 Open Access Article
Open Access ArticleDevelopment of a cellulose-based 96-well plate vertical flow pull-down assay†
Hoi Lok
Cheng‡
 a,
Huan
Jia‡
a,
Huan
Jia‡
 a,
Sing Mei
Lim
a,
Say Yong
Ng
a,
Patthara
Kongsuphol
a,
Megan E.
McBee
a and
Hadley D.
Sikes
a,
Sing Mei
Lim
a,
Say Yong
Ng
a,
Patthara
Kongsuphol
a,
Megan E.
McBee
a and
Hadley D.
Sikes
 *ab
*ab
aAntimicrobial Resistance Interdisciplinary Research Group (AMR-IRG), Singapore-MIT Alliance in Research and Technology (SMART), 1 CREATE Way, 138602, Singapore
bDepartment of Chemical Engineering, Massachusetts Institute of Technology (MIT), 25 Ames Street, Building 66, Cambridge, MA 02139, USA. E-mail: sikes@mit.edu
First published on 10th July 2023
Abstract
The abundance and low production cost of biomaterial cellulose paper have attracted attention for many applications. Point-of-care (PoC) diagnostic tests have been successfully developed using patterned cellulose paper. Although PoC diagnostic tests are rapid and simple to perform, their sample processing throughput is limited, allowing for only one sample to be evaluated at a time, which restricts potential applications. Thus, it was appealing to expand cellulose-based PoC tests to high-throughput versions to increase their applicability. Here, we present the development of a high-throughput cellulose-based 96-well plate vertical flow pull-down assay that can process 96 tests, is easy to prepare, and can be customized for different detection targets. The device has two key features: (i) patterned cellulose paper for 96 tests that do not require pre-immobilization of capturing reagents, and (ii) reusable sturdy housing. We believe that a variety of applications, including laboratory testing, population surveillance tests, and sizable clinical trials for diagnostic tests, can benefit from the adoption of this cellulose-based 96-well plate assay.
1. Introduction
Diagnostic tests greatly benefit from paper-based formats, particularly point-of-care (PoC) diagnostics.1,2 Among these, nitrocellulose paper and cellulose paper are the most frequently used types of paper.3,4 The most plentiful natural polymer resource on earth is cellulose, which is also used as the primary ingredient in the production of nitrocellulose paper.5 Although nitrocellulose is still the most utilized material for PoC tests, cellulose is garnering increased attention as it is widely available and has a straightforward production procedure at scale.6 A variety of strategies have been used to develop PoC testing using cellulose paper, including paper-based ELISA (P-ELISA),1–3 paper-based biosensors,7 and microfluidic paper-based analytical devices (μPADs).8 Several methods, including ink-jet etching, ink jet printing, wax printing, stamping, and toner laser printing, have been developed for prototyping of cellulose paper-based analytical tests.9,10 Manufacturing companies such as 3M have mature techniques utilizing UV-curable resin that allows rolls of cellulose paper to be printed with flow pathways for scaled production.6,11 Additionally, in order to improve the effectiveness of immobilizing capture reagents on paper, modifications of the cellulose surface have been explored.4 Recently, affinity coupling via a cellulose binding domain (CBD) genetically fused to a capture reagent was shown to be effective at immobilizing the reagent and increasing detection sensitivity.2,12,13COVID-19-related PoC diagnostic tests using cellulose paper have been successfully developed using the CBD fused to capture reagents including rcSso7d affinity proteins and the SARS-CoV-2 receptor binding domain (RBD).6,14–16 The development of a cellulose pull-down method, with cellulose paper as the support material to pull-down the ‘capture reagent – analyte – reporter reagent’ complex from a broad range of