Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Personalized tissue-engineered veins – long term safety, functionality and cellular transcriptome analysis in large animals†
Klas
Österberg
a,
Yalda
Bogestål
b,
Lachmi
Jenndahl
 c,
Tobias
Gustafsson-Hedberg
c,
Jane
Synnergren
de,
Gustav
Holmgren
d,
Eva
Bom
b,
Sarunas
Petronis
c,
Tobias
Gustafsson-Hedberg
c,
Jane
Synnergren
de,
Gustav
Holmgren
d,
Eva
Bom
b,
Sarunas
Petronis
 b,
Annika
Krona
f,
Jonna S.
Eriksson
g,
Jennifer
Rosendahl
b,
Annika
Krona
f,
Jonna S.
Eriksson
g,
Jennifer
Rosendahl
 b,
Veronica
Crisostomo
b,
Veronica
Crisostomo
 hij,
Francisco M.
Sanchez-Margallo
hij,
Claudia
Baez-Diaz
hij,
Raimund
Strehl
c and
Joakim
Håkansson
hij,
Francisco M.
Sanchez-Margallo
hij,
Claudia
Baez-Diaz
hij,
Raimund
Strehl
c and
Joakim
Håkansson
 *bk
*bk
aSahlgrenska Academy, Institution of Medicine, Wallenberg Laboratory, Department of Molecular and Clinical Medicine, Blå Stråket 5 B, 41345 Gothenburg, Sweden
bRISE Research Institutes of Sweden, Methodology, Textile and Medical Device, Brinellgatan 4, 50462 Borås, Sweden. E-mail: Joakim.hakansson@ri.se; Tel: +46 702172197
cVERIGRAFT AB, Arvid Wallgrensbacke 20, 41346, Göteborg, Sweden
dSystems Biology Research Center, School of Bioscience, University of Skövde, 54128 Skövde, Sweden
eGothenburg University, Department of Molecular and Clinical Medicine, Institute of Medicine, Sahlgrenska Academy, 41345 Gothenburg, Sweden
fRISE Research Institutes of Sweden, Agriculture and Food, Box 5401, 40229 Gothenburg, Sweden
gTATAA Biocenter AB, Gothenburg, Sweden
hJesús Usón Minimally Invasive Surgery Centre, Cáceres 10004, Spain
iCIBER de Enfermedades Cardiovasculares, CIBER CV, Madrid 28029, Spain
jRICORS-TERAV Network, ISCIII, Madrid 28029, Spain
kGothenburg University, Department of Laboratory Medicine, Institute of Biomedicine, Gothenburg, Sweden
First published on 13th April 2023
Abstract
Tissue engineering is a promising methodology to produce advanced therapy medicinal products (ATMPs). We have developed personalized tissue engineered veins (P-TEV) as an alternative to autologous or synthetic vascular grafts utilized in reconstructive vein surgery. Our hypothesis is that individualization through reconditioning of a decellularized allogenic graft with autologous blood will prime the tissue for efficient recellularization, protect the graft from thrombosis, and decrease the risk of rejection. In this study, P-TEVs were transplanted to vena cava in pig, and the analysis of three veins after six months, six veins after 12 months and one vein after 14 months showed that all P-TEVs were fully patent, and the tissue was well recellularized and revascularized. To confirm that the ATMP product had the expected characteristics one year after transplantation, gene expression profiling of cells from P-TEV and native vena cava were analyzed and compared by qPCR and sequencing. The qPCR and bioinformatics analysis confirmed that the cells from the P-TEV were highly similar to the native cells, and we therefore conclude that P-TEV is functional and safe in large animals and have high potential for use as a clinical transplant graft.
Introduction
Cardiovascular diseases cause high economical burdens on global healthcare systems as one of the leading causes of mortality and morbidity worldwide.1 Due to large efforts in research and development in the last decades, new treatments have improved the prognosis for arterial diseases.2 On the contrary, progress of treatments for venous diseases has been limited during the same period.3 The vast majority of venous pathology is of thrombogenic origin and is almost exclusively treated pharmacologically.4 But for some patients, the need for surgical reconstruction to restore the venous anatomy and function is of high priority to avoid severe clinical consequences.5 Efforts have been made to develop surgical methods to overcome long term symptoms after deep vein thrombosis, but results are discouraging due to graft failure.6 In present time, vascular graft procedures for venous pathology are mostly restricted to restore blood flow after traumatic injuries or extensive tumor resections.7,8 Autologous veins or synthetic grafts are mainly used, but conduits of different degree of tissue engineering have also been tried. Different graft substitutes, including cryopreserved or decellularized allogenic veins, vein conduits constructed of bovine pericardium or PTFE grafts have been evaluated, e.g., for chronic venous obstruction or deep vein insufficiency, but all have had issues with long-term patency.9–15We have recently, in a short-term in vivo pig safety study of vena cava transplantation shown that biological individualized vascular vein graft has potential to be a viable option for patients in need of restoration of crucial venous function.16 This technique involves decellularization of native blood vessels from human or animal donors followed by reconditioning with autologous peripheral blood to generate individualized tissue-engineered vascular grafts. Decellularization is conducted through chemical, enzymatical and/or physical methods to eliminate the donor cells from the extracellular matrix (ECM) scaffold.17 We believe that the use of native ECM structures has great potential for cell attachment, migration and proliferation, and our studies show efficient recellularization in vivo of both vein and artery grafts.16,18 Preservation of functional organs by perfusion with whole blood or blood-derived solutions as preparation for transplantation has been shown useful for maintaining the organ physiology and functionality.19–21 The ambition with this treatment is to prime the tissue for efficient recellularization in vivo, protect from exposure of the collagen to prevent thrombosis and personalize the tissue to thereby preclude rejection after transplantation.
VERIGRAFT is developing clinical-grade grafts under the trade name personalized tissue-engineered vein (P-TEV). The P-TEV protocols of decellularization and reconditioning have been used to produce porcine individualized tissue-engineered grafts for preclinical testing (referred to as P-TEV in this text).
To proceed from our previous successful short-term safety study of P-TEV in an in vivo model of vena cava transplantation, we here implanted P-TEV in pigs and analyzed their functionality and characteristics after up to one year in vivo. The study was conducted at two facilities in two different countries: one six months study in a facility classified to the good laboratory practice (GLP) quality system in Spain, and one one-year study in Sweden. The results showed full patency of all grafts throughout the whole study and that the characteristics of the cells repopulating the grafts were very similar with cells in native vein tissue.
Materials and methods
Preparation of P-TEV
Vena cava, a section between vena renalis and the bifurcation to vena iliaca communis, was excised from 12–16 weeks old female pigs (mixed breed of Yorkshire, Hampshire and Swedish Pigham) (bodyweight 45–69 kg) without any history of pregnancy, used in other research studies after euthanization, so no additional ethical permission was required. The blood vessel was dissected free from surrounding tissues and stored in ice-cold PBS and frozen at −80 °C. For decellularization, veins were thawed and decellularized using protocols previously described37,47 with or without perfusion. Vessels were agitated at 115 rpm, 37 °C for 24 h in 1% Triton X (Merck Millipore), followed by 8 h in 1% tri-n-butyl phosphate (TnBP, Merck Millipore) and 16 h in 40 U mL−1 DNase (Thermo Fisher Scientific), all containing 0.5% antibiotic-antimycotic (AA, Thermo Fisher Scientific). Between each step, the blood vessel was washed in H2O and the whole procedure was repeated. After two cycles, the vein segments were washed in 5 mM EDTA (Merck Millipore) for 48 h followed by PBS for 24 h. After decellularization, the vein segments were removed from the bioreactor, biopsies were taken for DNA quantification, histology, and immunohistochemistry before sterilization with peracetic acid followed by five washes in PBS during 24 h under sterile conditions. Sterilized decellularized vessels were frozen at −80 °C or used directly for reconditioning.From each pig to be transplanted with P-TEV, peripheral whole blood was collected in heparin vacutainers (BD) one week pre-surgery under sedation with tiletamine/zolazepam (Zoletil 50 mg ml−1 + 50 mg ml−1, 0.06 ml kg−1) and dexmedetomidin (Dexdomitor 2.5 mg). 25 mL blood was mixed immediately with 25 mL STEEN Solution (XVIVO Perfusion), 0.5% antibiotic-antimycotic mix (Thermo Fisher Scientific), 80 ng ml−1 VEGF (Cellgenics) and 10 ng ml−1 FGF (R&D Systems). 5 μg ml−1 acetylsalicylic acid (Sigma-Aldrich) was added to terminally inhibit all contained thrombocytes. The complete blood solution was added to the decellularized blood vessel inside a reconditioning bioreactor, which allowed circulation of the blood solution through and around the vessel in a vertical position at 2 mL min−1. This process was performed in a laminar flow hood at room temperature for seven days and the glucose level was measured with a Contour XT glucose meter (Bayer) and kept at 3–8 mM and adjusted with glucose solution (Life Technologies, USA) when needed. After perfusion, the graft was harvested, rinsed, biopsied for DNA quantification and histology and kept in PBS + AA until use. During blood procurement and production, a system was used to allow traceability of the autologous pig blood and the resulting graft to allow administration in an individualized, strictly autologous manner.
Transportation
This study was conducted at two facilities: one in Spain and one in Sweden. In Sweden, the animal facility was next door with the lab producing the P-TEV. The reconditioning process was started within 5 h from blood withdrawal and the time between harvest from reconditioning process and start of operation reached a maximum of two hours. For the study conducted in Spain, blood from the pigs was transported from Caceres to Gothenburg at 2–8 °C (DHL Same Day) and the reconditioning process was started in 36 h from blood withdrawal. The reconditioned vein segments were transported in 0.9% NaCl (B. Braun) from Gothenburg to Caceres at 2–8 °C transport temperature (DHL Same Day). The vein segments were transplanted to the pigs within 72 h from the time of ending the reconditioning process. Temperature was logged during the whole transport process.Pig in vivo model for vena cava transplantation of P-TEV
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Sweden and Spain according to the Directive 2010/63/EU of the European Parliament and have been approve by the local ethics committee for animal studies at the administrative court of appeals in Gothenburg, Sweden and Caceres, Spain. The model previously developed for P-TEV transplantation16 was used in this study with the exception that mini pigs were used for the 12 months setup.
