resistance to water between astrocytes (2.5–4.5 nm) | |||
| ■ Pressure gradient between consecutive PVS = 226 Pa | |||
| 103 | ■ Distance between vessels = 200 μm | ■ Pulsation induces 0 net flow in PVS; dispersion might be a better explanation of fluid movement. | |
| ■ Vessel radius = 10 μm | ■ Peak fluid velocity is small (<6 nm s−1) regardless of parameter change, indicative of mainly diffusion driven flow. | ||
| ■ Pulse frequency = 5 Hz | |||
| ■ Hydraulic conductivity = 5.63 × 10−12 m2 Pa−1 s−1 | |||
| ■ Vessel porosity = 0.2 | |||
| ■ Pore size = 60 nm | |||
| ■ Parenchyma as solid porous media | |||
| Tissue digital reconstruction | 104 | ■ 3D model from the electron microscopy of rat neuropil | ■ Extracellular flow velocity of 8.95–16 nm s−1. |
| ■ Pressure gradient = 1 mmHg mm−1 | ■ Solute movement is very constrained. Diffusion should be the main mechanism. | ||
| ■ Diffusion coefficients: | ■ Advection can take place only in the PVS. | ||
| (1) Potassium ion = 77 × 10−7 cm2 s−1 | |||
| (2) 3 kDa dextran = 5.3 × 10−7 cm2 s−1 | |||
| (3) 70 kDa dextran = 0.84 × 10−7 cm2 s−1 | |||
| ■ Permeability of the smallest element = 10.7 nm × 19 nm2 | |||
| 105 | ■ 2D model from the primate cerebral cortex | ■ AQP-4 affects only osmotically driven water transport. | |
| ■ Effective hydraulic conductance of ECM = 0.9 × 10−9 cm4 per (dyne per s) | ■ Diffusion is sufficient to explain results. | ||
| ■ Diffusion coefficients of solute A = 10−9 m2 s−1 and solute B = 0.2–5 × 10−10 m2 s−1 | ■ Advection in the parenchyma is unlikely. | ||
| ■ AC permeability = 0–0.4 cm s−1 | |||
| ■ Pressure periarterial space = 0–10 mmHg | |||
| ■ Pressure amplitude of pulsation = 0–100 mmHg |
Some issues arise with the use of in silico models when trying to approach specific experimental conditions, for example, AQP-4 depletion in the brain or disease-like conditions. Models tend to represent such states as a variation of some of their computational variables, such as trans-membrane water resistance or a decrease in the interfeet gaps of astrocytes.109,110 Meanwhile, in vivo experiments have shown that, in the case of AQP-4 depletion for example, the ECM volume fraction and diffusivity throughout the brain is also altered.111 This issue is less severe in localized in silico models, which can portray a more accurate representation of those phenomena thanks to being focused on a relatively small area of the brain physiology. Therefore, it should be of great interest to build computationally viable models that can incorporate the findings of both in vivo studies and other specific in silico models into a generalized parenchymal transport model.
Conclusions and future perspectives
In this review, we have summarized the scale and limitations of current solute brain transport studies, considering their reach and relevant parameters that can be obtained from them. Undoubtedly, in vivo studies are the most relevant of the brain clearance phenomena. However, studies in living humans fail to provide macroscopic readouts that definitely clarify the physical mechanisms of transport and clearance, although projects toward this goal are ongoing.112 There is a lack of sensitivity for probing interstitial flow. Therefore, other studies should support and seek an explanation of such behaviors at a smaller scale with high-resolution observations. To confirm the results of different models efficiently, several steps toward the standardization of measurements should be taken.On this review, we saw through the different sections how important correct brain tissue representation is when evaluating experimental results. Employment of ex vivo samples runs into the disadvantage of substantially modifying the brain tissue properties, due to terminal ischemia and fixation. In vitro models suffer of inaccurate ECM representation and poor control of the periodic gradients applied to the brain tissue by cardiac pulsations. In silico models lose detail when englobing different areas of the brain. The common issue among these is being distant from brain tissue physiology or the inaccurate representation of it in the case of in silico models. We think that relevant progress on the matter would be possible by including soft material physics into the equation, both on in vitro and in silico models.