sample solutions, was made possible by linking the CBD to capture reagents.6,12,14,17 Rapid chemical reactions in the aqueous phase are exploited in the cellulose pull-down method that eliminates the need for capture reagent pre-immobilization on cellulose paper.14–16 Additionally, the complexity of test device production has decreased due to the adoption of vertical flow technology in conjunction with the cellulose pull-down method.6,14–16 A small clinical study in Singapore has successfully used the vertical-flow cpVNT (cellulose pull-down viral neutralization test) to assess the efficacy of booster shots against SARS-CoV-2 strains.16
A PoC test can only process one sample per test, which restricts its use in high-throughput sample processing at clinical laboratories, population-level surveillance, and large-scale clinical trials.18 By creating hydrophobic borders and hydrophilic paths on cellulose paper, the Whitesides lab has shown that paper microzone plates were practical to allow multiple sample processing on cellulose-based tests.18,19 Paper microzone plates have also been used as a colorimetric sensor for Fritillariae cirrhosae bulbus and saccharide detection.20,21 However, the production procedure is complex as specially treated sheets with careful attention to registration between layers are required for the manufacture of microzone plates. On the other hand, the advances in 3D printing technology have provided a tool to fabricate prototypes for analytical devices, including immunosensor arrays, PoC tests and high-throughput spectroscopy.22–24 Here, we present an approach for creating a cellulose-based 96-well plate pull-down assay with two distinctive features: (i) patterned cellulose paper for 96 samples that does not require pre-immobilization of capturing reagents, and (ii) sturdy housing that can be re-used.
2. Results
2.1. Construction of a cellulose-based 96-well plate pull-down assay
The 96-well microplate housing was created using Greiner Bio-One Microplates' architecture as a reference.25 Two components make up the plate housing: a lid and a base. The two components were created using 3D printing with black polylactic acid (PLA) as the printing filament (Fig. 1A-1). The lid features 96 circular apertures that allows discrete opening of 96 test zones on the cellulose testing paper as well as to set additional physical separation between wells (Fig. S1A†). The base features three walls creating a 4 mm-deep housing area used for absorbent pads and cellulose testing papers (Fig. S1B†). Sliding tracks are included in the left and right walls of the base for ease of placement and securing of the lid.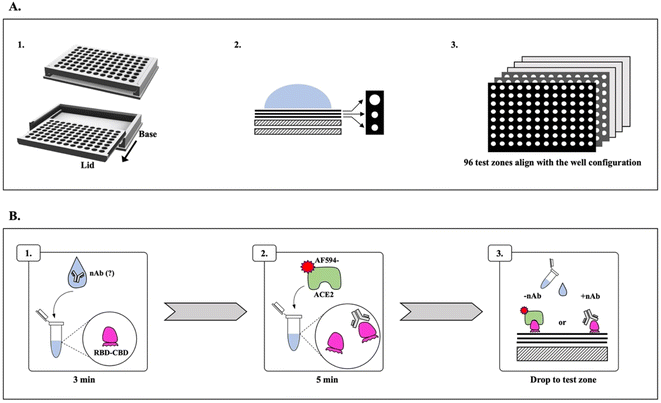 | ||
| Fig. 1 Schematic representation of the fabrication of a cellulose-based 96-well plate and cpVNT. (A) Fabricating cellulose-based 96-well plate involves three components, (1) 3D-printing drawing of 96-well plate housing with a lid and base; (2) illustration of an optimized cellulose test paper flow pathway (different types of flow pathways are depicted in Fig. 2A); and (3) optimized combination of cellulose test papers and absorbent pads for plate fabrication (different combinations of test papers and absorbent pads are depicted in Fig. 2B). (B) Schematic representation of cpVNT steps, (1) incubation of the RBD–CBD protein with the test sample that may contain neutralization antibodies (nAb) for 3 minutes; (2) incubation of the mixture from ‘1’ with ACE2-AF594 for 5 minutes; and (3) representation of the captured complex on cellulose test paper for samples with and without nAb, respectively, when the test sample is dropped onto the test zone. | ||