Briefly, 7 female minipigs around one year of age (ELLEGAARD, Denmark) (bodyweight 35–40 kg) were used for the setup up to 14 months in Sweden and 1 female and 2 males 4–5 months old pigs (Large white, CRISJEROPRA; EL PAGON° EXP, Spain) (bodyweight 40 kg ± 10% at inclusion) were used for the 6 months GLP setup in Spain. None of the female pigs had any history of pregnancy. The pigs were cared for in accordance with regulations for the protection of laboratory animals and the pigs were housed together before and after surgery. The rooms had bedding consisting of wood shavings and straw, and the pigs were fed twice daily and had free access to water. Acetylsalicylic acid (Trombyl, Pfizer) 160 mg was given orally once daily during the whole study starting six days before surgery and rivaroxaban (Xarelto, Bayer) 2 mg kg−1 was given orally twice daily during the whole study starting the day before surgery. All surgeries were performed under sterile conditions and under isoflurane anesthesia. Anesthesia was induced with intramuscular injections of dexmedetomidine 30 μg kg−1 (Dexdomitor, Orion Pharma Animal Health), tiletamine 3 mg kg−1 and zolazepam 3 mg kg−1 (Zoletil, Virbac). The pigs were intubated and given inhalation anesthesia with isoflurane (Attane vet, VM Pharma) (between 2.5–3.8% to keep 1.3–1.5 minimal alveolar concentration (MAC)). For pain relief, carprofen 4 mg kg−1 (Norocarp vet, N-vet) and buprenorphine 0.03 mg kg−1 (Vetergesic vet, Orion Pharma Animal Health) was given intra venously. An incision was made through linea alba and a retroperitoneal approach was performed to localize vena cava. On the right side, the peritoneum and the abdominal wall were separated down to the vena cava and thereby the intestines were left untouched. The section of vena cava between vena renalis and the bifurcation to vena iliaca communis was dissected free from surrounding tissue. At the time of implantation, heparin 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 IU (Leo Pharma) was administered intra venously before clamping the vein. P-TEV of average length 2.6 ± 0.3 cm and average luminal diameter 6.0 ± 1.2 mm was implanted using 6-0 Prolene (Ethicon) as end-to-end anastomoses with continuous suture. Detailed information of luminal diameter and length of each graft is noted in ESI Table I.† Metal clips were sutured at the anastomoses to facilitate orientation under X-ray imaging. The abdominal musculature was sutured with Maxon 0 (Ethicon) and the skin was sutured with a Monocryl 3-0 (Ethicon) intradermally. For post-surgery pain relief, carprofen 2 mg kg−1 (Norocarp, N-vet) was given orally twice daily for 3 days. Six of the pigs in Sweden were euthanized after 12 months and one 14 months after surgery. The results of these seven pigs were included in the same group. All 3 pigs in Spain were euthanized 6 months after surgery. At euthanization, angiography was performed on the operated area of vena cava under isoflurane anesthesia (as described above) by injecting contrast fluid (Urografin 76, Bayer) into a femoral vein or vena cava outside the graft. At harvest, the operated segment of vena cava, including adjacent area of native vena cava, was excised from the pigs under anesthesia before euthanization for further analysis. The animals were euthanized by an intravenous overdose of pentobarbital 100 mg kg−1 (Allfatal vet 100 mg ml−1). No extra anticoagulant or antiplatelet drugs were used.
000 IU (Leo Pharma) was administered intra venously before clamping the vein. P-TEV of average length 2.6 ± 0.3 cm and average luminal diameter 6.0 ± 1.2 mm was implanted using 6-0 Prolene (Ethicon) as end-to-end anastomoses with continuous suture. Detailed information of luminal diameter and length of each graft is noted in ESI Table I.† Metal clips were sutured at the anastomoses to facilitate orientation under X-ray imaging. The abdominal musculature was sutured with Maxon 0 (Ethicon) and the skin was sutured with a Monocryl 3-0 (Ethicon) intradermally. For post-surgery pain relief, carprofen 2 mg kg−1 (Norocarp, N-vet) was given orally twice daily for 3 days. Six of the pigs in Sweden were euthanized after 12 months and one 14 months after surgery. The results of these seven pigs were included in the same group. All 3 pigs in Spain were euthanized 6 months after surgery. At euthanization, angiography was performed on the operated area of vena cava under isoflurane anesthesia (as described above) by injecting contrast fluid (Urografin 76, Bayer) into a femoral vein or vena cava outside the graft. At harvest, the operated segment of vena cava, including adjacent area of native vena cava, was excised from the pigs under anesthesia before euthanization for further analysis. The animals were euthanized by an intravenous overdose of pentobarbital 100 mg kg−1 (Allfatal vet 100 mg ml−1). No extra anticoagulant or antiplatelet drugs were used.
DNA quantification
DNA was extracted from 10–25 mg wet tissue with the DNeasy Blood & Tissue kit (Qiagen) and quantified with the Qubit dsDNA HS assay kit (Life Technologies), according to the manufacturer's instructions. DNA concentration was calculated using a supplied DNA standard.Histology
4′,6-Diamidino-2-phenylindole (DAPI) and hematoxylin–eosin (H&E) staining was performed on paraffin embedded cross-sections of 5 μm thickness, following standard protocols for fixation, embedding, rehydration, and staining. Images were acquired using a fluorescence microscope (Zeiss Axiovert 40 CFL) for DAPI visualization, and in bright field for H&E visualization. Quantification of vascular wall thickness was performed using an electronic digital caliper on paraffinized samples from the native donor vessels and corresponding P-TEV after in vivo.Immunohistochemistry
Immunohistochemical staining was performed on paraffin sections by rehydrating samples following standard procedures and performing antigen retrieval by incubating samples for 15 min in a 95 °C water bath in a Tris-EDTA buffer (10 mM Tris base, 1 mM EDTA solution, 0.05% Tween 20, pH 9.0). The sections were blocked with SuperBlock Blocking Buffer (Thermo Scientific) for 1 h and incubated with primary antibodies against CD31 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, Abcam ab28364), CD45 (1
50, Abcam ab28364), CD45 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Abcam ab10558) or alpha smooth muscle actin (αSMA, 1
200, Abcam ab10558) or alpha smooth muscle actin (αSMA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Abcam ab7817) over night, followed by incubation with secondary antibodies, anti-mouse (1
100, Abcam ab7817) over night, followed by incubation with secondary antibodies, anti-mouse (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, A21203, Thermo Fisher) or anti-rabbit (1
200, A21203, Thermo Fisher) or anti-rabbit (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, A21207, Thermo Fisher), conjugated to Alexa Fluor 594 for 1 h. DAPI (Thermo Fisher Scientific) was used as a counterstain. Negative controls were performed without the primary antibody. Quantification of cellular (analyzing DAPI-staining) and capillary (analyzing CD31-staining) density was performed on images using the ImageJ 1.51R software.
200, A21207, Thermo Fisher), conjugated to Alexa Fluor 594 for 1 h. DAPI (Thermo Fisher Scientific) was used as a counterstain. Negative controls were performed without the primary antibody. Quantification of cellular (analyzing DAPI-staining) and capillary (analyzing CD31-staining) density was performed on images using the ImageJ 1.51R software.
Confocal microscopy
Biopsies were de-paraffinized in oven, 60 °C, until the wax had melted before rehydration. Microscopy slides and biopsies were rehydrated in xylene two times 10 min and 5 min as follows: 99% ethanol, 99% ethanol, 96% ethanol, 70% ethanol and ddH2O. Antigen retrieval was performed by heating the samples in Tris EDTA, pH 9, 0.5% Tween at 98 °C for 15 min. They were blocked with 5% BSA (Sigma-Aldrich, Germany) in PBS for 30 min followed by incubation overnight at 4 °C with primary antibody solution. Primary antibodies against αSMA (ab7817 mouse) and CD31 (ab28364 rabbit) were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 and 1
25 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, respectively, in PBS with 0.5% BSA, either one by one (single-labelling) or mixed together (double-labelling). Samples were rinsed 3 × 5 min in PBS and incubated in room temperature for 2 h in darkness with fluorescently labelled secondary antibodies, i.e., Alexa Fluor Plus 488 goat anti-mouse IgG (H + L) (Invitrogen, Carlsbad, USA) and Alexa Fluor 546 Goat anti-Rabbit IgG (H + L) (Invitrogen, Carlsbad, US) for aSMA and CD31 respectively, diluted 1
50, respectively, in PBS with 0.5% BSA, either one by one (single-labelling) or mixed together (double-labelling). Samples were rinsed 3 × 5 min in PBS and incubated in room temperature for 2 h in darkness with fluorescently labelled secondary antibodies, i.e., Alexa Fluor Plus 488 goat anti-mouse IgG (H + L) (Invitrogen, Carlsbad, USA) and Alexa Fluor 546 Goat anti-Rabbit IgG (H + L) (Invitrogen, Carlsbad, US) for aSMA and CD31 respectively, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS with 0.5% BSA. The samples were rinsed 3 × 5 min in PBS and stained with SYTOX™ Deep Red Nucleic Acid Stain (Invitrogen, Carlsbad, USA), washed in PBS and mounted with ProLong Diamond™ Antifade Mountant (Invitrogen, Carlsbad, USA). The samples were examined in a confocal laser scanning microscope (SP5, TSC Leica, Heidelberg, Germany) using a HCX PL APO lambda blue 20.0 × 0.70 IMM UV objective with zoom 1× and 4×. Excitation was performed with a 488 nm argon laser, a 543 nm HeNe laser and a 633 nm HeNe laser. Emission signals were collected at 500–530 nm (shown in green), 572–600 nm (shown in red) and 650–710 nm (shown in blue). Micrographs were acquired with 1024 × 1024 pixels, 8-line average.