When it comes to transport across the brain tissue, we propose diffusion coefficients as good standardizing candidates. Most in silico brain transport models include the diffusion coefficients of the solutes that they simulate as general calculation parameters. Similarly, ADCs or diffusive permeability englobe microscopic effects such as AC AQP-4 contributions to diffusive flow macroscopically. They can be calculated from MR images of living brains33 and in ex vivo tissue, even though the values obtained ex vivo are typically lower than that obtained from in vivo measurements.61,62 It would be interesting if the lowering ratio between in vivo and ex vivo ADCs remains in the same range in every direction of the brain tissue due to the conservation of diffusion anisotropy.61 This would allow the ex vivo ADC values to be taken more quantitatively. The measurement of effective diffusion coefficients in vitro (especially from 3D microfluidic cylindrical models) can also be a relatively simple task. Unfortunately, as discussed previously transport through the ECM is still not the main topic of brain in vitro models. Improving the in vitro ECM physiological relevance and allowing pressure control would be major milestones in the development of relevant glymphatics-on-a-chip models.
Because the flux by diffusion is conservative, brain-scale measurements can be connected to local insights on the BBB. In this context, the porosity of the barrier, or equivalently the diffusive permeability, are relevant to standardized validation. In vitro models allow BBB permeability measurement relatively easily through TEER measurements and tracer transport studies; however, most studies do not provide such measurements in their publications. On the other hand, BBB permeability measurement in vivo is yet a challenging task. Without observing the flow through the BBB at the microscopic scale in humans, only general fluid movement inside the brain can be used to infer BBB permeability. This could be solved with measurements on biopsied in vivo tissue in platforms such as BoC devices, and computational models can implement BBB permeability as one of their variables as the validation test for the permeability values obtained from other experiments.
In summary we discuss the necessity for cross-model validation parameters for brain clearance and solute transport studies. This review compiled the outputs from different techniques used to examine brain solute transport mechanisms. This is a complex process, involving the transport across the BBB, diffusion, and periodic pressure actuation by the cardiac pulses. A lack of attention to the coupling of the periodic pressure actuation and the soft, elastic brain tissue was observed. Interest in this matter is starting to rise, which can be seen in the depiction of dispersion fluid movements or poroelastic properties of the brain tissue implemented in some mathematical models. However, in vitro platforms are still behind and need to incorporate pressure controlling elements for a precise glymphatics-on-a-chip model development. We believe that this type of device would become a powerful tool in brain disease drug assessment and overall brain research. Finally, regarding the questions of data integration, we suggest that it is possible by defining simple and multiscale readouts such as ADCs when regarding parenchymal transport and cell barrier permeability in the case transport across the BBB, both representative of the overall brain tissue. However, we think that this analysis will be even more powerful after the effect of periodic pressure and ECM permeability/mechanical properties in the context of dispersion of fluid in the brain tissue are clarified first.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
D. A. acknowledges the doctoral fellowship from WINGS-QSTEP. J. C. acknowledges the JSPS for postdoctoral fellowship. The authors thank the LIMMS (CNRS-Institute of Industrial Science, University of Tokyo) for the financial support. This work was partially supported by the AMED-CREST (22gm1510009) and the JSPS Core-to-Core Program (grant no. JPJSCCA20190006).Notes and references
- W. G. Mayhan, Microcirculation, 2001, 8, 89–104 CAS.
- N. J. Abbott, J. Anat., 2002, 200, 523–534 CrossRef.
- M. M. A. Almutairi, C. Gong, Y. G. Xu, Y. Chang and H. Shi, Cell. Mol. Life Sci., 2016, 73, 57–77 CrossRef CAS PubMed.
- R. Daneman and A. Prat, Cold Spring Harbor Perspect. Biol., 2015, 7, a020412 CrossRef PubMed.
- K. Schoknecht and H. Shalev, Epilepsia, 2012, 53, 7–13 CrossRef CAS PubMed.
- E. Syková and C. Nicholson, Physiol. Rev., 2008, 88, 1277–1340 CrossRef PubMed.
- A. S. Verkman, Phys. Biol., 2013, 10, 045003 CrossRef CAS PubMed.
- J. J. Iliff, M. Wang, Y. Liao, B. A. Plogg, W. Peng, G. A. Gundersen, H. Benveniste, G. E. Vates, R. Deane, S. A. Goldman, E. A. Nagelhus and M. Nedergaard, Sci. Transl. Med., 2012, 4, 147ra111 Search PubMed.