Construction of the multi-well test used Whatman grade 1 chromatography (CHR) paper with hydrophilic wells made on cellulose paper by wax ink printing and baking to define the sample flow path. To control the sample flow rate, three layers of CHR sheets with patterned wax-ink printing were combined.6 Four different vertical flow pathways were evaluated to minimize crosstalk between samples loaded in different wells and to maximize the samples retained in the designated flow pathway: (a) all three layers of the cellulose papers patterned with 5 mm diameter circles; (b) circles measuring 5 mm on the 1st layer and 4 mm on the 2nd and 3rd layers; (c) circles measuring 5 mm, 4 mm, and 3 mm on the 1st, 2nd, and 3rd layer of the paper, respectively; and (d) circles measuring 5 mm on the 1st layer and 3 mm on the 2nd and 3rd layers (Fig. 2A). 4th and 5th layers consisted of absorbent pads (GB005 blotting paper). From all layers of cellulose test papers and absorbent pads, the crosstalk of loaded samples was evaluated. In the images of the 2nd layer and 3rd layer, obvious traces of liquid spreading outside of hydrophilic circles, which could lead to sample crosstalk to adjacent wells, were observed for type ‘a’, ‘b’ and ‘c’ but not for type ‘d’ (Fig. 2A). Additionally, sample traces on the first layer of the absorbent pad (4th layer) are more even with clear edges at each circular pattern in type ‘d’ as compared to the other three types. For the second layer of the absorbent pad (5th layer), circular sample traces were formed inadequately in type ‘a’, ‘b’ and ‘c’, which indirectly indicated sample loss through the flow pathway for these three types. With all three observations above, type ‘d’ shows the most optimized control of sample crosstalk and sample retention within its flow pathway. Therefore, type ‘d’ was chosen as the optimal vertical flow pathway for producing 96-well plate pull-down assays (Fig. 1A-2). Additionally, 5 alternate wells in a row were also tested for these four types of flow pathways, showing the same sample traces as aforementioned (Fig. S2†).
Next, the flow rate was evaluated on top of the optimized flow pathway. Combinations of test papers and absorbent pads of different thicknesses were assessed, while the type ‘d’ flow pathway was kept unchanged. As illustrated in Fig. 2B, combination 1 consisted of three layers of test paper and two layers of absorbent pad; combination 2 consisted of three layers of test paper and three layers of absorbent pad; and combination 3 consisted of five layers of test paper (no wax pattern on the last 2 layers) and three layers of absorbent pad. Out of the three tested combinations, combination 2 provided a consistently regulated slower flow rate at roughly 10 seconds per 40 μL of 1× PBS buffer (Fig. 2B).
Based on the optimizations, a cellulose-based test consisting of three layers of cellulose test paper with 5 mm circles on the 1st layer and 3 mm circles on the 2nd and 3rd layers followed by three layers of absorbent pad below the cellulose test paper was deemed optimal, as illustrated in Fig. 1A-3 and S4.† This configuration is referred to as a “cellulose-based 96-well plate”. One unique property to highlight is that the plate can easily be re-assembled for running 96 new tests by substituting new cellulose papers and absorbent pads, and we modified the single plex SARS-CoV-2 cpVNT that was previously developed in our lab to the cellulose-based 96-well plate to evaluate its performance using human serum samples.16 The cpVNT is depicted in Fig. 1B, where neutralizing antibodies, if present, inhibit binding of the fluorescent-conjugated human ACE2 (angiotensin-converting enzyme 2) receptor (ACE2-AF594) to the CBD-conjugated viral RBD (RBD–CBD) protein, leading to diminished fluorescent signal intensity.13,17 Upon increasing neutralizing antibody (nAb) concentrations, fluorescent signals will decrease. The assay's final readout is represented in % blocking.
