Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Physico-chemical properties of functionally adhesive spider silk nanofibres†
Anna-Christin
Joel
 *abc,
Aditya
Rawal
*abc,
Aditya
Rawal
 d,
Yin
Yao
d,
Andrew
Jenner
d,
Nicholas
Ariotti
de,
Margret
Weissbach
d,
Yin
Yao
d,
Andrew
Jenner
d,
Nicholas
Ariotti
de,
Margret
Weissbach
 c,
Lewis
Adler
d,
Jay
Stafstrom
bf and
Sean J.
Blamires
c,
Lewis
Adler
d,
Jay
Stafstrom
bf and
Sean J.
Blamires
 bdg
bdg
aDepartment of Biological Sciences, Macquarie University, Sydney, Australia. E-mail: joel@bio2.rwth-aachen.de
bSchool of Biological, Earth and Environmental Sciences, The University of New South Wales, Sydney, Australia
cInstitute of Zoology, RWTH Aachen University, Aachen, Germany
dMark Wainwright Analytical Centre, University of New South Wales, Sydney, Australia
eInstitute for Molecular Bioscience, The University of Queensland, Brisbane, Australia
fDepartment of Neurobiology and Behavior, Cornell University, Ithaca, New York, USA
gSchool of Mechanical and Mechatronic Engineering, University of Technology Sydney, Sydney, Australia
First published on 2nd February 2023
Abstract
Currently, synthetic fibre production focuses primarily on high performance materials. For high performance fibrous materials, such as silks, this involves interpreting the structure–function relationship and downsizing to a smaller scale to then harness those properties within synthetic products. Spiders create an array of fibres that range in size from the micrometre to nanometre scale. At about 20 nm diameter spider cribellate silk, the smallest of these silks, is too small to contain any of the typical secondary protein structures of other spider silks, let alone a hierarchical skin-core-type structure. Here, we performed a multitude of investigations to elucidate the structure of cribellate spider silk. These confirmed our hypothesis that, unlike all other types of spider silk, it has a disordered molecular structure. Alanine and glycine, the two amino acids predominantly found in other spider silks, were much less abundant and did not form the usual α-helices and β-sheet secondary structural arrangements. Correspondingly, we characterized the cribellate silk nanofibre to be very compliant. This characterization matches its function as a dry adhesive within the capture threads of cribellate spiders. Our results imply that at extremely small scales there may be a limit reached below which a silk will lose its structural, but not functional, integrity. Nano-sized fibres, such as cribellate silk, thus offer a new opportunity for inspiring the creation of novel scaled-down functional adhesives and nano meta-materials.
Since the invention of nylon and other synthetic fibres around 90 years ago, there has been an exponential increase in the use of petrochemical-based fibrous polymers. This has nevertheless come at substantial environmental costs. Research that aims to understand the functional limits of natural fibres and the barriers and limitations associated with their construction is thus being increasingly done.1–5
Spiders produce an array of silks, from extremely tough dragline silks, to silks used in web scaffolding, silks from which components of the egg sac are made, to glues and micro-adhesives for prey capture.6,7 For several decades, researchers have examined the remarkable properties of the different spider silks with an eye on the development of artificial fibres that mimic its performance. While an artificial fibre that performs as well as a natural spider silk counterpart has not yet been developed, recent advances in recombinant biotechnology, dry and wet spinning, and three-dimensional printing protocols, mean the production of a spider silk biomimetic fibre might not be very far away.8–11
The type of spider silk whose properties are most well-known is dragline, or major ampullate, silk. This silk has a hierarchical structure with a core that includes fibrous proteins (spidroins), a 40 to 100 nm wide skin that consists of highly oriented protein chains and a 10 to 20 nm wide glycoproteins and lipids outermost layer.6,12 The mechanical properties are mainly influenced by the spidroin secondary structure, where crystalline regions of β-sheets of approximately 6 nm diameter are embedded within an amorphous matrix of primarily α-helical structures. This combination of structures gives dragline silk its unique property of having high elasticity combined with exceptional strength.13
All silks studied so far are thought to attain a similar hierarchical structure and the skin-core model predicts that the secondary structures of the core proteins directly influence the functional properties of the silk.14 Most spider silks are circular at the micro-metre scale, while there are exceptions, like ribbon silks of brown recluse spider Loxosceles laeta, consisting of many aligned ∼20 nm diameter protein fibrils, or capture silks of cribellate spiders, being even smaller with single fibres of similar size compared to the protein fibrils of L. laeta.15–19 In Embiopteral nanofibrous silks, with 90 to 100 nm diameter slightly larger than the before mentioned spider silks, β-sheet secondary structures dominate the silk.20 Nonetheless, we expect silks with diameters close to the size of a single crystalline unit (6 nm) to be predominantly amorphous, which would henceforth be expected to render it functionally inert as a fibre. Intriguingly, however, such fibres exist and are functional, having the important task of catching and restraining prey within spider cribellate webs.
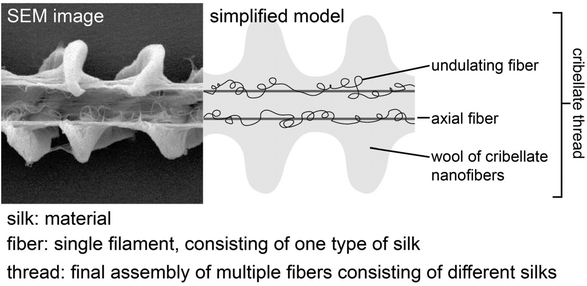
These extremely thin fibres are produced by cribellate web building spiders and function as adhesives within their capture threads.21,22 The hackled orb weavers Uloboridae or ogre-faced spiders Deinopidae belong to these cribellate spiders, but there are several more families comprising cribellate species. Such cribellate capture threads have a complex interwoven composition including at least three different types of silk (Fig. 1):22–25 The pseudoflagelliform silk forms the axial fibre, and either paracribellate fibres from the paracribellate glands or at least one undulating fibre from the minor ampullate gland is found next to several thousand eponymous cribellate nanofibres produced by the cribellate glands.21,22,24,26 Instead of glue droplets, typically used by ecribellate spiders like the well-studied golden orb-weaver Trichonephila spp., cribellate spiders use the viscous cuticle of the prey insect itself as the glue component, embedding the silken cribellate nanofibres in the waxy surface covering every insect.27 The cribellate silks thus appear to have highly hydrophobic components, which is necessary for its strong adhesion with the prey insect's cuticle. Nevertheless, they are spun in air and the adhesion force (on artificial wax-free surfaces) even increases with an elevation of humidity, though it decreases after contact with liquid water.28–30 This indicates that these fibres additionally must have hygroscopic, perhaps even hydrophilic, properties. The cribellate capture thread reacts as a dipole when exposed to positively and negatively charged objects. Accordingly, it is attracted to flying prey, which charges itself during flight as a result of triboelectrification.31,32 It is assumed that the cribellate fibres do not play an important role regarding the mechanical integrity of the fibres and most energy during prey capture is absorbed by the axial fibres.33 However, forces can be transferred by cribellate fibres to other fibres, at least in the threads of the lace sheet weaver Psechrus clavis and the Tasmanian cave spider Hickmania troglodytes, demonstrating that cribellate fibres have some degree of the mechanical strength for force absorption.30,34 Thus, multiple properties have to be combined in cribellate fibres for their functionality. While a few studies have attempted to clarify the genomic and/or proteomic compositions of cribellate nanofibres,35–41 the protein/molecular structure of cribellate silks is not well understood. The investigations have been especially hindered by the exceptionally small size of cribellate fibers and the difficulty of harvesting the threads in sufficiently large quantities.
