Open Access Article
Open Access ArticleUPLC-MS/MS method for quantitative determination of the advanced glycation endproducts Nε-(carboxymethyl)lysine and Nε-(carboxyethyl)lysine†
Lauren A.
Skrajewski-Schuler
ab,
Logan D.
Soule
 bc,
Morgan
Geiger
bc,
Morgan
Geiger
 bc and
Dana
Spence
bc and
Dana
Spence
 *bc
*bc
aDepartment of Chemistry, Michigan State University, East Lansing, MI 48824, USA
bInstitute for Quantitative Health, Michigan State University, East Lansing, MI 48824, USA. E-mail: spenceda@msu.edu; Tel: +1 517 353 1116
cDepartment of Biomedical Engineering, Michigan State University, East Lansing, MI 48824, USA
First published on 1st December 2023
Abstract
During blood storage, red blood cells (RBCs) undergo physical, chemical, and metabolic changes that may contribute to post-transfusion complications. Due to the hyperglycemic environment of typical solutions used for RBC storage, the formation of advanced glycation endproducts (AGEs) on the stored RBCs has been implicated as a detrimental chemical change during storage. Unfortunately, there are limited studies involving quantitative determination and differentiation of carboxymethyl-lysine (CML) and carboxyethyl-lysine (CEL), two commonly formed AGEs, and no reported studies comparing these AGEs in experimental storage solutions. In this study, CML and CEL were identified and quantified on freshly drawn blood samples in two types of storage solutions, standard additive solution 1 (AS-1) and a normoglycemic version of AS-1 (AS-1N). To facilitate detection of the AGEs, a novel method was developed to reliably extract AGEs from RBCs, provide Food and Drug Administration (FDA) bioanalytical guidance criteria, and enable acceptable selectivity for these analytes. Ultra-performance liquid chromatography with tandem mass spectrometry (UPLC-MS/MS) was utilized to identify and quantify the AGEs. Results show this method is accurate, precise, has minimal interferences or matrix effects, and overcomes the issue of detecting AGE byproducts. Importantly, AGEs can be detected and quantified in both types of blood storage solutions (AS-1 and AS-1N), thereby enabling long-term (6 weeks) blood storage related studies.
Introduction
Transfusion medicine is a critical component of modern healthcare, evident by the 10.7 million units of red blood cells (RBCs) transfused annually to patients.1 The collection of whole blood and subsequent storage of various blood components (such as the RBCs or plasma) into units ready for transfusion is relatively simple. Briefly, the process involves collection of ∼450 mL of whole blood from donors, centrifugation to separate the RBCs from the plasma and leukocytes, followed by storage in separate bags at 4 °C for various lengths of time depending on the component (plasma or RBC) and country regulations.2,3A key feature of current protocols for RBC storage is the collection solution into which the whole blood is drawn, and the solution into which the separated RBCs are stored (the latter also known as additive solution).4 The most popular collection solution is citrate–phosphate–dextrose (CPD), which contains citrate, phosphate, and dextrose (glucose) at concentrations shown in Table 1.5,6 Following centrifugation, the RBCs are then stored in one of multiple available additive solutions (e.g., AS-1, AS-3, AS-5, or AS-7).5,7,8 The contents of AS-1 are also shown in Table 1. While the current blood storage procedure has been in place since the 1970s, there are many reports showing adverse effects of storage over time on the RBC's chemical and physical properties.9–13 These adverse effects, collectively known as the RBC storage lesion, involve chemical, physical, and metabolic changes, as well as functional changes, to the RBC while in storage.14,15 Unfortunately, the exact origins of the storage lesion are not known, nor is the mechanism leading to the various changes to the stored RBC well understood.
| Constituent (mM) | CPD | CPD-N | AS-1 | AS-1N |
|---|---|---|---|---|
| Glucose | 129 | 5.5 | 111 | 5.5 |
| Sodium citrate | 89.4 | 89.4 | ||
| Monobasic sodium phosphate | 16.1 | 16.1 | ||
| Citric acid | 15.6 | 15.6 | ||
| Sodium chloride | 154 | 154 | ||
| Adenine | 2.0 | 2.0 | ||
| Mannitol | 41 | 41 | ||
| pH | 5.6 | 5.6 | 5.8 | 5.8 |
An interesting feature of the collection and additive solutions used in RBC storage is the high level of glucose in the CPD and the AS-1. Specifically, typical blood glucose concentration ranges from 4–6 mM in a healthy person.16 Currently, approved versions of CPD and AS-1 have glucose concentrations that exceed 110 mM; even after the RBCs are added, and the AS-1 is diluted due to mixing of the RBCs with the AS-1, the concentration of the glucose in the RBC/AS-1 solution is still in excess of 40 mM, a value much higher than that of healthy humans and humans with diabetes.10,17 It is noteworthy that after transfusion of the ∼280 mL of RBCs into a human, the glucose concentration in the storage bag will not affect the glucose levels in the human transfusion recipient (due to dilution of the 280 mL into a human who typically has a total blood volume of ∼5 L); rather, the concerning feature of the high glucose is the effect on the RBC properties during storage.
In continuance, people with high blood glucose levels, such as people with diabetes, have RBCs with increased levels of advanced glycation endproducts (AGEs), which are thought to be a negative determinant in overall cell health.18,19 Past work involving AGEs on the RBCs in the hyperglycemic bloodstream of people with diabetes provided the rationale to investigate the possible formation of AGEs on the RBCs in AS-1. A previous report20 suggests the formation of AGEs later in storage and provides motivation to (1) quantitatively determine the concentrations of the AGEs being formed and (2) evaluate these concentrations of RBC-bound AGEs from the beginning (day 1) to the end of storage duration (>42 days).15,20,21 Such time-based studies of AGE formation on stored RBCs are without precedent. Here, we describe novel mass spectrometric determination of two AGEs, Nε-carboxymethyl-lysine (CML) and Nε-carboxyethyl-lysine (CEL).22 The glyoxal mechanism produces CML, the first AGE discovered and the most widely studied AGE.23 Another important AGE, CEL, is associated with diabetes-related complications and derived from the methylglyoxal pathway.24 Measurement of the formation of these two AGEs (CML and CEL) on the stored RBC is the focus of this study to subsequently serve as a tool for determination of improved storage products in transfusion medicine.