 | ||
| Fig. 3 Optimized 96-well fluorescence readouts for high, mid and low signal plates. Data in the figure are mean ± s.d. with n = 96 readings, and each reading is represented as a dot. | ||
2.2. Signal readout optimization
The cellulose-based 96-well plate's fluorescence signal was read using a TECAN Microplate Reader Infinite® 200 PRO. To ensure even signal readouts through the 96 test wells, we changed the plate configuration settings in the TECAN plate reader software to fit the assembled cellulose-based plate dimensions. The parameters being configured include well diameter, plate size, and plate height. Eight plate configurations were created for evaluation (Table S1†). To evaluate signal readouts, the autofluorescence signal of cellulose test paper at an excitation wavelength of 590 nm and an emission wavelength of 618 nm was detected. The results showed that the eighth configuration provided the most consistent fluorescence readouts over 96 wells with the lowest standard deviation and CV (coefficient of variation) below 10%. Table S1† highlights the optimal plate configuration in green. To further evaluate the optimized configuration, samples in PBS buffer with a high AF594 (Alexa Fluor 594) fluorescence signal and mid AF594 fluorescence signal were applied to two assembled plates, respectively, along with one blank plate. As shown in Fig. 3, all high signal samples, mid signal samples, and the blank plate with a low signal presented homogeneous signals across the 96 wells, with CV values <10%. The raw data of fluorescence readouts are shown in Table S2.†2.3. Assay performance
Next, the cpVNT's performance on a 96-well cellulose-based plate was evaluated. Titrations of the fluorescence signal and the percentage of blocking were performed using a series of human serum samples spiked with serially diluted mouse SARS-CoV-2 nAbs. The range of nAb concentration was 0.25 nM to 128 nM. An IC50 value of 1.95 nM for the percentage of blocking was determined by nonlinear regression statistics (Fig. 4A). This assay sensitivity aligned with our reported single plex SARS-CoV-2 cpVNT with an IC50 value of 3.38 nM.26 To ensure good assay performance, blinded experiments were used to validate the 96-well cpVNT assay. Operator A generated nine samples with known doses of mouse SARS-CoV-2 nAbs and known % blocking that was calculated based on the titration curve shown in Fig. 4A. These samples were then sent to operator B for blinded testing. Three of the nine samples had concentrations below the IC50 value, while the remaining six samples had concentrations above the IC50 value. Fig. 4B illustrates the blind tested results. All % blocking obtained by operator B through blinded testing matched with the % blocking known to operator A. The 96-well cellulose-based plate cpVNT assay was shown to have a well-aligned linear correlation with the calculated % blocking values with a slope value of 0.9621 ± 0.0342 and a goodness of fit R2 value of 0.99 (Fig. 4B). Statistically, the blinded test gave 100% match at both the positive predictive value and negative predictive value.3. Discussion and conclusions
High throughput assays are necessary for batch analysis. However, they rely heavily on polystyrene plate platforms, which produce enormous amounts of plastic waste. A platform for substitution is thus highly needed.27 With the 96-well cellulose-based plate assay we designed, plastic waste is hugely substituted with cellulose paper waste that is readily biodegradable.28,29 Leveraging the cellulose-based PoC test for single samples, the cellulose-based 96-well plate assay can be widely applied in laboratory tests of biomarkers, clinical surveillance tests, and clinical trials for validating PoC tests utilizing the same test platform. Additionally, our test provides a much faster turn-around-time (TAT) as compared to commercial plate-based ELISA assays. Finally, both the reporter and the capture reagents are kept in a reaction tube, making it simple to produce the cellulose-based 96-well plate. With the advances in reagent lyophilization technologies, the capture and reporter reagents will be able to be stored at ambient temperature for a longer period of time as well.30,31In conclusion, the SARS-CoV-2 cpVNT assay served as an example for validating the cellulose-based 96-well pull-down assay that we developed. We believe that this cellulose-based plate assay can be converted into commercially available laboratory assays with further optimizations in production.