Here we have used cutting edge spectroscopic, microscopic, and mass spectrometry techniques to fill in some of the knowledge gaps pertaining to the protein composition and structures of nanofibrous cribellate spider silk. Our study thus explains how the exceptionally thin nanoscale cribellate spider silk fibres can sustain functionality, which provide insights into how nature is able to exceed the lower scaling limit, which could be used to promote production of an artificial nano-scale multifunctional spider silk analogue.
Results
Thread structure
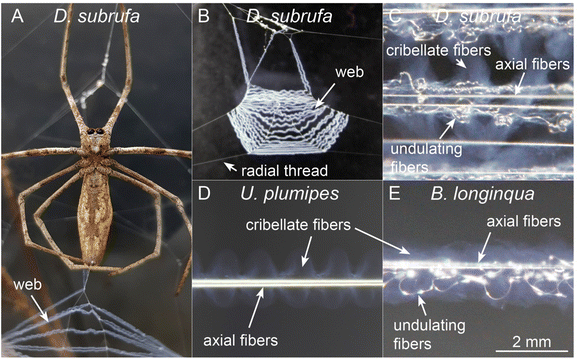
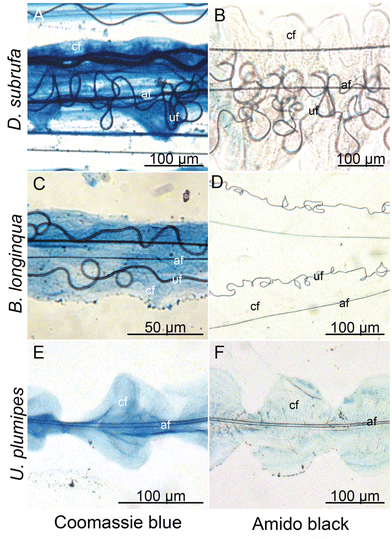
Cribellate threads are a composition of multiple silks. We examined here the threads from three species of cribellate silk producing spiders, the ogre-faced spider Deinopis subrufa, the feather-legged lace weaver Uloborus plumipes, as well as the grey house spider Badumna longinqua, each with differently structured cribellate threads. Fig. 2 shows optical images of the native orb web production and the finished web (Fig. 2A and B). Fig. 2C and D show the cribellate threads of the different species, highlighting the axial, undulating and cribellate fibres. Note that threads of all three species contain the eponymous cribellate nanofibres as a cloudy mass surrounding the other silks (Fig. 2C–E). Specific staining helps better image the distinct types of fibres (Fig. 3). The nanofibrous cribellate silk can be stained by Coomassie blue in all three species, while Amido black stains only the nanofibres of U. plumipes (Fig. 3F), but not those of the other spiders (Fig. 3B–D). Additionally, all species have axial fibres, which can be stained by both dyes. D. subrufa and B. longinqua also have additional undulating fibres, which are stained heavily by both dyes. Differences in stainability hint to differences in chemical composition of the different silks within one thread, but likewise between species (Fig. 3). Hence, we assumed, at least for B. longinqua and D. subrufa, that the chemical composition of cribellate silk were chemically different to other silks.The ultrastructure of cribellate fibres
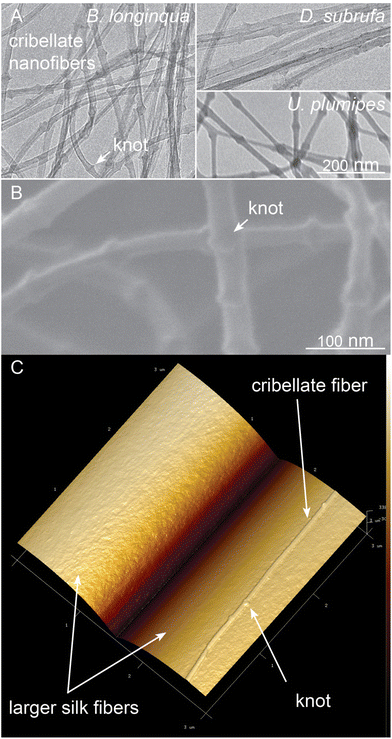
Electron Microscopy showed that cribellate fibres examined bear the characteristic knots described elsewhere.42 The function of these knots is not yet well understood, with indications they may enhance adhesion under increased humidity, or they may just be production artefacts.43,44 A more magnified Scanning Electron Microscopy image shows that the knots in U. plumipes cribellate fibres are not regularly shaped, but have a ‘saw tooth’ shape (Fig. 4B). This structure, found also in the cribellate fibres of B. longinqua and D. subrufa, made their nanofibres easily discriminable from other silk fibres when using Atomic Force Microscopy (AFM) (Fig. 4C).The cribellate fibres had a diameter of approximately 23 nm (Table 1), although D. subrufa's fibres had a slightly larger diameter compared to the other two species. The knots of the cribellate fibres enhanced their mean diameter to 35 nm (Table 1, Fig. 4). We were not able to perform thin-sections of the cribellate thread for TEM-analysis of the internal hierarchical structure (see material and methods).