100 in PBS with 0.5% BSA. The samples were rinsed 3 × 5 min in PBS and stained with SYTOX™ Deep Red Nucleic Acid Stain (Invitrogen, Carlsbad, USA), washed in PBS and mounted with ProLong Diamond™ Antifade Mountant (Invitrogen, Carlsbad, USA). The samples were examined in a confocal laser scanning microscope (SP5, TSC Leica, Heidelberg, Germany) using a HCX PL APO lambda blue 20.0 × 0.70 IMM UV objective with zoom 1× and 4×. Excitation was performed with a 488 nm argon laser, a 543 nm HeNe laser and a 633 nm HeNe laser. Emission signals were collected at 500–530 nm (shown in green), 572–600 nm (shown in red) and 650–710 nm (shown in blue). Micrographs were acquired with 1024 × 1024 pixels, 8-line average.
Scanning electron microscopy
Tissue samples were rinsed in PBS and submerged in 2.5% glutaraldehyde fixative at room temperature. After 2 hours, the samples were placed in a fresh fixative of the same concentration and left in a fridge at 5 °C for 24 hours. The samples were washed with buffer and submerged in 1% osmium tetroxide for 24 hours at room temperature for secondary fixation. Finally, the samples were rinsed in deionized water, plunge-frozen in liquid propane and freeze-dried overnight in VirTis Sentry 2.0 Benchtop Freeze Dryer (SP Scientific). Before the SEM analysis, samples were coated with 15 nm Au/Pd thin film using Gatan Model 682 PECS sputter-coater to prevent the charging under electron beam. The imaging was performed with a Zeiss Supra 40VP SEM, in secondary electrode image mode. The acceleration voltage and working distance were 4.03 kV and 9–12 mm, respectively.Isolation of RNA, quality control and reverse transcription
For qPCR assay validation, vena cava tissue was collected from pigs, added to RNALater and stored at −20 °C until extraction. RNA was extracted using RNeasy mini kit (Qiagen, cat. no. 74104) followed by a RNase-free DNase I treatment (Qiagen, cat. no. 1023460) according to the manufacturers’ protocols. Tissues were divided into equal pieces (∼20 mg) using a scalpel and placed in separate tubes together with one steel bead. To each tube, 600 μL RLT buffer and 10 μL β-mercaptoethanol were added. The tissues were homogenized using TissueLyser 85210 (Qiagen) with the following setting: 40′′ at 25 Hz and a second round for 60′′ at 25 Hz with the plates inverted. Each sample was eluted twice in separate tubes with 50 μL elution buffer (provided in the kit). RNA quantity and purity was measured using Nanodrop® ND-1000 spectrophotometer (Thermo Fisher Scientific). Only eluates with a OD260/280 ratio between 1.8 and 2.1 were accepted for downstream analysis. cDNA synthesis was performed using TATAA GrandScript cDNA FreePrime kit (TATAA Biocenter, cat. no. AF103b) according to the manufacturer's protocol with a mix of random hexamers and oligo(dT)'s in a final volume of 20 μL (∼500 ng total RNA) using a C1000 (Bio-Rad) cycler. After cDNA synthesis, the samples were diluted 5× with ultrapure distilled water (Invitrogen, cat. no. 10977-035) and stored at −20 °C.Primer and probe design
qPCR assays with hydrolysis probe were designed by TATAA Biocenter, Sweden. All primers were designed using NCBI Primer-Blast48–51 followed by selecting the assay specific probe using Beacon Designer™ 8.21 (PREMIER Biosoft). To avoid amplification of genomic DNA the assays were designed to span over at least one intron (the primers sit on an exon–intron–exon boundary). The assays were also designed to amplify all the isoforms (i.e., the choice of exon–intron–exon placement for primers had to exist for all isoforms). Primers and probes were evaluated in silico using OligoAnalyzer™ tool on IDT (Integrated DNA Technologies) – for Melt temperature, GC%, primer-dimer and hairpin formation – before ordering at IDT. Primers were selected with standard de-salting and as Lab Ready solution of 100 μM. Probes were labelled with FAM as reporter dye and ZEN Iowa Black® FQ as a quencher. The primers were validated using 1× TATAA SYBR® GrandMaster® mix (TATAA Biocenter, cat. no. TA01-625) and a final concentration of primer mix of 400 nM, with the following criteria for passing: r2 ≥ 0.995 on a seven-point standard curve (spanning 2E7-20 copies); efficiency between 80–110%; no amplification of genomic DNA or it must be distinguishable from the target amplification with at least 5 Cq; clean from NTC (no template control), other than primer-dimers (see ESI Table II†). Probes were evaluated in the same way as described for primers except for using 1× TATAA Probe GrandMaster® mix (TATAA Biocenter, cat. no. TA02-625) and a probe concentration of 200 nM.Gene specific pre-amplification
Single cell material from pigs, sorted in a 96-plate, were reverse transcribed into cDNA before gene-specific pre-amplification. Each cell was submerged in 5 μL CelluLyser Micro Lysis buffer (TATAA, #H104) and 2 μL 2× TATAA GrandScript cDNA Supermix (TATAA Biocenter, cat. no. AS103c) and 3 μL ultrapure water (Invitrogen, cat. no. 10977-035) were added to a final volume of 10 μL. The following temperature protocol was used: 25 °C for 5 min, followed by 42 °C for 30 min and 85 °C for 5 min, before hold at 4 °C. The 96-plate with 10 μL cDNA was immediately pre-amplified using 1× TATAA PreAmp GrandMaster® mix (TATAA Biocenter, cat. no. TA05-500), pooled primers with a final concentration of 50 nM of each primer and water for a final volume of 30 μL. The reaction was amplified for 22 cycles on C1000 (Bio-Rad) using the following protocol: 95 °C for 60 s; 22 cycles at 95 °C for 15 s, 60 °C for 120 s and 72 °C for 60 s; cooled using ice within 3 min after completion. A total of 21 assays were multiplexed and tested in this experiment. Pre-amplified cDNA was diluted prior to use in 6.6× water (final dilution in qPCR is 33×) and used immediately or stored at −20 °C until use.qPCR was performed using Bio-Rad CFX 384 system in singlicates for each assay. 10 μL reactions were run using 1× TATAA Probe GrandMaster® mix (TATAA Biocenter, cat. no. TA02-625), premixed 10 μM primers (final concentration of 400 nM), 200 nM probe, and 2 μL of 33× diluted pre-amplified cDNA. The reactions were run under the following thermal conditions: 96 °C for 60 s, 45 cycles at 95 °C for 5 s and 60 °C for 30 s. Data was processed using the CFX Manager 3.1 (Bio-Rad) with baseline subtracted curve fit correction and regression method for Cq determination. Quantitative PCR was performed in agreement with the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines.52
Gene expression data analysis
Data was exported and pre-processes using GenEx (MultiD). Samples and/or genes with >90% missing values were removed, and missing values were filled with column's max +1 resulting in data from 610 cells. The data was related to cells from native vena cava and log2 scale was applied. GenEx software was used to generate Principal Component Analysis (PCA) plots.Dissociation and FACS
Cut pieces from the proximal, central, and distal parts of P-TEV and normal tissue were dissociated in Hank's Balanced Salt Solution (HBSS, Gibco™ #14025092), 10 mM HEPES (Gibco™, #15630056) using 200 U ml−1 Collagenase Type II (Gibco™, #17101015), 37 °C, 100 rpm, for 4 hours. Cell suspensions were filtered through a 70 μm mesh and erythrocytes were lysed using 1× Red Blood cell lysis solution (Miltenyi, #130-094-183) according to the manufacturer's instructions. Cells were stained with Fixed viability Stain 450 (BD Biosciences, #562247, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), anti-pig CD31-PE (Bio-Rad, #MCA1746PE, 1
2000), anti-pig CD31-PE (Bio-Rad, #MCA1746PE, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20), anti-pig CD45-FITC (Bio-Rad, #MCA1222F, 1
20), anti-pig CD45-FITC (Bio-Rad, #MCA1222F, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and sorted on a BD FACS Melody using the ACDU system (Automated Cell Deposition Unit) into 96 well plates containing CelluLyser Micro Lysis buffer (TATAA, #H104) for single cell qRT-PCR.
10) and sorted on a BD FACS Melody using the ACDU system (Automated Cell Deposition Unit) into 96 well plates containing CelluLyser Micro Lysis buffer (TATAA, #H104) for single cell qRT-PCR.
Single-cell transcriptomics analysis
Prior to FACS sorting, a portion of the cell suspensions from graft and native tissue from each donor pig were removed and subject for single-cell full-length transcriptome analysis using SMART-Seq® ICELL8® system from Takara Bio (Takara Bio, #640220). To visualize cell nucleus for cell detection and to identify non-viable cells, cell suspensions were stained with ReadyProbes™ Cell Viability Imaging Kit, Blue/Red (Thermo Fisher Scientific, #R37610) before loading on the ICELL8® system. Cell suspensions from each individual donor pig were analysed using separate ICELL8® kits. Single cell sequencing libraries were prepared according to SMART-Seq® ICELL8® Application Kit User Manual (v.090619). In brief, single cell suspensions were dispensed into nanowells of the ICELL8® chip, followed by selection of wells for subsequent processing using CellSelect software (Takara Bio, v1.5). After dispenses of reagents for cDNA library preparation, the generated single cell cDNA libraries for each kit, were multiplexed together into one sequencing library. Each generated library was amplified and purified before the quality was assessed with Agilent 2100 Bioanalyzer and the High Sensitivity DNA Kit (Agilent, cat. no. 5067-4626), and the quantities were measured with a Qubit Fluorometer, and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, cat. no. Q32851). Each generated library was sequenced paired end 2 × 75 bp on one S4 lane on the NovaSeq 6000 system, using NovaSeq 6000 S4 Reagent Kit v1.5 (Illumina, #20028313).The data analysis was performed using the Seurat R-package for single cell analysis version v4.0.5.53 In total for all donor pigs, 2244 cells (including controls) were sequenced and of these 72 cells were flagged as low quality and removed from the dataset. During preprocessing, the dataset was filtered based on counts, mitochondrial signal and detected genes, and cells with less than 5000 counts and more than 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts were removed from the dataset. Moreover, cells with more than 5% mitochondrial signal, and cells with fewer than 500 or more than 20
000 counts were removed from the dataset. Moreover, cells with more than 5% mitochondrial signal, and cells with fewer than 500 or more than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 genes were filtered from the dataset. The latter is based on the assumption that too many detected genes are most likely the result from doublets of cells instead of single cells.54 To exclude blood derived cells remaining in the tissue, all CD45+ cells were removed. The dataset was then split into CD31+ and CD31− cells and these groups were analyzed separately.
