Open Access Article
Open Access ArticleLabel-free detection and mapping of graphene oxide in single HeLa cells based on MCR-Raman spectroscopy
Zuzana
Chaloupková
 a,
Ludmila
Žárská
a,
Jan
Belza
a,
Ludmila
Žárská
a,
Jan
Belza
 ab and
Kateřina
Poláková
ab and
Kateřina
Poláková
 *a
*a
aCATRIN – Regional Center of Advanced Technologies and Materials, Palacky University Olomouc, Olomouc, Czechia. E-mail: katerina.polakova@upol.cz
bDepartment of Physcial Chemistry, Faculty of Science, Palacky University Olomouc, Olomouc, Czechia
First published on 18th September 2023
Abstract
GO is a 2D nanomaterial that has attracted attention in many industries in recent years, such as the chemical industry, electronics or medicine. Due to its unique properties such as strength, hydrophilicity and large specific surface area with the possibility of functionalization, GO is a particularly attractive material in biomedicine as a candidate for use in targeted drug delivery. In such a case, we need information on whether graphene oxide penetrates into cells and whether we are able to detect and monitor GO in these cells during and also after the treatment to evaluate possible degradation process of GO and its interaction within the cell compartements. This work introduces the Raman spectroscopy as label-free detection method showing the advantages of combining Raman spectroscopy with MCR (Multivariate Curve Resolution) analysis for advanced detection of GO in cervical cancer (HeLa) cells. Our synthesized GO is characterized firstly by AFM, SEM and Raman spectroscopy and then MCR-Raman spectroscopy is used to detect internalized GO in individual HeLa cells. Moreover, by using our methodology, distribution of GO as well as its chemical stability inside the cell for up to six months is investigated without using any additional labeling or tracing the GO. Thus, MCR-Raman spectroscopy may become a new analytical tool in preclinical and clinical applications of graphene-based nanotheranostics.
Introduction
Graphene, which was first isolated in 2004,1 is one of the best known, explored and used 2D nanomaterials from that time on. For the application of graphene in other fields such as catalysis, detection of substances, carriers, adsorption, etc., the functionalization of graphene by chemical and physical methods is crucial to generate new properties of graphene derivatives according to the need of their application.2–4 An example of successful derivatization of graphene is the GO we have studied. Although GO, unlike graphene, has been known for a long time since it was first synthesized by oxidation of graphite by the chemist Brodie in 1859.5 However, until the isolation of graphene, GO was a relatively uninteresting and unused material. GO is an oxidized form of graphene that has better properties than graphene itself in certain parameters, is more reactive and has a large specific surface area with the possibility of further functionalization.6,7 It also exhibits excellent hydrophilicity and strength. All these features make it a promising material for use in catalysis, sensors, environment7 and also in biomedicine, since GO can non-covalently bind a variety of biomolecules, for example for targeted drug delivery.1,3,5,8 GO also exhibits antibacterial activity, mainly due to its high content of oxygen groups, which interact well with the polar membranes of bacteria.9,10 However, knowledge on the internalization of graphene oxide inside mammalian cells is still limited. The uptake of graphene oxide by cells is a complex phenomenon influenced by factors such as particle size and surface modification. In addition, nanomaterials introduced into living organisms interact with components of biological fluids. All these factors need to be taken into account when considering the uptake of potential drug carriers by cells.11 Thus, the fate of graphene oxide, biodegradation or active targeting in cells is extremely important from not only a safety perspective, but also in order to monitor the potential mechanism of action in targeted therapy.Raman spectroscopy has a number of advantages over other methods in biomedical applications. The low sample preparation requirements and the relatively non-destructive nature of Raman spectroscopy allow its use for tissue analysis and, in conjunction with the microscope, the analysis of individual cells and organelles can be easily performed. In addition, the absence of the need for sample staining, compared with other techniques such as fluorescence microscopy, allows its applications in drug transport and metabolism studies.10 In fact, dyes can significantly affect the interaction of the drugs used with the cell when investigating drug transport and metabolism.12,13 Raman microscopy is particularly frequently used, as it is sensitive enough to monitor changes in cells at the molecular level. In addition to the detection of cancer cells, Raman spectroscopy can also be used in the investigation of cancer treatment options14 or targeted drug transport.15,16 It has been published that 2D nanomaterials