Experimental methods and materials
RBC collection
Blood draw followed a protocol approved by the Institutional Review Board of Michigan State University. Blood was obtained from healthy humans and informed consent was obtained from all donors. All record keeping complied with Health Insurance Portability and Accountability Act regulations. Whole blood was collected via venipuncture from healthy consenting donors (140 mL total whole blood) into multiple 10 mL uncoated collection tubes (Thermo Fisher Scientific, Waltham, MA) that were prepared to contain either 1 mL of CPD or 1 mL of a normoglycemic version (CPD-N) (see Table 1). Each blood draw was divided into CPD or CPD-N solution tubes to maintain consistency between storage solution results; that is, each donor had 5 tubes of blood collected in CPD and 5 tubes collected in CPD-N. All whole blood tubes were centrifuged at 2000g for 10 minutes, the plasma and buffy coat (containing the white cells or leukocytes) were removed by aspiration, and packed RBCs (pRBCs) were kept in the tubes.Glucose concentration and hematocrit percentage
The pRBCs were added to either AS-1 or AS-1N in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio and mixed and stored at 4 °C for one hour before initial glucose concentration and percent hematocrit (the percentage of volume occupied by the RBCs) readings. The glucose concentration was determined with an Aimstrip Plus Blood Glucose Meter (VWR, Radnor, PA) using a 22-gauge needle and 1 mL syringe to collect and transfer a drop of RBC sample onto an Aimstrip Plus Blood Glucose Test Strip (VWR). The glucose reading was repeated, and the average glucose concentration (mg dL−1) was converted to a mM value. The glucose was adjusted to 5.5 mM after one hour for the AS-1N sample by adding an appropriate volume (typically between 50–400 μL) of a 100 mM glucose solution in 0.9% saline. The same volume of 0.9% saline was added to the AS-1 sample to maintain similarity in handling between the AS-1 and AS-1N. The RBC sample hematocrit was determined using a StatSpin MP microhematocrit centrifuge (Beckman Coulter, Brea, CA) and a hematocrit reader (StatSpin CritSpin). The remaining pRBCs were used for analysis either the same day or stored at 4 °C. Periodic feeding of the RBCs stored in AS-1N was achieved using a closed and automated feeding system. Stored samples were removed from bags on day 1, 8, 15, 22, 29, 36, and 43 for UPLC-MS/MS analysis following sample preparation techniques described below.
1 volume ratio and mixed and stored at 4 °C for one hour before initial glucose concentration and percent hematocrit (the percentage of volume occupied by the RBCs) readings. The glucose concentration was determined with an Aimstrip Plus Blood Glucose Meter (VWR, Radnor, PA) using a 22-gauge needle and 1 mL syringe to collect and transfer a drop of RBC sample onto an Aimstrip Plus Blood Glucose Test Strip (VWR). The glucose reading was repeated, and the average glucose concentration (mg dL−1) was converted to a mM value. The glucose was adjusted to 5.5 mM after one hour for the AS-1N sample by adding an appropriate volume (typically between 50–400 μL) of a 100 mM glucose solution in 0.9% saline. The same volume of 0.9% saline was added to the AS-1 sample to maintain similarity in handling between the AS-1 and AS-1N. The RBC sample hematocrit was determined using a StatSpin MP microhematocrit centrifuge (Beckman Coulter, Brea, CA) and a hematocrit reader (StatSpin CritSpin). The remaining pRBCs were used for analysis either the same day or stored at 4 °C. Periodic feeding of the RBCs stored in AS-1N was achieved using a closed and automated feeding system. Stored samples were removed from bags on day 1, 8, 15, 22, 29, 36, and 43 for UPLC-MS/MS analysis following sample preparation techniques described below.
Protein precipitation for free lysine quantification
To promote cell lysis, packed RBCs (100 μL) were frozen at −20 °C for one hour and then thawed for 10 minutes at room temperature before mixing with 300 μL of HPLC grade acetonitrile. The sample was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes at room temperature, and the supernatant was removed and stored at −80 °C until dried using a SpeedVac (Savant SpeedVac Concentrator, Thermo Fisher Scientific) with an acid vapor trap (Savant Refrigerated Vapor Trap, Thermo Fisher Scientific). The dried sample pellet was stored at −80 °C until prepared for measurement.
000g for 10 minutes at room temperature, and the supernatant was removed and stored at −80 °C until dried using a SpeedVac (Savant SpeedVac Concentrator, Thermo Fisher Scientific) with an acid vapor trap (Savant Refrigerated Vapor Trap, Thermo Fisher Scientific). The dried sample pellet was stored at −80 °C until prepared for measurement.
Preparation of RBC samples for UPLC-MS/MS
RBC samples (100 μL) for acid hydrolysis were frozen for one hour at −20 °C and thawed for 10 minutes at room temperature before centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes at 4 °C. The supernatant was removed, and pRBCs were collected for acid hydrolysis. The pRBCs were diluted to make a 1% RBC solution in constant boiling sequencing grade 6 M HCl (Thermo Fisher Scientific) in glass tubes and transferred to 10 mm, 6 mL vacuum hydrolysis tubes (Thermo Fisher Scientific). The samples were hydrolyzed at 110 °C for 16 hours using a 120 V Digital Dry Bath/Block Heater and Dry Bath Block Insert (Thermo Fisher Scientific). The samples were then removed using customized glassware (MSU Chemistry glass shop) designed to attach a 2 mL Pasteur rubber pipette bulb (Sigma Aldrich, St. Louis, MO) and extract the sample (see Fig. S1 in the ESI†). The glassware was designed using current Pasteur pipette models to extract a sample that would otherwise be unable to be removed cohesively. Using the glassware, the sample was placed into 1.7 mL tubes and frozen overnight at −80 °C. The following day, the samples were dried to completion using a SpeedVac at 75 °C for 4–5 hours and then kept at −80 °C until ready for measurement.