000 genes were filtered from the dataset. The latter is based on the assumption that too many detected genes are most likely the result from doublets of cells instead of single cells.54 To exclude blood derived cells remaining in the tissue, all CD45+ cells were removed. The dataset was then split into CD31+ and CD31− cells and these groups were analyzed separately.
To gain insights into potential sub-cell types present in the CD31+ and CD31− groups, the cells were clustered based on their expression profiles using the top 5 dimensions and resolution 0.4 in Seurat. Dimension reduction was performed with Uniform Manifold Approximation and Projection (UMAP). Differential expression analyses were performed of each cluster compared with all the other clusters, and markers that show distinct expression pattern in one group compared to all the other groups were identified using marker identification methods in Seurat. Heatmaps and dotplots were used to visualize the expression of these cell specific markers and compare their expression in both P-TEV and native samples. Heatmaps were generated based on the top significant biomarkers for each cluster to explore if large variations between clusters were present in the data or if the sorted CD31+ and CD31− populations were rather homogeneous. Dotplots of the three top cluster markers, for each of the five clusters, were used to investigate if the variance for these markers could be related to the treatment of the grafts or if the P-TEV and native samples show similar expression patterns for the 15 most significant cluster markers.
To investigate the similarities between P-TEV and native samples in terms of which genes are expressed, the average expression values (AEV) of genes in all cells in each group (P-TEV and native) were calculated and genes with AEV > −0.1 (normalized and scaled values) across all cells were considered expressed. Venn diagrams were used, applying functions in the ggvenn R-package, to visualize the overlap of expressed genes between P-TEV and native samples. The expression of a selection of typical biomarkers for endothelial cells in P-TEV and native cells are visualized with violin plots for the different cell clusters to explore if transcriptional deviations are present.
Statistical analysis
Statistical analysis of DNA-content was performed with Kruskal Wallis with Dunn's post hoc test, one-way ANOVA with Tukey post hoc test for gene expression data on blood vessel marker genes, Mann–Whitney U-test for cellular and capillary density as well as for vessel wall thickness and with Wilcoxon paired rank-sum test for vascular wall thickness. The software's SPSS or GraphPad Prism were used for calculations. P < 0.05 was considered statistically significant.Results
Preparation of P-TEV
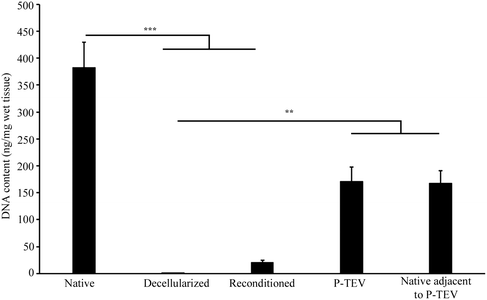
One vein was decellularized and reconditioned for each pig to be transplanted and the process was successful for all 10 animals (7 for the study in Sweden and 3 for the study in Spain).Lymphocyte count in the autologous peripheral whole blood, used for reconditioning, was between 4.0–8.0 × 106 lymphocytes per ml, which is in the lower range of published values from 6 months Gottingen pigs.22 DNA quantification of the decellularized vein showed almost complete removal (99.8%) of DNA from the native vessel, 383 ± 47 ng mg−1 tissue, compared with 0.7 ± 0.1 ng mg−1 tissue after decellularization. The reconditioning increased the DNA content to 20 ± 5 ng mg−1 tissue and the explanted graft, after 12–14 months in vivo, contained 171 ± 26 ng mg−1 wet tissue, which was equal to native vena cava adjacent to the P-TEV after 12–14 months in vivo (168 ± 23 ng mg−1 wet tissue) (Fig. 1).
Transplantation of P-TEV and patency after 12 months
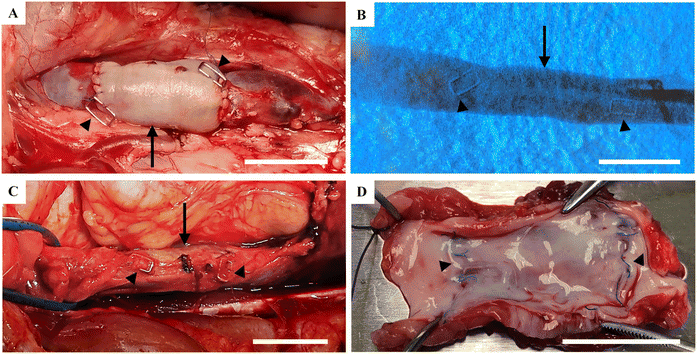
After perfusion of the decellularized veins with blood solution from the respective recipient pig, the reconditioned veins were transplanted to vena cava (Fig. 2A). To show the robustness of the technology, transplantations were performed on two breeds of pig, at two different facilities, in two different countries of which the study in Spain was performed under Good Laboratory Practice (GLP) quality conditions (the 6-months study). All animals had good health during the whole study without any contraindications. Angiography performed on the anaesthetized animals before harvest of the P-TEV, 6 and 12 months after surgery, showed that all grafts were fully patent (Fig. 2B). When examining vena cava macroscopically during dissection and after euthanization, no abnormalities were found on the vein tissue and there were no signs of clotting or thrombosis (Fig. 2C and D).Recellularization of P-TEV in vivo
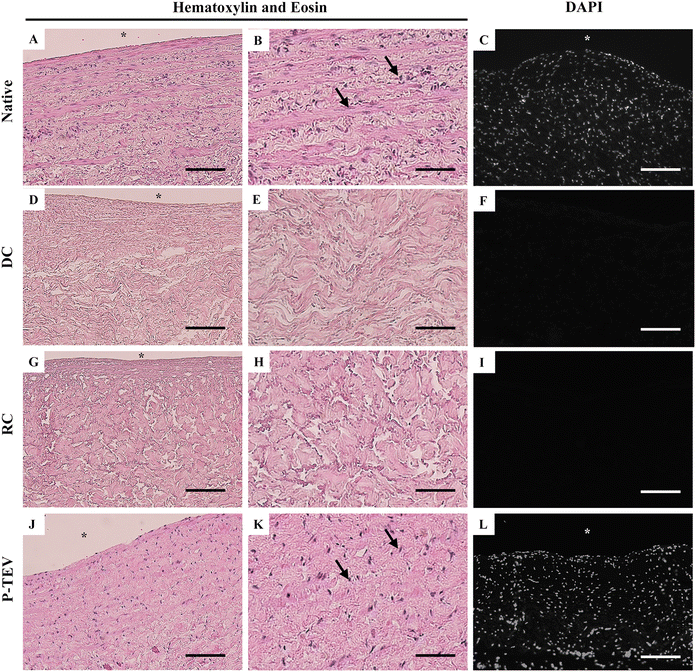
Histological analysis of vena cava shows the native morphology and cell density of the blood vessel tissue (Fig. 3A–C and ESI Fig. 1A†). Decellularization efficiently removed all cells from the tissue, leaving the intact extracellular matrix with no traces of cells (Fig. 3D–F). Reconditioning of the vein did not affect the blood vessel tissue morphology (Fig. 3G–I) and after 12 months in vivo, the vein tissue was recellularized with the same cell density and tissue morphology as the native vein (Fig. 3J–L and ESI Fig. 1B†). Histology analysis of the morphology was performed on proximal, central, and distal parts of the P-TEV as well on several native, decellularized and reconditioned veins with representative images shown in Fig. 3. Quantification of cellular density, on sections stained with DAPI, revealed no statistically significant difference between P-TEV and native tissue one year after transplantation (P-TEV 691 ± 48 cells per area unit (n = 12), native 794 ± 60 cells per area unit, p = 0.16 Mann Whitey U-test). The vascular wall thickness was quantified and compared pairwise between P-TEV and the corresponding native donor vessel, and there was no statistically significant difference between P-TEV and native (Native 1.2 ± 0.09 mm, P-TEV 1.3 ± 0.1 mm, p = 0.67 Wilcoxon paired rank-sum test).Luminal cellular coverage
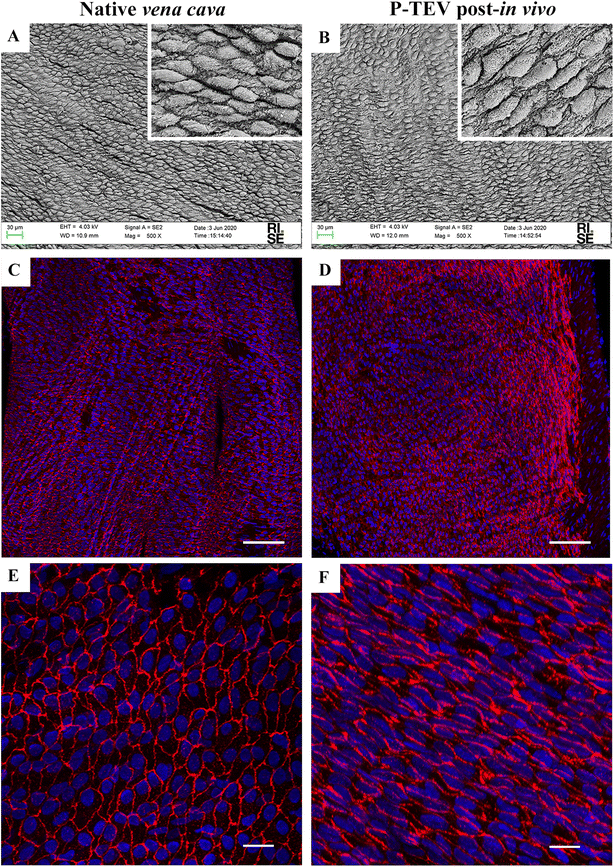
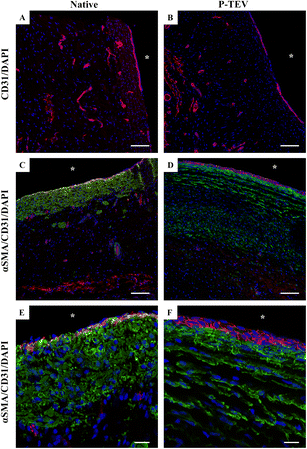
Scanning electron microscopy and immunohistochemistry was used to analyze the characteristics of the luminal surface. The recellularization in vivo had developed an endothelial cell layer covering the whole luminal surface with morphology indistinguishable to the native tissue (Fig. 4A and B). The endothelial cell marker CD31 was clearly expressed in all cells showing that the endothelial cells were tightly aligned to each other, and the expression pattern was equal between native tissue and P-TEV, as illustrated by confocal microscopy with en face images (Fig. 4C–F).Revascularization of the regenerated blood vessel tissue
Using confocal microscopy visualizing CD31 expression, we could clearly see that the blood vessel tissue of the P-TEV was revascularized with capillaries comparable with the native tissue (Fig. 5A and B). Quantitative analysis of capillary density showed no statistically significant difference between P-TEV tissue one year after transplantation and native tissue (percentage of tissue area covered with capillaries: P-TEV 1.0 ± 0.2 (n = 14), native 1.2 ± 0.3 (N = 5), p = 0.55; number of capillaries per area unit: P-TEV 5.1 ± 0.6 (N = 14), native 7.0 ± 1.2 (N = 5), p = 0.14). We could also observe that cells that had repopulated the P-TEV expressed the smooth muscle cell marker alpha smooth muscle actin (αSMA), but with a shifted pattern compared with the native tissue (Fig. 5C–F).The P-TEV's transplanted to the three pigs in the GLP facility, evaluated after 6 months, showed the same recellularization of the vascular tissue (ESI Fig. 2A and B†), even expression of the CD31 marker of endothelial cells (ESI Fig. 2C†) and expression pattern of αSMA (ESI Fig. 2D†) as the P-TEV's evaluated after 12–14 months.