Cribellate silk: structural composition
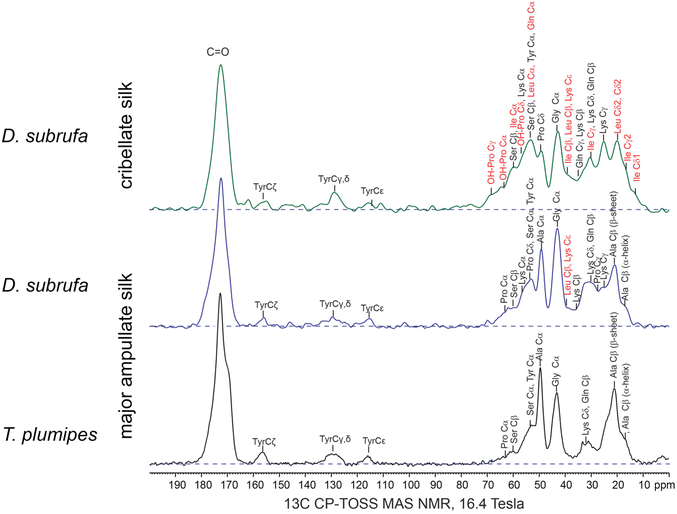
To analyse the molecular chemistry of cribellate silk, we simultaneously performed gas chromatography-mass spectroscopy (GC-MS) as well as solid-state nuclear magnetic resonance spectroscopy (ssNMR) on the cribellate threads (i.e. cribellate silk, pseudoflagelliform (axial) silk and, in case of B. longinqua and D. subrufa, also minor ampullate (undulating) silk) and compared them to major ampullate (dragline) silk from Trichonephila plumipes, U. plumipes and D. subrufa. For clarity, this cribellate thread composite will subsequently be called “cribellate silk”.The difference in molecular structure between the major ampullate (MA) silks and the cribellate silk is highlighted by the solid-state 13C MAS NMR spectra of the silk fibres (Fig. 5). The MA silk of D. subrufa is very similar to that dragline MA silk of T. plumipes with narrow lineshapes and dominant peak from the alanine and glycine residues. In comparison in the 13C spectrum of the D. subrufa cribellate silk, the peak widths are significantly broadened as highlighted in the 10–80 ppm region. This line broadening (in particular of the Cα peaks of the alanine and glycine residues) indicates that the cribellate silk is more amorphous than the MA silk. On the other hand, the greater number and more uniform distribution of spectral peaks found by the 13C ssNMR spectra point to a higher structural complexity in cribellate silk compared to the MA silk. In particular, the 13C ssNMR spectrum of the cribellate silk identifies a significant fraction of hydrophobic residues such as leucine/isoleucine as well as significant signals of hydroxyproline, an amino acid that has not been observed in any silk to date. Consistent with the reduction in order of the cribellate silk, there was no indication of the β-sheets motifs (formed from alanine residues) that are commonly observed in other spider and insect silks.45
Our ssNMR results were supported by our Gas Chromatography-Mass Spectrometry (GC-MS) analyses where the amino acid compositions of cribellate silk contrasted with those for MA silks, especially regarding its alanine and glycine content (Fig. 6, light blue and orange bars). In major ampullate silks, we found that the combined alanine and glycine represented 47.5% and 38.7% of the amino acids, whereas in cribellate silk the combined composition of alanine and glycine was reduced to 29.8%, 21.9%, respectively 32.2% for U. plumipes, B. longinqua, and D. subrufa respectively (Fig. 6). Especially the content of glycine was dramatically reduced (17.6% and 16.1% compared to 5.7%, 8.3%, respectively 13.7%). Alanine and glycine are the main components influencing the mechanical properties of spider silk. Our finding of high alanine and glycine compositions within the major ampullate silks suggests our GC-MS procedures were able to recover and accurately detect these amino acids.46
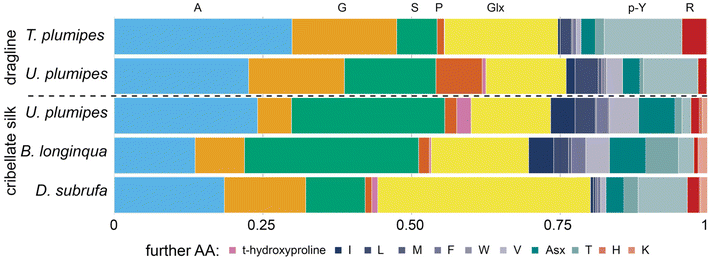 | ||
| Fig. 6 Amino acid composition of major ampullate silk (dragline) vs. cribellate silk of four spider species (GC-MS analysis). The acid hydrolysis may have caused some amino acids, i.e. asparagine and glutamine, to become deaminated to their respective acids during hydrolysis and thus could not be differentiated (named Glx and Asx). Data are presented in Table S1.† | ||
In contrast to MA silk, cribellate silk had a lower content of hydrophobic (mean: 40 ± 6% vs. 54 ± 2%) and higher contents of polar (assuming Asx and Glx are completely polar asparagine and glutamine: mean: 57 ± 6% vs. 43 ± 1%; excluding both: 30 ± 8% vs. 24 ± 3%) amino acids in the cribellate silk (Fig. 6, Table S1†). We found slightly elevated concentrations of the charged amino acid lysine (Fig. 6, pale red at the right hand side) in all cribellate silks, though an elevation of basic amino acids in general was not conspicuous (Fig. 6, red stripes at the right hand side). Fitting to the general elevation of polar amino acids, we found high compositions of serine (mean: 21.7 ± 10.3% vs. 11.1 ± 6.1%) and hydroxyproline (mean: 1.2 ± 1.1% vs. 0.4 ± 0.4%) in both our GC-MS data as well as ssNMR spectra (Fig. 5 and 6). The level of the hydrophobic isoleucine was also increased comparing cribellate silk (mean: 1.6 ± 2.1%) to MA silks (mean: 0.6 ± 0.8%).
The GC-MS analyses however, shows that there were species-dependent differences in the relative composition of the amino acids in the cribellate silks. For instance, isoleucine was in greater abundance in the cribellate silk of U. plumipes and B. longinqua compared to MA silks, but not in cribellate silk of D. subrufa (Fig. 6).