4. Materials and methods
4.1. Protein production and modification
The proteins of the soluble extracellular fragment of human ACE2 (residues 19–615) and wildtype (WT) SARS-CoV-2 RBD–CBD were expressed and purified according to previous publications.15,16 The ACE2 protein was conjugated to Alexa Fluor 594 with an Alexa Fluor® 594 Conjugation Kit (Fast)-Lightning-Link® (Abcam, ab269822, Singapore) following the manufacturer's protocol.4.2. Cellulose pull-down virus neutralization test (cpVNT)
To perform one cpVNT assay for each well, 10 μL of serum/plasma sample was added to 10 μL of 10 nM WT RBD–CBD in 1× PBS pH 7.6 with 1% BSA. After incubation for 3 minutes at ambient temperature, 20 μL of 5 nM ACE2-AF594 in 1× PBS pH 7.6 with 1% BSA was subsequently added and incubated for 5 minutes at ambient temperature. The mixture with a total volume of 40 μL was dropped onto the hydrophilic test zone of the cellulose test paper. This was followed by a one-time washing step with 40 μL of 1× PBS once the sample mixture was fully absorbed. The fluorescence signals were then detected, and the percent blocking (% blocking) could be calculated with this equation4.3. Fabrication of a cellulose-based 96-well plate
4.3.3.1. Four types of flow pathways for checking sample crosstalk and retention. In this section of plate fabrication, layers of absorbent pads were fixed to be 2 layers, and layers of test paper were fixed to be 3 layers. Types ‘a’ to ‘d’ of flow pathways as described in the Results section were assembled. Upon fabricating and assembling each type of cellulose test paper combination, 40 μL of diluted carmine fairbro batik dye (Fairbro Bairbreno, Malaysia) was loaded onto the top layer of test paper followed by washing with 40 μL of 1× PBS. Five consecutive wells (C4–C8) and five alternate wells (E3, E5, E7, E9 and E11) were used for testing in each type of test paper combination. Test papers and absorbent pads were extracted out to check cross talking of loaded sample on the lower layers of cellulose test papers and layers of absorbent pads (Fig. 2A and S2†).
4.3.3.2. Three combinations of cellulose test papers and absorbent pads for an optimized flow rate. To optimize the flow rate on the fabricated 96-well plate, we tested three combinations of cellulose test paper and absorbent pads as described in the Results section with a fixed type ‘d’ of flow pathway. Upon assembly of each combination, 12 wells at position C5–C7, D5–D7, E5–E7 and D5–D7 were selected to measure the flow time for 40 μL of 1× PBS (pH 7.6). When the sample loaded was not absorbed within 1 minute, the data point was recorded as an unmeasurable replicate (Fig. 2B).
4.6. Optimization of fluorescence readouts on the cellulose-based 96-well plate
The 96-well plate housing was designed to be compatible with most of the commercially available microplate readers and here we used a TECAN Microplate reader Infinite® 200 PRO for demonstration. During readout optimization, a blank plate was used for measurements. Briefly, the plate was placed in the microplate reader and the plate configuration was manually modified in plate reader software. Upon each plate definition setup, readings of all 96 wells were taken for analysis. The 8 plate definition settings are listed in Table S1.†Next, the optimized plate configuration was applied for reading fluorescence signals. The samples representing a high fluorescence signal were prepared by mixing and incubating 1 volume of 1× PBS, 1 volume of 10 nM RBD–CBD in 1× PBS pH 7.6 with 1% BSA, and 2 volumes of 5 nM ACE2-AF594 1× PBS pH 7.6 with 1% BSA. The samples representing a mid fluorescence signal were diluted from high fluorescence signal samples 2 fold with 1× PBS. 40 μL of the high intensity samples, or mid intensity samples was added to each of the 96 wells of the paper plate. The high intensity plate, the mid intensity plate, and a blank plate representing a low signal were measured, respectively, and evaluated for the homogeneity across the plate.