- N. A. Jessen, A. S. F. Munk, I. Lundgaard and M. Nedergaard, Neurochem. Res., 2015, 40, 2583–2599 CrossRef CAS PubMed.
- A. Louveau, I. Smirnov, T. J. Keyes, J. D. Eccles, S. J. Rouhani, J. D. Peske, N. C. Derecki, D. Castle, J. W. Mandell, K. S. Lee, T. H. Harris and J. Kipnis, Nature, 2015, 523, 337–341 CrossRef CAS PubMed.
- A. Aspelund, S. Antila, S. T. Proulx, T. V. Karlsen, S. Karaman, M. Detmar, H. Wiig and K. Alitalo, J. Exp. Med., 2015, 212, 991–999 CrossRef CAS PubMed.
- M. Nedergaard and S. A. Goldman, Science, 2020, 370, 50–56 CrossRef CAS PubMed.
- T. J. Lohela, T. O. Lilius and M. Nedergaard, Nat. Rev. Drug Discovery, 2022, 21, 763–779 CrossRef CAS PubMed.
- H. Benveniste, H. Lee, F. Ding, Q. Sun, E. Al-Bizri, R. Makaryus, S. Probst, M. Nedergaard, E. A. Stein and H. Lu, Anesthesiology, 2017, 127, 976–988 CrossRef CAS PubMed.
- A. Verkhratsky and M. Nedergaard, Physiol. Rev., 2018, 98, 239–389 CrossRef CAS PubMed.
- T. M. Mathiisen, K. P. Lehre, N. C. Danbolt and O. P. Ottersen, Glia, 2010, 58, 1094–1103 CrossRef PubMed.
- M.-J. Hannocks, M. E. Pizzo, J. Huppert, T. Deshpande, N. J. Abbott, R. G. Thorne and L. Sorokin, J. Cereb. Blood Flow Metab., 2018, 38, 669–686 CrossRef CAS PubMed.
- S. B. Hladky and M. A. Barrand, Fluids Barriers CNS, 2014, 11, 26 CrossRef PubMed.
- M. Nuriya, T. Shinotsuka and M. Yasui, Cereb. Cortex, 2013, 23, 2118–2126 CrossRef PubMed.
- S. Michinaga and Y. Koyama, Int. J. Mol. Sci., 2019, 20, 571 CrossRef CAS PubMed.
- N. Korogod, C. C. Petersen and G. W. Knott, eLife, 2015, 4, e05793 CrossRef PubMed.
- A. W. J. Morris, R. O. Carare, S. Schreiber and C. A. Hawkes, Front. Aging Neurosci., 2014, 6, 251 CAS.
- R. Hallmann, N. Horn, M. Selg, O. Wendler, F. Pausch and L. M. Sorokin, Physiol. Rev., 2005, 85, 979–1000 CrossRef CAS PubMed.
- N. J. Albargothy, D. A. Johnston, M. MacGregor-Sharp, R. O. Weller, A. Verma, C. A. Hawkes and R. O. Carare, Acta Neuropathol., 2018, 136, 139–152 CrossRef CAS PubMed.
- G. A. Gundersen, G. F. Vindedal, Ø. Skare and E. A. Nagelhus, Brain Struct. Funct., 2014, 219, 2181–2186 CrossRef CAS PubMed.
- V. A. Eidsvaag, R. Enger, H.-A. Hansson, P. K. Eide and E. A. Nagelhus, Glia, 2017, 65, 964–973 CrossRef PubMed.
- M. W. van der Helm, A. D. van der Meer, J. C. T. Eijkel, A. van den Berg and L. I. Segerink, Tissue Barriers, 2016, 4, e1142493 CrossRef PubMed.
- C. F. Ferris, Front. Neurosci., 2022, 16, 854377 CrossRef PubMed.
- D. Le Bihan, R. Turner, P. Douek and N. Patronas, AJR, Am. J. Roentgenol., 1992, 159, 591–599 CrossRef CAS PubMed.
- J. H. Burdette, D. D. Durden, A. D. Elster and Y. F. Yen, J. Comput. Assist. Tomogr., 2001, 25, 515–519 CrossRef CAS PubMed.
- R. Mills, J. Phys. Chem., 1973, 77, 685–688 CrossRef CAS.