can also be used as carriers to prevent premature degradation of the drug and minimize its non-specific toxicity.2 In addition to targeting tumor cells, the downstream action of the drug in the cell is also important. From a therapeutic and also safety point of view, affinity for specific organelles, particularly mitochondria, nucleus and lysosomes, is desirable. This is because if the drug is transported directly to the site where it is intended to act, it is less likely to be destroyed or rejected by the cell, thereby maximizing its efficacy.17 Therefore, 2D nanomaterials used for drug transport into cells must in particular meet the requirements of biocompatibility with the human body, biodegradability and low toxicity.18 A number of methods have been used to analyse spectra obtained by the Raman method.19 Mapping and multidimensional cellular resolution (MCR) are two well-known approaches that we have used recently to analyze Raman spectra.20 Unlike mapping, which scales intensity based on the acquired spectra, multidimensional curve resolution (MCR) is a statistical method that does not require reference spectra. The main difference between mapping and MCR is that mapping uses a single reference spectrum and applies it to the image, whereas MCR uses multiple reference spectra, spectral components and combines the experimental data.21 MCR can be used to obtain critical information about image composition, chemicals19,22 and map the surface based on composition, which is often presented through color coding. Correlation mapping can also extract the composition based on a single spectrum, but this could only be done in? One component at a time.23 MCR-based Raman spectroscopy can be used to detect GO penetrated inside HeLa cells, however, to study the fate of GO within the cells more deeply, here, we are introducing Raman spectroscopy with cell line mapping. The obtained background-adjusted multispectral data is analysed by using the MCR (Multivariate Curve Resolution) algorithm to identify individual components in the sample. MCR gives evolutionary analytical data originating from a process or analytical measurement24 and thus closely mimics the structure of an analytical measurement. The MCR algorithm is applied to the Raman spectra of GO inside the HeLa cells and their composition is quantitatively determined by comparison with the Raman spectra of each pure form. The aim of this research is the detection and imaging of GO distribution in single HeLa cells which is a promising approach to determining GO, its degradation products, or moreover interaction of GO with cellular components (lipids and proteins), organelles or cytoplasmic membrane, which shows great potential in expanding the knowledge of anticancer therapy and can be used in personalized diagnostics, therapy and drug transport in the future.
Experimental
Chemicals and reagents
Graphite powder (particle size below 20 μm), potassium permanganate (purity ≥99.0%), potassium nitrate (purity ≥99.0%), sulfuric acid (96%), hydrogen peroxide and sodium hydrogen phosphate dihydrate (p.a.) was purchased from Sigma-Aldrich. Phosphate buffer prepared from sodium chloride (purity ≥99.0%), potassium chloride (purity ≥99.0–100.5%) and potassium dihydrogen phosphate (purity ≥99.995%) from penta were used in the preparation of the cell samples. Cell fixation was performed with paraformaldehyde solution (4%) from penta. Ultrapure water was prepared in the laboratory using a Puris water system, from MiraeST, model Esse-UP analysis.Synthesis of GO
GO was prepared by oxidation of graphite powder using the Hummers' method.25 The procedure was as follows: 23 ml of sulphuric acid (dropwise) was added to a mixture of 1 g of graphite powder and 0.5 g of sodium nitrate. The reaction mixture was then cooled to 0 °C and 3 g of potassium permanganate was added successively during further cooling, keeping the temperature below 20 °C during this exothermic reaction. After the addition of the permanganate, the reaction mixture was heated to 35 °C and stirred for 30 minutes. In the next step, 46 ml of distilled water was slowly added and the reaction mixture was further stirred at 98 °C for 15 minutes. After 15 minutes, the mixture was cooled to room temperature, 1 ml of 30% hydrogen peroxide and 140 ml of distilled water were added. The resulting material was centrifuged at 6000 rpm for 7 minutes and washed with distilled water. This process was repeated several times to remove residual impurities. The obtained material was dried overnight in a vacuum oven and then GO powder was obtained. From the GO thus obtained, efficient size selection of GO powder dispersed in water was performed by sonication followed by centrifugation. 10 mg of GO powder was dispersed in 20 ml of distilled water and sonicated for 1 hour. Then the dispersion was stirred overnight on a magnetic stirrer. This was followed by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm for 20 min, and finally, decantation of the supernatant was performed, which contained GO particles of the desired size.