000g for 10 minutes at 4 °C. The supernatant was removed, and pRBCs were collected for acid hydrolysis. The pRBCs were diluted to make a 1% RBC solution in constant boiling sequencing grade 6 M HCl (Thermo Fisher Scientific) in glass tubes and transferred to 10 mm, 6 mL vacuum hydrolysis tubes (Thermo Fisher Scientific). The samples were hydrolyzed at 110 °C for 16 hours using a 120 V Digital Dry Bath/Block Heater and Dry Bath Block Insert (Thermo Fisher Scientific). The samples were then removed using customized glassware (MSU Chemistry glass shop) designed to attach a 2 mL Pasteur rubber pipette bulb (Sigma Aldrich, St. Louis, MO) and extract the sample (see Fig. S1 in the ESI†). The glassware was designed using current Pasteur pipette models to extract a sample that would otherwise be unable to be removed cohesively. Using the glassware, the sample was placed into 1.7 mL tubes and frozen overnight at −80 °C. The following day, the samples were dried to completion using a SpeedVac at 75 °C for 4–5 hours and then kept at −80 °C until ready for measurement.
Reagent materials and preparation
Immediately prior to measurement, the dried RBC sample used to detect free lysine sample pellet was reconstituted in 300 μL of 10 mM perfluoroheptanoic acid (PFHA) in water, and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes. The dried, hydrolyzed additive solution RBC samples were reconstituted in 500 μL of 10 mM PFHA in water, and centrifuged at 13
000g for 10 minutes. The dried, hydrolyzed additive solution RBC samples were reconstituted in 500 μL of 10 mM PFHA in water, and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes. All supernatant samples were removed and used for measurement or stored at −80 °C. The free lysine sample was diluted 1
000g for 10 minutes. All supernatant samples were removed and used for measurement or stored at −80 °C. The free lysine sample was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 using 10 mM PFHA in water. Each additive sample supernatant was divided into two categories: CML/CEL detection and lysine detection. The lysine detection samples were diluted 1
100 using 10 mM PFHA in water. Each additive sample supernatant was divided into two categories: CML/CEL detection and lysine detection. The lysine detection samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in 10 mM PFHA, and the CML/CEL detection samples were diluted 1
000 in 10 mM PFHA, and the CML/CEL detection samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in 10 mM PFHA. The sample supernatants were mixed 1
40 in 10 mM PFHA. The sample supernatants were mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) with an internal standard (IS) mixture containing: 0.1 μM Nε-(1-carboxymethyl)-L-lysine-d3 (Cayman Chemical, Ann Arbor, MI), 0.1 μM carboxyethyl-L-lysine-d4 (Toronto Research Chemicals, Toronto, ON), and 0.1 μM 13C6, 15N2 labelled L-Lysine (Sigma Aldrich). Nε-(1-carboxymethyl)-L-lysine (Cayman Chemical), Nε-(1-carboxyethyl)-L-lysine (Cayman Chemical), and 13C6, 15N2-lysine (Sigma Aldrich) are used to prepare calibrator samples by dissolving the lyophilized samples in water, diluting with 10 mM PFHA for a ten-point calibration curve, and mixing with the same internal standard mixture as above. The ten-point calibration curve (0, 4, 8, 16, 64, 128, 512, 1500, 2048, 5000 nM) was used to quantify CML, CEL, and lysine by adjusting analyte peak area relative to associated internal standards to illicit an overall response. During initial sample preparation, AS-1 and AS-1N solutions were also hydrolyzed to determine if diluting the samples in reconstituted sample solution would decrease matrix effects. Quality control (QC) samples were prepared using a stock of RBCs, from a healthy consenting donor, in AS-1 or AS-1N following the procedure above, preparing aliquots of the hydrolyzed sample stock into 1.7 mL vials, drying, and freezing at −80 °C until they were reconstituted for measurement.
1 (v/v) with an internal standard (IS) mixture containing: 0.1 μM Nε-(1-carboxymethyl)-L-lysine-d3 (Cayman Chemical, Ann Arbor, MI), 0.1 μM carboxyethyl-L-lysine-d4 (Toronto Research Chemicals, Toronto, ON), and 0.1 μM 13C6, 15N2 labelled L-Lysine (Sigma Aldrich). Nε-(1-carboxymethyl)-L-lysine (Cayman Chemical), Nε-(1-carboxyethyl)-L-lysine (Cayman Chemical), and 13C6, 15N2-lysine (Sigma Aldrich) are used to prepare calibrator samples by dissolving the lyophilized samples in water, diluting with 10 mM PFHA for a ten-point calibration curve, and mixing with the same internal standard mixture as above. The ten-point calibration curve (0, 4, 8, 16, 64, 128, 512, 1500, 2048, 5000 nM) was used to quantify CML, CEL, and lysine by adjusting analyte peak area relative to associated internal standards to illicit an overall response. During initial sample preparation, AS-1 and AS-1N solutions were also hydrolyzed to determine if diluting the samples in reconstituted sample solution would decrease matrix effects. Quality control (QC) samples were prepared using a stock of RBCs, from a healthy consenting donor, in AS-1 or AS-1N following the procedure above, preparing aliquots of the hydrolyzed sample stock into 1.7 mL vials, drying, and freezing at −80 °C until they were reconstituted for measurement.