4.7. Titration curve and blinded test of the cpVNT on the cellulose-based 96-well plate
Pre-COVID-19 serum (Sigma) was spiked with mouse anti-SARS-CoV-2 nAb (Sino Biological, 40591-MM43-100, USA) to generate serum samples containing 0.25 nM, 0.5 nM, 1 nM, 2 nM, 4 nM, 8 nM, 16 nM, 32 nM, 64 nM, and 128 nM nAb for a titration curve. Triplicate tests of each concentration were performed simultaneously on the same 96-well plate. The paper plate was also evaluated by blinded tests in which the assay operator was not informed about the nAb concentrations of the prepared serum samples prior to performing the assay and data analysis. The prepared nAb concentrations included 0, 0.25 nM, 4 nM, 8 nM, and 16 nM. The blinded test results were compared to the calculated % blocking values obtained from the titration curve.4.8. Statistical analysis
GraphPad Prism 9 (9.5.1) was utilized for all statistical analyses and graph preparation. Triplicate data were obtained for each group of experiments for analyzing the standard deviation that is represented as error bars in the figures. Simple linear regression was carried out to check the goodness of fit (R2) for the blinded test as shown in Fig. 3B.Author contributions
H. L. C. – conceptualization, performing experiments, data analysis, manuscript drafting and reviewing; H. J. – conceptualization, plate design, experimentation, data analysis, manuscript drafting and reviewing; S. M. L. – conceptualization, plate design and scientific inputs, S. Y. N., P. K. and M. E. M. – scientific inputs, and manuscript reviewing, H. D. S. – conceptualization, supervision, funding acquisition, and manuscript reviewing.Conflicts of interest
MIT (H. D. S.) and SMART (H. L. C., H. J., S. M. L., S. Y. N., P. K., M. E. M. and H. D. S.) declared that they have submitted a patent application in relation to this work.Acknowledgements
This work was supported by the National Research Foundation, Prime Minister's Office, Singapore, under its Campus for Research Excellence and Technological Enterprise (CREATE) programme through the Antimicrobial Resistance Interdisciplinary Research Group (AMR-IRG) of Singapore-MIT Alliance in Research and Technology (SMART). The authors thank the NTU Protein Production Platform especially Dr Chen Ming Wei for expression and purification of the SARS-CoV-2 RBD–CBD protein and ACE2 protein.References
- C. M. Cheng, A. W. Martinez, J. Gong, C. R. Mace, S. T. Phillips, E. Carrilho, K. A. Mirica and G. M. Whitesides, Angew. Chem., 2010, 122, 4881–4884 CrossRef.
- R. Pelton, TrAC, Trends Anal. Chem., 2009, 28, 925–942 CrossRef CAS PubMed.
- M. Costa, B. Veigas, J. Jacob, D. Santos, J. Gomes, P. Baptista, R. Martins, J. Inácio and E. Fortunato, Nanotechnology, 2014, 25, 094006 CrossRef CAS PubMed.
- C.-H. Yang, C.-A. Chen and C.-F. Chen, Sens. Actuators, B, 2018, 265, 506–513 CrossRef CAS.
- C. Saunders and L. Taylor, J. Energ. Mater., 1990, 8, 149–203 CrossRef CAS.
- S. Kim, E. Yee, E. A. Miller, Y. Hao, D. M. Tay, K.-J. Sung, H. Jia, J. M. Johnson, M. Saeed and C. R. Mace, ACS Appl. Mater. Interfaces, 2021, 13, 38990–39002 CrossRef CAS PubMed.
- K. Ratajczak and M. Stobiecka, Carbohydr. Polym., 2020, 229, 115463 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., 2007, 119, 1340–1342 CrossRef.
- L. Akyazi, Anal. Chim. Acta, 2018, 1001, 1 CrossRef PubMed.
- J. S. Ng and M. Hashimoto, RSC Adv., 2020, 10, 29797–29807 RSC.
- G. Zhu, J. Zhang, J. Huang, X. Yu, J. Cheng, Q. Shang, Y. Hu, C. Liu, L. Hu and Y. Zhou, Green Chem., 2021, 23, 5911–5923 RSC.
- E. A. Miller, S. Baniya, D. Osorio, Y. J. Al Maalouf and H. D. Sikes, Biosens. Bioelectron., 2018, 102, 456–463 CrossRef CAS PubMed.
- E. A. Miller, M. W. Traxlmayr, J. Shen and H. D. Sikes, Mol. Syst. Des. Eng., 2016, 1, 377–381 RSC.