- M. Holz, S. R. Heil and A. Sacco, Phys. Chem. Chem. Phys., 2000, 2, 4740–4742 RSC.
- L. M. Valnes, S. K. Mitusch, G. Ringstad, P. K. Eide, S. W. Funke and K.-A. Mardal, Sci. Rep., 2020, 10, 9176 CrossRef CAS PubMed.
- C.-H. Wu, J.-F. Lirng, Y.-H. Ling, Y.-F. Wang, H.-M. Wu, J.-L. Fuh, P.-C. Lin, S.-J. Wang and S.-P. Chen, Ann. Neurol., 2021, 89, 111–124 CrossRef CAS PubMed.
- P. K. Eide, S. A. S. Vatnehol, K. E. Emblem and G. Ringstad, Sci. Rep., 2018, 8, 7194 CrossRef PubMed.
- R. G. Thorne and C. Nicholson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5567–5572 CrossRef CAS PubMed.
- H. Hirase, L. Qian, P. Barthó and G. Buzsáki, PLoS Biol., 2004, 2, e96 CrossRef PubMed.
- M. T. Oberdier, J. F. Antaki, A. Kharlamov and S. C. Jones, Anim. Models Exp. Med., 2021, 4, 391–397 CrossRef CAS PubMed.
- H. Mestre, Y. Mori and M. Nedergaard, Trends Neurosci., 2020, 43, 458–466 CrossRef CAS PubMed.
- B. Bedussi, M. Almasian, J. de Vos, E. VanBavel and E. N. Bakker, J. Cereb. Blood Flow Metab., 2018, 38, 719–726 CrossRef PubMed.
- M. Moazen, A. Alazmani, K. Rafferty, Z.-J. Liu, J. Gustafson, M. L. Cunningham, M. J. Fagan and S. W. Herring, J. Biomech., 2016, 49, 123–126 CrossRef PubMed.
- S.-J. Guild, F. D. McBryde and S. C. Malpas, J. Appl. Physiol., 2015, 119, 576–581 CrossRef PubMed.
- O. Demeulenaere, A. Bertolo, S. Pezet, N. Ialy-Radio, B. Osmanski, C. Papadacci, M. Tanter, T. Deffieux and M. Pernot, EBioMedicine, 2022, 79, 103995 CrossRef PubMed.
- L. Xie, H. Kang, Q. Xu, M. J. Chen, Y. Liao, M. Thiyagarajan, J. O'Donnell, D. J. Christensen, C. Nicholson, J. J. Iliff, T. Takano, R. Deane and M. Nedergaard, Science, 2013, 342, 373–377 CrossRef CAS PubMed.
- P. G. Haydon, Curr. Opin. Neurobiol., 2017, 44, 28–33 CrossRef CAS PubMed.
- A. R. Mendelsohn and J. W. Larrick, Rejuvenation Res., 2013, 16, 518–523 CrossRef CAS PubMed.
- Ş. B. Demiral, D. Tomasi, J. Sarlls, H. Lee, C. E. Wiers, A. Zehra, T. Srivastava, K. Ke, E. Shokri-Kojori, C. R. Freeman, E. Lindgren, V. Ramirez, G. Miller, P. Bandettini, S. Horovitz, G.-J. Wang, H. Benveniste and N. D. Volkow, NeuroImage, 2019, 185, 263–273 CrossRef PubMed.
- J. J. Iliff, M. Wang, D. M. Zeppenfeld, A. Venkataraman, B. A. Plog, Y. Liao, R. Deane and M. Nedergaard, J. Neurosci., 2013, 33, 18190–18199 CrossRef CAS PubMed.
- V. Kiviniemi, X. Wang, V. Korhonen, T. Keinänen, T. Tuovinen, J. Autio, P. LeVan, S. Keilholz, Y.-F. Zang, J. Hennig and M. Nedergaard, J. Cereb. Blood Flow Metab., 2016, 36, 1033–1045 CrossRef CAS PubMed.
- M. Causemann, V. Vinje and M. E. Rognes, Fluids Barriers CNS, 2022, 19, 1–17 CrossRef PubMed.
- O. Balédent, in Adult Hydrocephalus, ed. D. Rigamonti, Cambridge University Press, Cambridge, 2014, pp. 121–138 Search PubMed.