To exclude any rejection reaction, P-TEV tissue was stained with antibodies against the leukocyte marker CD45. The only CD45 positive cells found were in the anastomosis in connection with the non-resorbable sutures, which is expected (data not shown). No damage or other visual sign could be found on the lumen indicating that an inflammatory response was initiated.
Gene expression profile of P-TEV equal to native vena cava after one year in vivo
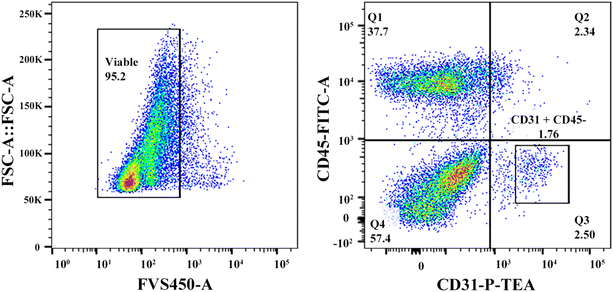
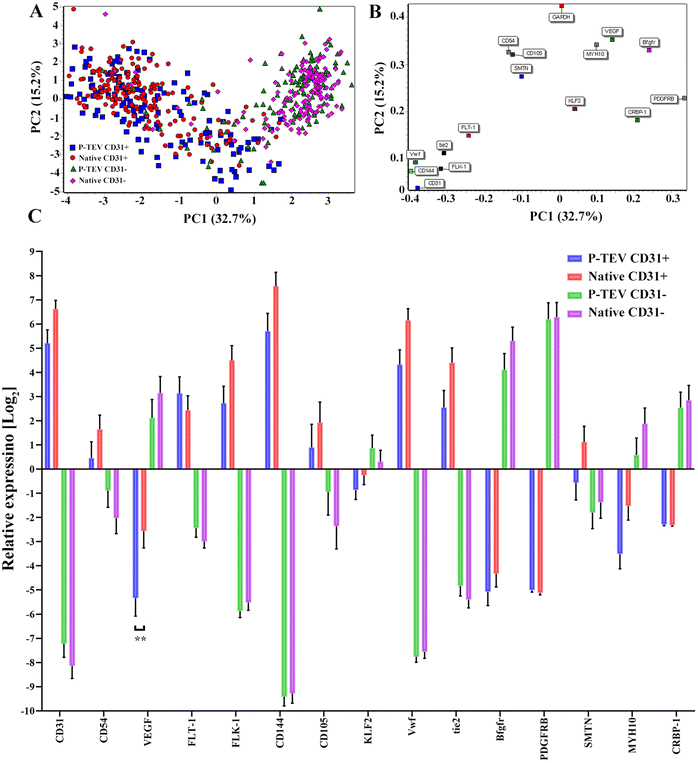
To evaluate if the cells in the regenerated P-TEV had differentiated to mature blood vessel cells and had adopted the expected characteristics, gene expression profiling was performed on P-TEV cells after one year in vivo and compared with cells in native vena cava tissue. Tissue from P-TEV and native vena cava was dissociated to a single cell suspension. Cells to be used for single cell qPCR analysis of a panel of blood vessel markers were FACS-sorted, first to discard the hematopoietic derived CD45-positive cells and then to separate CD31-positive endothelial cells from other blood vessel cells (CD31-negative). The gating used is illustrated in Fig. 6.The panel was chosen to include both highly and lowly expressed genes, in CD31+ and CD31− cells. Genes selected for CD31+ cells were CD31 (PECAM), CD54 (ICAM-1), Flt-1 (VEGFR1), FLK-1 (VEGFR2), CD144 (VE-cadherin), CD105 (Endoglin), vWF and Tie2. Genes chosen for CD31− cells were VEGF, KLF2 (Krüppel-like factor 2), bFGFR, PDGFRB, CEBP-1 and MYH10. A principal component analysis (PCA)-plot of the summarized gene expression from the single cell qPCR clearly illustrates, as expected, that the profile of the CD31+ cells separate from the CD31− cells. Importantly, the P-TEV (graft) cells from respective group overlapped well with cells from the native vena cava tissue. The expression level in native and P-TEV cells was statistically different only for one gene; VEGF in the CD31+ cells, Fig. 7A. The gene load is illustrated in Fig. 7B. Expression levels of the individual genes shows that the graft cells follow the native cells very well for all genes. One difference was for the gene SMTN. The SMTN protein is expressed in smooth muscle cells, and the gene expression level in graft and native CD31− cells was, as expected, very similar. In our analysis, the CD31+ cells also expressed this gene with a difference between the graft and native cells; however, the expression level is relatively low, the absolute difference between the groups is small and there is no statistically significant difference, Fig. 7C.
To perform an even more broad and deeper analysis of the gene expression characterization, in purpose to evaluate the similarities between the cells in the P-TEV and native cells, a portion of the dissociated cells were used for further large-scale single cell RNA sequencing.
Transcriptional characterization of the cells in the P-TEV one year after transplantation and cells in native vena cava
To understand transcriptional similarities and differences between the P-TEV transplants and the native blood vessels, single cell RNA-sequencing was applied. In total 2172 cells from P-TEV and native tissue were sequenced. After filtering on counts the number of cells was reduced to 1713. Furthermore, 834 cells were CD45 positive (hematopoietic cells) and thereby excluded. The remaining 879 cells were divided based on CD31 expression, to separate endothelial cells from other cell types in the blood vessel tissue, for further analyses. The number of cells from each filtering step is shown in ESI Fig. 3.†The expression signals from each group were scaled and normalized using appropriate methods from Seurat, and dimensional reduction was performed using a principal component analysis based on the 2000 most highly variable genes. The first 5 dimensions in the dataset were used for the UMAP visualization and cell type-marker analysis.
Results from the data analysis showed no distinct sub-cell types but more diffuse areas of cell clusters that connect to adjacent cell collections without forming distinct and separated UMAP clusters. Using the top five dimensions and a resolution of 0.4 in Seurat, the clustering identified five clusters (0–4) of cells that showed similar transcriptional profile (Fig. 8A and 9A). To highlight any overrepresentation of native or P-TEV cells in these clusters, they were coloured according to sample types in Fig. 8B and 9B. Overall, the native and P-TEV cells showed an even distribution among the clusters with a few exceptions. For the CD31+ data, an overrepresentation of P-TEV in cluster 0 (marked with an oval in Fig. 8B) was observed. For the CD31− data, there was an overrepresentation of P-TEV in cluster 0 (marked with an oval) and an over-representation of native cells in cluster 2 and 3 (marked with a dashed oval), see Fig. 9B. The heatmaps of the top marker genes for each of the five clusters from Fig. 8A and 9A confirmed that no large variation could be observed even for these top ranked marker genes in the specific clusters as the identified sub clusters showed a similar transcriptional profile for these marker genes (Fig. 8C and 9C). The expression levels of a selection of typical biomarkers for endothelial cells were investigated in the CD31+ data and results are shown in Fig. 8D where a strong signal of PECAM1 was observed in the majority of the cells in all five clusters. FLT1 was mainly expressed by cells in cluster 1, and KDR, TEK and CDH5 showed strong expression in P-TEV cells in cluster 2 and 3. FGFR1 showed strongest expression in cluster 4 for both the P-TEV and native cells. KLF4, ENG and ICAM1 showed to be highest expressed in cluster 2–4 and slightly stronger expression was observed for KLF4 by the native cells, while ENG and ICAM1 were more expressed in the P-TEV cells. VWF showed expression in all clusters except for cluster 4. In the CD31− cells, a strong expression of ICAM1 and FGFR1 were observed for cells in cluster 4, with high similarity between P-TEV and native cells (Fig. 9D). Moreover, KLF4 showed a strong expression in cells in cluster 2–4 and the transcription of this marker gene was highly similar in both the P-TEV and the native cells (Fig. 9D).