500 rpm for 20 min, and finally, decantation of the supernatant was performed, which contained GO particles of the desired size.
Cell preparation
The cell culture used for the experiment was human cervical epithelial tumour HeLa cells from ATCC, USA. The cells were cultured in DMEM (Dulbecco's Modified Eagle medium) with low glucose concentration from Life Technologies at 37 °C and 5% CO2 atmosphere. A calcium fluoride slide (substrate for Raman analysis) was inserted into a well in the culture plate. Standard trypsinization of the cells was performed and a sufficient number of cells (approximately 60% confluence) were seeded into the well on the substrate. The cells were kept to adhere to the surface of the substrate overnight. The next day, labelling of the cells with GO suspension was performed to achieve the desired concentration of GO in the volume of medium used. The cells with GO were then incubated for a further 24 hours. Control cells were cultured for 48 hours without labelling with GO suspension. After incubation, the culture medium was removed and the cells were gently washed with phosphate-buffered saline (PBS). Subsequently, 1 ml of 4% paraformaldehyde solution in PBS was added to the cells for fixation. The cells were left in the refrigerator for 10 min for fixation, after which the fixing agent was aspirated followed by washing with phosphate buffer and ultrapure water. The cell sample was let dry before measurement.Characterization of prepared GO
Raman spectroscopy
For the detection of Raman signal from GO, Raman spectral data were obtained on a DXR Raman microscopy (Thermos Scientific) with an excitation laser operating at 455 nm. Raman spectra were measured in the range from 500 to 3.500 cm−1. The spectral resolution of the applied combination of laser and optic was 1.0 cm−1. The step size of the X- and Y-axis displacement was no larger than 1 μm, allowing for high resolution. The range of motion in the Z-axis was not limited but allowed focusing on a sample with a maximum height of at least 5 mm. Experimental parameters were as follows: the number of exposures measured was 32 s and the length of exposure was 2 s, laser power 4 mW (on the sample surface). A lens with 50× magnification was chosen (with N.A. −0.55). Before the CCD detector, a 50 μm pinhole assembly with 1200 notches per mm grid was chosen. Fluorescence correction was also turned on to co-opt unwanted fluorescence from the cells. The spectra obtained were then adjusted to baseline using OMNIC 8.2.0.403 software (polynomial fit, where n = 3).The presence of GO in cells was confirmed by mapping. During the mapping, an area of 2–4 cells was mapped (usually 100 × 130 μm), in which a total of about 200 spectra were measured, corresponding to 200 sampling points within one map. Each spectrum was subjected to 32 s exposures and the exposure time was 2 s. A total of 512 exposures were made to collect the background. The measured maps were processed in OMNIC software for background editing and further processing. The obtained background-adjusted multispectral data were further analysed using the Multivariate Curve Resolution (MCR) algorithm to identify individual components in the sample. Two regions of expected components (GO and cell) and four components (only one region and three components for control cells without GO) were given for the calculation. The components corresponding to HeLa cells, GO and substrate were then selected from the algorithmically found components. From these, RGB maps were generated to show the spatial distribution of each component using the assigned colours.