- H. Jia, E. A. Miller, C. C. Chan, S. Y. Ng, M. Prabakaran, M. Tao, I. S.-Y. Cheong, S. M. Lim, M. W. Chen and X. Gao, Lab Chip, 2022, 22, 1321–1332 RSC.
- P. Kongsuphol, H. Jia, H. L. Cheng, Y. Gu, B. D. Shunmuganathan, M. W. Chen, S. M. Lim, S. Y. Ng, P. A. Tambyah and H. Nasir, Commun. Med., 2021, 1, 1–12 CrossRef PubMed.
- H. L. Cheng, S. M. Lim, H. Jia, M. W. Chen, S. Y. Ng, X. Gao, J. Somani, S. Sengupta, D. M. Y. Tay, P. W. L. Chua, A. R., S. Y. H. Ling, M. E. McBee, B. E. Yong, H. D. Sikes and P. R. Preiser, Microbiol. Spectrum, 2022, 10(5), e02257 Search PubMed.
- E. A. Miller, K.-J. Sung, P. Kongsuphol, S. Baniya, H. Q. Aw-Yong, V. Tay, Y. Tan, F. M. Kabir, K. Pang-Yeo and I. G. Kaspriskie, ACS Comb. Sci., 2019, 22, 49–60 CrossRef PubMed.
- E. Carrilho, S. T. Phillips, S. J. Vella, A. W. Martinez and G. M. Whitesides, Anal. Chem., 2009, 81, 5990–5998 CrossRef CAS PubMed.
- W. Zhao, M. M. Ali, S. D. Aguirre, M. A. Brook and Y. Li, Anal. Chem., 2008, 80, 8431–8437 CrossRef CAS PubMed.
- X. Lyu, V. Hamedpour, Y. Sasaki, Z. Zhang and T. Minami, Anal. Chem., 2020, 93, 1179–1184 CrossRef PubMed.
- L. Li, M. Yang, M. Zhang and M. Jia, Sens. Actuators, B, 2022, 368, 132210 CrossRef CAS.
- G. W. Bishop, J. E. Satterwhite-Warden, K. Kadimisetty and J. F. Rusling, Nanotechnology, 2016, 27, 284002 CrossRef PubMed.
- H. N. Chan, M. J. A. Tan and H. Wu, Lab Chip, 2017, 17, 2713–2739 RSC.
- M. Sharafeldin, K. Kadimisetty, K. S. Bhalerao, T. Chen and J. F. Rusling, Sensors, 2020, 20, 4514 CrossRef CAS PubMed.
- G. Bio-one, Microplate Dimensions Guide, https://www.gbo.com/fileadmin/media/GBO-International/01_Downloads_BioScience/SALES_Brochures/F073027_Microplate_Dimensions_Guide.pdf Search PubMed.
- S. M. Lim, H. L. Cheng, H. Jia, P. Kongsuphol, B. D. O. Shunmuganathan, M. W. Chen, S. Y. Ng, X. Gao, S. P. Turaga and S. P. Heussler, Bioeng. Transl. Med., 2022, e10293 CAS.
- A. E. Ongaro, Z. Ndlovu, E. Sollier, C. Otieno, P. Ondoa, A. Street and M. Kersaudy-Kerhoas, Lab Chip, 2022, 22, 3122–3137 RSC.
- B. Ai, L. Zheng, W. Li, X. Zheng, Y. Yang, D. Xiao, J. Shi and Z. Sheng, Front. Recent Dev. Plant Sci., 2021, 12, 625878 CrossRef PubMed.
- M. P. Motloung, V. Ojijo, J. Bandyopadhyay and S. S. Ray, Polymers, 2019, 11, 1270 CrossRef PubMed.
- S. J. Thorpe, B. Fox and D. Sands, Biologicals, 2002, 30, 315–321 CrossRef CAS PubMed.
- S. Ghosh and C. H. Ahn, Analyst, 2019, 144, 2109–2119 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ay00614j |
| ‡ H. L. C. and H. J. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |