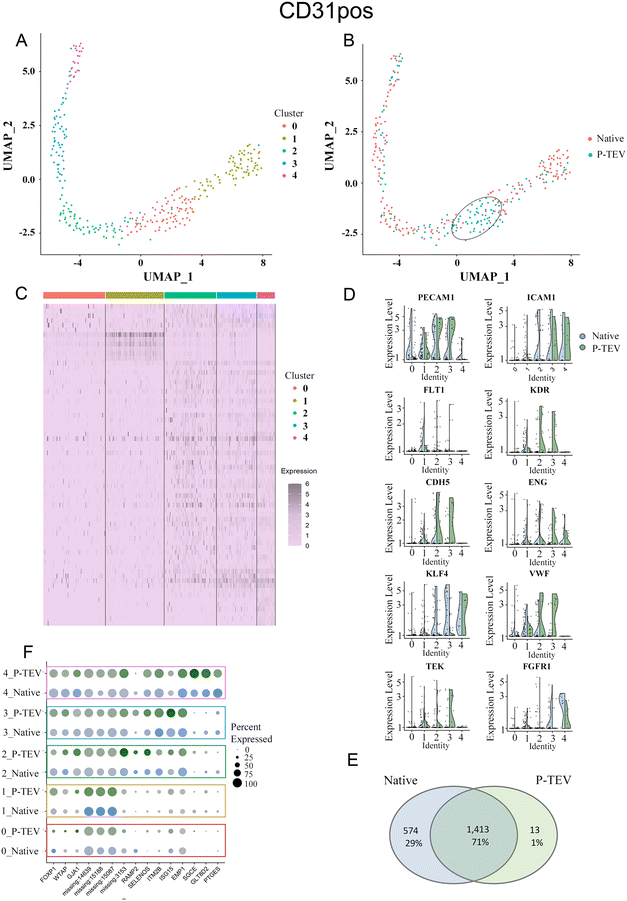 | ||
| Fig. 8 Single-cell RNA sequencing analysis of CD31 positive cells. (A) UMAP clustering of cells into five clusters. (B) Colour coding the UMAP on P-TEV and native cells demonstrated a similar distribution of P-TEV and native cells in the clusters except for cluster 0, marked with an oval, which has an overrepresentation of P-TEV cells. (C) Heatmap of the top markers for each of the five clusters shown in panel (A). Gene names in order from top to bottom are found in ESI Table III.† (D) Violin plots showing the expression of ten endothelial marker genes in the five identified clusters from panel (A). Blue represents native cells and green represents P-TEV cells. (E) Venn diagram showing the overlap of transcriptional activity between P-TEV and native samples. In total 1413 genes (70.7%) were expressed in both native and P-TEV samples, 574 genes (28.7%) were only expressed in the native samples and 13 genes (0.7%) were only expressed in the P-TEV samples. (F) The dotplot illustrates at a detailed level the similarities between the native and P-TEV samples for 15 top markers (three from each of the five identified clusters represented as rows in the plot). Similar dot diameter and colour intensity for the native (blue) and P-TEV (green) biomarkers indicate high similarity between these tissue samples. Colour intensities are relative to level of expression. The colour coded rectangles correspond to the clusters illustrated in panel (A). As shown in this plot, the variance of the identified marker genes is not attributed to the graft as the P-TEV and native samples show highly similar size and colour intensity for in principle all the top 15 marker genes. | ||
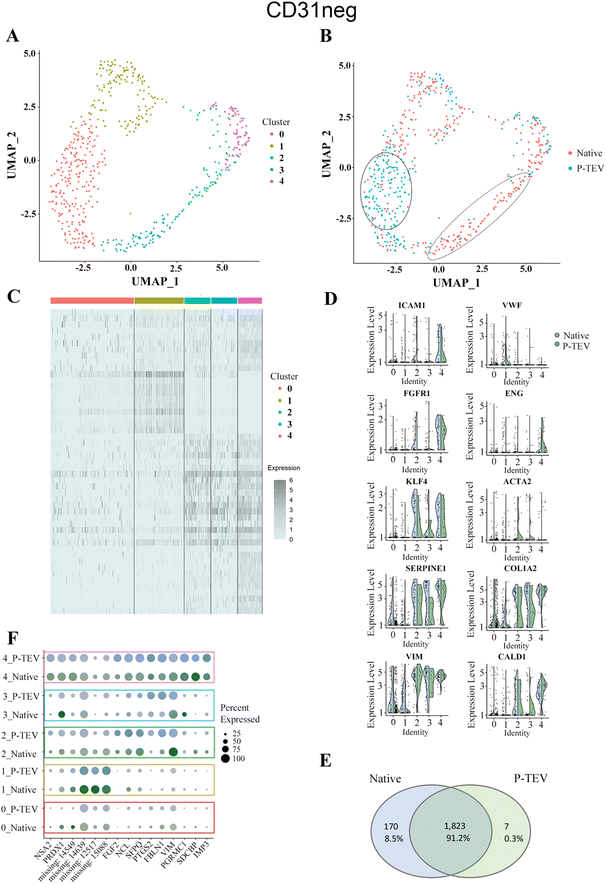 | ||
| Fig. 9 Single-cell RNA sequencing analysis of CD31 negative cells. (A) UMAP clustering of cells into five clusters. (B) Cells from both P-TEV and native samples were more or less equally represented in the clusters except for cluster 0 (marked with a solid oval line), which had an overrepresentation of P-TEV cells, and cluster 2 and 3 (marked with a dashed line), which were overrepresented with native cells. (C) Heatmap of the top markers for each of the five clusters shown in panel (A). Gene names in order from top to bottom are found in ESI Table IV.† (D) Violin plots showing the expression of ten typical blood vessel biomarker genes in the five identified clusters from panel (A). Blue represents native cells and green represents P-TEV cells. (E) Venn diagram showing the overlap of transcriptional activity between P-TEV and native samples. In total 1823 genes (91.2%) were expressed in both native and P-TEV samples, 170 genes (8.5%) were only expressed in the native samples and 7 genes (0.3%) were only expressed in the P-TEV samples. (F) The dotplot illustrates at a more detailed level the similarities between the native and P-TEV samples for 15 top markers (three from each of the five identified clusters represented as rows in the plot). Similar dot diameter and colour intensity for the native (blue) and P-TEV (green) biomarkers indicate high similarity between these tissue samples. Colour intensity represents level of expression. The colour coded rectangles correspond to the clusters illustrated in panel (A). Similarly, as for the CD31+ samples this plot the identified variance of the marker genes cannot be attributed to the graft, as the P-TEV and native samples show highly similar size and colour intensity for in principle all the top 15 marker genes. | ||
To further explore the transcriptional activity in P-TEV and native cells, the average expression values were calculated for each gene in the CD31+ and CD31− datasets, respectively, and the overall overlap of expressed genes between P-TEV and native cells in these datasets are shown in the Venn diagrams in Fig. 8E and 9E. For the CD31+ cells 71% of the transcripts were expressed in both P-TEV and native samples, and for the CD31− samples the overlap of transcriptional activity was as high as 91% between P-TEV and native. Moreover, the dotplots shown in Fig. 8F and 9F illustrate a high similarity between the P-TEV and the native samples for a selection of specific marker genes, shown by the comparable size of the dots (representing % of cells with expression of the specific marker gene) and the similar intensity of the colours (green for P-TEV and blue for native) which are relative to the level of gene expression.
Discussion
Tissue-engineered vascular grafts are promising alternatives to current vascular grafts. On the artery side, several studies have been performed on the use of decellularized grafts but with patency issues on small-diameter grafts (≤6 mm), reviewed in ref. 23–26. On the vein side, much less research has been performed to develop new vascular graft materials. Conduits for venous reconstructions have both early and late patency problems due to thrombosis.9–15 Even for autologous vein conduits, earlier experiences from treatments to overcome chronic venous obstruction or deep vein insufficiency, are associated with poor patency.11 Low flow velocity has been suggested to promote vein thrombosis but also vessel wall ischemia due to loss of the vasa vasorum circulation caused by transposition of the vein can explain graft failure. In previous safety and functionality short-term studies, we have shown that allogenic vascular grafts, decellularized and reconditioned with the recipient's peripheral blood, were efficiently recellularized in vivo, were patent and did not show any signs of thrombosis or significant stenosis due to intimal hyperplasia. This has been shown for both arteries and veins, and in two different species, pig, and sheep. Personalized tissue engineered veins (P-TEV) were implanted into pig, with the same model as in the current study, with a follow up time of one month,16 and personalized tissue engineered arteries (P-TEA) were inter-positioned carotid artery in a sheep model with a follow up time of four months.18Patency issues with decellularized grafts for artery surgery have been suggested to be due to thromboses initiated when the blood gets in contact with exposed collagen on the graft luminal surface.25,27,28 There are different potential ways of overcoming this problem: one is to immobilize modified peptides to the luminal side of the blood vessel to improve in vivo endothelialization.29,30 Another is recellularization of decellularized grafts prior to transplantation with different sources of endothelial cells, e.g. somatic ECs,31,32 endothelial progenitor cells,32–34 induced pluripotent stem cell derived ECs35 or embryonic stem cell derived ECs.36 However, growth and expansion of cells in vitro entails a potential risk with the introduction of genetic variation and mutations which brings regulatory hurdles in addition to higher costs and longer production time. In the present study, we used a decellularization method which preserves the biomechanical characteristics, such as maximum tensile strength and burst pressure, while the stiffness increases somewhat compared to the native vein. The amount of insoluble collagen in the ECM scaffold also remains unchanged whereas soluble collagen and glycosaminoglycans (GAGs), an important molecular target for organ rejection, decreases.37 Our aim is to protect the luminal surface from direct contact with the circulating blood after transplantation by a preconditioning process using the recipient's own blood to cover the collagen with a biolayer consisting of blood components.16 The theory is that this biolayer shields the graft from thrombosis and stimulates recruitment of cells for an efficient recellularization in vivo. One factor explaining the low predisposition for thrombosis could be that, compared with autologous grafts, the decellularized and reconditioned vessel wall contains no smooth muscle cells that could be induced to secrete tissue factor (TF) by the surgical transposition and blood contact and thereby cause a pro-coagulant response.38 Also, the personalization of the graft inhibits the immune system to react to any possible residual antigens and thereby protects from rejection of the implanted vessel.