Results & discussion
Cancer is among the leading causes of death worldwide. In 2018, there were 18.1 million new cases and 9.5 million cancer-related deaths worldwide. By 2040, the number of new cancer cases per year is expected to rise to 29.5 million and the number of cancer-related deaths to 16.4 million.26In the case of cancer, it is extremely important to make a diagnosis and classification of the cancer in the earliest phase to ensure on-time and appropriate treatment. However, tumor cells are still usually analyzed by H&E staining as a standard method for preliminary morphological diagnosis, which gives very low sensitivity and specificity, especially in the case of poorly differentiated cells.27 In this study we show the advantage of using Raman spectroscopy to study cells, where internal biochemical changes can be detected, to assess the physiological status and diagnose disease before any cellular morphological changes on one hand28 and on the other hand by the using of MCR-Raman spectroscopy we can study the GO chemical stability and its interaction within the cancer cells once it will be used as a nanoplatform for drug delivery purposes. Our study thus opens up the possibility of non-invasive and label-free diagnostics becoming an attractive and safe method for preclinical or clinical usage.29
Characterisation of prepared GO
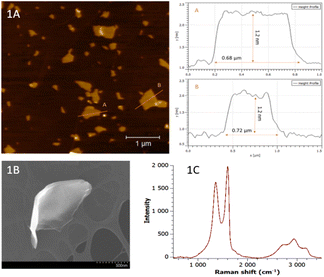
The prepared GO was characterized by AFM, SEM, ζ-potential measurements, Raman spectroscopy and D and G band intensity ratio. The AFM method was used to obtain the elevation topography of the GO. From Fig. 1A, it can be seen that the GO height profile reaches about 1.2 nm and shows a large amount of irregularities and is dissected. It can also be seen that the sample has a relatively large area and is mainly composed of areas with fewer layers. SEM images of GO with scales of 500 nm are shown in Fig. 1B. From the images, it can be seen that the surface is more indented, which is consistent with the AFM results. To characterize the properties of the layers in the dispersion, the zeta potential of GO was measured with a value of −30.5 mV. Negative values of zeta potential are due to the presence of electronegative functional groups formed on the graphite lattice during oxidation.30 It was thus demonstrated that GO exhibits moderate stability (according to the ASTM standard for stability of colloidal suspensions).31GO was then characterized by Raman spectroscopy. In the Fig. 1C is shown obtained spectrum typical for GO. The band with the highest intensity at a wavenumber of 1600 cm−1 corresponds to the G band, which is typical of both the Raman spectra of graphene and the spectra of its other forms. This band arises due to the vibration of the carbon bonds in the hexagonal lattice; the π electrons in this sp2 bond additionally cause resonance and the resulting band is therefore often very intense.32 The second typical but less intense band at a wavenumber of 1350 cm−1 belongs to the D band, which occurs in the region 1342–1356 cm−1 in GO spectra.33 The band is the result of distortions and deformations of the hexagonal sp2 lattice.34 In addition to these two intense bands D and G, less intense bands at wavenumbers 2715 cm−1, 2940 cm−1 and 3180 cm−1 can also be found in the spectra. These bands correspond to the overtones of the D and G bands. The most prominent of these (2940 cm−1) is referred to as the D + D′ band lying in the region 2920–2940 cm−1 and is a typical feature of GO spectra.32 The band at 2715 cm−1 is referred to as the 2D band (sometimes also G′) and is an overtone of the D band vibration.35,36 The sharp band is an indicator of single-layer graphene because it is sensitive to the thickness of the material being measured.36 The last mentioned band at a wavenumber of 3180 cm−1 is referred to as the 2D′ band and corresponds to the second-order vibration of the D and 2D bands.33