Elasticity of cribellate fibres
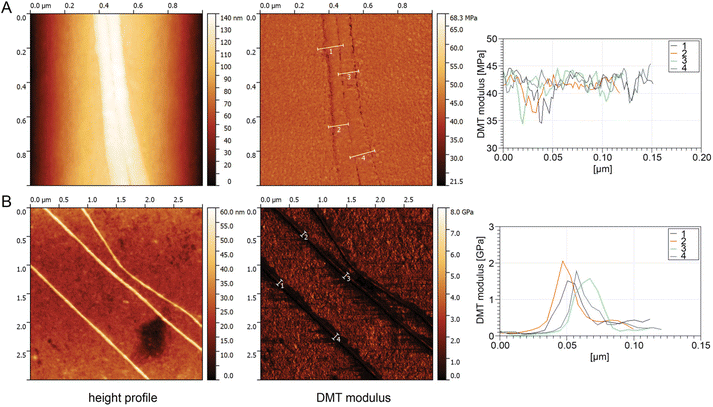
To determine the elasticity (resistance to being permanently deformed) of a single cribellate fibre, a single nanofibre was required to be separated from the bundle of nanofibres that typically build a cribellate thread. This was done by locating a single cribellate nanofibre of B. longinqua on top of a larger fibre via AFM. Once identified by its characteristic knot structure, the AFM peak force tapping mode was used following procedures outlined by Craig et al.47 The fibre was repeatedly probed at specific points along its surface to generate a series of AFM images, from which the Derjaguin–Muller–Toporov (DMT) modulus, a measure of elasticity, of the fibre was calculated. There was no discernible difference in the elasticity between the larger fibre and the cribellate fibre resting on it using this method, with both fibres being extremely compliant with a DMT modulus of about 40 to 45 MPa (Fig. 7A).For D. subrufa, we were able to separate the nanofibres by spreading the fibres over a clean glass cover slip and thus isolating single nanofibres directly on the glass cover slip. At 1.5–2 GPa, the DMT modulus of D. subrufa's nanofibres was stiffer compared to cribellate silk of B. longinqua, though still much lower than typical values of dragline spider silk (Fig. 7B). These low modular values correspond well with our molecular data, showing that cribellate silk lacks β-sheets structures, commonly associated with strength and stiffness in other silks.6
Discussion
A spider silk marginally greater than the size of single crystalline unit could be expected to have an irregular or amorphous protein secondary structure. Indeed, we found secondary structures typically associated with strength and extensibility in dragline and other spider silks (i.e. α-helices and β-sheets and associated elevated fraction of alanine and glycine)45 were not present in the nano-scaled cribellate silk fibres. We accordingly deduced that these silks are primarily amorphous in structure. The mechanical compliance measured for the cribellate silk fibres was consistent with the lack of high-strength forming protein structures. The molecular composition of cribellate spider silk consists of a significant fraction of both hydrophobic (leucine and isoleucine) as well as polar (hydroxyproline and serine) amino acid residues, which may enhance their adhesiveness to prey. Hence, significant physico-chemical changes of the silk come with scaling down spider silk fibres, limiting its toughness but adapting to its function as highly elastic and adhesive thread.Scaling down spider silk leads to unusual molecular composition
The evolutionary origin of the cribellum, the source of cribellate nanofibres, is still debated, though there are indications of them likely being modified spinnerets.48,49 Matching to these reports, recent investigations of cribellate spidroin sequences place these in the middle of the spidroin phylogenetic tree.38,40,41 Our results suggest that any downsizing of a spider silk to the nanometre scale to come with significant changes in its molecular composition: Many of the features, like the hierarchical core-skin structure as well as the β-sheet structures are unlikely to be present within silks with diameters under 100 nm. Indeed, genetic analyses of the Uloborids predicted their cribellate silks to be missing typical spider silk structures, like α-helices.40 Nevertheless, in the cribellate spidroin repetitive domain An regions were found, which are thought to be associated with the development of β-sheets structures.40,41,50–52 We herein confirmed the presence of alanine in the cribellate silks that we assessed. However, at least for the cribellate threads of Deinopis subrufa, our 13C NMR spectra failed to identify any associated β-sheet structures. Since cribellate threads are extremely fine and difficult to collect in large enough amounts, we could not ascertain a 13C NMR spectra for these silks from any other species. We recommend that the cribellate silks from other species be 13C enriched in order to gain a comparative ssNMR spectra, or indirect assessments methods such as FT-IR or Raman spectroscopy be used.As the evolutionary origin of the cribellate silk as well as all facets of its adhesive mechanism are still not fully understood, we cannot assess whether the decrease in fibre size is a secondary effect of molecular changes, necessary to increase the functionality of cribellate capture threads, or whether the reduced size itself is increasing the fibres functionality. If capillary forces, perhaps in an interplay with van der Waals forces, are increasing the preys’ adhesion, we would expect the second explanation to be the most likely.27,29,53 In any case, these extremely fine nanofibres have to balance the requirements of maximal adhesion with sufficient structural integrity as fibres, both to maintain their fibrillary form and to restrain prey. We found the silks to contain serine, hydroxyproline as well as charged lysine and polar amino acids, and these amino acids may be facilitating the structural integrity of the nanofibres.54
Serine is a typical component in the silk of the silkworm Bombyx mori with a function in stabilizing the silk by being involved in the formation of β-sheets, however, in combination with glycine and alanine.55,56 As glycine and alanine are reduced in the cribellate nanofibres, it is unlikely that serine forms similar structures for mechanical integrity here. For fibre formation in caddisworm silk, Ca2+ in combination with phosphoserines forms β-sheet structures leading to fibre formation after extraction.57–59 However, the aquatic caddisfly larvae take up their ions from the surrounding water. No similar mechanism of ion uptake has been reported for any kind of silk spun into air so far.
Hydroxyproline is rarely found in other proteins than collagen. However it is present in flagelliform spider silk (the core fibre of gluey capture threads), and is probably responsible for its high elasticity.54,60 In collagen molecules, hydroxyproline typically stabilizes a triple helix and thus it might have a stabilizing function of a helix here, too. However, at least in collagen, such helices require the presence of a repeated glycine-Xaa-Yaa sequence, with proline and hydroxyproline respectively occupying both positions most of the time.61–63 Signals pointing to the presence of such structures (i.e. sharp signals of proline or hydroxyproline and glycine, with both having the same intensity) were not found in our ssNMR spectra. The increased level of hydroxyproline additionally leaves open the question of the derivation of this molecule, as its synthesis has been reported to require vitamin C.
Promoting functionality of cribellate fibres
For the successful capture of prey, the mechanical properties of the deployed threads are particularly important and high extensibility ensures a better distribution of the energy after flying prey hits the web.64,65 The extreme extensibility of the axial flagelliform silk combined with the adhesiveness of the surrounding aggregate glue in viscous capture threads is presumed as a key element in the diversification of aranoid spiders. Flagelliform silks have a special molecular structure, which is linked to their mechanical properties: highly repetitive glycine-rich motifs form type II β-turns, again forming a β-spiral. Additionally, these silks are lacking alanine rich regions, thus forming no crystalline β-sheets.66 In the case of cribellate threads, the extremely pliant cribellate nanofibres should be eminently suitable for capturing prey, too, although they might be too pliant to capture prey all by themselves. Cribellate threads are, though, a composition of multiple fibres, all mechanically interacting together.24,33,34 Kono et al. predicted the amino acid composition of cribellate spidroins do be conserved and the repeating units of the cribellate spidroin characterized for the distant related species Stegodyphus mimosarum and Tengella perfuga shared a 56% identity at amino acid level.38,40 Correa-Garhwal et al. finally postulate differences in the arrangements of otherwise very similar repeat modules.41 Differences in amino acid composition as well as in-end mechanical properties as found in this study, however, would predict cribellate silk itself to be quite diverse when studied interspecifically. This diversity could be caused by genomic differences, but likewise by changed gene expression. Very likely, it reflects an adaption to the slightly different uses of cribellate silk, already apparent when comparing the structural diversity of cribellate threads.22,67–70 When interpreting our data, though, one has to keep in mind, that our amino acid analysis was performed on complete cribellate threads and not pure cribellate silk. Aside from pseudoflagelliform silk present in all cribellate threads, threads of the species D. subrufa and B. longinqua include minor ampullate silks (undulating fibres), but the threads of U. plumipes do not.Although cribellate fibres can distribute forces within the thread (at least in the species Psechrus clavis), the main task remains the adhesion to prey.27,34 The fibres thus have to (1) get in contact with the prey (e.g. by being electrostatically attracted to flying prey)31 (2) establish adhesion by van der Waals and hygroscopic forces29,44 and (3) embed the nanofibres into the cuticular hydrocarbons of the prey for enhancing adhesion.27,53 Our GC-MS analysis found elevated levels of charged amino acids that could be crucial for the attraction to prey as well as the establishment of van der Waals forces. Glutamine, on the other hand, has been described to interact with water and facilitate the supercontraction of major ampullate silk.51 Hence, an increased level of this polar amino acid might enhance the hygroscopic properties of the cribellate silk, described to influence adhesion, too.29
The molecular complexity that we observed in cribellate silk matches to the different end properties of spider silk: the major ampullate silk (i.e. dragline) has a purely mechanical function, while the cribellate silk functions to capture prey including a chemical function with a molecular driven adhesion. It remains unclear why we did not observe many non-polar amino acids. A higher proportion of non-polar amino acids should enhance the functionality of the cribellate silk, since hydrocarbons covering insects are non-polar. However, we found the exact opposite to be the case. Nonetheless, a high composition of nonpolar isoleucine was found, at least for U. plumipes and B. longinqua cribellate silk (consistent with other literature for Uloborids)35,36 and this amino acid was also found prominently in the ssNMR data of Deinopis subrufa. Additionally, we consistently found the cribellate silks to contain up to 4% of isoleucine composition, which is normally only found within spider silks in trace amounts. This amino acid might facilitate the hydrophobic interactions between cribellate silk and the hydrocarbons of the insect cuticle.27,53
Implications for technical nanofibre silk production
There is an increasing effort to produce and use spider silk biomimetic materials, and research is aiming also to explore the lower limits of this production. In particular, nano materials have gained lots of attention due to their high surface-to-volume ratio. However, studies exploring the structure–function relationship of silk fibres at the sub-micron scale are missing. We found that scaling down spider silk to ∼20 nm diameter, i.e. approximately twice the size of most of the secondary protein structure, the silk loses its molecular structure and, likewise, its high strength and toughness. Nevertheless new features are gained and this is matched to the functionality of the cribellate nanofibres as spiders adhesive. The cribellate fibres are extremely compliant and consisted of amino acids, that likely promote the adhesion to the preys’ surface. Accordingly, our results suggest that scaling down of the fibre size comes at a mechanical cost, but a new suit of functionalities can be gained.These results will influence the ambitions to scale down artificial spun spider silk, which currently reaches sizes about 90 nm, with some fibres having been produced with a diameter even below 50 nm.5 Spider silk with a diameter of ∼ 20 nm will not be able to combine high elasticity with exceptional strength, as characterized for other spider silk. With such dramatically different mechanical properties, such very small-scale spider silk will have other applications to those proposed for other silks, such as major ampullate silk.
Method section
Ethics
The species used in the experiments are not endangered or protected species. Export of Deinopis subrufa and Badumna longinqua was kindly granted by the Department of Agriculture, Water and the Environment (Australian Government, permission no. PWS2019-AU-000248). Other permits were not required. All applicable international, national, and institutional guidelines for the care and use of animals were followed.Study animals and silk collection
Deinopis subrufa (L. Koch, 1878) (Fig. 2A) were collected from local parks across Sydney, Australia, and housed at the University of New South Wales – Sydney campus (Kensington-Sydney, Australia). Spiders were kept in cylindrical, plastic enclosures (10 × 10 × 14 cm), and maintained at a temperature between 21 °C and 23 °C, and a relative humidity between 40% and 60%. Spiders were fed two small crickets (A. domesticus) twice a week and allowed water ad libitum. Spiders were maintained on a 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 LD cycle. This spider produces modified orb webs primarily out of cribellate threads with a few major ampullate frame threads for support. As we simply collected the completed “webs” upon construction and manually removed the major frame threads (Fig. 8), it was relatively easy for us to collect large enough samples of cribellate silk for our analysis. In our data, we will thus have impureness by having few other silks than cribellate included. However, the majority of extraneous silk will be the undulating and axial fibres, included in every capture thread of D. subrufa anyway (Fig. 2B and C). As a negative control, silk was taken which was placed randomly by the spiders in their container. As the ssNMR spectra of this silk did not notably differ from the major ampullate silk of Trichonephila plumipes, we assumed it to be mainly major ampullate silk.
12 LD cycle. This spider produces modified orb webs primarily out of cribellate threads with a few major ampullate frame threads for support. As we simply collected the completed “webs” upon construction and manually removed the major frame threads (Fig. 8), it was relatively easy for us to collect large enough samples of cribellate silk for our analysis. In our data, we will thus have impureness by having few other silks than cribellate included. However, the majority of extraneous silk will be the undulating and axial fibres, included in every capture thread of D. subrufa anyway (Fig. 2B and C). As a negative control, silk was taken which was placed randomly by the spiders in their container. As the ssNMR spectra of this silk did not notably differ from the major ampullate silk of Trichonephila plumipes, we assumed it to be mainly major ampullate silk.
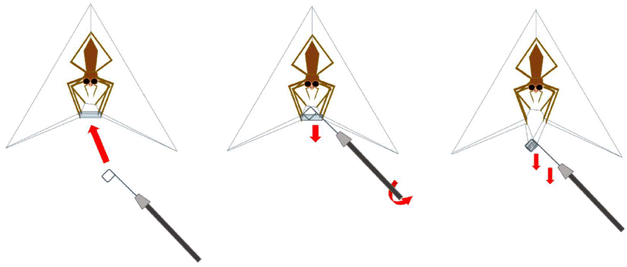 | ||
| Fig. 8 Schematic drawing of the cribellate silk collection by taking the complete web of Deinopis subrufa. This image was kindly provided by Nils Rosenkranz. | ||
We made additional analyses on the cribellate threads of the closely related Uloborus plumipes with simple cribellate threads lacking the undulating fibres (Fig. 2D) and of Badumna longinqua, a distantly related species with similar thread structure to D. subrufa (i.e. including undulating fibres) (Fig. 2E). Please note, that only for D. subrufa were we able to collect enough material to perform a solid-state nuclear magnetic resonance spectroscopy (described below).
B. longinqua (L. Koch, 1867) were collected in both Brisbane and Sydney (Australia) and returned to the laboratory at the University of New South Wales. In the lab, they were kept at 25 °C, 60% rH, light cycle within Perspex frames, wherein they built their capture webs. From which only the cribellate capture threads, without the frame threads, were removed for analysis.
U. plumipes (Lucas, 1846) were collected at garden centres across Germany. In the lab, they were kept at 20 °C with 10 h light. Only the capture threads were taken from the web for analysis. As negative control, the dragline of the spiders was harvested by forced silking.