- B. C. Reeves, J. K. Karimy, A. J. Kundishora, H. Mestre, H. M. Cerci, C. Matouk, S. L. Alper, I. Lundgaard, M. Nedergaard and K. T. Kahle, Trends Mol. Med., 2020, 26, 285–295 CrossRef CAS PubMed.
- F. Zhao, J. Deng, X. Xu, F. Cao, K. Lu, D. Li, X. Cheng, X. Wang and Y. Zhao, J. Neuroinflammation, 2018, 15, 157 CrossRef PubMed.
- Y. Ohene, I. F. Harrison, P. Nahavandi, O. Ismail, E. V. Bird, O. P. Ottersen, E. A. Nagelhus, D. L. Thomas, M. F. Lythgoe and J. A. Wells, NeuroImage, 2019, 188, 515–523 CrossRef PubMed.
- G. T. Manley, M. Fujimura, T. Ma, N. Noshita, F. Filiz, A. W. Bollen, P. Chan and A. S. Verkman, Nat. Med., 2000, 6, 159–163 CrossRef CAS PubMed.
- Z. Xu, N. Xiao, Y. Chen, H. Huang, C. Marshall, J. Gao, Z. Cai, T. Wu, G. Hu and M. Xiao, Mol. Neurodegener., 2015, 10, 58 CrossRef PubMed.
- Y. Huang, J. C. Williams and S. M. Johnson, Lab Chip, 2012, 12, 2103–2117 RSC.
- C. Rae and V. J. Balcar, in Brain Energy Metabolism, ed. J. Hirrlinger and H. S. Waagepetersen, Springer, New York, NY, 2014, pp. 217–241 Search PubMed.
- P. Herreros, S. Tapia-González, L. Sánchez-Olivares, M. F. Laguna Heras and M. Holgado, Int. J. Mol. Sci., 2022, 23, 2549 CrossRef CAS PubMed.
- S. Bang, S. Jeong, N. Choi and H. N. Kim, Biomicrofluidics, 2019, 13, 051301 CrossRef PubMed.
- H. E. D'Arceuil, S. Westmoreland and A. J. de Crespigny, NeuroImage, 2007, 35, 553–565 CrossRef PubMed.
- P. E. Thelwall, T. M. Shepherd, G. J. Stanisz and S. J. Blackband, Magn. Reson. Med., 2006, 56, 282–289 CrossRef PubMed.
- M. Nuriya and M. Yasui, J. Neurosci., 2013, 33, 3692–3698 CrossRef CAS PubMed.
- C. Giaume, A. Koulakoff, L. Roux, D. Holcman and N. Rouach, Nat. Rev. Neurosci., 2010, 11, 87–99 CrossRef CAS PubMed.
- P. Ezan, P. André, S. Cisternino, B. Saubaméa, A.-C. Boulay, S. Doutremer, M.-A. Thomas, N. Quenech'du, C. Giaume and M. Cohen-Salmon, J. Cereb. Blood Flow Metab., 2012, 32, 1457–1467 CrossRef CAS PubMed.
- D. M. Zeppenfeld, M. Simon, J. D. Haswell, D. D'Abreo, C. Murchison, J. F. Quinn, M. R. Grafe, R. L. Woltjer, J. Kaye and J. J. Iliff, JAMA Neurol., 2017, 74, 91–99 CrossRef PubMed.
- S. Saadoun, M. J. Tait, A. Reza, D. C. Davies, B. A. Bell, A. S. Verkman and M. C. Papadopoulos, Neuroscience, 2009, 161, 764–772 CrossRef CAS PubMed.
- M. C. Papadopoulos, G. T. Manley, S. Krishna and A. S. Verkman, FASEB J., 2004, 18, 1291–1293 CrossRef CAS PubMed.
- G. T. Manley, M. Fujimura, T. Ma, N. Noshita, F. Filiz, A. W. Bollen, P. Chan and A. S. Verkman, Nat. Med., 2000, 6, 159–163 CrossRef CAS PubMed.
- J. A. Kim, H. N. Kim, S.-K. Im, S. Chung, J. Y. Kang and N. Choi, Biomicrofluidics, 2015, 9, 024115 CrossRef PubMed.