GO penetration into HeLa cells
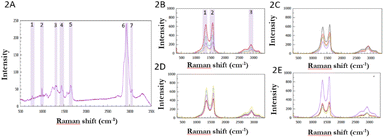
In the first step, control HeLa cells, i.e. cells without GO, were characterized by Raman spectroscopy. All measured control cells provided a spectrum typical of a cervical cancer cell line. One representative spectrum is shown in Fig. 2A. Bands are shown in the 800–1800 cm−1 wavenumber region, which is considered to be the fingerprint region in cells, corresponding to signals from nucleic acids and proteins.37 At 780 cm−1 wavenumber, signals from valence vibrations of the cytosine and uracil heterocycles were present, and a distinct narrow band of the amino acid phenylalanine was observed at 1000 cm−1 wavenumber. Around the 1100 cm−1 wavenumber, there was a band of symmetric vibration of the O–P–O bonds of the phosphate skeleton of nucleic acids. In addition to the vibrations from proteins and nucleic acids, a band of vibrations originating from C–C bonds in lipids was also discernible in this region at a wavenumber of 1130 cm−1. Other nucleotides showed bands in the wavenumber range of 1180–1400 cm−1 and a band originating from the vibration of amide III – at a wavenumber of 1310 cm−1 could be seen. At 1450 cm−1 there is a distinct band corresponding to the deformation vibrations of CH2 and CH3 bonds in proteins.38 At 1650 cm−1, a signal of amide I vibrations can be observed, which originates from valence vibrations of peptide bonds in proteins. In the 1800–2700 cm−1 wavenumber range, there was a region without significant signals. This is a typical feature of cell spectra and is referred to as the silent region. The dominant feature of the whole spectrum is a band at 2930 cm−1 with high intensity, which corresponds to the symmetric valence vibration of the CH3 phospholipid membrane. In the region 2854–2900 cm−1, a signal of lower intensity is evident, which corresponded to the asymmetric valence vibration of CH2 bonds in lipids and fatty acids. At a wavenumber of 3060 cm−1, there was a signal originating from vibrations of aromatic nuclei and CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH double bonds. In the region 3170–3500 cm−1 the signal originating from the low activity vibrations of water contained in the cell was found.
CH double bonds. In the region 3170–3500 cm−1 the signal originating from the low activity vibrations of water contained in the cell was found.
Subsequently, the presence of GO, at different concentrations during incubation, in HeLa cells was recorded by measuring individual Raman spectra. The results are shown in Fig. 2B–E. All the spectra obtained contained D and G bands of GO at the same wavenumbers as in the spectra of pure GO, i.e. at 1600 cm−1 and 1350 cm−1, respectively. In the region 2500–3300 cm−1, the overtone bands of the D band (i.e., 2D and 2D′ band) were visible, and the G′ band was obscured by the signal from the vibration of proteins in the cell at a wavenumber of 2930 cm−1.
The Raman spectra show that the intensity varies considerably in individual measurements (see the different colours of spectra in 2B, 2C, 2D and 2E) obtained in a single cell but also the spectra are different in each measured cell (2B–2E). This is due to the fact that the intensity in the case of cells is dependent on the cell profile in addition to the exact composition of the site, which varies from organelle to organelle. At sites where the cell is higher, its components are more abundant than at low-profile sites, since Raman spectroscopy allows focusing to different depths of the sample, which can cause significant differences in the intensity of the scattered radiation in high-profile samples. The intensity of the GO bands themselves also depends on the exact position of its particles within the cell. If a given GO particle is present within an organelle, its signal is attenuated relative to a GO particle located freely in the cytoplasm.
Raman mapping of GO distribution in cells
GO was further detected in the samples by using advanced mapping and analysed using multivariate curve resolution (MCR), Fig. 3A–D. The following maps were then generated from these results using RGB analysis.40 In these maps, the relative distribution of graphene oxide in HeLa cells is shown in colour. In all mapped samples, i.e., with concentrations of 18.75 μg ml−1, 25 μg ml−1, 37.5 μg ml−1 and 57.6 μg ml−1, GO was undoubtedly present in the resulting maps. As the concentration of GO used in sample incubation increased, the number of detected particles increased in individual cells.It can be seen from Fig. 3A that the map of the sample with the lowest mapped concentration (18.75 μg ml−1) showed a small amount of particles scattered throughout the cell cross-section. There was no clumping or accumulation at specific locations in the cell. In Fig. 3B and C, higher concentrations (25 μg ml−1 and 37.5 μg ml−1) showed a noticeable increase in the number of GO particles in the cells in the maps. There is also a noticeable clustering of more particles in both maps. In contrast, the map of the sample with the highest GO concentration, Fig. 3D, was significantly dominated by GO. The HeLa cell was completely overwhelmed by it and contained GO over almost the entire cross-section. There were large clusters of GO in the cells and the cells had significantly different morphology under the microscope from the low-concentration samples at first glance. The Raman spectrum of GO, shown in green on the RGB map, can be seen in Fig. 3E, and the Raman spectrum of the HeLa cell, shown in blue on the RGB map, can be seen in Fig. 3F.
From the obtained maps, it is noticeable that the number of particles that penetrated inside the cells increases with increasing concentration of GO used in cell incubation. At higher concentrations, GO particles cluster together and the maps show GO overwhelming the cells throughout the cross-section.
MCR-Raman imaging provided spectra of individual cells containing GO at 4 different concentrations and the spatial distribution of two chemical components (labeled GO and cell HeLa). Component 1 represents GO (green label) and a typical Raman spectrum of GO can be seen. For component 2, which represents the HeLa cell (blue label), a series of peaks can be observed, see Fig. 3A–D. At 2940 cm−1, a prominent C–H peak belonging to the lipids of the phospholipid membrane (symmetric CH2 stretch) can be observed.39 This peak can be observed at 2930 cm−1 in Fig. 2A, and is therefore shifted due to the presence of GO in the cell and its interaction with the cell. The same shift can be observed in the case of proteins present in the control cell and in cells with GO. These peaks are 1455 and 1660 cm−1, which represent the deformation vibrations of CH2 and CH3 bonds in the proteins,38 and the amide I vibration signal, which comes from the valence vibrations of peptide bonds in the proteins. Thus, it is clear that MCR-Raman imaging can image the intracellular distribution of lipids and proteins due to the improved resolution of overlapping Raman bands.41
Stability of cells with GO
The ratio of the intensities of the D and G bands – ID/IG (where the D peak indicates defects in the sample and the G peak indicates the vibration of sp2 carbon bonds) is used as an indicator of structure in Raman spectroscopy of graphene and its derivatives.42 In the characterization, the pure GO sample had an ID/IG ratio of 1.19. The higher the ratio, the more defects and irregularities are present in the structure of the measured graphene. Due to the disruption of sp2 bonds in the graphene structure during its oxidation to GO, the D band of GO is then intense. The ID/IG ratio is therefore higher for GO than for graphene and can be used as an indicator of the degree of its oxidation.The box plot shows the presence of significant changes in the ratio of bands D and G. A higher ID/IG ratio indicates more defects in the GO structure, which may be due to interaction with the cell. The change in these values suggests that chemical changes in GO within cells have occurred over time.
The stability of GO samples in cells was measured over a period of 6 months. To evaluate the stability, a sample with a concentration of 37.5 μg ml−1 was selected and measured, and this sample was then measured as mentioned above for 6 months, with several measurements taken at different points of the HeLa cell with GO each month. An overview of selected spectra from all months is shown in Fig. 4. Both GO and HeLa cell signals were present in the samples throughout the measurement period, but the samples were more sensitive during the last measurements. Changes in signal intensity were again observed during individual measurements due to a combination of differences in the profiles of individual cells, the precise location of GO in the cell, and the focus of the microscope itself. The intensity ratios of the D and G bands (ID/IG) were used to evaluate changes that have occurred in the sample over time. This approach would become very important in preclinical research from the long-term perspective to study possible changes in the structure of GO during the time within the cell, its interaction with cell compartments, and mechanisms of the degradation process of GO inside the individual single cells.