Since we know its amino acid composition and expected protein secondary structures,71,72 we used major ampullate silk from the orb web spider Trichonephila plumipes (Latreille, 1804) as a procedural control for the GC-MS and ssNMR methodologies outlined below. The silks used were taken from a silk repository at the Spider Silk Research Laboratory, University of New South Wales. The original collection methods are outlined by Blamires et al.72
Silk staining
An Amido black dye was prepared by dissolving 0.002 g Amido black 10B (Merck KGaA, Darmstadt, Germany) in 200 μL of 10% acetic acid (AA; 100%, Carl Roth GmbH & Co. KG, Karlsruhe, Germany) and 200 μL of 50% methanol (MeOH; 99.9%, Merck KGaA, Darmstadt, Germany). A volume of 30 μL of the staining solution was applied to the thread. The solution was left to react for at least 30 min. Decolourisation was done either with 50% methanol or deionized water (DI water). Any residues of the liquid or staining around the threads were removed with a filter paper. The samples were then air dried for a few minutes. Coomassie Brilliant Blue dye was prepared by dilution of 0.0025 g Serva Blue G250 (Serva Electrophoresis GmbH, Heidelberg, Germany) with 454 μL isopropyl alcohol (IPA; ≥99.5%, Carl Roth GmbH & Co. KG, Karlsruhe, Germany), 454 μL DI water and 92 μL acetic acid. 100 μL of the staining solution was added to the thread. Decolourisation took place after about 10 min by DI water.Electron microscopy
For cross-sectional analyses, ultramicromy of resin-embedded threads was performed. Threads under tension were mounted on a metal sample holder and treated to three different dehydration procedures; (i) a graded ethanol series with increasing percentages of ethanol (30%, 50%, 70%, 90%, 100%, 100%) (ii) a graded ethanol series with increasing percentages of acetone (30%, 50%, 70%, 90%, 100%, 100%), or (iii) threads were infiltrated with resins directly without dehydration. Cribellate threads from each procedure above were embedded into three different resins LX112 (Ladd Research, USA), Procure812 (Proscitech, Australia) or LR White resin (Proscitech, Australia) and polymerised to hardness at 60 °C. 70 nm ultrathin sections were collected and imaged on a JEOL1400 TEM at 100 kV. No discernible structure could be found.For transmission electron microscopy, thread samples of U. plumipes were picked up on a finder-grid (Plano GmbH) and observed without any further treatment (EM 10, Carl Zeiss AG). The nanofibres tend to rupture when exposed for longer time to the electron beam. Hence, images for measuring fibre diameter were quickly made to avoid or at least reduce artefacts. As the threads displayed significant dose-sensitivity, we undertook further TEM analysis of D. subrufa and B. longinqua silk using cryogenic-TEM (cryoTEM). Thread samples were adhered to a glow discharged quantifoil grid and rapidly plunge frozen in a Leica EM GP. Grids were imaged under low dose conditions on a 200 kV Talos Arctica fitted with a Falcon 3 direct electron detecting camera (Thermo Fisher Scientific).
Atomic force microscopy (AFM)
Cribellate capture threads of the webs of B. longinqua and D. subrufa were isolated by burning these from the web with hot needles and then transferred to freshly cleaned glass cover slips. To measure the elasticity of isolated nanofibres, threads were carefully pulled over the cover slip, thus single nanofibres could detach from the bundle as they adhered to the ground.All of the AFM measurements were performed on a Bruker Dimension ICON SPM with the SCANASYST-Air probe (Bruker AFM probes), respectively an OTESPA probe (Bruker AFM probes) at the Electron Microscopy Unit at the University of New South Wales. Derjaguin–Muller–Toporov (DMT) modulus was estimated along the fibre using peak force tapping mode. For accurate modulus measurements, the probe was calibrated according to the following procedure: Firstly, the probe was engaged on a hard surface (fused silica) with an engage set point of 0.05 V and a scan size of 0 nm. Then the probe was ramped on the surface with 0.1 V of deflection (trig threshold) to calculate the deflection sensitivity (∼77 nm V−1). Then the probe was withdrawn from the surface and lifted off the surface a few millimetres, so the spring constant could be calculated (∼ 0.3 N m−1) using thermal tuning. The tip radius of the probe was checked with a titanium roughness check (i.e. sample with sharp titanium flakes to check the tip radius) with a slow scan rate of 0.2 Hz, as the features of this sample are sharp and rough and can damage the probe apex if the scan rate is too high. After all the constants were determined, the probe was engaged on the silk sample surface. The scanning parameters, such as scan rate, peak force set point, feedback gain were optimized accordingly. The resolution of image was kept at 512 samples/line. Typical scan parameters were: Scan size: 3 μm; Scan rate: 0.25 Hz; Feedback gain: ∼12; Peak force set point: 1.5 nN; Poisson ratio: 0.5.73
AFM analyses were performed for D. subrufa as well as B. longinqua silk with both AFM probes. However, as quality of the data differed dramatically, analysis was only performed for the D. subrufa silk imaged with the OTESPA probe, and for B. longinqua, using the SCANASYST-Air probe. For data of the respective other tip, see Fig. S1.†
Solid-state nuclear magnetic resonance spectroscopy (ssNMR)
13C ssNMR was used to examine and compare the silk protein structures at the NMR Facility within the Mark Wainwright Analytical Centre, University of New South Wales. The cribellate web (15–30 mg) of D. subrufa were centre packed in 4 mm zirconia MAS rotors with Kel-f ® caps. The ssNMR measurements were carried out in a Bruker Avance III NMR spectrometer, with a 16.4 Tesla superconducting magnet operating at frequencies of 175 MHz and 700 MHz for 13C and 1H nuclei respectively. The 4 mm NMR rotor was spun to 8 kHz in a 4 mm double resonance probehead at the magic angle. The 13C NMR spectra were acquired with 1H to 13C cross-polarization with a 2 ms cross-polarization contact time, the TOSS scheme to remove the sinning sidebands, SPINAL-64 decoupling scheme for 1H decoupling with a field strength of 100 kHz and a recycle delay of 3 s. 4k−32k transients were co-added to minimize signal-to-noise.Gas chromatography-mass spectroscopy (GC-MS)
Silk samples (0.1–0.25 mg), containing the internal standards U-Glycine (13C2, 15N) and 4-chlorophenyalanine (50 nmol) were lyophilized in borosilicate glass, then placed into a mini-inert reaction vessel and evacuated using a vacuum pump. Samples were vapour phase acid hydrolysed in 6M HCl containing 1% phenol at 110 °C for 18 hours. Hydrolysates were lyophilized and then derivatised at 50 °C for 1 h with 15 μl acetonitrile plus 30 μl Nmethyl-N-(t-butyldimethylsilyl)trifluoroacetamide +1% t-butyldimethylchlorosilane (MtBSTFA +1% tBDMCS). After reconstitution in iso-octane t-butyldirimethylsilyl amino acid derivatives analysed by GC-MS (Agilent 5890II gas chromatograph and 5973A mass spectrometer). Derivatised samples (1 μL) were injected into a fused silica capillary column (25 m × 0.25 mm internal diameter) coated with cross-linked 5% phenylmethylsiloxane (film thickness 0.25 μm; Restek Rxi-5 ms) using a 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 split ratio and a Helium carrier gas flow rate of 1 mL min−1.
1 split ratio and a Helium carrier gas flow rate of 1 mL min−1.
Selected ion monitoring was performed by using the electron-ionization (EI) mode at 70 eV. Quantification of amino acids was performed by monitoring specific ions at standard retention times and calibrated using amino acid standard mixes analysed in the same batch. Data analysis was carried out using Agilent Masshunter software. For quality control, samples of bovine serum albumin and amino acid standard mixes containing internal standards were also hydrolysed, derivatised and analysed by GC-MS together with silk samples (see Table S1†). We put in blanks with no protein and observed no cross contamination of the samples during hydrolysis (i.e. blanks had no significant amino acids present).