It has earlier been suggested that growth factors in the preconditioning solution bind to the graft and potentially support regeneration of the tissue by promoting recellularization and differentiation of stem cells.39,40 Data from our previous ROTEM analysis showed increased clotting time, increased clot formation time and decreased alpha angle, indicating improved hemocompatibility and thereby a slower activation of the intrinsic coagulation system for the reconditioned P-TEV compared to native and decellularized tissue.16
In the current study, we present a tissue engineered vascular graft for venous reconstruction which become fully biologically integrated to the recipient animal with good short- and long-term patency. We used P-TEV, produced by decellularization and reconditioning, and evaluated safety and functionality of the grafts in a porcine in vivo vena cava transplantation model for up to 14 months. To further validate the concept, animal studies were performed in two different labs in two different countries (Sweden up to 14 months follow up and Spain up to 6 months follow up) on two different breeds of pig. Also, the study performed in Spain was conducted according to the good laboratory practice (GLP) quality system to complete the regulatory documentation in preparation of a clinical study. The decellularization process was efficient and limited the DNA content to a level well below the 50 ng DNA per mg dry ECM weight commonly used as the criterion for sufficient decellularization.41 The reconditioning protocol has been optimized with different perfusion time starting from two days. Binding of autologous components from the blood increased with time, but after seven days a decrease in the quality of the blood solution was apparent with increased hemolysis and methemalbumin, why seven days was chosen. The DNA content of the excised graft did not reach up to the DNA content of native tissue which is probably due to remaining scar tissue, formed during the surgery and healing, that was not possible to dissect from the P-TEV. This was supported by analyzing the DNA content of native tissue adjacent to the P-TEV excised after 12–14 months which contained the same DNA amount as the P-TEV (Fig. 1). At analysis, all P-TEVs were fully patent and did not show any signs of occlusion. We have previously shown that recellularization starts already three days post-surgery, and that the graft was well cellularized 17 days post-surgery.16 Here we could see that the patency (Fig. 2), cellular content and tissue morphology was consistent, and equal with native tissue up to 14 months post-surgery (Fig. 3–5 and ESI Fig. 1, 2†).
Intimal hyperplasia, where smooth muscle cells are abnormally integrated into the vascular tunica intima, inducing stenosis, is a common problem in blood vessel grafting in clinic. Despite the fact that pigs are known to be prone to develop intimal hyperplasia compared to other model animals,42 no intimal hyperplasia was observed in any of the P-TEV one-year post-surgery (data not shown).
Scanning electron microscopy and confocal microscopy en face revealed that the luminal surface of the P-TEV was completely covered with CD31-expressing endothelial cells with a morphology indistinguishable between native and P-TEV tissue and thereby shielding the luminal collagen from direct contact with the blood (Fig. 4). One key issue in regenerative medicine is that engineered tissue needs to be revascularized to be able to support the cells with oxygen and nutrition since the diffusion capacity is limited to approximately 150 μm.43–45 Cross section histology with confocal microscopy showed the important observation that the blood vessel tissue of the P-TEV was revascularized with capillaries comparable with the native tissue (Fig. 5). Also, no acellular regions or areas with necrosis were identified as possible signs of hypoxia or limited access to nutrient.
There is an ongoing essential discussion of what happens with an implanted ATMP-product over time in the body. Therefore, in addition to show safety and functionality of the P-TEV after one year in vivo, we here also wanted to investigate the characteristics of the regenerated P-TEV more deeply during long term conditions. This was performed using gene expression profiling, and to cover this in detail as comprehensive as possible, large-scale transcriptomic sequencing was performed on dissociated cells and specific analysis was performed on a panel of blood vessel markers with single cell qPCR. After excision of the P-TEV (one-year post-transplantation), P-TEV tissue was dissociated to get the cells into single cell suspension. A blood vessel is built up mainly by the three cell types; endothelial cells lining the luminal cell wall, perivascular mural cells which is a collection name for pericytes and vascular smooth muscle cells, and fibroblasts. Our analysis was based on a separation between endothelial cells in one group (CD31-positive cells) and the rest in the other group (CD31-negative cells). A portion of the dissociated cells were FACS sorted, initially to discard blood derived CD45-positive cells, and after that the cells were sorted for CD31-posivite and CD31-negative cells. From the single cell qPCR, we could draw the conclusions that the expression levels of the selected blood vessel marker genes showed highly similar expression levels in native and P-TEV derived cells with only VEGF expression in CD31+ cells as significantly different between native and P-TEV cells. Also, the expression levels of the genes were at the expected levels for markers related to CD31-positive and CD31-negative cells, respectively, except for SMTN with a lower expression in P-TEV CD31-positive cells compared to native. However, this gene is expressed by all blood vessel cells and the difference was not statistically significant between the groups (Fig. 7). The more comprehensive single cell transcriptomic analysis performed using RNA sequencing also revealed a high level of transcriptional similarity between the P-TEV and native tissue samples. The overlap of transcribed genes between P-TEV and native (ranging from 70–91%), shown in the Venn diagrams, demonstrates that the process for decellularization and reconditioning promotes the development and maturation of P-TEV with high similarity to the native tissue with respect to which genes that show transcriptional activity (Fig. 8E and 9E). However, the Venn diagram do not show the level of expression for each gene but instead the sets of genes that were detected as expressed in out data. The even distribution of both P-TEV and native cells in the UMAP clustering's also indicate high transcriptional similarity between the P-TEV and the native samples (Fig. 8B and 9B).
A known limitation of the study is that a decellularized scaffold was not included as a control group for the in vivo study. However, the aim of these long-term studies was to demonstrate safety as part of a preclinical safety package. The studies can further help to elucidate the functionality, safety and detailed gene expression characteristics relating the P-TEV to native tissue but not to compare mechanistical differences between decellularized scaffolds and P-TEV. This study was part of the final step before clinical trial, and the literature has already demonstrated several problems with decellularized scaffolds.9,24,27,46
Conclusion
In the present study, we have shown that decellularized blood vessels, reconditioned with the recipient's whole peripheral blood are safe and functional for vascular reconstruction without any signs of coagulation, thrombosis, intimal hyperplasia, or rejection. After analyzing 3 pigs 6 month post-surgery, 6 pigs 12 months post-surgery and one pig 14 months post-surgery, we conclude that all veins were fully patent and that the graft tissue got well recellularized and revascularized with a capillary network. The expected cell types were identified in the grafts and the transcriptional activity of the cells showed high overlap to cells from native vein tissue after one year in vivo. These results motivate to proceed with P-TEV for further validation in clinical studies.Author contributions
K. Ö., Y. B., L. J., T. H., J. S., G. H., E. B., J. S. E., R. S. and J. H. contributed to the study design. K. Ö., Y. B., L. J., T. H., J. S., G. H., E. B., S. P., A. K., J. S. E., J. R., V. C., F. M. S.-M., C. B.-D., R. S., J. H. performed experiments and analyzed results. All authors contributed to data production, statistical analysis, and interpretation. K. Ö., Y. B., L. J., T. H., J. S., G. H., R. S. and J. H. wrote the initial draft of the manuscript; all authors reviewed and revised the manuscript.Conflicts of interest
L. J., T. H. and R. S. are all employed by the company VERIGRAFT AB.Acknowledgements
This study was supported by Vinnova project CAMP (contract no. 2017-02130), a common call by VINNOVA and Vetenskapsrådet: Biologcal pharmaseuticals (Dnr 2017-02983), by University of Skövde under grants from the Swedish Knowledge Foundation [#2016-0330, #2020-0014] and Västra Götalandsregionen (consultant check). The company VERIGRAFT AB holds a patent on peripheral whole blood perfusion of decellularized tissues and did also finance the project. We want to acknowledge the staff at the Department of Experimental Biomedicine at Gothenburg University. The swine studies in Spain were conducted by the ICTS ‘NANBIOSIS’, specifically Units 21, 22, and 24 of the CCMIJU. Graphical Abstract image created with BioRender.com.References
- S. Mendis, P. Puska and B. Norrving, Global atlas on cardiovascular disease prevention and control, World Health Organization in Collaboration with the World Heart Federation and the World Stroke Organization, Geneva, 2011 Search PubMed.
- M. Ezzati, et al., Contributions of risk factors and medical care to cardiovascular mortality trends, Nat. Rev. Cardiol., 2015, 12(9), 508–530 CrossRef PubMed.
- S. Onida and A. H. Davies, Predicted burden of venous disease, Phlebology, 2016, 31(1 Suppl.), 74–79 CrossRef PubMed.
- M. G. De Maeseneer, et al., Editor's Choice – European Society for Vascular Surgery (ESVS) 2022 Clinical Practice Guidelines on the Management of Chronic Venous Disease of the Lower Limbs, Eur. J. Vasc. Endovasc. Surg., 2022, 63(2), 184–267 CrossRef PubMed.
- G. Plate, et al., Overcoming failure of venous vascular prostheses, Surgery, 1984, 96(3), 503–510 CAS.
- B. G. Eklof, R. L. Kistner and E. M. Masuda, Venous bypass and valve reconstruction: long-term efficacy, Vasc. Med., 1998, 3(2), 157–164 Search PubMed.