- L. A. Low, C. Mummery, B. R. Berridge, C. P. Austin and D. A. Tagle, Nat. Rev. Drug Discovery, 2021, 20, 345–361 CrossRef CAS PubMed.
- S. N. Bhatia and D. E. Ingber, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- R. Booth and H. Kim, Lab Chip, 2012, 12, 1784–1792 RSC.
- N. J. Abbott, D. E. M. Dolman, S. R. Yusof and A. Reichel, in Drug Delivery to the Brain: Physiological Concepts, Methodologies and Approaches, ed. M. Hammarlund-Udenaes, E. C. M. de Lange and R. G. Thorne, Springer, New York, NY, 2014, pp. 163–197 Search PubMed.
- T. Qian, S. E. Maguire, S. G. Canfield, X. Bao, W. R. Olson, E. V. Shusta and S. P. Palecek, Sci. Adv., 2017, 3, e1701679 CrossRef PubMed.
- X. Tian, O. Brookes and G. Battaglia, Sci. Rep., 2017, 7, 39676 CrossRef CAS PubMed.
- H. Cho, J. H. Seo, K. H. K. Wong, Y. Terasaki, J. Park, K. Bong, K. Arai, E. H. Lo and D. Irimia, Sci. Rep., 2015, 5, 15222 CrossRef CAS PubMed.
- G. Adriani, D. Ma, A. Pavesi, R. D. Kamm and E. L. K. Goh, Lab Chip, 2017, 17, 448–459 RSC.
- A. Wolff, M. Antfolk, B. Brodin and M. Tenje, J. Pharm. Sci., 2015, 104, 2727–2746 CrossRef CAS PubMed.
- M. E. Katt and E. V. Shusta, Curr. Opin. Chem. Eng., 2020, 30, 42–52 CrossRef PubMed.
- D. Baptista, L. Teixeira, C. van Blitterswijk, S. Giselbrecht and R. Truckenmüller, Trends Biotechnol., 2019, 37, 838–854 CrossRef CAS PubMed.
- R. Habibey, J. E. Rojo Arias, J. Striebel and V. Busskamp, Chem. Rev., 2022, 122, 14842–14880 CrossRef CAS PubMed.
- M. McRae, L. M. LaFratta, B. M. Nguyen, J. J. Paris, K. F. Hauser and D. E. Conway, Tissue Barriers, 2018, 6, e1405774 CrossRef PubMed.
- A. García-Salvador, A. Domínguez-Monedero, P. Gómez-Fernández, A. García-Bilbao, S. Carregal-Romero, J. Castilla and F. Goñi-de-Cerio, ATLA, Altern. Lab. Anim., 2020, 48, 184–200 CrossRef PubMed.
- L. M. Griep, F. Wolbers, B. de Wagenaar, P. M. ter Braak, B. B. Weksler, I. A. Romero, P. O. Couraud, I. Vermes, A. D. van der Meer and A. van den Berg, Biomed. Microdevices, 2013, 15, 145–150 CrossRef CAS PubMed.
- A. Herland, A. D. van der Meer, E. A. FitzGerald, T.-E. Park, J. J. F. Sleeboom and D. E. Ingber, PLoS One, 2016, 11, e0150360 CrossRef PubMed.
- E. Vandenhaute, E. Sevin, D. Hallier-Vanuxeem, M.-P. Dehouck and R. Cecchelli, Drug Discovery Today, 2012, 17, 285–290 CrossRef PubMed.
- M. Culot, S. Lundquist, D. Vanuxeem, S. Nion, C. Landry, Y. Delplace, M.-P. Dehouck, V. Berezowski, L. Fenart and R. Cecchelli, Toxicol. in Vitro, 2008, 22, 799–811 CrossRef CAS PubMed.
- S. I. Ahn, Y. J. Sei, H.-J. Park, J. Kim, Y. Ryu, J. J. Choi, H.-J. Sung, T. J. MacDonald, A. I. Levey and Y. Kim, Nat. Commun., 2020, 11, 175 CrossRef CAS PubMed.
- J.-K. Yoon, J. Kim, Z. Shah, A. Awasthi, A. Mahajan and Y. Kim, Adv. Healthcare Mater., 2021, 10, 2002285 CrossRef CAS PubMed.
- E. Ruoslahti, Glycobiology, 1996, 6, 489–492 CrossRef CAS PubMed.