The acid hydrolysis may have caused some amino acids, i.e. asparagine and glutamine, to become deaminated to their respective acids during hydrolysis and thus could not be differentiated by GC-MS. Amino acids with any value below 0.1% are most likely not present in the protein.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Schwedt and R. Harscheidt from the central facility for electron microscopy of the RWTH Aachen were so kind to provide the close-up picture of a carbon coated cribellate fibre (Fig. 4B). The scheme of the D. subrufa web collection was kindly provided by N. Rosenkranz (Fig. 8). The graphical table of content was designed by L. Kupferschläger. Many thanks to J. R. N. Linde and C. N. Creak for assistance in silk collection.This work was supported by the German national science foundation [JO 1464/1-1 and JO 1464/2-1 to A-CJ], the European Union's Horizon 2020 research and innovation programme within the project “BioCombs4Nanofibers” [grant agreement No. 862016], the Excellence Initiative of the German federal and state governments, the Rheinisch-Westfälische Technische Hochschule Aachen [RFwN program to MW] and by a Hermon Slade Foundation [HSF17/6] and Plus Alliance [PA21089] grant to SJB and AR.
References
- J. O. Wolff, D. Wells, C. R. Reid and S. J. Blamires, Bioinspiration Biomimetics, 2017, 12, 11 CrossRef PubMed
.
- S. J. Blamires, P. T. Spicer and P. J. Flanagan, Front. Mater., 2020, 7, 7 CrossRef
.
- M. Humenik, G. Lang and T. Scheibel, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10, 24 Search PubMed
.
- A. Luken, M. Geiger, L. Steinbeck, A. C. Joel, A. Lampert, J. Linkhorst and M. Wessling, Adv. Healthcare Mater., 2021, 10, 9 CrossRef PubMed
.
- F. Muller, S. Zainuddin and T. Scheibel, Molecules, 2020, 25, 12 Search PubMed
.
-
S. J. Blamires, T. A. Blackledge and I. M. Tso, in Annual Review of Entomology, ed. M. R. Berenbaum, Annual Reviews, Palo Alto, 2017, vol. 62, pp. 443–460 Search PubMed
.
-
C. L. Craig and L. Catherine, Spiderwebs and silk: tracing evolution from molecules to genes to phenotypes, Oxford University Press, 2003 Search PubMed
.
- A. Heidebrecht, L. Eisoldt, J. Diehl, A. Schmidt, M. Geffers, G. Lang and T. Scheibel, Adv. Mater., 2015, 27, 2189–2194 CrossRef CAS PubMed
.
- C. Holland, F. Vollrath, A. J. Ryan and O. O. Mykhaylyk, Adv. Mater., 2012, 24, 105 CrossRef CAS PubMed
.
- Y. X. Yang, Z. G. Qian, J. J. Zhong and X. X. Xia, Process Biochem., 2016, 51, 484–490 CrossRef CAS
.
- C. H. Bowen, B. Dai, C. J. Sargent, W. Q. Bai, P. Ladiwala, H. B. Feng, W. W. Huang, D. L. Kaplan, J. M. Galazka and F. Z. Zhang, Biomacromolecules, 2018, 19, 3853–3860 CrossRef CAS PubMed
.
- A. Sponner, W. Vater, S. Monajembashi, E. Unger, F. Grosse and K. Weisshart, PLoS One, 2007, 2, 8 CrossRef PubMed
.
- Y. Termonia, Macromolecules, 1994, 27, 7378–7381 CrossRef CAS
.
- M. Heim, D. Keerl and T. Scheibel, Angew. Chem., Int. Ed., 2009, 48, 3584–3596 CrossRef CAS PubMed
.
- Q. Wang and H. C. Schniepp, ACS Macro Lett., 2018, 7, 1364–1370 CrossRef CAS PubMed
.
- S. R. Koebley, F. Vollrath and H. C. Schniepp, Mater. Horiz., 2017, 4, 377–382 RSC
.
- H. C. Schniepp, S. R. Koebley and F. Vollrath, Adv. Mater., 2013, 25, 7028–7032 CrossRef CAS PubMed
.
- R. Lehmensick and E. Kullmann, Zool. Anz., 1957, 20, 123–129 Search PubMed
.
- V. Friedrich and R. M. Langer, Am. Zool., 1969, 9, 91–96 CrossRef
.
- J. B. Addison, T. M. Osborn Popp, W. S. Weber, J. S. Edgerly, G. P. Holland and J. L. Yarger, RSC Adv., 2014, 4, 41301–41313 RSC
.
- H. M. Peters, Zoomorphology, 1984, 104, 96–104 CrossRef
.
- A.-C. Joel, P. Kappel, H. Adamova, W. Baumgartner and I. Scholz, Arthropod Struct. Dev., 2015, 44, 568–573 CrossRef PubMed
.
- A.-C. Joel, I. Scholz, L. Orth, P. Kappel and W. Baumgartner, R. Soc. Open Sci., 2016, 3, 150617 CrossRef PubMed
.
- C. C. F. Grannemann, M. Meyer, M. Reinhardt, M. J. Ramírez, M. E. Herberstein and A.-C. Joel, Sci. Rep., 2019, 9, 17273 CrossRef PubMed
.
- M. Weissbach, M. Neugebauer and A.-C. Joel, J. Comp. Physiol., A, 2021, 207, 127–139 CrossRef PubMed
.
- H. M. Peters, Zoomorphology, 1992, 112, 27–37 CrossRef
.
- R. A. Bott, W. Baumgartner, P. Bräunig, F. Menzel and A.-C. Joel, Proc. R. Soc. London, Ser. B, 2017, 284, 20170363 CrossRef PubMed
.
- H. Elettro, S. Neukirch, A. Antkowiak and F. Vollrath, Denkschr. Schweiz. Naturforsch. Ges., 2015, 102, 41 Search PubMed
.
- A. C. Hawthorn and B. D. Opell, J. Exp. Biol., 2003, 206, 3905–3911 CrossRef PubMed
.
- D. Piorkowski, C.-P. Liao, A.-C. Joel, C.-L. Wu, N. Doran, S. J. Blamires, N. M. Pugno and I. M. Tso, J. Mech. Behav. Biomed. Mater., 2021, 114, 104200 CrossRef CAS PubMed
.
- A.-C. Joel and W. Baumgartner, J. Exp. Biol., 2017, 220, 2243–2249 Search PubMed
.
- V. M. Ortega-Jimenez and R. Dudley, Sci. Rep., 2013, 3, 4 Search PubMed
.
- T. A. Blackledge and C. Y. Hayashi, J. Exp. Biol., 2006, 209, 3131–3140 CrossRef PubMed
.
- D. Piorkowski, T. A. Blackledge, C.-P. Liao, A.-C. Joel, M. Weissbach, C.-L. Wu and I. M. Tso, J. Exp. Biol., 2020, 223, jeb215269 CrossRef PubMed
.