- F. S. Lozano, M. C. Estevan and J. R. Gónzalez-Porras, Femoral vein injury and transposition techniques: a new approach to venous reconstruction in the setting of trauma, J. Trauma, 2009, 67(4), E118–E120 Search PubMed.
- D. Kleive, et al., Portal vein reconstruction using primary anastomosis or venous interposition allograft in pancreatic surgery, J. Vasc. Surg.: Venous Lymphatic Disord., 2018, 6(1), 66–74 Search PubMed.
- R. L. Madden, et al., Experience with cryopreserved cadaveric femoral vein allografts used for hemodialysis access, Ann. Vasc. Surg., 2004, 18(4), 453–458 CrossRef PubMed.
- M. Jara, et al., Bovine pericardium for portal vein reconstruction in abdominal surgery: a surgical guide and first experiences in a single center, Dig. Surg., 2015, 32(2), 135–141 CrossRef PubMed.
- C. J. Jost, et al., Surgical reconstruction of iliofemoral veins and the inferior vena cava for nonmalignant occlusive disease, J. Vasc. Surg., 2001, 33(2), 320–327 CrossRef CAS PubMed , discussion 327–328.
- R. T. Faulkner, G. S. Cowan, Jr. and L. Rothouse, Early failure of expanded polytetrafluoroethylene in femoral vein replacement, Arch. Surg., 1979, 114(8), 939–943 CrossRef CAS PubMed.
- A. J. Kovalic, D. K. Beattie and A. H. Davies, Outcome of ProCol, a bovine mesenteric vein graft, in infrainguinal reconstruction, Eur. J. Vasc. Endovasc. Surg., 2002, 24(6), 533–534 CrossRef CAS PubMed.
- S. Manduz, et al., Early thrombosis in bovine mesenteric vein grafts after infrainguinal reconstruction, Int. J. Angiol., 2008, 17(1), 37–39 CrossRef PubMed.
- J. Schmidli, et al., Bovine mesenteric vein graft (ProCol) in critical limb ischaemia with tissue loss and infection, Eur. J. Vasc. Endovasc. Surg., 2004, 27(3), 251–253 CrossRef CAS PubMed.
- J. Håkansson, et al., Individualized tissue-engineered veins as vascular grafts: A proof of concept study in pig, J. Tissue Eng. Regener. Med., 2021, 15(10), 818–830 CrossRef PubMed.
- T. W. Gilbert, T. L. Sellaro and S. F. Badylak, Decellularization of tissues and organs, Biomaterials, 2006, 27(19), 3675–3683 CAS.
- L. Jenndahl, et al., Personalized tissue-engineered arteries as vascular graft transplants: a safety study in sheep, Regener. Ther., 2022, 21, 331–341 CrossRef CAS PubMed.
- S. A. Hosgood and M. L. Nicholson, First in Man Renal Transplantation After Ex Vivo Normothermic Perfusion, Transplantation, 2011, 92(7), 735–738 CrossRef PubMed.
- S. Steen, et al., Safe orthotopic transplantation of hearts harvested 24 hours after brain death and preserved for 24 hours, Scand. Cardiovasc. J., 2016, 50(3), 193–200 CrossRef PubMed.
- M. L. Nicholson and S. A. Hosgood, Renal transplantation after ex vivo normothermic perfusion: the first clinical study, Am. J. Transplant., 2013, 13(5), 1246–1252 CrossRef CAS PubMed.
- L. Ellegaard, et al., Haematologic and Clinical Chemical values in 3 and 6 months old Göttingen minipigs, Scand. J. Lab. Anim. Sci., 1995, 22(03), 239–248 Search PubMed.
- B. C. Isenberg, C. Williams and R. T. Tranquillo, Small-diameter artificial arteries engineered in vitro, Circ. Res., 2006, 98(1), 25–35 CrossRef CAS PubMed.
- C.-H. Lin, et al., In Vivo Performance of Decellularized Vascular Grafts: A Review Article, Int. J. Mol. Sci., 2018, 19(7), 2101 CrossRef PubMed.
- S. Pashneh-Tala, S. MacNeil and F. Claeyssens, The Tissue-Engineered Vascular Graft-Past, Present, and Future, Tissue Eng., Part B, 2016, 22(1), 68–100 CrossRef CAS PubMed.
- E. C. Scott and M. H. Glickman, Conduits for hemodialysis access, Semin. Vasc. Surg., 2007, 20(3), 158–163 CrossRef PubMed.
- L. Gui, et al., Development of decellularized human umbilical arteries as small-diameter vascular grafts, Tissue Eng., Part A, 2009, 15(9), 2665–2676 CrossRef CAS PubMed.
- J. W. Heng, et al., Coatings in Decellularized Vascular Scaffolds for the Establishment of a Functional Endothelium: A Scoping Review of Vascular Graft Refinement, Front. Cardiovasc. Med., 2021, 8, 677588 CrossRef CAS PubMed.
- A. Mahara, et al., Tissue-engineered acellular small diameter long-bypass grafts with neointima-inducing activity, Biomaterials, 2015, 58, 54–62 CrossRef CAS PubMed.
- H. Bai, et al., Hyaluronic acid-heparin conjugated decellularized human great saphenous vein patches decrease neointimal thickness, J. Biomed. Mater. Res., Part B, 2020, 108(6), 2417–2425 CrossRef CAS PubMed.
- N. Dahan, et al., Dynamic Autologous Reendothelialization of Small-Caliber Arterial Extracellular Matrix: A Preclinical Large Animal Study, Tissue Eng., Part A, 2017, 23(1–2), 69–79 CrossRef CAS PubMed.
- C. Quint, et al., Decellularized tissue-engineered blood vessel as an arterial conduit, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(22), 9214–9219 CrossRef CAS PubMed.
- A. J. Melchiorri, et al., In Vitro Endothelialization of Biodegradable Vascular Grafts Via Endothelial Progenitor Cell Seeding and Maturation in a Tubular Perfusion System Bioreactor, Tissue Eng., Part C, 2016, 22(7), 663–670 CrossRef CAS PubMed.
- S. Kaushal, et al., Functional small-diameter neovessels created using endothelial progenitor cells expanded ex vivo, Nat. Med., 2001, 7(9), 1035–1040 CrossRef CAS PubMed.
- K. H. Nakayama, et al., Bilayered vascular graft derived from human induced pluripotent stem cells with biomimetic structure and function, Regener. Med., 2015, 10(6), 745–755 CrossRef CAS PubMed.
- Q. Shi, et al., Ex vivo reconstitution of arterial endothelium by embryonic stem cell-derived endothelial progenitor cells in baboons, Stem Cells Dev., 2013, 22(4), 631–642 CrossRef CAS PubMed.
- R. Simsa, et al., Systematic in vitro comparison of decellularization protocols for blood vessels, PLoS One, 2018, 13(12), e0209269 CrossRef PubMed.
- S. A. Smith, R. J. Travers and J. H. Morrissey, How it all starts: Initiation of the clotting cascade, Crit. Rev. Biochem. Mol. Biol., 2015, 50(4), 326–336 CrossRef CAS PubMed.
- I. Ullah, et al., VEGF – Supplemented extracellular matrix is sufficient to induce endothelial differentiation of human iPSC, Biomaterials, 2019, 216, 119283 CrossRef CAS PubMed.
- Y. Ikada, Challenges in tissue engineering, J. R. Soc., Interface, 2006, 3(10), 589–601 CrossRef CAS PubMed.
- P. M. Crapo, T. W. Gilbert and S. F. Badylak, An overview of tissue and whole organ decellularization processes, Biomaterials, 2011, 32(12), 3233–3243 CrossRef CAS PubMed.
- R. H. Liu, et al., Review of Vascular Graft Studies in Large Animal Models, Tissue Eng., Part B, 2018, 24(2), 133–143 CrossRef CAS PubMed.
- P. L. Olive, C. Vikse and M. J. Trotter, Measurement of oxygen diffusion distance in tumor cubes using a fluorescent hypoxia probe, Int. J. Radiat. Oncol., Biol., Phys., 1992, 22(3), 397–402 CrossRef CAS PubMed.
- I. F. Tannock, Oxygen diffusion and the distribution of cellular radiosensitivity in tumours, Br. J. Radiol., 1972, 45(535), 515–524 CrossRef CAS PubMed.
- R. H. Thomlinson and L. H. Gray, The histological structure of some human lung cancers and the possible implications for radiotherapy, Br. J. Cancer, 1955, 9(4), 539–549 CrossRef CAS PubMed.
- N. Das, et al., Results of a seven-year, single-centre experience of the long-term outcomes of bovine ureter grafts used as novel conduits for haemodialysis fistulas, Cardiovasc. Intervention Radiol., 2011, 34(5), 958–963 CrossRef PubMed.
- R. Simsa, et al., Effect of fluid dynamics on decellularization efficacy and mechanical properties of blood vessels, PLoS One, 2019, 14(8), e0220743 CrossRef CAS PubMed.
- A. Untergasser, et al., Primer3—new capabilities and interfaces, Nucleic Acids Res., 2012, 40(15), e115 CrossRef CAS PubMed.
- J. Ye, et al., Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction, BMC Bioinf., 2012, 13, 134 CrossRef CAS PubMed.
- T. Koressaar and M. Remm, Enhancements and modifications of primer design program Primer3, Bioinformatics, 2007, 23(10), 1289–1291 CrossRef CAS PubMed.
- T. Kõressaar, et al., Primer3_masker: integrating masking of template sequence with primer design software, Bioinformatics, 2018, 34(11), 1937–1938 CrossRef PubMed.
- S. A. Bustin, et al., The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments, Clin. Chem., 2009, 55(4), 611–622 CrossRef CAS PubMed.
- T. Stuart, et al., Comprehensive Integration of Single-Cell Data, Cell, 2019, 177(7), 1888–1902 CrossRef CAS PubMed .e21.
- M. D. Luecken and F. J. Theis, Current best practices in single-cell RNA-seq analysis: a tutorial, Mol. Syst. Biol., 2019, 15(6), e8746 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm02011d |
| This journal is © The Royal Society of Chemistry 2023 |