 | ||
| Fig. 4 Graph showing the change in intensity ratios of bands D and G measured in one sample over the six months (I–VI). | ||
Conclusions
This work shows for the first time the use of Raman spectroscopy to study the presence of graphene oxide in cervical cancer cells, the fate of GO within a single cell and its evaluation by MCR analysis. MCR-Raman spectroscopy could be used as highly complementary to fluorescence labeling-based methods and MCR imaging can become a tool for exploratory single-cell studies,43 as MCR analysis faithfully mimics the structure of the analytical measurement and the MCR-Raman method offers a promising approach to determine GO within single cells at the same time. Since MCR-Raman spectroscopy can be used to observe peak shifts within a cell as a result of GO interaction with single-cell components, there is a great potential for studying changes in the consequences of this GO interaction within cell compartments. Based on the mapping, graphene oxide was confirmed in all mapped samples in cells, at various concentrations. It was evident from the obtained maps that the number of particles that penetrated inside each of the cells increased with increasing concentration of GO used during the cell incubation. At the highest concentrations, clustering of GO particles was observed and the maps showed GO overwhelming the cells throughout the cross-section. Raman spectroscopy proved to be a promising label-free method that, together with MCR analysis, provides information on GO detection even in individual single HeLa cells which is a beneficial complementary analytical method in drug delivery treatment studies using 2D graphene-based nanomaterials.Author contributions
Conceptualization, ZC, LŽ, JB and KP; investigation, ZC, LŽ and KP; writing – preparation of original draft, ZC, and KP; writing – review and editing, ZC and KP; all authors have read and agree to the published version of the manuscript.Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
The work was supported by the HORIZON-EIC-2021-PATHFINDERCHALLENGES-01:101070865 and by the Internal Grant Agency of the Palacký University Olomouc (grant No. IGA_PrF_2023_024).Notes and references
- K. S. Novoselov, et al. , Science, 2004, 306(5696), 666–669 CrossRef CAS PubMed.
- H. Zhang, T. Fan, W. Chen, Y. Li and B. Wang, Bioact. Mater., 2020, 5(4), 1071–1086 CrossRef PubMed.
- G. Hongcai and D. Hongwei, Biosens. Bioelectron., 2015, 65(16), 404–419 Search PubMed.
- V. B. Mohan, K. Lau, D. Hui and D. Bhattacharyya, Composites, Part B, 2018, 142(16), 200–220 CrossRef CAS.
- B. C. Brodie XIII, Philos. Trans. R. Soc. London, 1859, 149(4), 249–259 Search PubMed.
- S. Georgitsopolou, N. D. Stola, A. Bakadritsos and V. Georgakilas, Surf. Interfaces, 2021, 26, 101320 CrossRef.
- A. Bakandritsos, et al. , ACS Nano, 2017, 11(3), 2982–2991 CrossRef CAS PubMed.
- T. Malina, et al. , Carbon, 2023, 118093 CrossRef CAS.
- T. Malina, A. Lamczová, E. Maršálková, R. Zbořil and B. Maršálek, Chemosphere, 2022, 291(1), 132739 CrossRef CAS PubMed.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6(3), 1879–1889 RSC.
- B. Dabrowski, et al. , Chem.-Biol. Interact., 2023, 376, 110444 CrossRef CAS PubMed.
- T. Chernenko, Drug Delivery Transl. Res., 2013, 3, 575–586 CrossRef CAS PubMed.
- M. Eliášová Sohová, et al. , Analyst, 2018, 143, 3686–3692 RSC.
- V. Ranc, J. Srovnal, L. Kvítek and M. Hajduch, Analyst, 2013, 138, 5983–5988 RSC.
- G. W. Auner, S. K. Koya, Ch. Huang and B. Broadbent, et al. , Cancer Metastasis Rev., 2018, 37, 691–717 CrossRef CAS PubMed.
- Z. Liu, Z. Guo, H. Zhong, X. Qin, M. Wan and B. Yang, Phys. Chem. Chem. Phys., 2013, 15, 2961–2966 RSC.
- T. Chernenko, et al. , Mol. Pharm., 2012, 9, 930–936 CrossRef CAS PubMed.
- A. Balzerová, K. Poláková, T. Malina, J. Belza, V. Ranc and R. Zbořil, Conference Proceedings Nanocon 2018, 2018, pp. 566–571 Search PubMed.
- L. Zhang, M. J. Henson and S. S. Sekulic, Anal. Chim. Acta, 2005, 545, 262–278 CrossRef CAS.
- Z. Chaloupková, et al. , Front. Anal. Sci., 2022, 2, 847730 CrossRef.
- J. P. Smith, F. C. Smith and K. S. Booksh, Appl. Spectrosc., 2017, 72, 404–419 CrossRef PubMed.
- G. P. S. Smith, C. M. McGoverin, S. J. Fraser and K. C. Gordon, Adv. Drug Delivery Rev., 2015, 89, 21–41 CrossRef CAS PubMed.
- Y. Shen, F. Hu and W. Min, Annu. Rev. Biophys., 2019, 48, 347–369 CrossRef CAS PubMed.
- De A. Juan, et al. , Anal. Chim. Acta, 2021, 59, 1145 Search PubMed.
- W. S. Hummers, S. William, R. E. Offeman and E. Richard, J. Am. Chem. Soc., 1958, 80(6), 1339 CrossRef CAS.
- International Agency for Research on Cancer, https://www.iarc.who.int, accessed on September 2020.
- S. Cui, S. Zhang and S. Yue, J. Healthc. Eng., 2018, 8619642 Search PubMed.
- A. Sahu, N. Nandakumar, S. Sawant and C. M. Krishna, Analyst, 2015, 140(7), 2294–2301 RSC.
- C. W. Hsu and C. C. Huang, et al. , J. Biomed. Opt., 2016, 21(7), 075006 CrossRef PubMed.
- I. Jung, D. A. Dikin, R. D. Pinera and R. S. Ruoff, Nano Lett., 2008, 8, 4283–4287 CrossRef CAS PubMed.
- ASTM, Zeta Potential of Colloids in Water and Waste Water, ASTM Standard D 4187–82, American Society for Testing and Materials, 1985 Search PubMed.
- S. Claramunt, A. Varea, D. López-Díaz, M. M. Velázquez, A. Cornet and A. Cirera, J. Phys. Chem. C, 2015, 119(18), 10123–10129 CrossRef CAS.
- A. C. Ferrari, Solid State Commun., 2007, 143, 47–57 CrossRef CAS.
- E. Aliyev, et al. , Nanomaterials, 2019, 9, 1180 CrossRef CAS PubMed.
- K. Krishnamoorthy, M. Veerapandian, K. Yun and S. J. Kim, Carbon, 2013, 53, 38–49 CrossRef CAS.
- E. Hahriary, A. Njali and A. Material, Science, 2014, 2(1), 58–63 Search PubMed.
- B. Kann, H. L. Offerhaus, M. Windbergs and C. Otto, Adv. Drug Delivery Rev., 2015, 89, 71–90 CrossRef CAS PubMed.
- K. Moto, H. Inadome, Y. Kanaho, S. Narumiya and M. Umeda, J. Biol. Chem., 2005, 280, 37901–37907 CrossRef PubMed.
- Ch. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., 2009, 121(26), 4879–4881 CrossRef.
- Z. Liu, J. T. Robinson, X. Sun and H. Dai, J. Am. Chem. Soc., 2008, 130(33), 10876–10877 CrossRef CAS PubMed.
- S. Goenka, V. Sant and S. Sant, J. Controlled Release, 2014, 173, 75–88 CrossRef CAS PubMed.
- A. Jorio, et al. , Phys. Status Solidi B, 2010, 247, 2980–2982 CrossRef CAS.
- J. P. Smith, E. C. Holahan, F. C. Smith, V. Marrero and K. S. Booksh, Analyst, 2019, 144, 5425–5438 RSC.
| This journal is © The Royal Society of Chemistry 2023 |