- X. Liao, G. Yin, Z. Huang, Y. Yao, J. Gu and D. Han, Mater. Sci. Eng., C, 2011, 31, 128–133 CrossRef CAS
.
- Z. B. Huang, D. H. Yan, M. Yang, X. M. Liao, Y. Q. Kang, G. F. Yin, Y. D. Yao and B. Q. Hao, J. Colloid Interface Sci., 2008, 325, 356–362 CrossRef CAS PubMed
.
- Z. Huang, X. Liao, G. Yin, Y. Kang and Y. Yao, J. Phys. Chem. B, 2009, 113, 5092–5097 CrossRef CAS PubMed
.
- S. M. Correa-Garhwal, R. C. Chaw, T. H. Clarke III, L. G. Alaniz, F. S. Chan, R. E. Alfaro and C. Y. Hayashi, PLoS One, 2018, 13, e0203563 CrossRef PubMed
.
- S. M. Correa-Garhwal, T. H. Clarke, M. Janssen, L. Crevecoeur, B. N. McQuillan, A. H. Simpson, C. J. Vink and C. Y. Hayashi, Sci. Rep., 2019, 9, 12 CrossRef PubMed
.
- N. Kono, H. Nakamura, M. Mori, M. Tomita and K. Arakawa, Sci. Rep., 2020, 10, 15721 CrossRef CAS PubMed
.
- S. M. Correa-Garhwal, R. H. Baker, T. H. Clarke, N. A. Ayoub and C. Y. Hayashi, BMC Ecol. Evol., 2022, 22, 89 CrossRef CAS PubMed
.
- C. E. Griswold, M. J. Ramirez, J. A. Coddington and N. I. Platnick, Proc. Calif. Acad. Sci., 2005, 56, 1 Search PubMed
.
- K. Kronenberger and F. Vollrath, Biol. Lett., 2015, 11, 20140813 CrossRef PubMed
.
- A. C. Hawthorn and B. D. Opell, Biol. J. Linn. Soc., 2002, 77, 1–8 CrossRef
.
-
S. Blamires, Silk: Exploring Nature’s Superfibre, Xlibris Corporation, 2022 Search PubMed
.
- S. Kambhampati, J. Li, B. S. Evans and D. K. Allen, Plant Methods, 2019, 15, 46 CrossRef PubMed
.
- H. C. Craig, Y. Yao, N. Ariotti, M. Setty, R. Remadevi, M. M. Kasumovic, R. Rajkhowa, A. Rawal and S. J. Blamires, J. Mater. Chem. B, 2022, 10, 5561–5570 RSC
.
- M. Pechmann, S. Khadjeh, F. Sprenger and N.-M. Prpic, Arthropod Struct. Dev., 2010, 39, 453–467 CrossRef PubMed
.
- M. R. Gray, Rec West Aust Mus., 1995, 52, 79–89 Search PubMed
.
- J. D. van Beek, S. Hess, F. Vollrath and B. H. Meier, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10266–10271 CrossRef CAS PubMed
.
- J. M. Gosline, P. A. Guerette, C. S. Ortlepp and K. N. Savage, J. Exp. Biol., 1999, 202, 3295–3303 CrossRef CAS PubMed
.
- A. H. Simmons, C. A. Michal and L. W. Jelinski, Science, 1996, 271, 84–87 CrossRef CAS PubMed
.
- A.-C. Joel, D. Schmitt, L. Baumgart and F. Menzel, J. Exp. Biol., 2022, 225, jeb242514 CrossRef PubMed
.
- H. C. Craig, S. J. Blamires, M. A. Sani, M. M. Kasumovic, A. Rawal and J. M. Hook, Chem. Commun., 2019, 55, 4687–4690 RSC
.
- K. Okushita, A. Asano, M. P. Williamson and T. Asakura, Macromolecules, 2014, 47, 4308–4316 CrossRef CAS
.
- T. Asakura, K. Okushita and M. P. Williamson, Macromolecules, 2015, 48, 2345–2357 CrossRef CAS
.
- N. N. Ashton and R. J. Stewart, FASEB J., 2019, 33, 572–583 CrossRef CAS PubMed
.
- J. B. Addison, W. S. Weber, Q. Mou, N. N. Ashton, R. J. Stewart, G. P. Holland and J. L. Yarger, Biomacromolecules, 2014, 15, 1269–1275 CrossRef CAS PubMed
.
- P. B. Frandsen, M. G. Bursell, A. M. Taylor, S. B. Wilson, A. Steeneck and R. J. Stewart, Philos. Trans. R. Soc., B, 2019, 374, 20190206 CrossRef CAS PubMed
.
- J. R. A. dos Santos-Pinto, H. A. Arcuri, F. G. Esteves, M. S. Palma and G. Lubec, Sci. Rep., 2018, 8, 14674 CrossRef PubMed
.
- S. Xu, M. Gu, K. Wu and G. Li, J. Phys. Chem. B, 2019, 123, 7754–7763 CrossRef CAS PubMed
.
- K. Mizuno, T. Hayashi, D. H. Peyton and H. P. Bächinger, J. Biol. Chem., 2004, 279, 38072–38078 CrossRef CAS PubMed
.
- C. A. Miles, L. Knott, I. G. Sumner and A. J. Bailey, J. Mol. Biol., 1998, 277, 135–144 CrossRef CAS PubMed
.
- T. A. Blackledge and C. Y. Hayashi, J. Exp. Biol., 2006, 209, 2452–2461 CrossRef PubMed
.
- T. Kohler and F. Vollrath, J. Exp. Zool., 1995, 271, 1–17 CrossRef
.
- C. Y. Hayashi and R. V. Lewis, BioEssays, 2001, 23, 750–756 CrossRef CAS PubMed
.
- A.-C. Joel and M. Weissbach, Integr. Comp. Biol., 2019, 59, 1673–1680 CrossRef PubMed
.
- B. D. Opell, Biol. J. Linn. Soc., 1999, 68, 593–612 CrossRef
.
- B. D. Opell, J. Arachnol., 2002, 30, 10–19 CrossRef
.
- P. Michalik, D. Piorkowski, T. A. Blackledge and M. J. Ramírez, Sci. Rep., 2019, 9, 9092 CrossRef PubMed
.
- S. J. Blamires, M. Nobbs, P. J. Martens, I. M. Tso, W. T. Chuang, C. K. Chang and H. S. Sheu, PLoS One, 2018, 13, 23 CrossRef PubMed
.
- S. J. Blamires, M. M. Kasumovic, I. M. Tso, P. J. Martens, J. M. Hook and A. Rawal, Int. J. Mol. Sci., 2016, 17, 13 CrossRef PubMed
.
- S. P. Patil, B. Markert and F. Grater, Biophys. J., 2014, 106, 2511–2518 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Additional AFM data and the data of the GC-MS analysis. See DOI: https://doi.org/10.1039/d2bm01599d |
| This journal is © The Royal Society of Chemistry 2023 |