Chromatography and MRM mass spectrometry
To determine if CML, CEL, and lysine were present, the reconstituted hydrolyzed RBC samples were analyzed by UPLC-MS/MS using a Waters Xevo TQ-S Micro QqQ system interfaced with a Waters Acquity UPLC. A 10 μL aliquot was injected onto a reverse phase column (High Strength Silica (HSS) T3 2.1 × 100 mm) and the compounds were separated using ion-pair chromatography. Mobile phase A was 10 mM PFHA in water and mobile phase B consisted of 10 mM ammonium formate in a solution consisting of 45% acetonitrile, 45% methanol, and 10% water. The analytes were eluted using the following gradient: initial conditions were 90% mobile phase A and 10% mobile phase B for 5 minutes and then changed to 35% mobile phase A and 65% mobile phase B before switching to 1% mobile phase A and 99% mobile phase B between 5.01 and 6 minutes. Finally, at 6.01 minutes, mobile phase A was 90% and mobile phase B was 10%, and these conditions were held until the 8 minute run was complete. The column was maintained at 40 °C and a flow rate of 0.3 mL min−1. Prior to sample injection, a needle wash was utilized containing 80/20 isopropyl alcohol (IPA)/water with 10 mM ammonium formate. For multiple reaction monitoring mass spectrometry (MRM MS) of 9 channels, the sample was ionized by electrospray operating in positive ion mode (ES+), with a capillary voltage of 1.00 kV (Table 2).| Analyte compound | Precursor ion mass | Product ion mass | Cone voltage (V) | Collision energy (eV) |
|---|---|---|---|---|
| 13C6, 15N2 lysine | 147.1 | 84.0 | 19.0 | 14.0 |
| N-CML | 205.0 | 84.0 | 15.0 | 22.0 |
| N-CEL | 219.0 | 84.0 | 15.0 | 28.0 |
| 13C6, 15N2 labelled L-lysine | 155.1 | 90.1 | 19.0 | 14.0 |
| CML-d3 | 208.0 | 87.0 | 15.0 | 28.0 |
| CEL-d4 | 223.0 | 134.0 | 15.0 | 15.0 |
The source temperature was 150 °C, desolvation temperature was 350 °C, desolvation gas flow was 800 L h−1, and cone gas flow was 40 L h−1. CML, CEL, and lysine concentrations were calculated based on the integrated areas relative to the internal standard peak areas. The total AGE concentrations were calculated from measured CML and CEL and adding them together for each sample. The total protein-bound lysine helps evaluate total protein concentration over time, and thus the number of lysines that can be glycated.
Carry-over
For each analyte, there was significant carry-over of analyte signal (>20%), but less than 5% of IS signal. Therefore, after each standard curve, at least 5 blanks (water only) were run before continuing sample analysis. After analysis, 5 blanks were sufficient to remove any remaining analyte eluting (<0.1%). Also, a minimum of two blank samples were run in between each analyte sample (see Fig. S2†).Data analysis
Calibration plots of analyte/IS peak area ratio versus CML, CEL, and lysine concentrations were constructed, and a linear regression was used for all analytes. The peak area ratio of sample versus associated IS was used to produce a response to determine the concentrations from the calibration line. SigmaPlot (Systat Software Inc.) was used to plot results and R software (Rstudio version 4.2.2) was used to perform statistical testing in the “rstatix” package (0v.7.2; Kassambara, A. 2023). One-way ANOVA tests for each analyte were conducted to examine differences between storage solution types over 43 days. Days were treated as a repeated measure independent variable and the storage solutions were treated as a between-subjects dependent variable. Prior to conducting the ANOVA test, the assumptions of normality and homogeneity of variances were assessed using Shapiro–Wilk and Levene's tests, respectively. If these assumptions were met, the ANOVA test was performed. In the case of a significant ANOVA result, post hoc Bonferroni tests were conducted to determine which specific solution types and/or days in storage exhibited statistically significant differences on the 4 analytes.Results
Intra- and inter-assay accuracy and precision
Intra- and inter-assay variations were assessed utilizing quality control (QC) reconstituted hydrolyzed RBC samples spiked with known concentrations of CML, CEL, or lysine for each batch analysis. The assay variation was determined by reading the plate samples (n = 6) for over 20 hours kept at 4 °C in the sample manager of the autoinjector. Inter-assay variation was determined by measuring the analytes on 4 different samples (n = 4) over a 2 day period at 4 °C in the sample manager. Both intra- and inter-assay variations were prepared for the analytes and the results are reported in Table 3.| Analyte | Intra-assay (n = 6)a | Inter-assay (n = 4)b | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy (% of target) | Precision (RSD%) | Accuracy (% of target) | Precision (RSD%) | |||||
| Low | High | Low | High | Low | High | Low | High | |
| a Intra-assay (six repeated analyses within one experiment during 20 h with samples stored in sample manager). b Inter-assay (three independent experiments within 2 days) validation results. Concentration low: ten-times endogenous levels, high: forty-times endogenous levels for all analytes. | ||||||||
| CML | 113 | 116 | 4.26 | 4.47 | 92.8 | 102 | 12.69 | 12.2 |
| CEL | 97.4 | 93.7 | 14.3 | 1.87 | 93.5 | 94.4 | 5.25 | 3.58 |
| Lysine | 94.2 | 102 | 3.36 | 1.27 | 93.4 | 98.8 | 8.14 | 7.06 |
As seen in eqn (1), the stock QC sample expected concentration (spike concentration) was determined and either diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 for CML/CEL or 1
40 for CML/CEL or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for lysine. The two spiked concentrations (concentration of spiking solution) relate to a low (10 times the exogenous concentration), and high (40 times the exogenous concentration) concentration of the analytes. Percent recovery (% of target) in eqn (2) was used to determine the accuracy and subsequent percent relative standard deviation (RSD%) in eqn (3) for precision.
000 for lysine. The two spiked concentrations (concentration of spiking solution) relate to a low (10 times the exogenous concentration), and high (40 times the exogenous concentration) concentration of the analytes. Percent recovery (% of target) in eqn (2) was used to determine the accuracy and subsequent percent relative standard deviation (RSD%) in eqn (3) for precision.
 | (1) |
 | (2) |
 | (3) |
The sample accuracy and precision were evaluated at two different concentrations of spiked analyte (low and high) as reported in Table 3. Intra-assay accuracy ranged between 113% and 116% for CML, 93.7% and 97.4% for CEL, and 94.2% and 102% for lysine. All analyte intra-assay accuracy results are in the 80–120% range, which is acceptable for EMA and US FDA bioanalytical guidelines. Each analyte precision was below the EMA and US FDA threshold of 15%. Inter-assay accuracy ranged between 92.8% and 102% for CML, 93.5% and 94.4% for CEL, and 93.4% and 98.8% for lysine. These intra-assay accuracy results are also in the acceptable range for EMA and US FDA bioanalytical guidelines, as is the inter-assay precision.
Linearity, detection limit, and quantification limit
To assess linearity of the developed method, nine calibration standards and a zero standard were measured for each analyte in three experiments; specifically, each standard was diluted using either 10 mM PFHA in water, AS-1, or AS-1N reconstituted supernatant. To test the effect of acid hydrolysis on the reconstitution solution, AS-1 and AS-1N solution containing no sample were hydrolyzed and dried under vacuum. After initial analysis, it was determined that the hydrolyzed sample solutions did not statistically alter the peak shape, retention time, or matrix effects. Therefore, to simplify sample processing, all standards were simply diluted in 10 mM PFHA in water. The correlation coefficient for standard curves using 10 mM PFHA in water (R2) are reported in Table 4.| Analyte | Linearity (R2) | LOD (nM) | LOQ (nM) | Matrix effects (%) |
|---|---|---|---|---|
| a Validated method utilized (n = 6). CML: Nε-carboxymethyl-lysine (CML); CEL: Nε-carboxyethyl-lysine (CEL). LOQ: limit of quantification (10 × standard deviationblankresponse/slope). LOD: limit of detection (3 × standard deviationblankresponse/slope). Matrix effects calculated from ratio of ((IS area in matrix/IS area in blank sample) − 1) × 100. IS: internal standard. | ||||
| CML | 0.999 | 1.07 | 3.58 | 7.48 |
| CEL | 0.999 | 0.494 | 1.65 | 8.99 |
| Lysine | 0.999 | 0.345 | 1.15 | 6.33 |
The limit of detection (LOD) and limit of quantification (LOQ) were calculated based on the signal-to-noise ratio of the blank signal and the sample signal as reported in Table 3 for each analyte. The calibration curve of all three analytes using stable IS was measured and indicated good linearity within the concentration range selected (see Table 4). The relative peak area versus injected relative concentration to IS was found to be linear with a regression coefficient R2 = 0.999 for all three analytes. The LOD and LOQ were determined using standard deviation of the blank response divided by the slope of the calibration curve multiplied by 3 or 10, respectively.
Matrix effects
There are no commercially available RBC samples in AS-1/AS-1N that are free of the three analytes, so it is impossible to obtain analyte-free biological samples for method validation. However, to adhere to the bioanalytical method guidelines, the hydrolyzed RBC samples were prepared and analyzed to determine the overall extent of suppression or enhancement of signal. The matrix effects were determined using only the IS mix in either sample or blank (water). The matrix effects were calculated using eqn (4). | (4) |
According to bioanalytical method validation, less than 13% matrix effects are acceptable criteria. Due to presence of endogenous concentrations of the three analytes tested, the matrix effects were evaluated using IS peak areas. Matrix effects were found to be in all analytes tested (Table 4). All matrix effects were below 13%, which shows there is no significant enhancement or suppression of chromatogram signal due to the blood component matrix. Therefore, the chromatograms can be used to quantify analytes during further analysis.
Absolute quantification of CML, CEL, and lysine
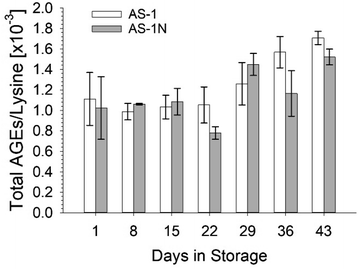
The AGEs under investigation, CML and CEL, were successfully detectable in all blood samples from five healthy controls assayed in biological duplicate, as shown in Fig. 1. While there appeared to be no statistically significant differences in AGE formation based on storage solution, there was a significant difference in AGE formation as a function of storage duration for both CML and CEL. The CML concentration in AS-1 on day 1 of storage (774 ± 113 nM) was not statistically different when stored in AS-1N (869 ± 61 nM). There was also no statistically significant difference for CML at day 43 of storage between AS-1 (1210 ± 120 nM) and AS-1N (1052 ± 149 nM). However, there was a statistically significant increase in AGE formation for both storage solutions across the 43 day storage period (see analysis in Fig. 1 caption). In fact, there was a 53% increase in CML concentration for the RBCs stored in AS-1, a value much higher than the 21% increase in CML formation on RBCs stored in AS-1N. Similar results were measure for the CML, which had a concentration in AS-1 of 565 ± 53 nM on day 1 and a concentration of 953 ± 65 nM on day 43. The CML formation on the RBCs stored in AS-1N increased from 643 ± 49 nM on day 1 to 901 ± 161 nM on day 43. The percent increase for the CEL in AS-1 was higher (69%) than the AS-1N stored cells (40%).The data in Fig. 2 represent the total increase in AGEs for each storage solution during the entire storage duration (43 days) relative to the lysine concentration. Similar to the individual determinations of CML and CEL, the storage solutions did not seem to have an effect on the AGE formation. Furthermore, although the total AGEs clearly increased as a function of storage duration, there was not a large difference in AGE formation from day 1 to day 43 (total AGE change was 54% for AS-1 and 49% for AS-1N). However, an increase in AGEs (relative to day 1 of storage) were measured after 3 weeks of storage.
Discussion
This study includes a new protocol with high accuracy and precision for the quantification of two types of AGEs using UPLC-MS/MS. In the present study, UPLC-MS/MS with MRM was highly useful for the measurement of AGEs in blood samples. Specifically, we were able to report the concentrations of CML, CEL, and lysine in two different blood samples on day 1 through day 43 of storage. This study focused on CML and CEL without the addition of common byproducts because these are the most abundant and widely studied AGEs related to complications associated with diabetes. Due to the hyperglycemic storage conditions used in RBC storage for transfusion medicine, it was anticipated that the stored RBCs would also exhibit AGE formation. There is limited research for detecting and quantifying both CML and CEL, and almost no present research involving stored blood. Current commercial kit assays are not selective enough for only CML or CEL without other glycation mechanism byproducts. Current methods of detecting and quantifying AGEs include non-selective enzyme linked immunosorbent assay (ELISAs), which use antibodies and are often limited to reliable types of antibodies and decreased sensitivity.22 Other types of analytical methods include time-consuming immunohistochemical detection, fluorescence spectroscopy and size exclusion chromatography with fluorescence detection.25–27 Thus, we developed a reliable, sensitive, specific (see ESI Fig. S3†), and reproducible method to detect CML, CEL and lysine in human blood samples.Other challenges when developing a robust method for detection and quantification of AGEs, include the ten-thousand-fold difference between the concentration of protein-lysine on RBC membranes versus modified lysine adducts (CML and CEL); only 1% of RBC proteins that contain lysine are glycated and form AGEs.23,28 Acid hydrolysis is a well-known technique that can be used to extract proteins and amino acids on the RBC membrane to adequately prepare various biological samples (urine, pRBCs, plasma) for high throughput analysis such as UPLC-MS/MS.29,30 There are many drawbacks with previous methodology reports, such as biological interferences and matrix effects enhancing or suppressing chromatographic signal.31–33 There are several methods that report CML, CEL, and/or lysine detection for AGEs studies utilizing various analytical techniques.34–36
The bioanalytical method validation by the European Medicines Agency (EMA) provides guidance and recommendations for bioanalytical assays, which can be seen as the “gold standard” in other types of method development.37 By using the FDA approved guideline M10 by the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH), this study used sample analysis recommended chemical, biological, and metabolite drug guidelines to explore various analytical parameters, such as LOD, LOQ, matrix effects, intra- and inter-assay accuracy and precision, and matrix interferences. The EMA bioanalytical method validation guidance provides clear acceptance and reliability for biological assays and analytical results. Through this process, our results showed acceptable linearity, LOD, and LOQ. The matrix effects did not contribute to chromatographic signals, which is a major concern for biological samples. There was clear specificity, requisite peak resolution, and distinguishable IS correlation to the analyte detected. The intra- and inter-assay results showed EMA and US FDA bioanalytical guideline acceptable accuracy and precision. Overall, the results showed this methodology to detect and quantify CML, CEL, and lysine in stored blood solutions can be used in other analyses involving suspected AGE formation on blood components.
To date, most reports reporting AGE formation have primarily focused on AGEs in food, biological plasma or serum, or human tissues (retina, kidney, endothelial and smooth muscle cells).24,27,38–45 Increased protein glycation has been reported in clinical studies involving people with diabetes, and it is linked to various complications associated with increased AGEs and oxidative stress.19,46–48 Glycation involves the Maillard reaction, a non-enzymatic process where the carbonyl group of a reducing sugar reacting with the amino group of a protein, lipid or nucleic acid, generates Schiff bases to produce Amadori products.22 These Amadori products, as well as other byproducts and intermediates during glucose oxidation, lead to the formation of various AGEs, such CML and CEL.22 The mechanism that produces CML was the first discovered and is the most widely studied AGE.23 Another important AGE, CEL, is associated with diabetic complications and derived from the methylglyoxal pathway (see ESI Fig. S4†).24 Although glycation occurs on most cell types, AGE detection of bloodstream such bloodstream components as proteins and intact cells may provide insight into increased pathologic conditions.28,49 For example, glycated albumin in the bloodstream has been shown to influence delivery of biologically active peptides to healthy RBCs, while that same delivery was reduced in RBCs from people with diabetes.50,51
Here, there were no statistically significant differences in the two blood storage solutions on a given day, although there were some interesting features of the collected data. For example, while there was no difference in AGE formation between the AS-1 and AS-1N storage solutions on any particular day, there was a significant increase in AGE formation on the RBCs during storage duration for both storage solution strategies. Another interesting feature of the data shown in Fig. 2 is the increase in total AGE formation after 2–3 weeks in storage. While the concept of “fresh” stored RBCs do not perform any better than “older” stored RBCs is debatable, it is clear in the literature that fresh RBCs are preferred in certain situations (e.g., infant transfusions).52,53 Furthermore, some reports suggest a reduction in post-transfusion complications when using RBCs stored for less than 2 weeks.54,55 The data in Fig. 2 show that the covalent AGE formation experiences an increase in formation rate around this 14–21 day period. These results correspond closely to our previous results showing irreversible damage to the RBCs stored in AS-1 after ∼2 weeks.56
Conclusions
AGEs are known to be related to the pathomechanism of diabetes and other degenerative disorders.48,57,58 This study involved utilizing blood from healthy donors to explore methodology that can reliably and accurately detect and measure AGEs, while maintaining sensitivity and bioanalytical merits to other reports using different instruments or assays. We were able to confirm the detection and quantification of CML, CEL, and lysine present on stored RBCs, without significant issues of matrix effects and with analyte recovery. The purpose of this small initial study was to validate the robustness of the biomarkers and provide a pilot study for future blood banking AGE research. The data encourages further investigation of the accumulation of AGEs.Author contributions
The idea of the experiment belongs to M. G. and L. A. S. S.; M. G. and L. A. S. S. planned and organized the work. L. D. S. performed the blood collection and L. D. S. and L. A. S. S. performed blood preparation; L. A. S. S. and M. G. developed TQ-S Micro sample preparation methods and protocol; L. A. S. S. conducted the experiments, developed the quantitative method, and interpreted the UPLC-MS/MS data; L. A. S. S. performed calculations and revisions; L. A. S. S., L. D. S., M. G., A. S., and D. S., helped in the writing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the volunteers for their blood donation. The authors also thank Dr Anthony Schilmiller and others at MSU RTSF Mass Spectrometry and Metabolomics Core for assisting with measurements and experimental development, Dr Liangliang Sun for his instrumentation and continued assistance with the project, and Scott Bankroff for designing the customized scientific glassware. The financial support from the National Institute of Health (5R01HL56440-02) is gratefully acknowledged.References
- M. R. P. Sapiano, J. M. Jones, A. A. Savinkina, K. A. Haass, J. J. Berger and S. V. Basavaraju, Transfusion, 2020, 60, 17–37 CrossRef PubMed.
- D. W. Greening, K. M. Glenister, R. L. Sparrow and R. J. Simpson, J. Proteomics, 2010, 73, 386–395 CrossRef CAS PubMed.
- Association for Advancement of Blood and Biotherapies, AABB User Guide, 2021 Search PubMed.
- G. L. Moore and J. G. Batsakis, Crit. Rev. Clin. Lab. Sci., 1987, 25, 211–229 CrossRef CAS PubMed.
- M. A. Meledeo, G. C. Peltier, C. S. McIntosh, J. A. Bynum and A. P. Cap, Transfusion, 2019, 59, 1549–1559 CrossRef CAS PubMed.
- E. K. Meyer, D. F. Dumont, S. Baker and L. J. Dumont, Transfusion, 2011, 51, 1574–1579 CrossRef CAS PubMed.
- C. Robert Valeri, L. E. Pivacek, G. P. Cassidy and G. Ragno, Transfusion, 2000, 40, 1341–1345 CrossRef PubMed.
- J. A. Cancelas, L. J. Dumont, L. A. Maes, N. Rugg, L. Herschel, P. H. Whitley, Z. M. Szczepiokowski, A. H. Siegel, J. R. Hess and M. Zia, Transfusion, 2015, 55, 491–498 CrossRef CAS PubMed.
- G. M. D'Amici, C. Mirasole, A. D'Alessandro, T. Yoshida, L. J. Dumont and L. Zolla, Blood Transfus., 2012, 2, 46–54 Search PubMed.
- G. Moroff and D. Dende, Transfusion, 1983, 23, 484–489 CrossRef CAS PubMed.
- T. Yoshida, M. Prudent and A. D'Alessandro, Blood Transfus., 2019, 17, 27–52 Search PubMed.
- Y. Liu, C. Chen, S. Summers, W. Medawala and D. M. Spence, Integr. Biol., 2015, 7, 534 CrossRef CAS PubMed.
- A. D'Alessandro, G. Liumbruno, G. Grazzini and L. Zolla, Blood Transfus., 2010, 8, 82–88 Search PubMed.
- J. R. Hess, Transfus. Apher. Sci., 2010, 43, 51–59 CrossRef PubMed.
- E. Bennett-Guerrero, T. H. Veldman, A. Doctor, M. J. Telen, T. L. Ortel, T. S. Reid, M. A. Mulherin, H. Zhu, R. D. Buck, R. M. Califf and T. J. McMahon, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17063–17068 CrossRef CAS PubMed.
- R. L. Sparrow, Blood Transfus., 2012, 10, 7–11 Search PubMed.
- N. S. R. Maluf, J. Hist. Med. Allied Sci., 1954, 9, 59–107 CrossRef CAS PubMed.
- A. L. Y. Tan, J. M. Forbes and M. E. Cooper, Semin. Nephrol., 2007, 27, 130–143 CrossRef CAS PubMed.
- M. Hellwig and T. Henle, Angew. Chem., Int. Ed., 2014, 53, 10316–10329 CrossRef CAS PubMed.
- N. S. Mangalmurti, S. Chatterjee, G. Cheng, E. Andersen, A. Mohammed, D. L. Siegel, A. M. Schmidt, S. M. Albelda and J. S. Lee, Transfusion, 2010, 50, 2353–2361 CrossRef CAS PubMed.
- C. Oyet, B. Okongo, R. Onyuthi Apecu and E. Muwanguzi, J. Blood Med., 2018, 9, 111–115 CrossRef CAS PubMed.
- K. Nowotny, T. Jung, A. Höhn, D. Weber and T. Grune, Biomolecules, 2015, 5, 194–222 CrossRef CAS PubMed.
- C. Delgado-Andrade, Food Funct., 2016, 7, 46–57 RSC.
- M. U. Ahmed, E. B. Frye, T. P. Degenhardt, S. R. Thorpe and J. W. Baynes, Biochem. J., 1997, 324, 565–570 CrossRef CAS PubMed.
- W. Schwab, U. Friess, U. Hempel, E. Schulze, Z. Makita, M. Kasper and H.-G. Simank, Histochem. Cell Biol., 2002, 117, 541–546 CrossRef CAS PubMed.
- S. Jaisson and P. Gillery, Curr. Opin. Clin. Nutr. Metab. Care, 2021, 24, 411–415 CrossRef CAS PubMed.
- T. Henle, R. Deppisch, W. Beck, O. Hergesell, G. M. Hänsch and E. Ritz, Nephrol., Dial., Transplant., 1999, 14, 1968–1975 CrossRef CAS PubMed.
- J. Xue, V. Rai, D. Singer, S. Chabierski, J. Xie, S. Reverdatto, D. S. Burz, A. M. Schmidt, R. Hoffmann and A. Shekhtman, Structure, 2011, 19, 722–732 CrossRef CAS PubMed.
- M. Fountoulakis and H.-W. Lahm, J Chromatogr A, 1998, 826, 109–134 CrossRef CAS PubMed.
- A. Tsugita and J.-J. Scheffler, Eur. J. Biochem., 1982, 124, 585–588 CrossRef CAS PubMed.
- S. L. Thomas, J. B. Thacker, K. A. Schug and K. Maráková, J. Sep. Sci., 2021, 44, 211–246 CrossRef CAS PubMed.
- Y. V. Karpievitch, A. D. Polpitiya, G. A. Anderson, R. D. Smith and A. R. Dabney, Ann. Appl. Stat., 2010, 4, 1797–1823 Search PubMed.
- A. M. Rowan, P. J. Moughan and M. N. Wilson, J. Agric. Food Chem., 1992, 40, 981–985 CrossRef CAS.
- N. J. Rankin, K. Burgess, S. Weidt, G. Wannamethee, N. Sattar and P. Welsh, Ann. Clin. Biochem, 2019, 56, 397–407 CrossRef CAS PubMed.
- E. Tareke, A. Forslund, C. H. Lindh, C. Fahlgren and E. Östman, Food Chem., 2013, 141, 4253–4259 CrossRef CAS PubMed.
- N. Shoji, K. Nakagawa, A. Asai, I. Fujita, A. Hashiura, Y. Nakajima, S. Oikawa and T. Miyazawa, J. Lipid Res., 2010, 51, 2445–2453 CrossRef CAS PubMed.
- L. Schwieler, A. Trepci, S. Krzyzanowski, S. Hermansson, M. Granqvist, F. Piehl, T. Venckunas, M. Brazaitis, S. Kamandulis, D. Lindqvist, A. D. Jones, S. Erhardt and L. Brundin, Bioanalysis, 2020, 12, 379–392 CrossRef CAS PubMed.
- V. P. Singh, A. Bali, N. Singh and A. S. Jaggi, Korean J. Physiol. Pharmacol., 2014, 18, 1–14 CrossRef CAS PubMed.
- M. W. Poulsen, J. M. Andersen, R. V. Hedegaard, A. N. Madsen, B. N. Krath, R. Monošík, M. J. Bak, J. Nielsen, B. Holst, L. H. Skibsted, L. H. Larsen and L. O. Dragsted, Br. J. Nutr., 2016, 115, 629–636 CrossRef CAS PubMed.
- Y. Kim, J. B. Keogh, P. Deo and P. M. Clifton, Nutrients, 2020, 12, 1–11 CAS.
- A. Twarda-Clapa, A. Olczak, A. M. Białkowska and M. Koziołkiewicz, Cells, 2022, 11, 1312 CrossRef CAS PubMed.
- Y. Nomi, H. Kudo, K. Miyamoto, T. Okura, K. Yamamoto, H. Shimohiro, S. Kitao, Y. Ito, S. Egawa, K. Kawahara, Y. Otsuka and E. Ueta, Biochimie, 2020, 179, 69–76 CrossRef CAS PubMed.
- P. J. Thornalley, S. Battah, N. Ahmed, N. Karachalias, S. Agalou, R. Babaei-Jadidi and A. Dawnay, Biochem. J., 2003, 375, 581–592 CrossRef CAS PubMed.
- J. Uribarri, S. Woodruff, S. Goodman, W. Cai, X. Chen, R. Pyzik, A. Yong, G. E. Striker and H. Vlassara, J. Am. Diet. Assoc., 2010, 110, 911–916 CrossRef PubMed.
- C. Nevin, L. McNeil, N. Ahmed, C. Murgatroyd, D. Brison and M. Carroll, Sci. Rep., 2018, 8, 9002 CrossRef PubMed.
- M. A. Mengstie, E. Chekol Abebe, A. Behaile Teklemariam, A. Tilahun Mulu, M. M. Agidew, M. Teshome Azezew, E. A. Zewde and A. Agegnehu Teshome, Front. Mol. Biosci., 2022, 9, 1–11 Search PubMed.
- J. W. Baynes, Diabetes, 1991, 40, 405–412 CrossRef CAS PubMed.
- J. Peyroux and M. Sternberg, Pathol. Biol., 2006, 54, 405–419 CrossRef CAS PubMed.
- J. Wautier, M. Wautier, A. Schmidt, G. M. Anderson, C. Zoukourian, L. Capron, S. Yan, J. Breyf, P. Guillausseauii and D. Sterni, Proc. Natl. Acad. Sci. U.S.A., 1994, 91, 7742–7746 CrossRef CAS PubMed.
- M. J. Jacobs, M. K. Geiger, S. E. Summers, C. P. DeLuca, K. R. Zinn and D. M. Spence, ACS Meas. Sci. Au, 2022, 2, 278–286 CrossRef CAS PubMed.
- M. Geiger, T. Janes, H. Keshavarz, S. Summers, C. Pinger, D. Fletcher, K. Zinn, M. Tennakoon, A. Karunarathne and D. Spence, Sci. Rep., 2020, 10, 17493 CrossRef CAS PubMed.
- D.-H. Kim, Korean J. Pediatr., 2018, 61, 265–270 CrossRef PubMed.
- M. Burke, P. Sinha, N. L. C. Luban and N. G. Posnack, Front. Pediatr., 2021, 9, 765306 CrossRef PubMed.
- P. Maruti, S. Musyoki and A. Amayo, Pan Afr. Med. J., 2021, 38, 280 Search PubMed.
- C. G. Koch, L. Li, D. I. Sessler, P. Figueroa, G. A. Hoeltge, T. Mihaljevic and E. H. Blackstone, N. Engl. J. Med., 2008, 358, 1229–1239 CrossRef CAS PubMed.
- Y. Liu, L. E. Hesse, M. K. Geiger, K. R. Zinn, T. J. McMahon, C. Chen and D. M. Spence, Lab Chip, 2022, 22, 1310–1320 RSC.
- A. Damasiewicz-Bodzek, B. Łabuz-Roszak, B. Kumaszka and K. Tyrpień-Golder, Arch. Med. Sci., 2020, 95654 Search PubMed.
- H. Vlassara and J. Uribarri, Curr. Diabetes Rep., 2014, 14, 453 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ay01817b |
| This journal is © The Royal Society of Chemistry 2023 |