- A. L. Placone, P. M. McGuiggan, D. E. Bergles, H. Guerrero-Cazares, A. Quiñones-Hinojosa and P. C. Searson, Biomaterials, 2015, 42, 134–143 CrossRef CAS PubMed.
- P. A. Soden, A. R. Henderson and E. Lee, Adv. Biol., 2022, 6, 2200027 CrossRef CAS PubMed.
- J. Wan, S. Zhou, H. J. Mea, Y. Guo, H. Ku and B. M. Urbina, Chem. Rev., 2022, 122, 7142–7181 CrossRef CAS PubMed.
- M. Coloma, J. D. Schaffer, P. Huang and P. R. Chiarot, Biomicrofluidics, 2019, 13, 024103 CrossRef PubMed.
- U. Blache, E. M. Ford, B. Ha, L. Rijns, O. Chaudhuri, P. Y. W. Dankers, A. M. Kloxin, J. G. Snedeker and E. Gentleman, Nat. Rev. Methods Primers, 2022, 2, 1–22 CrossRef.
- A. D. Martinac and L. E. Bilston, Biomech. Model. Mechanobiol., 2020, 19, 781–800 CrossRef PubMed.
- D. H. Kelley and J. H. Thomas, Annu. Rev. Fluid Mech., 2023, 55, 237–264 CrossRef.
- R. T. Kedarasetti, P. J. Drew and F. Costanzo, Fluids Barriers CNS, 2022, 19, 34 CrossRef PubMed.
- V. Vinje, E. N. T. P. Bakker and M. E. Rognes, Sci. Rep., 2021, 11, 16085 CrossRef CAS PubMed.
- C. Daversin-Catty, I. G. Gjerde and M. E. Rognes, Front. Phys., 2022, 10, 882260 CrossRef.
- M. Asgari, D. de Zélicourt and V. Kurtcuoglu, Sci. Rep., 2015, 5, 15024 CrossRef CAS PubMed.
- J. Rey and M. Sarntinoranont, Fluids Barriers CNS, 2018, 15, 20 CrossRef PubMed.
- B.-J. Jin, A. J. Smith and A. S. Verkman, J. Gen. Physiol., 2016, 148, 489–501 CrossRef CAS PubMed.
- K. E. Holter, B. Kehlet, A. Devor, T. J. Sejnowski, A. M. Dale, S. W. Omholt, O. P. Ottersen, E. A. Nagelhus, K.-A. Mardal and K. H. Pettersen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9894–9899 CrossRef CAS PubMed.
- A. K. Diem, R. O. Carare, R. O. Weller and N. W. Bressloff, PLoS One, 2018, 13, e0205276 CrossRef PubMed.
- M. Asgari, D. de Zélicourt and V. Kurtcuoglu, Sci. Rep., 2016, 6, 38635 CrossRef CAS PubMed.
- C. A. Hawkes, N. Jayakody, D. A. Johnston, I. Bechmann and R. O. Carare, Brain Pathol., 2014, 24, 396–403 CrossRef CAS PubMed.
- D. K. Binder, M. C. Papadopoulos, P. M. Haggie and A. S. Verkman, J. Neurosci., 2004, 24, 8049–8056 CrossRef CAS PubMed.
- X. Yao, S. Hrabětová, C. Nicholson and G. T. Manley, J. Neurosci., 2008, 28, 5460–5464 CrossRef CAS PubMed.
- E. A. Nagelhus and O. P. Ottersen, Physiol. Rev., 2013, 93, 1543–1562 CrossRef CAS PubMed.
- S. Y. Huang, T. Witzel, B. Keil, A. Scholz, M. Davids, P. Dietz, E. Rummert, R. Ramb, J. E. Kirsch, A. Yendiki, Q. Fan, Q. Tian, G. Ramos-Llordén, H.-H. Lee, A. Nummenmaa, B. Bilgic, K. Setsompop, F. Wang, A. V. Avram, M. Komlosh, D. Benjamini, K. N. Magdoom, S. Pathak, W. Schneider, D. S. Novikov, E. Fieremans, S. Tounekti, C. Mekkaoui, J. Augustinack, D. Berger, A. Shapson-Coe, J. Lichtman, P. J. Basser, L. L. Wald and B. R. Rosen, NeuroImage, 2021, 243, 118530 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |